Abstract
Natural killer (NK) cells are effectors of the innate immune system and recognize cells transformed by viruses or neoplasia. Their response to “missing self” signals was described 3 decades ago, but the recent discovery of a panoply of activating receptors has made it clear that NK cell reactivity arises from a combination of inhibitory and activating signals. Successful clinical exploitation of NK cell reactivity was demonstrated in allogeneic transplantation for acute myelogenous leukemia from HLA-haploidentical donors when matched donors were not available. Multiple clinical studies have since attempted to use NK reactivity in the setting of both HLA-matched and -mismatched transplantation, with varying results. This review summarizes the heterogeneous clinical results and explains them based on a succinct description of NK cell biology.
Keywords: Natural killer, NK cells, Haploidentical transplantation, KIR, NKG2D, Missing self, Missing ligand
INTRODUCTION
Alternative donors are required for allogeneic hematopoietic cell transplantation (HCT) when suitable matched donors are not available in a timely fashion. Early studies investigating unmanipulated bone marrow (BM) grafts from mismatched or haploidentical related or unrelated donors demonstrated a high rate of nonrelapse mortality (NRM) resulting from graft failure, graft-versus-host disease (GVHD), or delayed immune reconstitution, and showed an association between increasing HLA disparity and worse prognosis [1-5]. Approaches designed to circumvent these hurdles included increasing peritransplantation immunosuppression, increasing the dose of hematopoietic stem cells [6,7], and manipulating the graft through T cell depletion (TCD) [8] or positive selection for CD34+ hematopoietic stem cells [9,10]. A recent review of these studies concluded that GVHD in haploidentical transplant recipients can be ameliorated by TCD, but at the cost of increased relapse and delayed immune reconstitution [5]. Using a reduced intensity conditioning regimen before mismatched or haploidentical transplantation was found to be associated with high rates of engraftment and low treatment-related mortality (TRM) when combined with in vivo or ex vivo TCD [11-13]; however, these approaches were again complicated by relapse of malignancy and delayed immune reconstitution.
Pioneering work from Perugia, Italy, highlighted a remarkable effect of donor natural killer (NK) cells in reducing relapse after TCD haploidentical transplant for acute myelogenous leukemia (AML) [14-16]. This group reported that allogeneic, alloreactive NK cells promote engraftment and the graft-versus-tumor effect, whereas they reduce GVHD. Relapse-free and event-free survival outcomes were significantly better in patients exhibiting killer immunoglobulin-like receptor (KIR) ligands that were mismatched with those from their donor. The publication of these provocative data spurred numerous studies attempting to document the beneficial effects of NK cell alloreactivity; however, these studies'design, methods, and thus results have been very heterogeneous. Examination of a nonexhaustive list of these studies (Table 1) suggests that the following factors may be important in maximizing NK cell alloreactivity: high stem cell dose, extensive TCD, no GVHD prophylaxis, and myelogenous malignancy as the target. Other possible reasons for the disparate findings include the differences in the definition of NK cell alloreactivity (phenotypic vs genotypic; mismatch algorithm), donor source (related vs unrelated; haploidentical), disease state at transplantation, and ethnicity. Grafts from both BM or peripheral blood (PB) sources have been effective, but KIR-mismatched grafts may be associated with worse survival in nonmyeloablative transplants from unrelated cord blood (UCB) [17]. Recent evidence also suggests that mothers may be a superior haploidentical donor source [18].
Table 1.
Selected Clinical Trials Investigating NK Alloreactivity
| Reference | Disease | Number of patients | Conditioning | TCD | Graft source and composition | NK alloreactivity* | GVHD prophylaxis | Engraftment failure | Acute GVHD grade II +/Chronic GVHD | Infection | TRM | RFS | OS | Benefit from NK alloreactivity? |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haploidentical | ||||||||||||||
| Ruggeri et al., 2007 [16] | AML | 112 | MA | Ex vivo | PB; 15 × 106/kg CD34, 3 × 104/kg CD3 | 1, 2, 3 | 0 | 6% vs 10% | 10%, NS/NR | 38% fatal infections | 43% | 67% vs 18% | NR | Yes |
| Leung et al., 2004 [86] | AML, ALL; pediatric | 36 | NR | CD34+ selection | PB; < 3 × 104/kg CD3 | 1, 2, 4 | 0 | NR | NR/NR | NR | NR | 13% vs 54% relapse rate | NR | Yes, for relapse, “missing ligand” model |
| Lang 2004 [134] | Various | 63 | MA | CD34 or CD133 selected + ATG | PB; 19.5 × 106/kg MNC, < 2.5 × 104/kg CD3 | 1 | 0 | 17% | 7%/13% | 17% fatal infections | 27% | NR | 42% | No (equivalent to historical matched unrelated donor) |
| HLA-identical related | ||||||||||||||
| Hsu et al., 2005 [47] | AML, CML, ALL, MDS | 178 | MA | Ex vivo | BM; 9× 105/kg CD3 | 2 | Yes | 0% | NS/NS | NR | NR | × 0.41 relapse (AML, MDS) | × 0.52 risk | Yes |
| Cook 2004 [135] | Various | 220 | MA/RIC | NR | NR | 1, 2 | Yes | NR | NS/NR | NR | NR | NS | 31.6%) vs 56.1% (4 years) | No; worse survival for myeloid patients with C2/C2 and KIR2DS2 donor |
| Unrelated | ||||||||||||||
| Giebel 2003 [136] | Various | 130 | MA | ATG | BM; 4.3 × 108/kg MNC | 1 | Yes | 0% vs 4% | 0% vs 15% (grade III-IV/NS | NR | 6% vs 40% | Relapse 6% vs 21% | 87% vs 48% (4.5 years | Yes |
| Kroger et al., 2006 [104] | AML, CML, ALL, MDS | 142 | MA | ATG | PB/BM | 1, 2 | Yes | 0% | NS/NS | Increased | × 2.2 risk if alloreactive | × 3 relapse risk (activating KIR) | × 0.5 unless donors are KIR haplotype A | No; ligand/ligand model |
| Cooley et al., 2008 [59] | AML | 448 | MA | No | PB/BM | 1, 2 | NR | NR | NS/× 1.5 risk if activating haplotype | NR | NS | × 2 RFS (activating KIR) | × 1.5 with higher number of activating KIR (3 years) | Y for donors with group B KIR haplotype |
| Davies 2002 [137] | Various | 175 | MA | Ex vivo, minority | BM; 2 × 108/kg MNC | 1 | Yes | NS | NS/NR | NR | NR | 9%-12% at 5 years (NS) | NS (whole group); × 0.5 (myeloid) | No |
| Farag 2006 [138] | AML, CML MDS | 1571 | MA | Minority ex vivo TCD | BM | 1 | NR | NS | NS/NS | NR | × 1.95 risk | NS | × 0.5 risk | No |
| Yabe et al., 2008 [132] | Various | 1489 | MA | ATG, minority | BM | 1, 2 | Yes | NR | × 1.7 risk; increased with 2DS2 gene in donor/NR | NR | NR | NS | × 1.93 risk | No (worse GVHD) |
| Miller 2007 [139] | AML, CML MDS | 2062 | NR | NR | NR | 1 | NR | NR | 44% vs 30% (late-phase CML)/NR | NR | NR | × 0.54 relapse in early disease | NR | Yes, for early myeloid disease |
| Bornhauser et al, 2004 [99] | AML, CML, MDS | 118 | MA | ATG | PB/BM; 4 × l06/kg CD34 | 1 | NR | 10%, NS | 46% vs 69%, NS/NS | NR | NR | Relapse 60% vs 35% | NS | No |
RFS indicates relapse-free survival; OS, overall survival; MA, myeloablative conditioning; RIC, reduced-intensity conditioning; ATG, antithymocyte globulin; MNC, mononuclear cell; haploA, KIR haplotype A; C2, HLA-C2 alleles; NS, no significant difference; NR, not reported.
Model used to define NK alloreactivity:
1, ligand/ligand (HLA typing of donor and recipient).
2, receptor/ligand (KIR genotyping or phenotyping of donor).
3, specific cytotoxicity assay of donor NK cells against recipient cells.
4, nonspecific cytotoxicity assay of donor NK cells against NK-susceptible targets (eg, K562 cell line).
The aforementioned approaches rely on the development of donor hematopoietic stem cell–derived NK cells in the host. Adoptive therapy using mature NK cells from haploidentical donors has been attempted, but a significant antileukemia effect has been difficult to demonstrate [19-22].
In an attempt to explain the heterogeneous outcomes of studies documenting NK alloreactivity, in this review we present a brief update of NK cell biology, describing NK cell development, activation, receptor types, effector functions, and models of alloreactivity. We then summarize the current understanding of the role of NK cells in mediating and modulating engraftment as well as the graft-versus-host and graft-versus-tumor responses.
OVERVIEW OF NK CELL BIOLOGY
NK cells circulate in the PB and lymphogenous organs, licensed and ready to engage target cells. After a short engagement, they are able to kill unless stopped by an inhibitory signal. NK cells develop in the BM and migrate to the PB, spleen, lymph nodes, and other tissues (eg, lung, liver, or uterus). As a component of the innate immune system, NK cells play a role in surveillance against transformed and virally infected cells [23]. NK cells compose 2% to 18% of the mononuclear cells in human PB [23] and have a turnover rate of approximately 14 days [24]. In humans, NK cells have traditionally been defined as expressing CD56 with or without CD16, without expressing T cell markers (CD3, T cell receptors). Level of CD56 expression further subdivides human NK cells into 2 broad groups, the CD56dimCD16bright subset, which composes 90% of PB NK cells and has cytotoxic function, and the CD56brightCD16- subset, which cooperates with dendritic cells (DCs) and T cells in lymph nodes to secrete interferon (IFN)-γ and promote adaptive immune responses. The CD56dimCD16bright subset expresses major histocompatibility complex (MHC) class I allele–specific KIR, as well as the CXCR1 and CX3CR1 chemokine receptors, whereas the CD56brightCD16- subset expresses the CD94/NKG2A receptors along with lymphogenous organ homing markers, such as CCR7, CD62L, and CXCR3. Recent evidence suggests that the CD56bright differentiates into the CD56dim subset [25]. In the mouse, NK cells are defined as CD3- NK1.1+ or DX5+ cells, and mature NK cells are further subdivided into functionally disparate CD11b+CD27bright and CD11b+CD27dim subsets [26]. The CD11b+CD27bright subset is highly cytotoxic, localized in the lymph nodes, and interacts with dendritic cells, whereas the CD11b+CD27dim subset has a higher stimulatory threshold and is found in the spleen and PB. Recent evidence suggests that NKp46 may be a unifying marker of NK cells across both species [27].
NK Cell Development
NK cells arise from common lymphogenous progenitors in the BM in a process requiring signaling through the Flt3-, c-kit–, and gamma chain–associated receptors [28]. Further development, peripheral expansion, and survival depends on cytokine stimulation through the interleukin (IL)-2/IL-15 receptor [29,30]; an indirect effect through osteopontin in the microenvironment also has been postulated [31]. The transcription factors Ets-1 and PU.1 are important in early NK cell development, whereas Gata-2 and T-bet play a role in maturation, and CEPγ, MEF, and MITF are responsible for cytotoxicity and cytokine production in mature NK cells [28].
An NK cell's ability to respond to stimulation is related to the strength of the inhibitory signal received during its development, a process termed “licensing” or “education” [32,33]. Thus, NK cells without self-specific KIR are likely to be hyporesponsive, although evidence from mouse studies suggests that they still may retain the ability to react robustly when stimulated by cytokines or by a suitably potent antigen [34,35]. The acquisition of Ly49 receptors in mice and KIR in humans may be regulated by the cytokines IL-15 and IL-2, respectively [36,37].
NK Cell Activation
After infection or inflammation, NK cells are recruited to tissues under the control of the chemokine receptors CCR2, CCR5, CX3CR1, and CXCR3 [28]. Resting human PB or mouse splenic NK cells have poor cytotoxic potential and require activation, either by direct cell-to-cell contact and receptor recognition or by the action of cytokines. Type I interferon secretion by plasmacytoid and myelogenous DCs [38], DC trans-presentation of IL-15 [39], and CD4+ T cell production of IL-2 in the lymph node [40] activate NK cells, leading to translation of a preexisting pool of granzyme B and perforin mRNA [41] and secretion of these effector molecules, whereas IL-12 and IL-18 lead to increased IFN-γ secretion by NK cells [42]. In contrast, transforming growth factor-β secreted by regulatory T cells [43] is a negative regulator of NK homeostasis and induces down-regulation of natural cytotoxicity receptor (NCR) families [28,43].
NK Receptors
NK cells recognize their targets through inhibitory and activating cell surface receptors. Three main receptor families have been described: KIR, C-type lectin receptors (including NKG2A-E and Ly49 in mice), and NCR (see Table 2). The observation that NK cytotoxicity is triggered by tumor (and other) cells lacking expression of self MHC class I molecules first led to the “missing self” hypothesis and the discovery of the inhibitory receptors [44]; however, it is now apparent that it is the balance of signaling through the different receptors that leads to the final “decision” on NK cell reactivity [45].
Table 2.
Selected Human NK Cell Receptors
| Receptor/Gene | Target (Where Known) | Allele Frequency* |
|---|---|---|
| Inhibitory | ||
| KIR2DL2/3 | HLA-C group 1 (also recognize some HLA-C group 2) | 2DL2: 40%-60% 2DL3: 80%-95% |
| KIR2DL1 | HLA-C group 2 | 90%-100% |
| KIR3DL1 | HLA-A and B with Bw4 motifs at position 77-83 (but not HLA-B1301 or B1302) | 90%-95% |
| CD94/NKG2A | HLA-E | 100% |
| KIR3DL2 | HLA-A3/A11 | 100% |
| LAIR-1 | Collagen | |
| KIR2DL4 | HLA-G | 100% |
| Activating | ||
| KIR2DS4 | 85%-95% | |
| KIR2DS1 | HLA-C group 2 | 30%-50% |
| KIR2DS2 | 40%-50% | |
| KIR2DS3 | 0-30% | |
| KIR2DS5 | 20%-40% | |
| KIR3DS1 | 20%-40% | |
| NKG2D | MICA/B, ULBP | |
| CD94/NKG2C | HLA-E | |
| DNAM-1 (CD226) | CD112, CD155 | |
| NKp30 | BAT3 | |
| NKp44 | Viral hemagglutinin | |
| NKp46 (CD335) | Viral hemagglutinin | |
| 2B4 (CD244) | CD48 |
From http://www.allelefrequencies.net; accessed December 23, 2008.
The KIR are type I transmembrane molecules belonging to the immunoglobulin superfamily that are encoded on chromosome 19q13.4. They are expressed on γδ CD8 T cells as well as NK cells, and recognize amino acids in the carboxyl terminal domain of the MHC class I α1 helix in specific groups of HLA-A, -B, or C alleles. The specificity of KIR for HLA-C allotypes is determined by allelic dimorphism at residues 77 and 80 of the HLA-C molecule [46]. KIR genetics are of importance for transplantation, because significant diversity occurs at the population level both within and between ethnicities [47]. Furthermore, KIR genes segregate independently of the HLA genes (encoded on chromosome 6), and thus 2 HLA-matched individuals (even if related) may still be KIR-mismatched. An individual's HLA class I genotype dictates which KIR or NKG2A receptor combination occurs as inhibitory receptors on the surface of that person's NK cells, however. Thus, functionally mature NK cells express at least 1 inhibitory receptor for self-HLA, and occasionally as many as 3 or 4 such receptors. Some KIR are more often used as single receptors [48,49].
KIRs may be inhibitory or activating. Recognition of the MHC class I target by inhibitory KIRs lead to phosphorylation of an immunoreceptor tyrosine-based inhibitory motif in their cytoplasmic tail, followed by an inhibitory downstream signal. Activating KIR contain the same extracellular and transmembrane domains as the related inhibitory KIR, but lack cytoplasmic tails. Thus, target recognition by activating KIR leads to an interaction with an adapter protein and activation of alternative downstream signaling pathways [45]. The activating KIR bind MHC class I molecules more weakly than the respective inhibitory KIR [50,51], although the ligands for activating KIR are not well characterized, and it is possible that asyet unidentified non-HLA proteins serve as their true ligands [52,53]. Based on murine data, the inhibitory signal has been considered the dominant one [54], although this notion has been challenged by recent human data [55].
The mouse homologs of the human KIR are the Ly49 family of receptors, which are structurally distinct type II lectin–related homodimers. Like KIR, this family of receptors recognizes classical MHC class Ia molecules, such as H-2d and H-2k in mice, and also occurs in both immunoreceptor tyrosine-based inhibitory and stimulatory forms.
A particular NK cell in mice or humans may express anywhere from 0 to 4 Ly49 receptors or KIR, respectively. The expression of the inhibitory NKG2A receptor varies inversely with the number of KIR genes coexpressed on the cell surface, being highest in cells with no KIR expression [49,56]. Each KIR has a different affinity for its target, although some redundancy exists, such that a single KIR can recognize epitopes shared between different HLA alleles [57]; see Table 2. Further complexity is added by the fact that each MHC class I molecule may be recognized by both inhibitory and activating receptors on the same NK cell.
Inhibition is not solely MHC class I–mediated. The target of the CD94/NKG2A, B, or C receptor is the nonclassical MHC HLA-E molecule [58]; other inhibitory receptors include NKR-P1A and LAIR-1, which bind to LLT-1 and collagen, respectively [23,51].
The complement of KIR genes on one chromosome comprises a KIR haplotype. KIR haplotypes may vary in terms of gene number and gene content, [51]. Haplotype B is defined as encoding at least one of KIR2DL5, KIR2DS1, KIR 2DS2, KIR2DS3, KIR2DS5, or KIR3DS1, and haplotype A is defined as having none of these loci [59]. Homozygosity for haplotype A is seen in 25% to 30% of Caucasians and 80% of Japanese [59,60], whereas the remainder are heterozygous or homozygous for haplotype B and thus have combinations of activating and inhibitory KIR. Furthermore, KIR genotype correlates with phenotype in only about 75% of cases, because of allelic polymorphism and epigenetic silencing [61-64].
NK cells also may recognize MHC class I–negative targets using the NCR, including NKp30, NKp44, and NKp46, as well as NKG2D, CD16, and DNAM-1. Whereas NKp30 and NKp46 are constitutively expressed by all PB NK cells [65,66], NKp44 is up-regulated in IL-2–activated NK cells [67]. The ligands for the NCR in humans are not well characterized, but NKp30 may recognize BAT3, an intracellular ligand released in exosomes from tumor cells and DCs [68], and also may play a role in regulating DC lysis and maturation [69]. NKG2D plays a role in tumor immunosurveillance [70,71]; its ligands are rarely expressed by normal cells, but are up-regulated in response to cellular stress signals from transformation, viral infection, heat shock, and DNA damage. The ligands for NKG2D include MHC I–related genes A and B (MICA and MICB), as well as UL16-binding proteins (ULBPs) in humans and Rae-1 and H-60 in mice [72,73]. Another receptor with a recently identified role in immunosurveillance is DNAM-1, which recognizes the CD112 and CD155 in both mice and humans [74]. Human NK cells also express CD16, the FCγRIII receptor, which binds the Fc portion of IgG and thus mediates antibody-dependent cellular cytotoxicity [75].
NK Cell Effector Functions
On recognition, an immunologic synapse forms between the NK cell and its target, allowing direct cytotoxicity that is mediated through the perforin, granzyme, Fas/FasL, and TRAIL pathways [76-78], as well as by production of IFN-γ [23]. In addition, it has become clear that the original description of NK cells as “natural killers” spontaneously lysing transformed and virally infected cells encompasses only part of their effector mechanism; NK cells also promote DC maturation through tumor necrosis factor-α and IFN-γ secretion [79] and enhance Th1 polarization in secondary lymphogenous organs [80]. In addition to their viral or tumor targets, NK cells also may kill activated autologous CD4+ cells [81] and thus provide a link between the innate and adaptive immune responses [82,83], with a possible additional role in protecting the host from excessive immune response to pathogens [23].
ALGORITHMS OF NK CELL ALLOREACTIVITY
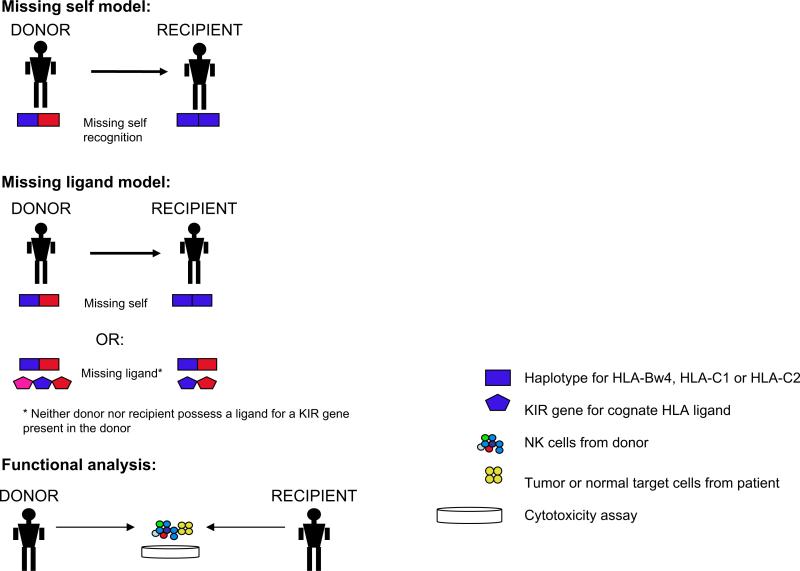
The 2 major models used to predict NK cell alloreactivity are the “missing self” and “missing ligand” models (see Figure 1). According to the “missing self” model, NK alloreactivity is stimulated when the recipient lacks one or more HLA class I alleles present in the donor [84,85]. This was the model used by the Perugia group to predict NK alloreactivity in their transplantation studies. According to the alternative “missing ligand” (also known as the “receptor-ligand”) model, NK cell alloreactivity also may occur in donor–recipient pairs matched for KIR and KIR ligands: when there is an extra KIR in the donor for which neither the donor nor the recipient has a ligand, the donor's potentially self-reactive NK cells are anergic in situ, but can trigger an alloreactive effect in the recipient. This finding is based on the observation that most individuals have 3 inhibitory KIR (for HLA-C1 and -C2 and for HLA-Bw4 alleles), but only 1 or 2 HLA KIR ligands on their own cells [47,64,86,87]. This latter model was found to be a better predictor of risk of leukemia relapse by some groups [47,86] but not by others [16], with the discrepancy likely related to differences in age, disease, and conditioning regimens.
Figure 1.
Models used to predict NK cell alloreactivity.
Functional analysis may resolve the differences between predictive models and in some cases reveal unexpected findings, as exemplified by recent data reported by several groups. Ruggeri et al. [16] performed functional analysis to identify and quantitate the frequency of alloreactive NK clones against HLA-C–mismatched targets. Alloreactive clones were present in all donors, at a frequency of 8% (± 6%) for HLA-C group 2 mismatches and a frequency of 5% (± 3%) for HLA-C group 1 mismatches. Alloreactive clones were present in only 2/3 of HLA-Bw4–mismatched donors, however. As predicted by their model, Ruggeri et al. [16] found no NK alloreactivity in donor–recipient pairs that were not KIR ligand–mismatched. Fauriat et al. [49] evaluated the frequency of the alloreactive repertoire in donors with the A haplotype (consisting only of inhibitory KIR and KIR2DS4) and found that the use of KIR and HLA genotyping alone to predict alloreactivity may significantly overestimate the size of the alloreactive repertoire. They found a wide variability (0 to 62%) in the alloreactivity of the total NK pool in donor–recipient pairs (all of which would have been predicted to be alloreactive by use of genotyping) depending on the extent of KIR ligand expression on theoretical recipients; the alloreactive subset was larger for recipients lacking more than one KIR ligand [49]. Finally, Foley et al. [88] demonstrated that some Bw4 alleles (HLA-B*1301 and HLA*1302) failed to protect targets from KIR3DL1-dependent lysis but, unexpectedly, HLA-A*2402 and HLA-A*3201 were protective against lysis; these results have implications for donor selection. Up to now, the 1/3 of individuals expressing all 3 KIR ligands (HLA-C1, HLA-C2, and HLA-Bw4) were thought to inhibit NK cells from all donors and thus be unable to benefit from NK alloreactivity [16]. Thus, the aforementioned findings, along with recent work suggesting that activating receptors may occasionally predominate, have important functional ramifications and suggest the need for caution when relying on genotypic predictive models alone.
ENGRAFTMENT AND IMMUNE RECONSTITUTION
Many studies have documented NK cells’ ability to mediate rejection of allogeneic BM in murine models, as initially described in the “hybrid resistance” model [89]. This phenomenon has been ascribed to classical “missing self” recognition, but activating receptors, such as NKG2D, may play a role as well [90].
In an extension of these observations, infusion of alloreactive NK cells (in the graft-versus-host direction) into haploidentical mice led to ablation by the NK cells of host hematopoiesis and antigen-presenting cells (14). This may explain the ability of alloreactive NK transplants to facilitate engraftment, as first described by the Perugia group in 2002. Interestingly, a recent update of this data no longer showed a significant impact of NK alloreactivity on rejection [16]. NK cells are relatively radioresistant [91], and the presence of host-versus-graft alloreactive NK cells may increase the risk of graft failure or incomplete chimerism [92,93].
NK cells are the first lymphogenous cells to repopulate after engraftment [94]. Early after HLA-matched HCT, NK cells are NKG2A+ and KIR-; reconstitution kinetics are variable, and acquisition of a donor-type KIR repertoire may take anywhere from 3 months to 3 years [86,95]. Alloreactive NK cells of donor origin were detectable from 1 to 3 months up until at least 12 months posttransplantation in some studies of haploidentical transplantation [16,55], although another study found that NK cells reconstituting in the haploidentical setting had an immature CD56bright KIR- NKG2A+ phenotype, and that even putatively alloreactive cells had poor effector function against primary leukemia cells [96]. Reconstitution is adversely affected in the setting of T cell–replete grafts [97,98], and peritransplantation immunosuppression affects NK cell subsets and function [99,100]. In contrast, several groups have reported an association between NK recovery after HLA-identical HCT and improved relapse-free and overall survival (RFS, OS) [101,102].
Increased incidences of infection and infection-related mortality have been noted in several studies [16,103,104]. Impaired immune function is likely related to the extensive TCD required to prevent GVHD, as well as to the contribution of an NK-mediated attack on host antigen-presenting cells. Finally, cytomegalovirus (CMV), a common pathogen in severely immunocompromised patients, has been shown to shape the NK receptor repertoire in healthy donors [105]. Conversely, donor KIR genotype was found to have an effect on CMV reactivation in HCT in some studies [106], but not in others [104].
ANTITUMOR EFFECT
NK cell–mediated rejection of tumor cells occurs through MHC class I–dependent and –independent mechanisms. In vitro cytotoxicity has been demonstrated against many tumor types, including AML and chronic myelogenous leukemia (CML), chronic lymphocytic leukemia (CLL), non-Hodgkin lymphoma (NHL), multiple myeloma (MM), T-cell acute lymphoblastic leukemia (ALL), melanoma, renal cell carcinoma (RCC), and neural tumors [107-109]. Preclinical models have convincingly demonstrated efficacy against human AML [14]. Tumor cells exhibit differential sensitivity to NK cytotoxicity because of variegated expression of inhibitory and activating receptors on NK cells [23,71], thus NK-mediated clearance of leukemia cells can be augmented by the blockade of inhibitory receptors [110,111].
Clinical data on NK cell antitumor efficacy is limited mainly to hematologic malignancies. The most impressive and frequently quoted data was published in 2002 and most recently updated in 2007. In total, 112 patients with high-risk AML, of whom 61 were in remission and 51 in relapse, were transplanted with HLA-haploidentical grafts from 51 NK alloreactive or 61 nonalloreactive related donors. In this case, NK alloreactivity was defined by the presence of KIR ligands in the donor, which were absent in the recipient, KIR gene for missing self recognition in the recipient, and alloreactive NK clones against recipient targets. Transplantation from NK alloreactive donors led to a remarkably low relapse rate in patients transplanted in remission (3% vs 47%), and to a superior EFS for patients whether transplanted in remission (67% vs 18%) or in relapse (34% vs 6%). Disease status and transplantation from an NK alloreactive donor were the only independent prognostic factors [16]. A lower relapse rate in patients transplanted in the setting of potential NK alloreactivity has been demonstrated by some groups, but not by others (see Table 1).
Although ALL cells are generally held to be less susceptible to NK cell mediated attack than AML [14,112], MLL-rearranged ALL cells have been shown to be susceptible to NK alloreactivity, with a sensitivity which is proportional to the extent of KIR-ligand mismatch [86]; this effect was also found in AML and myelodysplastic syndrome (MDS) [47]. Clearly, inhibitory KIR are not the only factor permitting antitumor NK alloreactivity. The presence of certain activating KIR genes in the donor may be associated with a lower relapse rate [113], and NKG2D plays a role in AML and CML [114,115].
Barriers to successful NK antitumor activity include tumor bulk and immunoevasion mechanisms [116,117]. Tumor escape from NK attack has been shown to occur through down-regulation on the tumor of activating receptor ligands, such as MICA/B, ULBP, CD112, CD155, and CD48 [112,115,118,119]. Direct contact with AML cells leads to down-regulation of NCR on the NK cells, correlating with worse survival [120]. NKG2D may be down-regulated after exposure to high levels of soluble tumor-derived MICA/MICB [121]. Finally, an NK cell–suppressive NK subset may be induced by tumor cells [122].
Attempts to counter tumor immunoevasion mechanisms by manipulation and enhancement of NK cell function can be achieved using cytokines or ligation or modulation of inhibitory or activating receptors, as has been reviewed recently [116,123]. Examples of this include blockade of the interaction between KIR2DL1/2/3 and HLA-C molecules as postremission therapy in patients with AML and MM [124], use of bispecific antibodies to direct effectors to lyse otherwise-refractory target cells [125], genetic modification of NK or T cells to express a chimeric NKG2D receptor [126], and down-regulation of myeloma MHC class I molecules using bortezomib [127].
EFFECTS ON GVHD
In a preclinical model, infusion of alloreactive NK cells into haplodentical mouse recipients as part of the conditioning regimen was protective against GVHD after a T cell–replete transplantation [14]. A rationale for this observation may be provided by NK-mediated ablation of host DCs [14], lysis of donor T cells [128-130], and the absence of activating NK receptor ligands on normal nonhematopoietic cells [131].
Clinical results vary, however. Although the initial report from the Perugia group suggested a favorable effect of NK alloreactivity on the incidence of acute GVHD (aGVHD), the updated results no longer showed a significant difference [16]. As can be seen in Table 1, most groups report no significant difference in GVHD rates, and in fact GVHD may be worsened by KIR mismatch or in the presence of some donor-activating KIR genes [21,93,132,133]. Because KIR mismatch correlated with HLA mismatch in some of these studies, T cell–induced GVHD clearly is a major confounder.
CONTROVERSIES AND CONCLUSIONS
Alloreactive NK cells in transplantation can have remarkably favorable effects on relapse and survival, as well as adverse outcomes related to relapse, infection, and GVHD. These discrepancies relate to differences in donor selection, conditioning regimens, extent of TCD, hematopoietic stem cell dose, disease state at transplantation, nature of disease, and algorithm of NK alloreactivity. In broad brushstrokes, it seems the most favorable conditions include a myeloablative conditioning regimen, maximal stem cell and minimal T cell doses, lack of interference with NK expansion by posttransplantation GVHD prophylaxis, and selection of patients with myelogenous disease in remission.
Selecting the most appropriate NK alloreactivity model is of vital importance. Preclinical and clinical results point variously to the “missing self ” or “missing ligand” model as the most predictive. NK cell receptor recognition is a complex and incompletely elucidated process, and tests of donor NK cell function against leukemia and patient target cells have sometimes revealed unexpected results [49,55,88]. Thus, functional analysis, including quantitation of the alloreactive educated NK cell pool, should be incorporated in real time into the donor selection process, as should KIR genotyping of the donor. Proper donor selection may be only part of the answer, however; perhaps “tumor selection”—an individualized assessment of tumor cell expression of NK receptor ligands or strategies to increase expression of ligands—could be incorporated into these models.
Hopefully, these major issues will be addressed with well-designed clinical trials. Nonetheless, several unanswered questions will require further biological input:
Is the observed reduction in relapse because of a persistence of NK immunosurveillance?
How are donor stem cell–derived NK cells educated in the recipient, and do hyporesponsive NK cells acquire effector function after transplantation?
What are the ideal conditions for in vivo persistence of NK cells after transplantation?
Should a particular NK cell subset be preferentially expanded, and if so, by what means?
Despite these unanswered questions, the use of alloreactive NK cells in transplantation continues to be an exciting example of the translation of basic biological principles to clinical medicine.
ACKNOWLEDGMENTS
Financial disclosure: The authors have nothing to disclose.
REFERENCES
- 1.Anasetti C, Amos D, Beatty PG, et al. Effect of HLA compatibility on engraftment of bone marrow transplants in patients with leukemia or lymphoma. N Engl J Med. 1989;320:197–204. doi: 10.1056/NEJM198901263200401. [DOI] [PubMed] [Google Scholar]
- 2.Beatty PG, Clift RA, Mickelson EM, et al. Marrow transplantation from related donors other than HLA-identical siblings. N Engl J Med. 1985;313:765–771. doi: 10.1056/NEJM198509263131301. [DOI] [PubMed] [Google Scholar]
- 3.Szydlo R, Goldman JM, Klein JP, et al. Results of allogeneic bone marrow transplants for leukemia using donors other than HLA-identical siblings. J Clin Oncol. 1997;15:1767–1777. doi: 10.1200/JCO.1997.15.5.1767. [DOI] [PubMed] [Google Scholar]
- 4.Kanda Y, Chiba S, Hirai H, et al. Allogeneic hematopoietic stem cell transplantation from family members other than HLA-identical siblings over the last decade (1991-2000). Blood. 2003;102:1541–1547. doi: 10.1182/blood-2003-02-0430. [DOI] [PubMed] [Google Scholar]
- 5.Koh LP, Rizzieri DA, Chao NJ. Allogeneic hematopoietic stem cell transplantation using mismatched/haploidentical donors. Biol Blood Marrow Transplant. 2007;13:1249–1267. doi: 10.1016/j.bbmt.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Aversa F, Tabilio A, Terenzi A, et al. Successful engraftment of T-cell–depleted haploidentical “three-loci” incompatible transplants in leukemia patients by addition of recombinant human granulocyte colony-stimulating factor–mobilized peripheral blood progenitor cells to bone marrow inoculum. Blood. 1994;84:3948–3955. [PubMed] [Google Scholar]
- 7.Bachar-Lustig E, Rachamim N, Li HW, et al. Megadose of T cell–depleted bone marrow overcomes MHC barriers in sublethally irradiated mice. Nat Med. 1995;1:1268–1273. doi: 10.1038/nm1295-1268. [DOI] [PubMed] [Google Scholar]
- 8.Soiffer RJ, Mauch P, Tarbell NJ, et al. Total lymphoid irradiation to prevent graft rejection in recipients of HLA non-identical T cell–depleted allogeneic marrow. Bone Marrow Transplant. 1991;7:23–33. [PubMed] [Google Scholar]
- 9.Aversa F, Tabilio A, Velardi A, et al. Treatment of high-risk acute leukemia with T-cell–depleted stem cells from related donors with one fully mismatched HLA haplotype. N Engl J Med. 1998;339:1186–1193. doi: 10.1056/NEJM199810223391702. [DOI] [PubMed] [Google Scholar]
- 10.Kato S, Yabe H, Yasui M, et al. Allogeneic hematopoietic transplantation of CD34+ selected cells from an HLA haploidentical related donor: a long-term follow-up of 135 patients and a comparison of stem cell source between the bone marrow and the peripheral blood. Bone Marrow Transplant. 2000;26:1281–1290. doi: 10.1038/sj.bmt.1702707. [DOI] [PubMed] [Google Scholar]
- 11.Ogawa H, Ikegame K, Yoshihara S, et al. Unmanipulated HLA 2-3 antigen–mismatched (haploidentical) stem cell transplantation using nonmyeloablative conditioning. Biol Blood Marrow Transplant. 2006;12:1073–1084. doi: 10.1016/j.bbmt.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Handgretinger R, Chen X, Pfeiffer M, et al. Feasibility and outcome of reduced-intensity conditioning in haploidentical transplantation. Ann N Y Acad Sci. 2007;1106:279–289. doi: 10.1196/annals.1392.022. [DOI] [PubMed] [Google Scholar]
- 13.Rizzieri DA, Koh LP, Long GD, et al. Partially matched, non-myeloablative allogeneic transplantation: clinical outcomes and immune reconstitution. J Clin Oncol. 2007;25:690–697. doi: 10.1200/JCO.2006.07.0953. [DOI] [PubMed] [Google Scholar]
- 14.Ruggeri L, Capanni M, Urbani E, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295:2097–2100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 15.Aversa F, Terenzi A, Tabilio A, et al. Full haplotype-mismatched hematopoietic stem-cell transplantation: a phase II study in patients with acute leukemia at high risk of relapse. J Clin Oncol. 2005;23:3447–3454. doi: 10.1200/JCO.2005.09.117. [DOI] [PubMed] [Google Scholar]
- 16.Ruggeri L, Mancusi A, Capanni M, et al. Donor natural killer cell allorecognition of missing self in haploidentical hematopoietic transplantation for acute myeloid leukemia: challenging its predictive value. Blood. 2007;110:433–440. doi: 10.1182/blood-2006-07-038687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunstein C, Wagner JE, Weisdorf D, et al. Negative effect of KIR alloreactivity in recipients of umbilical cord blood transplantation depends on transplantation conditioning intensity [abstract]. ASH annual meeting abstracts. 2008;112:152. doi: 10.1182/blood-2008-12-197467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stern M, Ruggeri L, Mancusi A, et al. Survival after T cell–depleted haploidentical stem cell transplantation is improved using the mother as donor. Blood. 2008;112:2990–2995. doi: 10.1182/blood-2008-01-135285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koehl U, Sorensen J, Esser R, et al. IL-2–activated NK cell immunotherapy of three children after haploidentical stem cell transplantation. Blood Cells Mol Dis. 2004;33:261–266. doi: 10.1016/j.bcmd.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Passweg JR, Tichelli A, Meyer-Monard S, et al. Purified donor NK-lymphocyte infusion to consolidate engraftment after haploidentical stem cell transplantation. Leukemia. 2004;18:1835–1838. doi: 10.1038/sj.leu.2403524. [DOI] [PubMed] [Google Scholar]
- 21.Cooley S, Gada P, McKenna D, et al. Successful haploidentical hematopoietic cell engraftment using a non-myeloablative preparative regimen including natural killer (NK) cells. ASH annual meeting abstracts. Blood. 2008;112:827. [Google Scholar]
- 22.Miller JS, Soignier Y, Panoskaltsis-Mortari A, et al. Successful adoptive transfer and in vivo expansion of human haploidentical NK cells in patients with cancer. Blood. 2005;105:3051–3057. doi: 10.1182/blood-2004-07-2974. [DOI] [PubMed] [Google Scholar]
- 23.Vivier E, Tomasello E, Baratin M, et al. Functions of natural killer cells. Nat Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Wallace DL, de Lara CM, et al. In vivo kinetics of human natural killer cells: the effects of ageing and acute and chronic viral infection. Immunology. 2007;121:258–265. doi: 10.1111/j.1365-2567.2007.02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan A, Hong DL, Atzberger A, et al. CD56bright human NK cells differentiate into CD56dim cells: role of contact with peripheral fibroblasts. J Immunol. 2007;179:89–94. doi: 10.4049/jimmunol.179.1.89. [DOI] [PubMed] [Google Scholar]
- 26.Hayakawa Y, Smyth MJ. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J Immunol. 2006;176:1517–1524. doi: 10.4049/jimmunol.176.3.1517. [DOI] [PubMed] [Google Scholar]
- 27.Walzer T, Blery M, Chaix J, et al. Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc Natl Acad Sci USA. 2007;104:3384–3389. doi: 10.1073/pnas.0609692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Santo JP. Natural killer cell developmental pathways: a question of balance. Ann Rev Immunol. 2006;24:257–286. doi: 10.1146/annurev.immunol.24.021605.090700. [DOI] [PubMed] [Google Scholar]
- 29.Cooper MA, Bush JE, Fehniger TA, et al. In vivo evidence for a dependence on interleukin 15 for survival of natural killer cells. Blood. 2002;100:3633–3638. doi: 10.1182/blood-2001-12-0293. [DOI] [PubMed] [Google Scholar]
- 30.Ranson T, Vosshenrich CA, Corcuff E, et al. IL-15 is an essential mediator of peripheral NK-cell homeostasis. Blood. 2003;101:4887–4893. doi: 10.1182/blood-2002-11-3392. [DOI] [PubMed] [Google Scholar]
- 31.Chung JW, Kim MS, Piao ZH, et al. Osteopontin promotes the development of natural killer cells from hematopoietic stem cells. Stem Cells. 2008;26:2114–2123. doi: 10.1634/stemcells.2008-0370. [DOI] [PubMed] [Google Scholar]
- 32.Kim S, Poursine-Laurent J, Truscott SM, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 33.Brodin P, Lakshmikanth T, Johansson S, et al. The strength of inhibitory input during education quantitatively tunes the functional responsiveness of individual natural killer cells. Blood. doi: 10.1182/blood-2008-05-156836. Prepublished online Oct 30, 2008; doi 10.1182/blood-2008-05-156836. [DOI] [PubMed] [Google Scholar]
- 34.Sun JC, Lanier LL. Cutting edge: viral infection breaks NK cell tolerance to “missing self.”. J Immunol. 2008;181:7453–7457. doi: 10.4049/jimmunol.181.11.7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortaldo JR, Young HA. Expression of IFN-gamma upon triggering of activating Ly49D NK receptors in vitro and in vivo: costimulation with IL-12 or IL-18 overrides inhibitory receptors. J Immunol. 2003;170:1763–1769. doi: 10.4049/jimmunol.170.4.1763. [DOI] [PubMed] [Google Scholar]
- 36.Lian RH, Chin RK, Nemeth HE, et al. A role for lymphotoxin in the acquisition of Ly49 receptors during NK cell development. Eur J Immunol. 2004;34:2699–2707. doi: 10.1002/eji.200425394. [DOI] [PubMed] [Google Scholar]
- 37.Ferlazzo G, Munz C. NK cell compartments and their activation by dendritic cells. J Immunol. 2004;172:1333–1339. doi: 10.4049/jimmunol.172.3.1333. [DOI] [PubMed] [Google Scholar]
- 38.Strowig T, Brilot F, Munz C. Noncytotoxic functions of NK cells: direct pathogen restriction and assistance to adaptive immunity. J Immunol. 2008;180:7785–7791. doi: 10.4049/jimmunol.180.12.7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lucas M, Schachterle W, Oberle K, et al. Dendritic cells prime natural killer cells by trans-presenting interleukin 15. Immunity. 2007;26:503–517. doi: 10.1016/j.immuni.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fehniger TA, Cooper MA, Nuovo GJ, et al. CD56bright natural killer cells are present in human lymph nodes and are activated by T cell–derived IL-2: a potential new link between adaptive and innate immunity. Blood. 2003;101:3052–3057. doi: 10.1182/blood-2002-09-2876. [DOI] [PubMed] [Google Scholar]
- 41.Fehniger TA, Cai SF, Cao X, et al. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity. 2007;26:798–811. doi: 10.1016/j.immuni.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 42.Orange JS, Biron CA. Characterization of early IL-12, IFN-alphabeta, and TNF effects on antiviral state and NK cell responses during murine cytomegalovirus infection. J Immunol. 1996;156:4746–4756. [PubMed] [Google Scholar]
- 43.Smyth MJ, Teng MW, Swann J, et al. CD4+CD25+ T regulatory cells suppress NK cell–mediated immunotherapy of cancer. J Immunol. 2006;176:1582–1587. doi: 10.4049/jimmunol.176.3.1582. [DOI] [PubMed] [Google Scholar]
- 44.Karre K, Ljunggren HG, Piontek G, et al. Selective rejection of H-2–deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 45.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colonna M, Brooks EG, Falco M, et al. Generation of allospecific natural killer cells by stimulation across a polymorphism of HLA-C. Science. 1993;260:1121–1124. doi: 10.1126/science.8493555. [DOI] [PubMed] [Google Scholar]
- 47.Hsu KC, Keever-Taylor CA, Wilton A, et al. Improved outcome in HLA-identical sibling hematopoietic stem-cell transplantation for acute myelogenous leukemia predicted by KIR and HLA genotypes. Blood. 2005;105:4878–4884. doi: 10.1182/blood-2004-12-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parham P. Taking license with natural killer cell maturation and repertoire development. Immunol Rev. 2006;214:155–160. doi: 10.1111/j.1600-065X.2006.00462.x. [DOI] [PubMed] [Google Scholar]
- 49.Fauriat C, Andersson S, Bjorklund AT, et al. Estimation of the size of the alloreactive NK cell repertoire: studies in individuals homozygous for the group A KIR haplotype. J Immunol. 2008;181:6010–6019. doi: 10.4049/jimmunol.181.9.6010. [DOI] [PubMed] [Google Scholar]
- 50.Vales-Gomez M, Reyburn HT, Erskine RA, et al. Differential binding to HLA-C of p50-activating and p58-inhibitory natural killer cell receptors. Proc Natl Acad Sci USA. 1998;95:14326–14331. doi: 10.1073/pnas.95.24.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrow AD, Trowsdale J. The extended human leukocyte receptor complex: diverse ways of modulating immune responses. Immunol Rev. 2008;224:98–123. doi: 10.1111/j.1600-065X.2008.00653.x. [DOI] [PubMed] [Google Scholar]
- 52.Smith HR, Heusel JW, Mehta IK, et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci USA. 2002;99:8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arase H, Mocarski ES, Campbell AE, et al. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 54.Ortaldo JR, Winkler-Pickett R, Willette-Brown J, et al. Structure/function relationship of activating Ly-49D and inhibitory Ly-49G2 NK receptors. J Immunol. 1999;163:5269–5277. [PubMed] [Google Scholar]
- 55.Pende D, Marcenaro S, Falco M, et al. Anti-leukemia activity of alloreactive NK cells in KIR ligand-mismatched haploidentical HSCT for pediatric patients: evaluation of the functional role of activating KIR and re-definition of inhibitory KIR specificity. Blood. doi: 10.1182/blood-2008-06-164103. Prepublished online Oct 22, 2008; doi:10.1182/blood-2008-06-164103. [DOI] [PubMed] [Google Scholar]
- 56.Yawata M, Yawata N, Draghi M, et al. MHC class I–specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Farag SS, Fehniger TA, Ruggeri L, et al. Natural killer cell receptors: new biology and insights into the graft-versus-leukemia effect. Blood. 2002;100:1935–1947. doi: 10.1182/blood-2002-02-0350. [DOI] [PubMed] [Google Scholar]
- 58.Braud VM, Allan DS, O'Callaghan CA, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 59.Cooley S, Trachtenberg E, Bergemann TL, et al. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood. 2008;113:726–732. doi: 10.1182/blood-2008-07-171926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yawata M, Yawata N, McQueen KL, et al. Predominance of group A KIR haplotypes in Japanese associated with diverse NK cell repertoires of KIR expression. Immunogenetics. 2002;54:543–550. doi: 10.1007/s00251-002-0497-x. [DOI] [PubMed] [Google Scholar]
- 61.Santourlidis S, Graffmann N, Christ J, et al. Lineage-specific transition of histone signatures in the killer cell Ig-like receptor locus from hematopoietic progenitor to NK cells. J Immunol. 2008;180:418–425. doi: 10.4049/jimmunol.180.1.418. [DOI] [PubMed] [Google Scholar]
- 62.Santourlidis S, Trompeter HI, Weinhold S, et al. Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J Immunol. 2002;169:4253–4261. doi: 10.4049/jimmunol.169.8.4253. [DOI] [PubMed] [Google Scholar]
- 63.Chan HW, Miller JS, Moore MB, et al. Epigenetic control of highly homologous killer Ig-like receptor gene alleles. J Immunol. 2005;175:5966–5974. doi: 10.4049/jimmunol.175.9.5966. [DOI] [PubMed] [Google Scholar]
- 64.Leung W, Iyengar R, Triplett B, et al. Comparison of killer Ig-like receptor genotyping and phenotyping for selection of allogeneic blood stem cell donors. J Immunol. 2005;174:6540–6545. doi: 10.4049/jimmunol.174.10.6540. [DOI] [PubMed] [Google Scholar]
- 65.Sivori S, Vitale M, Morelli L, et al. p46, a novel natural killer cell–specific surface molecule that mediates cell activation. J Exp Med. 1997;186:1129–1136. doi: 10.1084/jem.186.7.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pende D, Parolini S, Pessino A, et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J Exp Med. 1999;190:1505–1516. doi: 10.1084/jem.190.10.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vitale M, Bottino C, Sivori S, et al. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non–major histocompatibility complex–restricted tumor cell lysis. J Exp Med. 1998;187:2065–2072. doi: 10.1084/jem.187.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simhadri VR, Reiners KS, Hansen HP, et al. Dendritic cells release HLA-B–associated transcript-3–positive exosomes to regulate natural killer function. PLoS ONE. 2008;3:e3377. doi: 10.1371/journal.pone.0003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferlazzo G, Tsang ML, Moretta L, et al. Human dendritic cells activate resting natural killer (NK) cells and are recognized via the NKp30 receptor by activated NK cells. J Exp Med. 2002;195:343–351. doi: 10.1084/jem.20011149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guerra N, Tan YX, Joncker NT, et al. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 2008;28:571–580. doi: 10.1016/j.immuni.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Diefenbach A, Jensen ER, Jamieson AM, et al. Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature. 2001;413:165–171. doi: 10.1038/35093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Steinle A, Li P, Morris DL, et al. Interactions of human NKG2D with its ligands MICA, MICB, and homologs of the mouse RAE-1 protein family. Immunogenetics. 2001;53:279–287. doi: 10.1007/s002510100325. [DOI] [PubMed] [Google Scholar]
- 73.Cosman D, Mullberg J, Sutherland CL, et al. ULBPs, novel MHC class I–related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14:123–133. doi: 10.1016/s1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- 74.Iguchi-Manaka A, Kai H, Yamashita Y, et al. Accelerated tumor growth in mice deficient in DNAM-1 receptor. J Exp Med. 2008;205:2959–2964. doi: 10.1084/jem.20081611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lanier LL, Ruitenberg JJ, Phillips JH. Functional and biochemical analysis of CD16 antigen on natural killer cells and granulocytes. J Immunol. 1988;141:3478–3485. [PubMed] [Google Scholar]
- 76.Screpanti V, Wallin RP, Ljunggren HG, et al. A central role for death receptor–mediated apoptosis in the rejection of tumors by NK cells. J Immunol. 2001;167:2068–2073. doi: 10.4049/jimmunol.167.4.2068. [DOI] [PubMed] [Google Scholar]
- 77.Trapani JA, Smyth MJ. Killing by cytotoxic T cells and natural killer cells: multiple granule serine proteases as initiators of DNA fragmentation. Immunol Cell Biol. 1993;71(Pt 3):201–208. doi: 10.1038/icb.1993.22. [DOI] [PubMed] [Google Scholar]
- 78.Smyth MJ, Takeda K, Hayakawa Y, et al. Nature's TRAIL: on a path to cancer immunotherapy. Immunity. 2003;18:1–6. doi: 10.1016/s1074-7613(02)00502-2. [DOI] [PubMed] [Google Scholar]
- 79.Gerosa F, Baldani-Guerra B, Nisii C, et al. Reciprocal activating interaction between natural killer cells and dendritic cells. J Exp Med. 2002;195:327–333. doi: 10.1084/jem.20010938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martin-Fontecha A, Thomsen LL, Brett S, et al. Induced recruitment of NK cells to lymph nodes provides IFN-gamma for T(H)1 priming. Nat Immunol. 2004;5:1260–1265. doi: 10.1038/ni1138. [DOI] [PubMed] [Google Scholar]
- 81.Lu L, Ikizawa K, Hu D, et al. Regulation of activated CD4+ T cells by NK cells via the Qa-1-NKG2A inhibitory pathway. Immunity. 2007;26:593–604. doi: 10.1016/j.immuni.2007.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raulet DH. Interplay of natural killer cells and their receptors with the adaptive immune response. Nat Immunol. 2004;5:996–1002. doi: 10.1038/ni1114. [DOI] [PubMed] [Google Scholar]
- 83.Yoshida N, Ino K, Ishida Y, et al. Overexpression of indoleamine 2,3-dioxygenase in human endometrial carcinoma cells induces rapid tumor growth in a mouse xenograft model. Clin Cancer Res. 2008;14:7251–7259. doi: 10.1158/1078-0432.CCR-08-0991. [DOI] [PubMed] [Google Scholar]
- 84.Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 85.Karre K. Immunology. A perfect mismatch. Science. 2002;295:2029–2031. doi: 10.1126/science.1070538. [DOI] [PubMed] [Google Scholar]
- 86.Leung W, Iyengar R, Turner V, et al. Determinants of antileukemia effects of allogeneic NK cells. J Immunol. 2004;172:644–650. doi: 10.4049/jimmunol.172.1.644. [DOI] [PubMed] [Google Scholar]
- 87.Hsu KC, Gooley T, Malkki M, et al. KIR ligands and prediction of relapse after unrelated donor hematopoietic cell transplantation for hematologic malignancy. Biol Blood Marrow Transplant. 2006;12:828–836. doi: 10.1016/j.bbmt.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 88.Foley BA, De Santis D, Van Beelen E, et al. The reactivity of Bw41 HLA-B and HLA-A alleles with KIR3DL1: implications for patient and donor suitability for haploidentical stem cell transplantations. Blood. 2008;112:435–443. doi: 10.1182/blood-2008-01-132902. [DOI] [PubMed] [Google Scholar]
- 89.Barao I, Murphy WJ. The immunobiology of natural killer cells and bone marrow allograft rejection. Biol Blood Marrow Transplant. 2003;9:727–741. doi: 10.1016/j.bbmt.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 90.Ogasawara K, Benjamin J, Takaki R, et al. Function of NKG2D in natural killer cell–mediated rejection of mouse bone marrow grafts. Nat Immunol. 2005;6:938–945. doi: 10.1038/ni1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Clave E, Socie G, Cosset JM, et al. Multicolor flow cytometry analysis of blood cell subsets in patients given total body irradiation before bone marrow transplantation. Int J Radiat Oncol Biol Phys. 1995;33:881–886. doi: 10.1016/0360-3016(95)00213-6. [DOI] [PubMed] [Google Scholar]
- 92.Beelen DW, Ottinger HD, Ferencik S, et al. Genotypic inhibitory killer immunoglobulin-like receptor ligand incompatibility enhances the long-term antileukemic effect of unmodified allogeneic hematopoietic stem cell transplantation in patients with myeloid leukemias. Blood. 2005;105:2594–2600. doi: 10.1182/blood-2004-04-1441. [DOI] [PubMed] [Google Scholar]
- 93.De Santis D, Bishara A, Witt CS, et al. Natural killer cell HLA-C epitopes and killer cell immunoglobulin-like receptors both influence outcome of mismatched unrelated donor bone marrow transplants. Tissue Antigens. 2005;65:519–528. doi: 10.1111/j.1399-0039.2005.00396.x. [DOI] [PubMed] [Google Scholar]
- 94.Small TN, Papadopoulos EB, Boulad F, et al. Comparison of immune reconstitution after unrelated and related T-cell–depleted bone marrow transplantation: effect of patient age and donor leukocyte infusions. Blood. 1999;93:467–480. [PubMed] [Google Scholar]
- 95.Shilling HG, McQueen KL, Cheng NW, et al. Reconstitution of NK cell receptor repertoire following HLA-matched hematopoietic cell transplantation. Blood. 2003;101:3730–3740. doi: 10.1182/blood-2002-08-2568. [DOI] [PubMed] [Google Scholar]
- 96.Nguyen S, Dhedin N, Vernant JP, et al. NK-cell reconstitution after haploidentical hematopoietic stem-cell transplantations: immaturity of NK cells and inhibitory effect of NKG2A override GvL effect. Blood. 2005;105:4135–4142. doi: 10.1182/blood-2004-10-4113. [DOI] [PubMed] [Google Scholar]
- 97.Giebel S, Locatelli F, Lamparelli T, et al. Survival advantage with KIR ligand incompatibility in hematopoietic stem cell transplantation from unrelated donors. Blood. 2003;102:814–819. doi: 10.1182/blood-2003-01-0091. [DOI] [PubMed] [Google Scholar]
- 98.Cooley S, McCullar V, Wangen R, et al. KIR reconstitution is altered by T cells in the graft and correlates with clinical outcomes after unrelated donor transplantation. Blood. 2005;106:4370–4376. doi: 10.1182/blood-2005-04-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bornhauser M, Schwerdtfeger R, Martin H, et al. Role of KIR ligand incompatibility in hematopoietic stem cell transplantation using unrelated donors. Blood. 2004;103:2860–2862. doi: 10.1182/blood-2003-11-3893. [DOI] [PubMed] [Google Scholar]
- 100.Wang H, Grzywacz B, Sukovich D, et al. The unexpected effect of cyclosporin A on CD56+CD16- and CD56+CD16+ natural killer cell subpopulations. Blood. 2007;110:1530–1539. doi: 10.1182/blood-2006-10-048173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dunbar EM, Buzzeo MP, Levine JB, et al. The relationship between circulating natural killer cells after reduced-intensity conditioning hematopoietic stem cell transplantation and relapse-free survival and graft-versus-host disease. Haematologica. 2008;93:1852–1858. doi: 10.3324/haematol.13033. [DOI] [PubMed] [Google Scholar]
- 102.Savani BN, Mielke S, Adams S, et al. Rapid natural killer cell recovery determines outcome after T-cell–depleted HLA-identical stem cell transplantation in patients with myeloid leukemias but not with acute lymphoblastic leukemia. Leukemia. 2007;21:2145–2152. doi: 10.1038/sj.leu.2404892. [DOI] [PubMed] [Google Scholar]
- 103.Schaffer M, Malmberg KJ, Ringden O, et al. Increased infection-related mortality in KIR ligand–mismatched unrelated allogeneic hematopoietic stem-cell transplantation. Transplantation. 2004;78:1081–1085. doi: 10.1097/01.tp.0000137103.19717.86. [DOI] [PubMed] [Google Scholar]
- 104.Kroger N, Binder T, Zabelina T, et al. Low number of donor activating killer immunoglobulin-like receptors (KIR) genes but not KIR-ligand mismatch prevents relapse and improves disease-free survival in leukemia patients after in vivo T-cell–depleted unrelated stem cell transplantation. Transplantation. 2006;82:1024–1030. doi: 10.1097/01.tp.0000235859.24513.43. [DOI] [PubMed] [Google Scholar]
- 105.Guma M, Angulo A, Vilches C, et al. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 2004;104:3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- 106.Cook M, Briggs D, Craddock C, et al. Donor KIR genotype has a major influence on the rate of cytomegalovirus reactivation following T-cell–replete stem cell transplantation. Blood. 2006;107:1230–1232. doi: 10.1182/blood-2005-03-1039. [DOI] [PubMed] [Google Scholar]
- 107.Ruggeri L, Mancusi A, Burchielli E, et al. NK cell alloreactivity and allogeneic hematopoietic stem cell transplantation. Blood Cells Mol Dis. 2008;40:84–90. doi: 10.1016/j.bcmd.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 108.Sivori S, Parolini S, Marcenaro E, et al. Involvement of natural cytotoxicity receptors in human natural killer cell–mediated lysis of neuroblastoma and glioblastoma cell lines. J Neuroimmunol. 2000;107:220–225. doi: 10.1016/s0165-5728(00)00221-6. [DOI] [PubMed] [Google Scholar]
- 109.Igarashi T, Wynberg J, Srinivasan R, et al. Enhanced cytotoxicity of allogeneic NK cells with killer immunoglobulin-like receptor ligand incompatibility against melanoma and renal cell carcinoma cells. Blood. 2004;104:170–177. doi: 10.1182/blood-2003-12-4438. [DOI] [PubMed] [Google Scholar]
- 110.Koh CY, Blazar BR, George T, et al. Augmentation of antitumor effects by NK cell inhibitory receptor blockade in vitro and in vivo. Blood. 2001;97:3132–3137. doi: 10.1182/blood.v97.10.3132. [DOI] [PubMed] [Google Scholar]
- 111.Koh CY, Raziuddin A, Welniak LA, et al. NK inhibitory-receptor blockade for purging of leukemia: effects on hematopoietic reconstitution. Biol Blood Marrow Transplant. 2002;8:17–25. doi: 10.1053/bbmt.2002.v8.pm11846352. [DOI] [PubMed] [Google Scholar]
- 112.Pende D, Spaggiari GM, Marcenaro S, et al. Analysis of the receptor–ligand interactions in the natural killer–mediated lysis of freshly isolated myeloid or lymphoblastic leukemias: evidence for the involvement of the poliovirus receptor (CD155) and Nectin-2 (CD112). Blood. 2005;105:2066–2073. doi: 10.1182/blood-2004-09-3548. [DOI] [PubMed] [Google Scholar]
- 113.Verheyden S, Schots R, Duquet W, et al. A defined donor-activating natural killer cell receptor genotype protects against leukemic relapse after related HLA-identical hematopoietic stem cell transplantation. Leukemia. 2005;19:1446–1451. doi: 10.1038/sj.leu.2403839. [DOI] [PubMed] [Google Scholar]
- 114.Sconocchia G, Lau M, Provenzano M, et al. The antileukemia effect of HLA-matched NK and NK-T cells in chronic myelogenous leukemia involves NKG2D–target cell interactions. Blood. 2005;106:3666–3672. doi: 10.1182/blood-2005-02-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Verheyden S, Demanet C. NK cell receptors and their ligands in leukemia. Leukemia. 2008;22:249–257. doi: 10.1038/sj.leu.2405040. [DOI] [PubMed] [Google Scholar]
- 116.Ljunggren HG, Malmberg KJ. Prospects for the use of NK cells in immunotherapy of human cancer. Nat Rev Immunol. 2007;7:329–339. doi: 10.1038/nri2073. [DOI] [PubMed] [Google Scholar]
- 117.Albertsson PA, Basse PH, Hokland M, et al. NK cells and the tumour microenvironment: implications for NK-cell function and anti-tumour activity. Trends Immunol. 2003;24:603–609. doi: 10.1016/j.it.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 118.Romanski A, Bug G, Becker S, et al. Mechanisms of resistance to natural killer cell–mediated cytotoxicity in acute lymphoblastic leukemia. Exp Hematol. 2005;33:344–352. doi: 10.1016/j.exphem.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 119.Nowbakht P, Ionescu MC, Rohner A, et al. Ligands for natural killer cell–activating receptors are expressed upon the maturation of normal myelomonocytic cells but at low levels in acute myeloid leukemias. Blood. 2005;105:3615–3622. doi: 10.1182/blood-2004-07-2585. [DOI] [PubMed] [Google Scholar]
- 120.Fauriat C, Just-Landi S, Mallet F, et al. Deficient expression of NCR in NK cells from acute myeloid leukemia: evolution during leukemia treatment and impact of leukemia cells in NCRdull phenotype induction. Blood. 2007;109:323–330. doi: 10.1182/blood-2005-08-027979. [DOI] [PubMed] [Google Scholar]
- 121.Groh V, Wu J, Yee C, et al. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. 2002;419:734–738. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- 122.Ullrich E, Terme M, Aymeric L, et al. IL-18–elicited suppressor NK cells with immunoregulatory functions [abstract]. ASH annual meeting abstracts. 2008;106:112. [Google Scholar]
- 123.Terme M, Ullrich E, Delahaye NF, et al. Natural killer cell–directed therapies: moving from unexpected results to successful strategies. Nat Immunol. 2008;9:486–494. doi: 10.1038/ni1580. [DOI] [PubMed] [Google Scholar]
- 124.Wagtmann N, Andre P, Zahn S, et al. Anti-KIR (1-7F9): a fully human monoclonal antibody (mAb) that blocks KIR2DL1, 2, and 3, promoting natural killer (NK) cell–mediated lysis of tumor cells in vitro and in vivo. ASH annual meeting abstracts. Blood. 2007;110:582. [Google Scholar]
- 125.Shahied LS, Tang Y, Alpaugh RK, et al. Bispecific minibodies targeting HER2/neu and CD16 exhibit improved tumor lysis when placed in a divalent tumor antigen–binding format. J Biol Chem. 2004;279:53907–53914. doi: 10.1074/jbc.M407888200. [DOI] [PubMed] [Google Scholar]
- 126.Zhang T, Barber A, Sentman CL. Generation of antitumor responses by genetic modification of primary human T cells with a chimeric NKG2D receptor. Cancer Res. 2006;66:5927–5933. doi: 10.1158/0008-5472.CAN-06-0130. [DOI] [PubMed] [Google Scholar]
- 127.Shi J, Tricot GJ, Garg TK, et al. Bortezomib down-regulates the cell-surface expression of HLA class I and enhances natural killer cell–mediated lysis of myeloma. Blood. 2008;111:1309–1317. doi: 10.1182/blood-2007-03-078535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Olson JA, Leveson-Gower DB, Baker J, et al. NK cells suppress GVHD by directly inhibiting activated alloreactive T cells through an NKG2D-mediated mechanism [abstract]. ASH annual meeting abstracts. 2008;112:61. [Google Scholar]
- 129.Rabinovich BA, Li J, Shannon J, et al. Activated, but not resting, T cells can be recognized and killed by syngeneic NK cells. J Immunol. 2003;170:3572–3576. doi: 10.4049/jimmunol.170.7.3572. [DOI] [PubMed] [Google Scholar]
- 130.Cerboni C, Zingoni A, Cippitelli M, et al. Antigen-activated human T lymphocytes express cell-surface NKG2D ligands via an ATM/ATR-dependent mechanism and become susceptible to autologous NK-cell lysis. Blood. 2007;110:606–615. doi: 10.1182/blood-2006-10-052720. [DOI] [PubMed] [Google Scholar]
- 131.Bottino C, Castriconi R, Moretta L, et al. Cellular ligands of activating NK receptors. Trends Immunol. 2005;26:221–226. doi: 10.1016/j.it.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 132.Yabe T, Matsuo K, Hirayasu K, et al. Donor killer immunoglobulin-like receptor (KIR) genotype–patient cognate KIR ligand combination and antithymocyte globulin preadministration are critical factors in outcome of HLA-C-KIR ligand–mismatched T cell–replete unrelated bone marrow transplantation. Biol Blood Marrow Transplant. 2008;14:75–87. doi: 10.1016/j.bbmt.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 133.Lima MD, Rodrigues M, Cano P, et al. Donor–recipient mismatches in MHC class I chain-related gene A (MICA) in unrelated donor (UD) transplantation [abstract]. ASH annual meeting abstracts. 2008;112:58. [Google Scholar]
- 134.Lang P, Greil J, Bader P, et al. Long-term outcome after haploidentical stem cell transplantation in children. Blood Cells Mol Dis. 2004;33:281–287. doi: 10.1016/j.bcmd.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 135.Cook MA, Milligan DW, Fegan CD, et al. The impact of donor KIR and patient HLA-C genotypes on outcome following HLA-identical sibling hematopoietic stem cell transplantation for myeloid leukemia. Blood. 2004;103:1521–1526. doi: 10.1182/blood-2003-02-0438. [DOI] [PubMed] [Google Scholar]
- 136.Giebel S, Locatelli F, Lamparelli T, et al. Survival advantage with KIR ligand incompatibility in hematopoietic stem cell transplantation from unrelated donors. Blood. 2003;102:814–819. doi: 10.1182/blood-2003-01-0091. [DOI] [PubMed] [Google Scholar]
- 137.Davies SM, Ruggieri L, DeFor T, et al. Evaluation of KIR ligand incompatibility in mismatched unrelated donor hematopoietic transplants. Blood. 2002;100:3825–3827. doi: 10.1182/blood-2002-04-1197. [DOI] [PubMed] [Google Scholar]
- 138.Farag SS, Bacigalupo A, Eapen M, et al. The effect of KIR ligand incompatibility on the outcome of unrelated donor transplantation: a report from the center for international blood and marrow transplant research, the European blood and marrow transplant registry, and the Dutch registry. Biol Blood Marrow Transplant. 2006;12:876–884. doi: 10.1016/j.bbmt.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 139.Miller JS, Cooley S, Parham P, et al. Missing KIR ligands are associated with less relapse and increased graft-versus-host disease (GVHD) following unrelated donor allogeneic HCT. Blood. 2007;109:5058–5061. doi: 10.1182/blood-2007-01-065383. [DOI] [PMC free article] [PubMed] [Google Scholar]