Abstract
Objective
To examine the association between renin-angiotensin-aldosterone system (RAAS) genes and salt-sensitivity of blood pressure (BP).
Methods
A 7-day low-sodium followed by a 7-day high-sodium dietary intervention was conducted among 1,906 participants living in a rural region of north China where habitual sodium-intake is high. BP measurements were obtained at baseline and following each intervention using a random-zero sphygmomanometer.
Results
Diastolic BP (DBP) and mean arterial pressure (MAP) responses increased with the number of rs4524238 A alleles in the angiotensin II receptor, type 1 (AGTR1) gene. For example, mean DBP responses (95% CI) among those with genotypes G/G, G/A, and A/A were −2.53 (−2.89, −2.18), −3.49 (−4.13, −2.86), and −5.78 (−9.51, −2.06) mmHg, respectively, following the low-sodium intervention (p=0.0008). Carriers of the rare A allele of rs5479 in the hydroxysteroid (11-beta) dehydrogenase 2 (HSD11B2) gene had decreased DBP responses to low-sodium (p-value=0.00004). Those with the C/A and C/C genotypes had DBP responses of −0.70 (−6.62, 5.22) and −2.71 (−4.88, −0.54) mmHg, respectively. X-chromosome renin-binding protein (RENBP) gene markers rs1557501 and rs2269372 were associated with systolic BP (SBP) response to low-sodium in men (p=0.00004 and 0.0001, respectively). SBP responses (95% CI) were −6.13 (−6.68, −5.58) versus −4.07 (−4.88, −3.26) and −6.04 (−6.57, −5.52) versus −3.94 (−4.90, −2.99) mmHg among men with major versus those with minor alleles of rs1557501 and rs2269372, respectively. Haplotype analyses of these genes supported our single marker findings.
Conclusions
We identified RAAS variants that were predictive of salt-sensitivity in a Han population with habitually high-sodium intake.
Keywords: blood pressure, genetics, polymorphism, dietary sodium, salt sensitivity, renin-angiotensin-aldosterone system
INTRODUCTION
Salt-sensitivity of blood pressure (BP) has been associated with an increased risk of hypertension, cardiovascular disease (CVD) and premature death [1,2]. Examining the genetic determinants of salt-sensitivity will enhance our understanding of the mechanisms underlying hypertension and thus facilitate the development of targeted prevention and treatment strategies for reducing BP and the related CVD burden. High salt-sensitivity is a phenomenon that can be described as an increased BP response to sodium consumption and is heterogeneous within populations. Previous studies have suggested that an individual’s genetic profile may play a large role in their BP responses to salt intake [3–7]. For example, family studies, conducted by us and others, have documented a moderately high heritability of salt-sensitivity [3–5]. Likewise, candidate gene studies have identified a significant association between genetic variants and salt-sensitivity [6,7].
The renin-angiotensin-aldosterone system (RAAS) plays a central role in BP regulation by maintaining sodium and water homeostasis and vascular tone [8]. Low-renin hypertension is associated with an increased BP response to high salt intake, and salt-sensitive individuals exhibit a blunted response of the RAAS when switching from low to high salt intake, compared to that of salt-resistant subjects [9,10]. In addition, a few polymorphisms in the RAAS candidate genes have been associated with salt-sensitive hypertension [7,11,12]. Most previous studies, however, have had relatively small sample sizes and have exhibited inconsistent findings as well as substantial variation in genetic and phenotypic measurements [7].
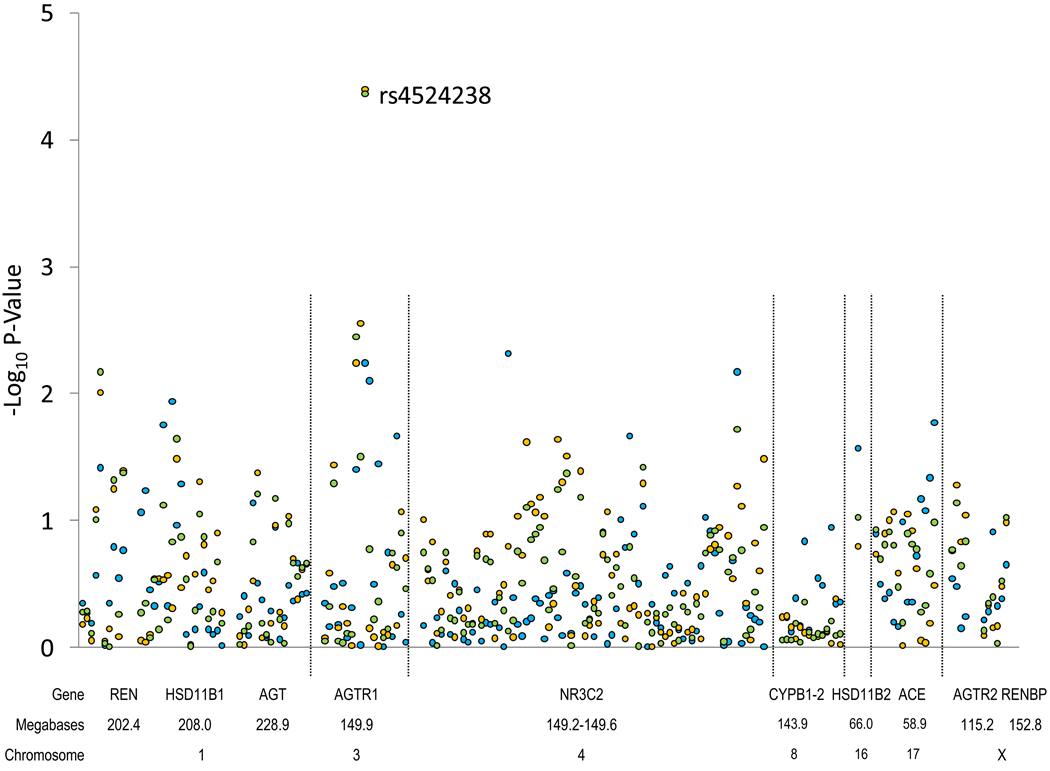
The current study was conducted in a large, homogeneous sample of Han Chinese families who took part in the Genetic Epidemiology Network of Salt Sensitivity (GenSalt). The objective of this analysis was to examine the association between RAAS candidate genes [renin (REN); hydroxysteroid (11-beta) dehydrogenase 1 (HSD11B1); angiotensinogen (AGT); angiotensin II type 1 receptor (AGTR1); nuclear receptor subfamily 3, group C, member 2 (NR3C2); cytochrome P450, family 11, subfamily B, polypeptide 1 (CYP11B1); cytochrome P450, family 11, subfamily B, polypeptide 2 (CYP11B2); hydroxysteroid (11-beta) dehydrogenase 2 (HSD11B2); angiotensin converting enzyme (ACE); angiotensin II type 1 receptor 2 (AGTR2); and renin-binding protein (RENBP)] and systolic (SBP), diastolic (DBP), and mean arterial (MAP) pressure responses to a dietary sodium intervention.
METHODS
Study Population
The GenSalt study was conducted in a Han Chinese population living in rural areas of northern China where habitual salt-intake is high (13, 14). A community-based BP screening was conducted among persons aged 18–60 years in the study villages to identify potential probands and their families. Those with a mean SBP between 130–160 mmHg and/or a DBP between 85–100 mmHg and no use of antihypertensive medications and their spouses, siblings and offspring were recruited as volunteers for the dietary intervention study. Detailed eligibility criteria for the probands and siblings/spouses/offspring have been presented elsewhere [15]. Individuals who had stage-2 hypertension, secondary hypertension, a history of clinical CVD or diabetes, used antihypertensive medications, or were pregnant, heavy alcohol drinkers or currently on a low-sodium diet were excluded from the study. Among the 1,906 eligible participants, 1,871 (98.2%) and 1,860 (97.6%) completed the low-sodium and high-sodium interventions, respectively, and were included in the current analysis.
Institutional Review Boards at all of the participating institutions approved the GenSalt study. Written informed consents for the baseline observation and for the intervention program were obtained from each participant.
Dietary Intervention
The study participants received a 7-day low-sodium diet (3 grams of sodium chloride or 51.3 mmol of sodium per day) followed by a 7-day high-sodium diet (18 grams of sodium chloride or 307.8 mmol of sodium per day). During the period of sodium intervention, dietary potassium intake remained unchanged. Total energy intake was varied according to each participant’s baseline energy intake. All study foods were cooked without salt, and pre-packaged salt was added to the individual study participant’s meal when it was served by the study staff. To ensure study participants’ compliance to the intervention program, they were required to have their breakfast, lunch and dinner at the study kitchen under supervision of the study staff during the entire study period. The study participants were instructed to avoid consuming any foods that were not provided by study personnel. Three timed urinary specimens (one 24–hour and two overnight) were collected at baseline and at the end of each phase of intervention (days 5, 6, and 7) to monitor each participants’ compliance with their dietary sodium intervention. For each urinary specimen, the exact urine volume, electrolyte concentrations and duration of collection were recorded. Two-hundred thirty-eight participants collected a 24-hour specimen from each intervention phase in two separate containers, one for the overnight 8-hours and one for the remaining 16-hours. Eight-hour overnight measures for each intervention phase were extrapolated to 24-hour measures for all GenSalt intervention participants based on an adjustment using the two-part 24-hour collections of the 238 participants. The mean of three 24-hour measures (one collected 24-hour measure and two adjusted 24-hour measures) were used to estimate each participant’s average 24-hour urinary excretion for each intervention phase. The results from the 24-hour urinary excretions of sodium showed excellent compliance with the study diet. The mean (standard deviation) 24-hour urinary excretions of sodium and potassium were 242.4 (66.7) mmol and 36.9 (9.6) mmol at baseline, 47.5 (16.0) and 31.4 (7.7) during the low-sodium intervention, and 244.3 (37.7) and 35.7 (7.5) during the high-sodium intervention, respectively. Baseline 24-hour urinary sodium excretions were not significantly different from those of the high-sodium intervention phase, suggesting that sodium intake during the high-sodium diet was similar to the habitual sodium intake of this population.
Phenotype Measurement
A standard questionnaire was administered by trained staff at the baseline examination to collect information on family structure, demographic characteristics, personal and family medical history, and lifestyle risk factors including alcohol consumption, cigarette smoking, and physical activity. Three morning BP measurements were obtained according to a standard protocol during each of the 3-days of baseline observation and on days 5, 6 and 7 of each intervention period. All BP readings were measured by trained and certified observers using a random–zero sphygmomanometer [16]. BP was measured with the participant in the sitting position after 5 minutes of rest. In addition, participants were advised to avoid alcohol, cigarette smoking, coffee/tea, and exercise for at least 30 minutes prior to their BP measurements. All BP observers were blinded to the participant’s dietary intervention. Body weight and height were measured twice in light indoor clothing without shoes during the baseline examination. Body mass index (BMI) was calculated as kilograms per meters squared (kg/m2).
Mean BP response to low-sodium was calculated as the mean of 9 measurements on days 5, 6 and 7 during the low-sodium intervention minus the mean of 9 measurements at baseline; and response to high-sodium as the mean of 9 measurements on days 5, 6 and 7 during the high-sodium intervention minus the mean of 9 measurements on days 5, 6 and 7 during the low-sodium intervention.
Candidate Gene and SNP Selection and Genotyping
Eleven candidate genes in the RAAS were selected based on their potential biological effect on BP regulation, and included REN, HSD11B1, AGT, AGTR1, NR3C2, CYP11B1, CYP11B2, HSD11B2, ACE, AGTR2, and RENBP. We used Tagger software to identify 149 tagSNPs in these candidate genes based on the empirical patterns of linkage disequilibrium (LD) structure in the Chinese population, as determined by the Chinese HapMap [17,18]. In addition, functional SNPs were chosen based on previously reported associations with BP. SNP genotyping was performed using SNPlex assays (Applied Biosystems) based on oligonucleotide ligation assay for capillary electrophoresis on an automated DNA Sequencer (ABI 3700 DNA Analyzer). These SNPs were supplemented with an additional 60 SNPs (27 tagSNPs and 33 functional SNPs) genotyped as part of the Affymetrix 6.0 platform to provide better coverage of the candidate genes. Functional SNPs were identified via Washington University’s SNPseek website [19].
Data quality control revealed 13 monomorphic SNPs, 14 SNPs with a low genotyping call rate (<85%), and 1 tagSNP with a MAF<0.01. After exclusion of these 28 SNPs, a total of 181 SNPs remained (please see Supplemental Table).
Statistical Analysis
Quality control, including checks of Mendelian consistency, genotyping call rate, MAF, and HWE, was performed using PLINK software [20].
The mean of each baseline characteristic and BP response variable was calculated for each study participant. Additive associations between single SNPs and BP responses to low and high-sodium interventions were assessed using a mixed linear regression model. We conducted haplotype analyses to follow-up on genes with significant results. Linkage disequilibrium (LD) was calculated using the pairwise r2 correlation between SNPs, and LD block structure was estimated using the method of Gabriel et. al., as implemented in Haploview [21]. Haplotypes were inferred based on the most likely pattern of gene flow using Merlin software [22]. Similar to single marker analysis, haplotype associations were estimated using a mixed linear regression model. A sandwich estimator was used to account for the non-independence of family members. This method assumes the same degree of dependency among family members. Age, gender, BMI, BP measurement room temperature, and study site were adjusted in multivariable analyses. A similar, gender-stratified analysis was conducted for those SNPs located on the X-chromosome. To adjust for multiple comparisons, the false discovery rate (FDR) q-value was calculated for all SNPs and haplotypes [23]. For SNPs with a q-value<0.05 and all haplotypes, we estimated the mean effect size and 95% confidence interval (CI) for each genotype using a mixed linear regression model. These analyses were conducted using SAS statistical software (version 9.1; SAS Institute Inc).
RESULTS
On average, study participants were 38.7 years old and had a mean BMI of 23.3 kg/m2, mean SBP of 116.9 and mean DBP of 73.7 mmHg. Approximately 53% of participants were male. BP levels were similar during baseline and the high-sodium intervention, and BP responses to sodium intervention were significantly different from zero (Table 1).
Table 1.
Characteristics of 1,906 study participants.
| Variable | Mean ± SD or Percentage |
Median (Inter-quartile Range) |
|---|---|---|
| Age, yrs | 38.7 ± 9.6 | 39.0 (33.0, 46.0) |
| Male, % | 53.0 | … |
| BMI, kg/m2 | 23.3 ± 3.2 | 22.9 (21.1, 25.2) |
| Baseline BP, mm Hg | ||
| Systolic | 116.9 ± 14.2 | 115.8 (106.4, 127.1) |
| Diastolic | 73.7 ± 10.3 | 73.3 (66.7, 80.7) |
| Mean arterial pressure | 88.1 ± 10.9 | 87.7 (80.0, 95.4) |
| BP during low salt, mm Hg | ||
| Systolic | 111.4 ± 12.2 | 110.0 (102.7, 119.3) |
| Diastolic | 71.0 ± 9.7 | 70.7 (64.2, 77.3) |
| Mean arterial pressure | 84.5 ± 9.7 | 84.1 (77.4, 90.6) |
| BP response to low salt, mm Hg | ||
| Systolic | −5.5 ± 7.0* | −4.4 (−8.9, −1.3) |
| Diastolic | −2.8 ± 5.5* | −2.7 (−5.6, 0.4) |
| Mean arterial pressure | −3.7 ± 5.3* | −3.3 (−6.6, −0.6) |
| BP during high salt, mm Hg | ||
| Systolic | 116.3 ± 13.6 | 114.4 (106.7, 124.6) |
| Diastolic | 72.9 ± 10.3 | 72.4 (66.0, 79.1) |
| Mean arterial pressure | 87.4 ± 10.6 | 86.7 (79.6, 94.3) |
| BP response to high salt, mm Hg | ||
| Systolic | 4.9 ± 6.0* | 4.7 (0.6, 8.2) |
| Diastolic | 1.9 ± 5.4* | 1.8 (−1.6, 5.3) |
| Mean arterial pressure | 2.9 ± 5.0* | 2.7 (−0.4, 5.9) |
P-value<0.0001 when compared to no BP change during sodium interventions.
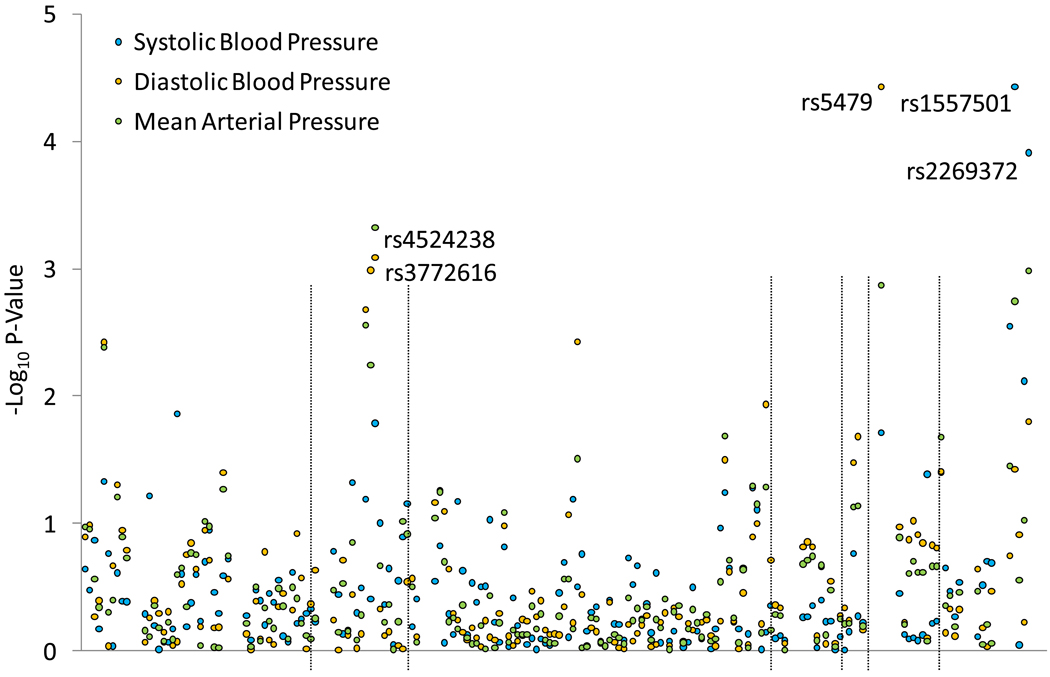
Figure 1 shows the association between each SNP and absolute SBP, DBP, and MAP responses to the low (upper) and high (lower) sodium interventions. Several SNPs in the AGTR1 (rs4524238 and rs3772616), HSD11B2 (rs5479) and RENBP (rs1557501 and rs2269372) genes were associated with BP responses to low-sodium, while only AGTR1 SNP rs4524238 was associated with BP response to high-sodium.
Figure 1.
Log p-values for the association between 181 SNPs in 11 candidate genes and systolic blood pressure, diastolic blood pressure, and mean arterial pressure responses to low (top) and high (bottom) sodium interventions. Labeled SNPs had a q-value<0.05.
Mean BP responses to the high- and low-sodium interventions, by genotype, for AGTR1 SNPs are shown in Table 2. DBP and MAP responses to sodium intervention increased with the number of A alleles of marker rs4524238. For example, mean DBP responses (95% CI) for genotypes G/G, G/A, and A/A were −2.53 (−2.89, −2.18), −3.49 (−4.13, −2.86), and −5.78 (−9.51, −2.06) mmHg, respectively, during the low-sodium intervention (p-value=0.0008), and 1.41 (1.09, 1.72), 2.59 (1.95, 3.23), and 5.24 (2.06, 8.42) mmHg, respectively, during the high-sodium intervention (p-value=0.00004). A similar trend was observed for mean SBP responses to sodium intervention. Additionally, the number of A alleles of rs3772616 was associated with an increased mean DBP response to low-sodium intervention (p-value=0.001).
Table 2.
BP response to dietary sodium intervention according to AGTR1 genotypes
| SNP | Genotype | Response to Low Salt |
Response to High Salt |
||||
|---|---|---|---|---|---|---|---|
| Absolute Change (95% CI) |
P-value for Additive Model |
Q-value* | Absolute Change (95% CI) |
P-value for Additive Model |
Q-value* | ||
| Systolic Blood Pressure | |||||||
| rs4524238 | G/G | −5.54 (−5.95, −5.13) | 4.37 (4.03, 4.71) | ||||
| A/G | −6.57 (−7.39, −5.75) | 0.02 | 0.37 | 5.41 (4.64, 6.18) | 0.006 | 0.29 | |
| A/A | −6.97 (10.61, −3.33) | 6.61 (3.32, 9.90) | |||||
| rs3772616 | G/G | −5.65 (−6.09, −5.20) | 4.58 (4.22, 4.94) | ||||
| A/G | −5.87 (−6.56, −5.18) | 0.40 | 0.91 | 4.50 (3.87, 5.14) | 0.97 | 1.00 | |
| A/A | −6.59 (−9.13, −4.05) | 5.05 (2.79, 7.30) | |||||
| Diastolic Blood Pressure | |||||||
| rs4524238 | G/G | −2.53 (−2.89, −2.18) | 1.41 (1.09, 1.72) | ||||
| A/G | −3.49 (−4.13, −2.86) | 0.0008 | 0.05 | 2.59 (1.95, 3.23) | 0.00004 | 0.004 | |
| A/A | −5.78 (−9.51, −2.06) | 5.24 (2.06, 8.42) | |||||
| rs3772616 | G/G | −2.49 (−2.87, −2.11) | 1.42 (1.09, 1.75) | ||||
| A/G | −3.31 (−3.87, −2.75) | 0.001 | 0.05 | 2.33 (1.76, 2.90) | 0.003 | 0.17 | |
| A/A | −4.61 (−6.60, −2.62) | 2.52 (1.03, 4.01) | |||||
| Mean Arterial Pressure | |||||||
| rs4524238 | G/G | −3.54 (−3.87, −3.20) | 2.39 (2.11, 2.68) | ||||
| A/G | −4.53 (−5.14, −3.91) | 0.0005 | 0.08 | 3.52 (2.92, 4.13) | 0.00004 | 0.008 | |
| A/A | −6.19 (−9.19, −3.19) | 5.71 (2.88, 8.53) | |||||
| rs3772616 | G/G | −3.55 (−3.91, −3.18) | 2.47 (2.17, 2.77) | ||||
| A/G | −4.17 (−4.71, −3.63) | 0.006 | 0.13 | 3.05 (2.51, 3.58) | 0.03 | 0.61 | |
| A/A | −5.28 (−7.07, −3.50) | 3.38 (1.89, 4.86) | |||||
FDR Q-Value
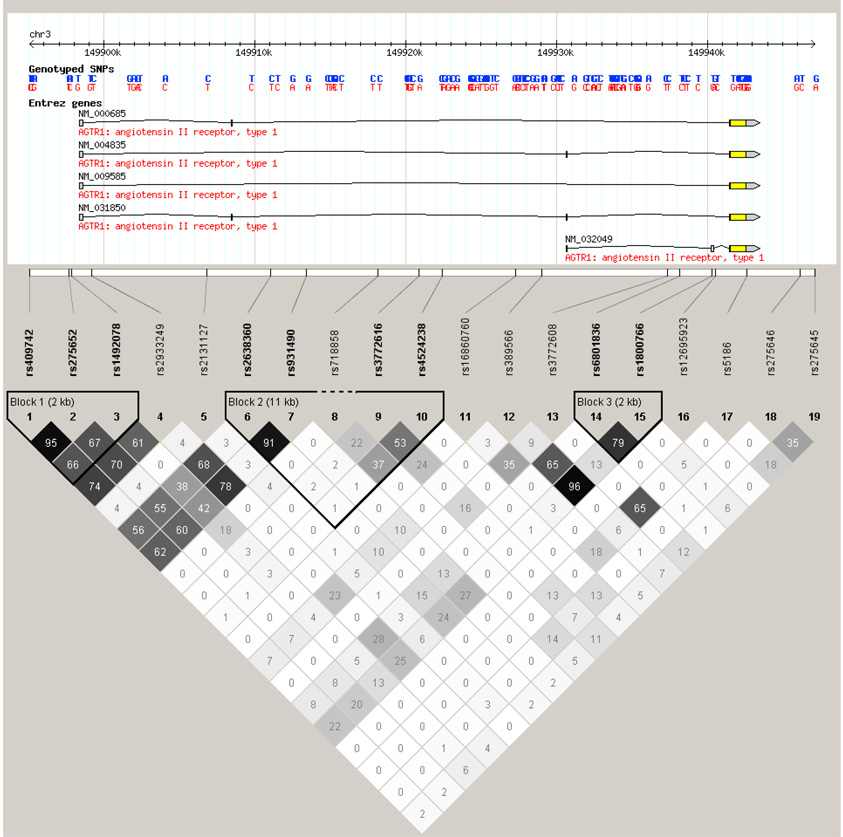
Three distinct LD blocks were identified in the AGTR1 gene (Figure 2). LD block 1 (rs409742-rs275652-rs1492078) contained three common haplotypes (frequency>1%) including: C-C-A (13.4%), T-A-A (4.9%), and T-A-G (74.7%). We identified four common haplotypes in LD block 2 (rs2638360-rs931490-rs3772616-rs4524238), which included: C-G-G-G (9.0%), T-A-A-A (8.9%), T-A-A-G (6.5%), and T-A-G-G (62.9%); and four common haplotypes in LD block 3 (rs6801836-rs1800766): C-C (7.4%), C-T (3.1%), T-C (8.2%), and T-T (51.6%). Similar to findings from single marker analysis, haplotype T-A-A-A of LD block 2, which contained the A variant of the rs4524238 SNP, was significantly associated with DBP and MAP responses to low- and high-sodium. DBP and MAP responses increased with the number of copies of the T-A-A-A haplotype (Table 5). In addition, haplotype T-A-A of LD block 1 was associated with DBP and MAP responses to both low and high-sodium interventions.
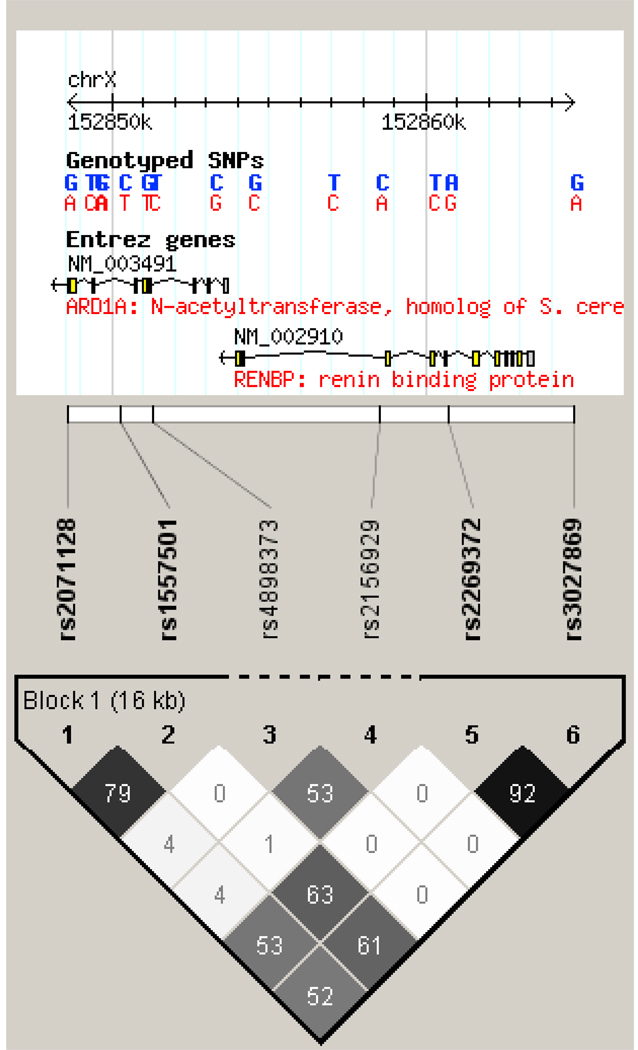
Figure 2.
Linkage disequilibrium structure of the AGTR1 (top) and RENBP (bottom) genes.
Table 5.
BP responses to dietary sodium intervention according to the number of copies of AGTR1 haplotypes.
| Haplotype | Response to Low Salt |
Response to High Salt |
||||||
|---|---|---|---|---|---|---|---|---|
| Absolute Change (95% CI) | P-Value | Absolute Change (95% CI) | P-Value | |||||
| 0 | 1 | 2 | 0 | 1 | 2 | |||
| Systolic Blood Pressure | ||||||||
| rs409742-rs275652-rs1492078 | ||||||||
| C-C-A | −5.84 (−6.27, −5.42) | −5.68 (−6.33, −5.03) | −3.94 (−5.52, −2.37) | 0.17 | 4.61 (4.25, 4.97) | 4.46 (3.92, 5.00) | 3.17 (1.67, 4.66) | 0.23 |
| T-A-A | −5.67 (−6.07, −5.27) | −6.56 (−7.74, −5.38) | −11.08 (−19.82, −2.33) | 0.07 | 4.43 (4.10, 4.77) | 5.49 (4.44, 6.54) | 10.50 (4.50, 16.50) | 0.02 |
| T-A-G | −6.18 (−8.19, −4.18) | −5.69 (−6.29, −5.10) | −5.78 (−6.22, −5.35) | 0.97 | 4.54 (3.23, 5.85) | 4.68 (4.17, 5.19) | 4.48 (4.11, 4.86) | 0.58 |
| rs2638360-rs931490-rs3772616-rs4524238 | ||||||||
| C-G-G-G | −5.81 (−6.23, −5.39) | −5.37 (−6.17, −4.57) | −5.07 (−5.91, −4.23) | 0.22 | 4.57 (4.23, 4.92) | 4.33 (3.70, 4.97) | 4.50 (2.75, 6.25) | 0.52 |
| T-A-A-A | −5.57 (−6.00, −5.15) | −6.30 (−7.14, −5.46) | −6.78 (−10.10, 3.45) | 0.09 | 4.32 (3.99, 4.66) | 5.29 (4.54, 6.04) | 7.32 (4.19, 10.45) | 0.004 |
| T-A-A-G | −5.75 (−6.17, −5.33) | −5.71 (−6.70, −4.73) | 0.94 (−4.42, 2.53) | 0.42 | 4.65 (4.32, 4.99) | 3.79 (2.90, 4.68) | 2.99 (−0.28, 6.26) | 0.06 |
| T-A-G-G | −5.92 (−7.09, −4.75) | −5.63 (−6.22, −5.04) | −5.76 (−6.23, −5.28) | 0.95 | 5.57 (4.54, 6.61) | 4.25 (3.74, 4.75) | 4.61 (4.21, 5.00) | 0.70 |
| rs6801836-rs1800766 | ||||||||
| C-C | −5.78 (−6.20, −5.37) | −5.21 (−6.07, −4.34) | −5.91 (−8.20, −3.62) | 0.40 | 4.59 (4.26, 4.92) | 4.51 (3.72, 5.30) | 3.58 (1.65, 5.51) | 0.47 |
| C-T | −5.64 (−6.04, −5.23) | −6.29 (−7.41, −5.17) | −8.31 (−16.75, 0.13) | 0.21 | 4.53 (4.20, 4.86) | 4.91 (3.74, 6.08) | 2.23 (−0.21, 4.67) | 0.67 |
| T-C | −5.69 (−6.11, −5.28) | −5.75 (−6.55, −4.95) | −4.77 (−8.38, −1.17) | 0.95 | 4.58 (4.24, 4.92) | 4.41 (3.76, 5.07) | 5.95 (2.85, 9.04) | 0.95 |
| T-T | −6.12 (−7.08, −5.15) | −5.29 (−5.95, −4.63) | −5.79 (−6.23, −5.35) | 0.88 | 4.65 (3.77, 5.54) | 4.35 (3.77, 4.94) | 4.63 (4.26, 4.99) | 0.72 |
| Diastolic Blood Pressure | ||||||||
| rs409742-rs275652-rs1492078 | ||||||||
| C-C-A | −2.77 (−3.14, −240) | −2.73 (−3.33, −2.13) | −1.18 (−2.69, 0.34) | 0.34 | 1.62 (1.29, 1.95) | 1.72 (1.19, 2.25) | 0.60 (−1.27, 2.48) | 0.79 |
| T-A-A | −2.59 (−2.94, −2.25) | −4.00 (−4.96, −3.05) | −3.43 (−9.33, −2.48) | 0.004* | 1.47 (1.17, 1.78) | 3.08 (2.14, 4.01) | 0.09 (−4.78, 4.96) | 0.002* |
| T-A-G | −2.64 (−3.02, −2.27) | −2.91 (−3.45, −2.36) | −2.64 (−3.02, −2.27) | 0.43 | 1.36 (0.03, 2.68) | 2.05 (1.55, 2.54) | 1.44 (1.10, 1.78) | 0.10 |
| rs2638360-rs931490-rs3772616-rs4524238 | ||||||||
| C-G-G-G | −2.86 (−3.23, −2.50) | −2.38 (−3.07, −1.69) | −2.53 (−4.56, −1.69) | 0.20 | 1.70 (1.38, 2.02) | 1.47 (0.87, 2.08) | 1.81 (−0.42, 4.03) | 0.58 |
| T-A-A-A | −2.58 (−2.94, −2.22) | −3.45 (−4.07, −2.82) | −5.75 (−9.21, −2.29) | 0.002* | 1.42 (1.11, 1.73) | 2.51 (1.86, 3.15) | 5.44 (2.47, 8.42) | 8.82E-05* |
| T-A-A-G | −2.76 (−3.12, −2.41) | −2.98 (−3.74, −2.23) | 1.05 (−1.66, 3.76) | 0.79 | 1.69 (1.38, 2.00) | 1.56 (0.80, 2.32) | −0.83 (−3.89, 2.22) | 0.41 |
| T-A-G-G | −2.87 (−3.92, −1.83) | −2.95 (−3.41, −2.50) | −2.61 (−3.04, −2.19) | 0.31 | 2.38 (1.52, 3.24) | 1.77 (1.32, 2.22) | 1.47 (1.11, 1.84) | 0.05 |
| rs6801836-rs1800766 | ||||||||
| C-C | −2.70 (−3.05, −2.36) | −3.00 (−3.74, −2.25) | −1.44 (−3.27, 0.39) | 0.85 | 1.64 (1.33, 1.94) | 1.90 (1.20, 2.60) | 1.71 (0.17, 3.25) | 0.54 |
| C-T | −2.70 (−3.05, −2.35) | −2.93 (−3.81, −2.04) | −3.58 (−5.08, −2.07) | 0.57 | 1.74 (1.44, 2.05) | 0.99 (−0.00, 1.98) | 0.79 (−2.49, 4.06) | 0.13 |
| T-C | −2.72 (−3.05, −2.34) | −2.77 (−3.32, −2.21) | −1.72 (−3.87, −2.21) | 0.89 | 1.73 (1.42, 2.05) | 1.50 (0.93, 2.07) | 0.71 (−1.39, 2.81) | 0.31 |
| T-T | −2.74 (−3.43, −2.05) | −2.70 (−3.25, −2.14) | −2.72 (−3.11, −2.33) | 0.99 | 1.37 (0.63, 2.11) | 1.68 (1.14, 2.21) | 1.74 (1.40, 2.07) | 0.41 |
| Mean Arterial Pressure | ||||||||
| rs409742-rs275652-rs1492078 | ||||||||
| C-C-A | −3.80 (−4.15, −3.45) | −3.71 (−4.26, −3.17) | −2.11 (−3.48, −0.74) | 0.21 | 2.61 (2.31, 2.92) | 2.64 (2.16, 3.11) | 1.46 (−0.13, 3.05) | 0.52 |
| T-A-A | −3.62 (−3.95, −3.30) | −4.87 (−5.77, −3.96) | −5.97 (−9.99, −1.95) | 0.003* | 2.46 (2.18, 2.74) | 3.88 (3.01, 4.76) | 3.54 (1.56, 5.53) | 0.001* |
| T-A-G | −3.96 (−5.41, −2.50) | −3.84 (−4.34, −3.34) | −3.69 (−4.05, −3.34) | 0.55 | 2.42 (1.32, 3.52) | 2.92 (2.47, 3.37) | 2.45 (2.14, 2.76) | 0.15 |
| rs2638360-rs931490-rs3772616-rs4524238 | ||||||||
| C-G-G-G | −3.85 (−4.19, −3.51) | −3.38 (−4.03, −2.73) | −3.39 (−4.83, −1.94) | 0.15 | 2.65 (2.36, 2.95) | 2.43 (1.89, 2.98) | 2.71 (0.93, 4.50) | 0.52 |
| T-A-A-A | −3.58 (−3.92, −3.24) | −4.40 (−5.01, −3.80) | −6.10 (−8.90, −3.29) | 0.002* | 2.39 (2.82, 4.03) | 3.42 (2.82, 4.03) | 6.09 (3.42, 8.77) | 0.00006* |
| T-A-A-G | −3.76 (−4.10, −3.43) | −3.90 (−4.65, −3.16) | 0.40 (−1.91, 2.70) | 0.59 | 2.67 (2.39, 2.96) | 2.31 (1.59, 3.02) | 0.42 (−2.18, 3.02) | 0.17 |
| T-A-G-G | −3.89 (−4.81, −2.97) | −3.85 (−4.30, −3.40) | −3.67 (−4.06, −3.27) | 0.48 | 3.45 (2.66, 4.24) | 2.59 (2.17, 3.01) | 2.51 (2.19, 2.84) | 0.11 |
| rs6801836-rs1800766 | ||||||||
| C-C | −3.73 (−4.06, −3.40) | −3.74 (−4.45, −3.02) | −2.92 (−4.73, −1.11) | 0.62 | 2.62 (2.35, 2.90) | 2.77 (2.10, 3.43) | 2.33 (0.90, 3.76) | 0.90 |
| C-T | −3.68 (−4.01, −3.35) | −4.04 (−4.88, −3.19) | −5.15 (−7.07, −3.23) | 0.34 | 2.67 (2.39, 2.95) | 2.30 (1.35, 3.25) | 1.28 (−0.09, 2.65) | 0.38 |
| T-C | −3.71 (−4.05, −3.37) | −3.77 (−4.34, −3.20) | −2.75 (−4.87, −0.63) | 0.91 | 2.68 (2.39, 2.97) | 2.48 (1.95, 3.00) | 2.47 (0.52, 4.42) | 0.48 |
| T-T | −3.86 (−4.56, −3.17) | −3.57 (−4.10, −3.04) | −3.75 (−4.11, −3.39) | 0.93 | 2.47 (1.78, 3.17) | 2.56 (2.07, 3.06) | 2.70 (2.39, 3.00) | 0.48 |
FDR Q-value < 0.05.
Mean BP responses to the low- and high-sodium interventions by genotype of HSD11B2 SNP rs5479 are shown in Table 3. While there were only 12 heterozygotes and no homozygotes for the A allele (MAF=0.0009), participants with one copy of the A allele had decreased DBP in response to the low-sodium intervention (p-value=0.00004). A similar trend was observed for all other BP responses to the low and high-sodium interventions. Because we genotyped only 1 SNP in the HSD11B2 gene, haplotype analysis was not possible.
Table 3.
BP response to dietary sodium intervention according to HSD11B2 genotype
| SNP | Genotype | Response to Low Salt |
Response to High Salt |
||||
|---|---|---|---|---|---|---|---|
| Absolute Change (95% CI) |
P-value for Additive Model |
Q-value* | Absolute Change (95% CI) |
P-value for Additive Model |
Q-value* | ||
| Systolic Blood Pressure | |||||||
| rs5479 | C/C | −5.74 (−8.24, −3.23) | 0.02 | 0.36 | 4.63 (2.63, 6.63) | 0.03 | 0.45 |
| C/A | −2.67 (−19.19, 13.85) | 0.53 (−22.91, 23.98) | |||||
| Diastolic Blood Pressure | |||||||
| rs5479 | C/C | −2.71 (−4.88, −0.54) | 0.00004 | 0.003 | 1.69 (−0.18, 3.57) | 0.16 | 0.57 |
| C/A | −0.70 (−6.62, 5.22) | −1.33 (−28.70, 26.03) | |||||
| Mean Arterial Pressure | |||||||
| rs5479 | C/C | −3.73 (−5.78, −1.67) | 0.001 | 0.08 | 2.67 (0.97, 4.38) | 0.10 | 0.61 |
| C/A | −1.41 (−10.40, 7.57) | −0.69 (−26.21, 24.83) | |||||
FDR Q-Value
In the RENBP gene, SNPs rs1557501 and rs2269372 were associated with mean SBP response to low-sodium in men, and the associations remained significant after adjustment for multiple comparisons (Table 4). Male participants with the variant alleles had smaller absolute decreases in SBP in response to low-sodium compared to those with the more common alleles (p-values=0.00004 and 0.0001 for rs1557501 and rs2269372, respectively). These SNPs were not significantly associated with BP responses to high-sodium, and the associations did not persist among women (data not shown).
Table 4.
BP response to dietary sodium intervention according to RENBP allele in males
| SNP | Allele | Response to Low Salt |
Response to High Salt |
||||
|---|---|---|---|---|---|---|---|
| Absolute Change (95% CI) |
P-value for Additive Model |
Q-value* | Absolute Change (95% CI) |
P-value for Additive Model |
Q-value* | ||
| Systolic Blood Pressure | |||||||
| rs1557501 | T | −6.13 (−6.68, −5.58) | 0.00004 | 0.003 | 4.35 (3.88, 4.83) | 0.53 | 0.90 |
| C | −4.07 (−4.88, −3.26) | 4.08 (3.37, 4.80) | |||||
| rs2269372 | G | −6.04 (−6.57, −5.52) | 0.0001 | 0.007 | 4.39 (3.93, 4.85) | 0.42 | 0.87 |
| A | −3.94 (−4.90, −2.99) | 4.03 (3.25, 4.82) | |||||
| Diastolic Blood Pressure | |||||||
| rs1557501 | T | −2.70 (−3.19, −2.21) | 0.04 | 0.45 | 1.33 (0.87, 1.79) | 0.47 | 0.87 |
| C | −1.84 (−2.53, −1.15) | 1.06 (0.45, 1.67) | |||||
| rs2269372 | G | −2.71 (−3.19, −2.23) | 0.02 | 0.29 | 1.35 (0.90, 1.79) | 0.33 | 0.72 |
| A | −1.64 (−2.41, −0.87) | 0.95 (0.26, 1.64) | |||||
| Mean Arterial Pressure | |||||||
| rs1557501 | T | −3.85 (−4.31, −3.39) | 0.002 | 0.08 | 2.34 (1.93, 2.75) | 0.45 | 0.87 |
| C | −2.59 (−3.24, −1.93) | 2.07 (1.50, 2.64) | |||||
| rs2269372 | G | −3.83 (−4.27, −3.38) | 0.001 | 0.08 | 2.36 (1.97, 2.76) | 0.30 | 0.69 |
| A | −2.41 (−3.16, −1.67) | 1.98 (1.34, 2.61) | |||||
FDR Q-Value
The RENBP gene contained one LD block, comprised of SNPs rs2071128-rs1557501-rs2269372-rs3027869, with 4 common haplotypes: A-T-G-A (61.1%), G-C-A-G (21.2%), G-C-G-A (7.2%), and G-T-G-A (3.5%). Men with the A-T-G-A haplotype had significantly increased SBP and MAP responses to low-sodium intervention, while those with the G-C-A-G haplotype had decreased SBP and MAP responses to low-sodium (Table 6).
Table 6.
BP responses to dietary sodium intervention according to the number of copies of RENBP rs2071128-rs1557501-rs2269372-rs3027869 haplotypes in men.
| Haplotype | Response to Low Salt |
Response to High Salt |
||||
|---|---|---|---|---|---|---|
| Absolute Change (95% CI) | P-Value | Absolute Change (95% CI) | P-Value | |||
| 0 | 1 | 0 | 1 | |||
| Systolic Blood Pressure | ||||||
| A-T-G-A | −4.41 (−5.15, −3.67) | −6.01 (−6.57, −5.45) | 0.0005* | 4.02 (3.38, 4.65) | 4.36 (3.90, 4.83) | 0.37 |
| G-C-A-G | −5.95 (−6.47, −5.43) | −3.78 (−4.70, −2.86) | 0.00004* | 4.40 (3.95, 4.84) | 3.73 (2.98, 4.48) | 0.12 |
| G-C-G-A | −5.53 (−6.02, −5.03) | −4.56 (−5.95, −3.17) | 0.17 | 4.22 (3.81, 4.63) | 4.57 (2.93, 6.22) | 0.66 |
| G-T-G-A | −5.35 (−5.87, −4.84) | −7.73 (−9.58, −5.88) | 0.008 | 4.23 (3.79, 4.66) | 4.61 (2.69, 6.53) | 0.68 |
| Diastolic Blood Pressure | ||||||
| A-T-G-A | −1.91 (−2.56, −1.26) | −2.59 (−3.08, −2.09) | 0.09 | 1.04 (0.48, 1.61) | 1.25 (0.79, 1.71) | 0.55 |
| G-C-A-G | −2.60 (−3.06, −2.14) | −1.51 (−2.27, −0.74) | 0.01 | 1.30 (0.87, 1.73) | 0.76 (0.06, 1.46) | 0.18 |
| G-C-G-A | −2.34 (−2.77, −1.91) | −2.49 (−3.87, −1.11) | 0.82 | 1.16 (0.76, 1.55) | 1.48 (0.12, 2.84) | 0.63 |
| G-T-G-A | −2.32 (−2.78, −1.86) | −3.09 (−4.94, −1.24) | 0.39 | 1.15 (0.73, 1.57) | 1.80 (0.25, 3.35) | 0.39 |
| Mean Arterial Pressure | ||||||
| A-T-G-A | −2.75 (−3.36, −2.14) | −3.73 (−4.19, −3.27) | 0.01* | 2.04 (1.52, 2.56) | 2.29 (1.88, 2.70) | 0.45 |
| G-C-A-G | −3.72 (−4.15, −3.29) | −2.27 (−2.99, −1.54) | 0.0006* | 2.33 (1.95, 2.72) | 1.75 (1.11, 2.38) | 0.11 |
| G-C-G-A | −3.41 (−3.81, −3.01) | −3.20 (−4.49, −1.92) | 0.75 | 2.17 (1.82, 2.52) | 2.53 (1.21, 3.85) | 0.59 |
| G-T-G-A | −3.33 (−3.76, −2.91) | −4.65 (−6.39, −2.90) | 0.12 | 2.17 (1.80, 2.55) | 2.75 (1.26, 4.25) | 0.42 |
FDR Q-value < 0.05.
DISCUSSION
Our study identified several novel genetic variants in the RAAS which may play a critical role in salt-sensitivity of BP. It is the first investigation to observe a strong, dose-response, and consistent relationship between the number of copies of the A allele variant of rs4524238 in the AGTR1 gene and DBP and MAP responses to low and high-sodium interventions. AGTR1 marker rs3772616 was also associated with DBP and MAP responses to low-sodium intervention in our analysis. Furthermore, two AGTR1 haplotypes, one containing rs4524238 and rs3772616 and one in the promoter region of this gene, were associated with DBP and MAP responses to salt-intake. In the HSD11B2 gene, carriers of the rare A allele of rs5479 had a decreased DBP response to low-sodium. Given the growing awareness that rare genetic variants may also contribute to the inheritance of common chronic diseases [24], this finding may be of particular relevance. Furthermore, we described a potential role for the X-linked RENBP gene in salt-sensitivity, identifying a strong association between markers rs1557501 and rs2269372 and SBP responses to low-sodium intervention in men. Haplotype analysis of the RENBP gene supported these results. In aggregate, these findings may have important clinical and public health implications. Discovery of novel genetic variants that determine an individual’s susceptibility to dietary sodium intervention is crucial for the identification of individuals who will benefit the most from a low-sodium diet. Moreover, these study findings will help in our understanding of the genetic mechanisms underlying hypertension.
To date, the GenSalt study is the largest dietary intervention study to examine the association between genetic variants in the RAAS and BP response to dietary sodium intervention. Study attributes, including the recruitment of all Han Chinese participants, should make the analysis robust to population stratification. The study participants were also similar with respect to lifestyle and environmental risk factors, including high habitual sodium intake, low BMI and high physical activity levels. While these similarities may limit the generalizability of study findings to Western populations, confounding of genetic associations due to these exposures should be minimized. The majority of participants completed the dietary intervention (98.2% and 97.6% for the low- and high-sodium interventions, respectively). Of those who withdrew from the dietary intervention, 1 (2%) started BP medication, 17 (38%) refused to continue, 3 (7%) had a health problem which prohibited them from continuing on in the study and 25 (54%) withdrew for other reasons. Compliance with the study interventions, as assessed by urinary excretion of sodium during each intervention period, was excellent. We have selected multiple tag-SNPs and previously reported SNPs which provided full coverage of these candidate genes. Finally, stringent quality control procedures were employed during measurement of BP and the other study covariables, conduct of the dietary interventions, genotyping, and marker data cleaning.
The AGTR1 gene mediates the vasoconstrictive and salt-conserving actions of the RAAS, via its action with angiotensin II, and may be an important regulator of BP response to salt intake [25]. A few previous studies have examined the association between the A1166C polymorphism of AGTR1 and BP salt-sensitivity, with mostly negative findings [7,11,12,26]. The current study also examined the association between rs5186 and BP responses to sodium intervention, and, similar to previous reports, no significant relationship was observed. Other variants and haplotypes in the AGTR1 gene have been related to hypertension [27]. Similarly, we found that SNP markers rs4524238 and rs3772616 were associated with both DBP and MAP responses to sodium intervention. Results of haplotype analysis suggested that rs3772616 was likely related to BP responses due to its correlation with rs4524238, as the rs3772616 variant was only associated with increased BP response in the presence of the rs4524238 variant. Furthermore, rs4524238 was more strongly associated with BP responses than its corresponding haplotype, indicating that this SNP might be in stronger LD with the causative loci than the haplotype itself. It is also possible that the results of the haplotype analysis are diluted by the imprecision of the haplotype inference procedure. In addition, haplotype T-A-A in block 1 of the AGTR1 gene was associated with BP responses to sodium and consisted of SNPs that were not identified in single marker analysis. This LD block lies in the promoter region of the AGTR1 gene, suggesting that it may harbor a causative locus that regulates gene transcription.
The HSD11B2 gene also plays an important role in sodium homeostasis [28]. Compromised function of the HSD11B2 enzyme has been shown to result in overstimulation of the mineralocorticoid receptor by cortisol, leading to sodium retention, hypokalemia and hypertension [28,29]. HSD11B2 has been associated with salt-sensitivity in previous studies [12,29,30]. Similarly, we observed a significant association between marker rs5479 of the HSD11B2 gene and BP salt-sensitivity, with the variant A allele of this SNP providing a protective effect on BP response to salt-intake. Because rs5479 had a low MAF in the GenSalt study, a word of caution regarding our result is warranted.
Although the relationship between the RENBP gene and salt-sensitivity is not well studied, this gene has been shown to increase prorenin levels in men and has been implicated as a potential regulator of BP [31]. In the current analysis, we identified strong associations between rs1557501 and rs2269372 and SBP responses to the low-sodium intervention in men. A haplotype containing the minor alleles of rs1557501 and rs2269372 decreased BP responses to sodium and was more strongly associated with BP response than either variant alone. This could suggest that both SNPs are in LD with separate causal loci, or that the haplotype is in greater LD with the causal loci than either variant alone. Synergistic effects of the two loci were not detected in a test of interaction.
Several studies have identified an association between the ACE insertion/deletion (I/D), AGT M235T (rs699), and CYP11B2 C-344T polymorphisms with salt-sensitivity [7, 11, 12, 32–35]. While the current study did not examine the ACE I/D polymorphism specifically, two studies have suggested that the ACE I/D polymorphism is in complete LD (D’ and r2=1.0) with SNPs rs4343 and rs4331 in Caucasian and Asian populations, respectively [36,37]. The current study examined SNPs rs4343 and rs4331, but did not find an association between either of the SNPs and BP salt-sensitivity. These findings were similar to those of three other studies, including one conducted in a different Asian population [26,32,38]. We also failed to replicate the AGT M235T findings, with results similar to those of three other studies [11,12,39]. Furthermore, Iwai and colleagues reported that the CYP11B2 C-344T polymorphism (rs1799998) was associated with salt sensitivity in a Japanese population [40]. However, the current study did not find any significant associations with this SNP, and our results are similar to those of other studies [7,12,26,41]. Several possibilities may explain the discrepancies between studies. Our study was conducted in a Han Chinese population, where LD structure may be different to that of Western populations. If the identified variant is in LD with the true causal variant, then it may not be replicated in populations with a distinct ethnic background. Furthermore, the same genetic variants may have distinct BP effects due to their interactions with genetic and environmental factors that are unique to certain populations. Phenotypic heterogeneity between studies also exists. Salt-sensitivity has been defined by BP response to acute sodium loading, dietary interventions or diuretic administration, depending on the study [7]. In addition, salt-sensitivity has been measured as a continuous BP response in some studies and as a dichotomous trait in others. Phenotypic standardization and replication in different populations will be critical for identification of true genetic variants that increase susceptibility to high salt-sensitivity.
We have described novel genetic variants in the AGTR1, HSD11B2 and RENBP genes that are significantly associated with BP responses to changes in sodium intake in a Han Chinese population known for its habitual high-salt diet. These findings may have important clinical and public health implications, but some additional work is needed. While these genes have been implicated in previous studies for their association with salt-sensitivity and related phenotypes, replication of the loci identified in the current analysis are needed. Studies aimed at identification of the causal loci along with their functions are also warranted.
Supplementary Material
Acknowledgments
Funding sources: The Genetic Epidemiology Network of Salt Sensitivity (GenSalt) is supported by research grants (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, Maryland.
APPENDIX
GenSalt Collaborative Research Group
Tulane University Health Sciences Center, New Orleans, USA: Jiang He (PI), Lydia A. Bazzano, Chung-Shiuan Chen, Jing Chen, Tanika N. Kelly, L. Lee Hamm, Paul Muntner, Kristi Reynolds, Wenjie Yang, and Qi Zhao.
Washington University School of Medicine, St. Louis, USA: DC Rao (PI), Matthew Brown, Charles Gu, Treva Rice, Karen Schwander, and Shiping Wang.
University of Texas Health Sciences Center, Houston, USA: James E. Hixson (PI) and Lawrence C. Shimmin.
Loyola University Health System and Medical Center, Chicago, USA: Paul K. Whelton
National Heart, Lung, and Blood Institute, Bethesda, USA: Cashell E. Jaquish
Chinese Academy of Medical Sciences, Beijing, China: Dongfeng Gu (PI), Jie Cao, Jichun Chen, Jingping Chen, Zhenhan Du, Jianfeng Huang, Hongwen Jiang, Jianxin Li, Xiaohua Liang, Depei Liu, Xiangfeng Lu, Donghua Liu, Qunxia Mao, Dongling Sun, Hongwei Wang, Qianqian Wang, Xigui Wu, Ying Yang, and Dahai Yu.
Shandong Academy of Medical Sciences, Shandong, China: Fanghong Lu (PI), Zhendong Liu, Shikuan Jin, Yingxin Zhao, Shangwen Sun, Shujian Wang, Qengjie Meng, Baojin Liu, Zhaodong Yang, and Chuanrui Wei.
Shandong Center for Diseases Control and Prevention, Shandong, China: Jixiang Ma (PI), Jiyu Zhang, and Junli Tang.
Zhengzhou University, Henan, China: Dongsheng Hu, Hongwei Wen, Chongjian Wang, Minghui Shen, Jingjing Pan, and Liming Yang.
Xinle Traditional Chinese Medicine Hospital, Hebei, China: Xu Ji (PI), Rongyan Li, Haijun Zu, and Junwei Song.
Ganyu Center for Disease Control and Prevention: Delin Wu (PI), Xushan Wang, and Xiaofeng Zhang.
Xi’an Jiaotong University, Shanxi, China: Jianjun Mu (PI), Enrang Chen, Fuqiang Liu, and Guanji Wu.
Chinese National Human Genome Center, Beijing, China:
Zhi-Jian Yao (PI), Shufeng Chen, Dongfeng Gu, Hongfan Li, Laiyuan Wang, and Penghua Zhang.
Footnotes
Conflict of interest statement: We declare that we have no conflict of interest.
REFERENCES
- 1.Morimoto A, Uzu T, Fujii T, Nishimura M, Kuroda S, Nakamura S, Inenaga T, Kimura G. Sodium sensitivity and cardiovascular events in patients with essential hypertension. Lancet. 1997;350:1734–1737. doi: 10.1016/S0140-6736(97)05189-1. [DOI] [PubMed] [Google Scholar]
- 2.Weinberger MH, Fineberg NS, Fineberg SE, Weinberger M. Salt sensitivity, pulse pressure, and death in normal and hypertensive humans. Hypertension. 2001;37:429–432. doi: 10.1161/01.hyp.37.2.429. [DOI] [PubMed] [Google Scholar]
- 3.Svetkey LP, McKeown SP, Wilson AF. Heritability of salt sensitivity in black Americans. Hypertension. 1996;28:854–858. doi: 10.1161/01.hyp.28.5.854. [DOI] [PubMed] [Google Scholar]
- 4.Miller JZ, Weinberger MH, Christian JC, Daugherty SA. Familial resemblance in the blood pressure response to sodium restriction. Am J Epidemiol. 1987;126:822–830. doi: 10.1093/oxfordjournals.aje.a114719. [DOI] [PubMed] [Google Scholar]
- 5.Gu D, Rice T, Wang S, Yang W, Gu C, Chen CS, et al. Heritability of blood pressure responses to dietary sodium and potassium intake in a Chinese population. Hypertension. 2007;50:116–122. doi: 10.1161/HYPERTENSIONAHA.107.088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Svetkey LP, Chen YT, McKeown SP, Preis L, Wilson A. Preliminary evidence of salt sensitivity in black Americans at the beta2-adrenergic receptor locus. Hypertension. 1997;29:918–922. doi: 10.1161/01.hyp.29.4.918. [DOI] [PubMed] [Google Scholar]
- 7.Beeks E, Kessels AG, Kroon AA, van der Klauw MM, de Leeuw PW. Genetic predisposition to salt-sensitivity: a systematic review. J Hypertens. 2004;22:1243–1249. doi: 10.1097/01.hjh.0000125443.28861.0d. [DOI] [PubMed] [Google Scholar]
- 8.Ames RP. The role of the renin-angiotensin-aldosterone system in blood pressure regulation. Am J Hypertens. 2002;15(7 Pt 1):653–654. doi: 10.1016/s0895-7061(02)02935-7. [DOI] [PubMed] [Google Scholar]
- 9.Weinberger MH, Miller JZ, Luft FC, Grim CE, Fineberg NS. Definitions and characteristics of sodium sensitivity and blood pressure resistance. Hypertension. 1986;8 suppl II:II-127–II-134. doi: 10.1161/01.hyp.8.6_pt_2.ii127. [DOI] [PubMed] [Google Scholar]
- 10.De la Sierra A, Lluch MM, Coca A, Aguilera MT, Giner V, Bragulat E, Urbano-Márquez A. Fluid, ionic and hormonal changes induced by high salt intake in salt-sensitive and salt-resistant hypertensive patients. Clin Sci. 1996;91:155–161. doi: 10.1042/cs0910155. [DOI] [PubMed] [Google Scholar]
- 11.Giner V, Poch E, Bragulat E, Oriola J, Gonzalez D, Coca A, De La Sierra A. Renin-angiotensin-aldosterone system genetic polymorphisms and salt sensitivity in essential hypertension. Hypertension. 2000;35:512–517. doi: 10.1161/01.hyp.35.1.512. [DOI] [PubMed] [Google Scholar]
- 12.Poch E, Gonzalez D, Giner V, Bragulat E, Coca A, de La Sierra A. Molecular basis of salt sensitivity in human hypertension. Evaluation of renin-angiotensin-aldosterone system gene polymorphisms. Hypertension. 2001;38:1204–1209. doi: 10.1161/hy1101.099479. [DOI] [PubMed] [Google Scholar]
- 13.Elliot P, Dyer A, Stamler R. The INTERSALT study: results for 24 hour sodium and potassium, by age and sex. J Hum Hypertens. 1989;3:331–407. [PubMed] [Google Scholar]
- 14.Zhao L, Stamler J, Yan LL, Zhou B, Wu Y, Liu K, Daviglus ML, Dennis BH, Ellieot P, Ueshima H, Yang J, Zhu L, Guo D. Blood pressure differences between northern and southern Chinese: Role of dietary factors. Hypertension. 2004;43:1332–1337. doi: 10.1161/01.HYP.0000128243.06502.bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.GenSalt Collaborative Research G. GenSalt: rationale, design, methods and baseline characteristics of study participants. J Human Hypertens. 2007;21:639–646. doi: 10.1038/sj.jhh.1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perloff D, Grim C, Flack JM, Frohlich ED, Hill M, McDonald M, Morgenstern BZ. Writing Group. Human blood pressure determination by sphygmomanometry. Circulation. 1993;88(5 Part 1) doi: 10.1161/01.cir.88.5.2460. 2460–1270. [DOI] [PubMed] [Google Scholar]
- 17.The International HapMap Consortium. The International HapMap Project. Nature. 2005;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 18.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 19.Washington University in St. Louis School of Medicine. SNPSEEK. Retrieved June 2009 from: A web based interface to human SNPs, frequencies and annotation data. Web site: http://snp.wustl.edu/cgi-bin/SNPseek/index.cgi.
- 20.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 23.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995;57:289–300. [Google Scholar]
- 24.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmid C, Castrop H, Reitbauer J, Della Bruna R, Krutz A. Dietary salt intake modulates angiotensin II type 1 receptor gene expression. Hypertension. 1997;29:923–929. doi: 10.1161/01.hyp.29.4.923. [DOI] [PubMed] [Google Scholar]
- 26.Pamies-Andreu E, Ramirez-Lorca R, Garcia-Junco PS, Muniz-Grijalbo O, Vallejo-Maroto I, Morillo SG, et al. Renin-angiotensin-aldosterone system and G-protein beta-3 subunit gene polymorphisms in salt-sensitive essential hypertension. J Human Hypertens. 2003;17:187–191. doi: 10.1038/sj.jhh.1001534. [DOI] [PubMed] [Google Scholar]
- 27.Zhu X, Chang YC, Yan D, Weder A, Cooper R, Luke A, et al. Associations between hypertension and genes in the renin-angiotensin system. Hypertension. 2003;41:1027–1034. doi: 10.1161/01.HYP.0000068681.69874.CB. [DOI] [PubMed] [Google Scholar]
- 28.White PC. Disorders of aldosterone biosynthesis and action. New Engl J Med. 1994;331:250–258. doi: 10.1056/NEJM199407283310408. [DOI] [PubMed] [Google Scholar]
- 29.Lovati E, Ferrari P, Dick B, Jostarndt K, Frey BM, Frey FJ, et al. Molecular basis of human salt sensitivity: the role of the 11beta-hydroxysteroid dehydrogenase type 2. J Clin Endocr Metab. 1999;84:3745–3749. doi: 10.1210/jcem.84.10.6098. [DOI] [PubMed] [Google Scholar]
- 30.Agarwal AK, Giacchetti G, Lavery G, Palermo M, Ricketts M, McTernan C, et al. CA-Repeat polymorphism in intron 1 of HSD11B2 : effects on gene expression and salt sensitivity. Hypertension. 2000;36:187–194. doi: 10.1161/01.hyp.36.2.187. [DOI] [PubMed] [Google Scholar]
- 31.Knoll A, Schunkert H, Reichwald K, Danser AHJ, Bauer D, Platzer M, et al. Human renin binding protein: complete genomic sequence and association of an intronic T/C polymorphism with the prorenin level in males. Hum Mol Genet. 1997;6:1527–1534. doi: 10.1093/hmg/6.9.1527. [DOI] [PubMed] [Google Scholar]
- 32.Johnson AG, Nguyen TV, Davis D. Blood pressure is linked to salt intake and modulated by the angiotensinogen gene in normotensive and hypertensive elderly subjects. J Hypertens. 2001;19:1053–1060. doi: 10.1097/00004872-200106000-00009. [DOI] [PubMed] [Google Scholar]
- 33.Hunt SC, Geleijnse JM, Wu LL, Witteman JC, Williams RR, Grobbee DE. Enhanced blood pressure response to mild sodium reduction in subjects with the 235T variant of the angiotensinogen gene. Am J Hypertens. 1999;12:460–466. doi: 10.1016/s0895-7061(99)00014-x. [DOI] [PubMed] [Google Scholar]
- 34.Hiraga H, Oshima T, Watanabe M, Ishida M, Ishida T, Shingu T, et al. Angiotensin I-converting enzyme gene polymorphism and salt sensitivity in essential hypertension. Hypertension. 1996;27:569–572. doi: 10.1161/01.hyp.27.3.569. [DOI] [PubMed] [Google Scholar]
- 35.Dengel DR, Brown MD, Ferrell RE, Supiano MA. Role of angiotensin converting enzyme genotype in sodium sensitivity in older hypertensives. Am J Hypertens. 2001;14:1178–1184. doi: 10.1016/s0895-7061(01)02204-x. [DOI] [PubMed] [Google Scholar]
- 36.Glenn KL, Du Z-Q, Eisenmann JC, Rothschild MF. An alternative method for genotyping of the ACE I/D polymorphism. Mol Biol Rep. 2009;36:1305–1310. doi: 10.1007/s11033-008-9313-5. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka C, Kamide K, Takiuchi S, Miwa Y, Yoshii M, Kawano Y, Miyata T. An alternative fast and convenient genotyping method for the screening of angiotensin converting enzyme gene polymorphisms. Hyperten Res Clin Exper. 2003;26:301–306. doi: 10.1291/hypres.26.301. [DOI] [PubMed] [Google Scholar]
- 38.Kojima S, Inenaga T, Matsuoka H, Kuramochi M, Omae T, Nara Y, Yamori Y. The association between salt sensitivity of blood pressure and some polymorphic factors. J Hypertens. 1994;12:797–801. [PubMed] [Google Scholar]
- 39.Schorr U, Blaschke K, Beige J, Distler A, Sharma AM. Angiotensinogen M235T variant and salt sensitivity in young normotensive Caucasians. J Hypertens. 1999;17:475–479. doi: 10.1097/00004872-199917040-00004. [DOI] [PubMed] [Google Scholar]
- 40.Iwai N, Kajimoto K, Tomoike H, Takashima N. Polymorphism of CYP11B2 determines salt sensitivity in Japanese. Hypertension. 2007;49:825–831. doi: 10.1161/01.HYP.0000258796.52134.26. [DOI] [PubMed] [Google Scholar]
- 41.Brand E, Schorr U, Ringel J, Beige J, Distler A, Sharma AM. Aldosterone synthase gene (CYP11B2) C-344T polymorphism in Caucasians from the Berlin Salt-Sensitivity Trial (BeSST) J Hypertens. 1999;17:1563–1567. doi: 10.1097/00004872-199917110-00009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.