Abstract
In eukaryotes, mRNA turnover and translational repression represent important regulatory steps in gene expression. Curiously, when under cellular stresses, factors involved in these processes aggregate into cytoplasmic foci known as Processing bodies (P-bodies) and Stress Granules (SGs). In animals, CCCH Tandem Zinc Finger (TZF) proteins play important roles in mRNA decay within P-bodies. TTP, a P-body localized mammalian TZF, can bind to the 3'UTRs of mRNAs containing AU-rich elements (AREs) and recruit and activate RNA decay machineries. In plants, the function of CCCH TZF proteins is still unclear. We previously showed that AtTZF1, a member of the Arabidopsis CCCH TZF protein family, could bind both RNA and DNA in vitro, traffic between the nucleus and cytoplasmic foci, and co-localize with P-body markers AGO1, DCP2 and XRN4. In this report, we show that the other 10 members of the AtTZF gene family can similarly localize in cytoplasmic foci. Together these 11 genes, though similar in TZF sequence and sub-cellular localization, appear to be differentially expressed in response to developmental and environmental cues. We propose that AtTZF1 and its gene family members may each play distinct roles at different developmental stages and environmental conditions.
Key words: CCCH, TZF, AtTZF, zinc finger, P-body, stress granule
The precise control over gene expression in eukaryotes is a multi-step regulatory process. In addition to the control of transcript production, RNAs are subjected to the regulation of RNA processing, quality control, transport and turnover that ultimately dictate the availability of transcript for translation.1 Thus mRNAs exported to the cytoplasm can be either translated or subjected to translational repression caused by various silencing mechanisms that can dissociate mRNAs from active polysomes and aggregate them into P-bodies (PBs).2,3 In yeast and animals, RNA turnover via nonsense mediated decay (NMD), AU-rich element mediated decay (AMD), and RNAi can all take place in PBs.2,4,5 As suggested by their functions, animal PBs are largely composed of mRNAs and RNA decay machineries including: decapping and deadenylation complexes, exonucleases, RNA-induced silencing complexes (RISCs) and various other RNA-binding proteins.6 Similarly, SGs are closely related cytoplasmic bodies that can also form in response to cellular stresses. Though compositionally and functionally linked, there are distinct components that distinguish PBs from SGs. Whereas PBs are primarily composed of mRNA decay machineries, SGs are enriched with stalled preinitiation complexes and translation regulators.7 Of the PB and SG-localized proteins, CCCH TZF proteins such as TTP and BRF1 can bind AU-rich elements (AREs) in the 3’UTR of mRNAs and recruit and activate RNA decay proteins.82-10
CCCH TZF proteins are conserved in multi-cellular eukaryotes and participate in various cellular processes. For example, C. elegans PIE-1 OMA-1 and OMA-2 are involved in germline specification, and they are associated with cytoplasmic RNPs termed P-granules.11,12 In the nucleus, PIE1 acts as a transcriptional inhibitor promoting germline development. When shuttled to the cytoplasm, PIE-1 plays a genetically separable function by enhancing maternal NOS2 RNA stability and translation. Although CCCH TZF genes can also be found in green organisms from algae to higher vascular plants, the plant TZF motif is distinct from the mammalian TZFs in the CCCH spacing pattern. In Arabidopsis, SOMNUS (AtTZF4) is involved in light dependent seed germination, partly by regulating the expression of ABA/GA anabolic/catabolic genes.13 AtSZF1 and AtSZF2 (AtTZF11 and AtTZF10 respectively) both regulate salt stress and confer salt resistance when overexpressed.14 Besides Arabidopsis, rice and cotton CCCH TZF genes are also involved in stress responses.15,16 It is intriguing, however, none of the plant CCCH TZF proteins have been found in either PBs or SGs.13–16
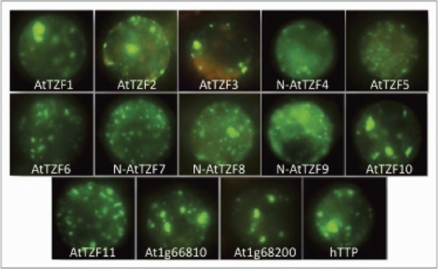
Recently, we showed that AtTZF1 could localize in cytoplasmic foci, similar to those described for CCCH TZF proteins in animals and yeast.17 We further demonstrated that AtTZF1 could traffic between the nucleus and cytoplasmic foci, and could co-localize with both P-body (DCP2, XRN4 and AGO1) and SG markers (PABP) when localized in the cytoplasm.17 Although the 11 AtTZF genes show high homologies within the TZF and upstream 50 aa regions, it cannot be assumed that all of them can be localized in cytoplasmic foci. To determine the subcellular localization patterns for the other 10 AtTZF genes, we used maize protoplast transient expression analyses.18 Each AtTZF-GFP fusion construct was individually expressed and then visualized under a fluorescent microscope. All AtTZF gene family members, including previously characterized AtTZF 4, 10 and 11,13,14 localized to cytoplasmic foci similar to those of AtTZF1 and http (Fig. 1). The localization patterns of two other CCCH TZF genes (At1g66810 and At1g68200), not part of the AtTZF gene family, were also found to be within cytoplasmic foci. Though the size and number of foci formed by overexpression varied between each AtTZF gene, owing to the experimental variations introduced by the DNA and cell qualities, it is difficult to draw any conclusions. However, as seen in Figure 1, AtTZF5, 7, 8, 9, 11 and At1g66810 all seemed to show a greater number of fine cytoplasmic foci, as opposed to fewer but larger foci of the other genes.
Figure 1.
Arabidopsis CCC H TZFs are localized in cytoplasmic foci that resemble P-bodies. GFP fusions of the AtTZF gene family, two other related TZF genes, At1g66810 and At1g68200, and a human TZF (hTT P) were expressed in etiolated maize mesophyll protoplasts. All were C-terminal GFP fusions unless otherwise noted. Scale bar: 10 µm. AtTZF1-At2g25900, AtTZF2-At2g19810, AtTZF3-At4g29190, AtTZF4-At1g03790, AtTZF5-At5g44260, AtTZF6-At5g07500, AtTZF7- At2g41900, AtTZF8-At5g12850, AtTZF9-At5g58620, AtTZF10-At2g40140, AtTZF11-At3g55980, hTT P-ZFP36.
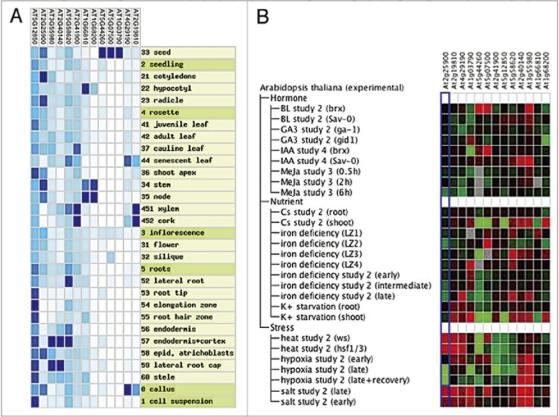
Compared to mammals, plants seem to have unusually high number of CCCH TZF genes. The Genevestigator metaprofile analyses19 revealed that despite similarities in sequence and subcellular localization patterns, the expression of each of the AtTZF genes varies widely, and is defined by developmental stages (Fig. 2A). Furthermore, their expression levels are affected by various cues, such as hormones, nutrients and environmental stresses (Fig. 2B). Interestingly, some AtTZFs’ expression patterns correlate with their reported functions. For example, AtTZF4 and AtTZF6, which have been reported to regulate germination, are preferentially expressed in seeds and embryos. AtTZF10 and AtTZF11 are involved in salt stress, and they are upregulated in the presence of salt (Fig. 2B).13,14
Figure 2.
Temporal, spatial and stimulus-induced expression of AtTZF gene family and two close related TZF, At1g66810 and At1g68200. Shown are heat-maps of meta-profile analyses using Genevestigator programs (www.genevestigator.com/gv/index.jsp). The level of gene expression is indicated by the intensity of the blue color in the left panel. The level of stimulus-induced gene expression change is indicated by the brightness of the red (induction) and green (repression) color.
The distinct gene expression patterns and subtle differences in the morphology of the cytoplasmic foci of the AtTZF gene family members have raised many questions regarding to the potential functions of each gene. So far, only a handful of the AtTZF gene family members have been characterized, and they seem to play distinct roles in plant development and environmental responses. However, preliminary observations in our laboratory have revealed considerable phenotypic similarities among AtTZF genes when ubiquitously expressed in transgenic plants. Because AtTZF genes share high sequence homologies in TZF region and similar subcellular localization patterns, our findings suggest that each AtTZF gene may have a similar molecular function—e.g., targeting various genes for RNA stability control. We thus propose that each AtTZF gene may function via a similar mechanism but will play a distinct role in response to various developmental cues and environmental conditions.
Acknowledgements
We would like to thank Dr. Perry J. Blackshear for providing http clone, Cyrus Hah for helping with the experiments, and Arabidopsis Biological Resource Center for providing DNA clones. This work was supported by the National Science Foundation grant IOB- 0543751 to J.-C.J. Salaries and research support also provided by state and federal funds appropriated to The Ohio State University, Ohio Agricultural Research and Development Center.
Abbreviations
- P-bodies, PBs
processing bodies
- SGs
stress granules
- TZF
tandem zinc finger
- mRNA
messenger RNA
- UTR
untranslated region
- AREs
AU-rich elements
- NMD
nonsense mediated decay
- AMD
AU-rich element mediated decay
- RNAi
RNA interference
- RISC
RNA induced silencing complex
- RNP
ribonucleoprotein
- GFP
green fluorescent protein
- AGO1
argonaute 1
- DCP2
decapping 2
- XRN4
exoribunuclease 4
- PABP
polyadenylate-binding protein (At1g49760)
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10988
References
- 1.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 2.Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 3.Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007;21:719–735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newbury SF, Muhlemann O, Stoecklin G. Turnover in the Alps: an mRNA perspective. Workshops on mechanisms and regulation of mRNA turnover. EMBO Rep. 2006;7:143–148. doi: 10.1038/sj.embor.7400628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 9.Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- 10.Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 11.Mello CC, Schubert C, Draper B, Zhang W, Lobel R, Priess JR. The PIE-1 protein and germline specification in C. elegans embryos. Nature. 2006;382:710–712. doi: 10.1038/382710a0. [DOI] [PubMed] [Google Scholar]
- 12.Shimada M, Yokosawa H, Kawahara H. OMA-1 is a P granules-associated protein that is required for germline specification in Caenorhabditis elegans embryos. Genes Cells. 2006;11:383–396. doi: 10.1111/j.1365-2443.2006.00945.x. [DOI] [PubMed] [Google Scholar]
- 13.Kim DH, Yamaguchi S, Lim S, Oh E, Park J, Hanada A, et al. SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell. 2008;20:1260–1277. doi: 10.1105/tpc.108.058859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun J, Jiang H, Xu Y, Li H, Wu X, Xie Q, et al. The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 2007;48:1148–1158. doi: 10.1093/pcp/pcm088. [DOI] [PubMed] [Google Scholar]
- 15.Kong Z, Li M, Yang W, Xu W, Xue Y. A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 2006;141:1376–1388. doi: 10.1104/pp.106.082941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo YH, Yu YP, Wang D, Wu CA, Yang GD, Huang JG, et al. GhZFP1, a novel CCCH-type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol. 2009;183:62–75. doi: 10.1111/j.1469-8137.2009.02838.x. [DOI] [PubMed] [Google Scholar]
- 17.Pomeranz MC, Hah C, Lin PC, Kang SG, Finer JJ, Blackshear PJ, et al. The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol. 2009;152:151–165. doi: 10.1104/pp.109.145656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jang JC, Sheen J. Sugar sensing in higher plants. Plant Cell. 1994;6:1665–1679. doi: 10.1105/tpc.6.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, et al. Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinform. 2008. p. 420747. [DOI] [PMC free article] [PubMed]