Abstract
Protein S has an established role in the protein C anticoagulant pathway, where it enhances the factor Va (FVa) and factor VIIIa (FVIIIa) inactivating property of activated protein C (APC). Despite its physiological role and clinical importance, the molecular basis of its action is not fully understood. To clarify the mechanism of the protein S interaction with APC, we have constructed and expressed a library of composite or point variants of human protein S, with residue substitutions introduced into the Gla, thrombin-sensitive region (TSR), epidermal growth factor 1 (EGF1), and EGF2 domains. Cofactor activity for APC was evaluated by calibrated automated thrombography (CAT) using protein S–deficient plasma. Of 27 variants tested initially, only one, protein S D95A (within the EGF1 domain), was largely devoid of functional APC cofactor activity. Protein S D95A was, however, γ-carboxylated and bound phospholipids with an apparent dissociation constant (Kdapp) similar to that of wild-type (WT) protein S. In a purified assay using FVa R506Q/R679Q, purified protein S D95A was shown to have greatly reduced ability to enhance APC-induced cleavage of FVa Arg306. It is concluded that residue Asp95 within EGF1 is critical for APC cofactor function of protein S and could define a principal functional interaction site for APC.
Introduction
Protein S, a vitamin K–dependent plasma anticoagulant protein, functions as an enhancing cofactor to activated protein C (APC) in the inactivation of activated factors V (FVa) and VIII (FVIIIa).1 Protein S also has an APC-independent activity that has recently been attributed to its ability to enhance tissue factor pathway inhibitor (TFPI).2,3 Protein S has an important role in vivo, as is shown clinically by infants with complete deficiency, who suffer purpura fulminans, and by heterozygous carriers of PROS1 gene deletions and point mutations, who are at enhanced risk of venous thromboembolism.4,5 The importance of protein S has also been demonstrated in murine knockouts, which fail to survive development.6,7
Protein S binds phospholipids and helps to localize APC to the membrane surface, in proximity to FVa and FVIIIa. In doing so, rather than the preferential cleavage at Arg506 of FVa by APC, Arg306 is then favored, with cleavage enhanced approximately 20-fold.8 Although this effect has generally been accepted to be due to protein S repositioning of the active site of APC,9 this has recently been questioned.10 Cleavage at Arg506 and Arg306 fully inactivates FVa, thereby efficiently down-regulating coagulation.11 APC also inactivates FVIIIa by cleaving first at Arg336 and thereafter at Arg562. In the presence of protein S, it has been shown that the efficiency of cleavage after Arg336 by APC is enhanced approximately 3-fold.12
Protein S is a 635–amino acid glycoprotein, circulating at a plasma concentration of approximately 350nM. It is composed of an N-terminal Gla domain (amino acids 1-45), a thrombin-sensitive region (TSR; amino acids 46-75), 4 epidermal growth factor (EGF)–like domains (EGF1, amino acids 76-116; EGF2, amino acids 117-160; EGF3, amino acids 161-202; EGF4, amino acids 203-242), and a C-terminal sex hormone–binding globulin (SHBG)–like domain (amino acids 243-635).13,14 Approximately 60% of protein S circulates as a noncovalent, high-affinity complex with β-chain containing C4b binding protein.15 This interaction is mediated through the SHBG domain of protein S. Although it has been generally understood that the free form (∼ 40%) of protein S alone is important in anticoagulant function, this has recently been questioned.16
Whereas the functions of the domains of protein S have been extensively studied, the APC cofactor function of protein S remains poorly understood at the molecular level. Several basic and clinical studies have confirmed that alteration of protein S can disrupt phospholipid binding and thereby impair function of protein S.17,18 Saller et al prepared protein S/prothrombin chimeras and, on the basis of functional studies supported by molecular modeling, identified a cluster of protein S Gla domain residues (termed Face2), which may provide an APC interaction site.19
Additional studies have focused on the possible role of TSR and EGF domains. Several studies have reported that cleavage in the TSR of protein S affects its APC cofactor activity.20–22 Dahlbäck et al23 and Giri et al24 used domain-specific monoclonal antibodies directed against protein S and their Fab fragments and suggested that the TSR and EGF1 domains may be involved in APC cofactor activity. The TSR and EGF1 domains of protein S have also been reported to be responsible for species specificity.25 Using EGF-like, modules Stenberg et al concluded the EGF1 module to be crucial for interaction with APC.26 Hackeng et al demonstrated that the isolated EGF1 domain of protein S could inhibit full-length protein S anticoagulant enhancing function against APC: direct binding of the domain to APC was also demonstrated.27 Mille-Baker et al prepared EGF2 domain deletion and substitution mutants of protein S and demonstrated reduced anticoagulant activity, in the presence of normal phospholipid binding.28 Collectively, these investigators have suggested Gla, TSR, EGF1, and EGF2 domains may each contain recognition elements on protein S needed for APC cofactor function.
Because these diverse approaches have not yet fully defined the functional interaction site(s) on protein S for APC, we have investigated the role of individual and groups of residue substitutions in Gla, TSR, EGF1, and EGF2 domains of protein S. Using a library of 27 protein S variants, we performed thrombin generation assays in protein S–deficient plasma to which APC and these protein S variants were added. We report here that EGF1 has a critical role in anticoagulant function of protein S, provided specifically by Asp95, the mutation of which almost completely abolishes APC cofactor activity.
Methods
Generation and expression of protein S and its variants
A previous reported pcDNA6/protein S vector containing the cDNA for wild-type (WT) protein S29 was used to generate recombinant full-length WT protein S and as a template to produce protein S mutants by polymerase chain reaction site-directed mutagenesis (QuikChange XL Site-Directed Mutagenesis Kit; Stratagene) using mutagenic oligonucleotide primers (Thermo Scientific). Single and composite residue variants produced are shown in Table 1.
Table 1.
Protein S variants generated by site-directed mutagenesis
| GLA1 | D38N/V46G/K97Q |
|---|---|
| GLA2/Face2 | L21T/N23S/K24Y/R28F/D34S/Y41W/L45T |
| TSR1 | R49Q/R60Q/D68N/R70Q/D78N |
| EGF1 single-residue variants | K94A, D95A, K97A, S99A, T101A, T103A, K105A, P106A, W108A, Q109A, E111A, K112A |
| EGF2 single-residue variants | D135A, N136A, T137A, Y141A, H142A, S144A, K146A, S153A, N154A, K155A, K156A, D157A |
Residues to mutate were selected based on nonconserved residues in domains of other human coagulation factors (Gla mutants) and charge (TSR mutant). Residues in the B and C regions (between the third/fourth and fifth/sixth Cys residues, respectively) of both EGFs were mutated, as these regions on the EGF-like domains are likely to be involved in protein interaction as suggested by (1) the highly divergent sequence of the B region,30 (2) the association of dysfunctional proteins with natural PROS1 mutations in these regions,4 and (3) the location in these regions of many mutations associated with dysfunctional proteins in the other coagulation genes coding for EGF domains, such as FVII, FIX, and protein C.
WT and mutant protein S expression vectors were either transiently or stably transfected into HEK293T or HEK293 cells (ATCC), respectively. Stably transfected cells were selected with 5 μg/mL Blasticidin-HCl (Invitrogen) for 4 weeks. Protein S was expressed in OptiMem I (Invitrogen) with 10 μg/mL vitamin K (Roche Products Ltd) for 3 days. Media containing protein S was harvested, centrifuged, filtered, dialyzed in 20 mM Tris (tris(hydroxymethyl)aminomethane)–HCl (pH 7.5), 140 mM NaCl (TBS), and concentrated, as required. All variants were successfully expressed into the media and could be readily detected by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blotting and by enzyme-linked immunosorbent assay (ELISA). We have previously reported that the protein S expressed in this way is functionally active in APC cofactor assays and can be fully γ-carboxylated.17,31
Purification of protein S
WT protein S and selected variants were either partially purified by barium citrate precipitation as previously described32 for use in phospholipid binding analysis or extensively purified by chromatography for use in APC-dependent functional assays. Chromatography was performed using an ÄKTA purifier UPC-10 and the Unicorn 5.11 (Build 407) software, strategy Version 1.00 (both from GE Healthcare). Concentrated and dialyzed conditioned media containing protein S supplemented with 4 mM EDTA (ethylenediaminetetraacetic acid) was applied to an anionic Q Sepharose Fast Flow (5 mL of HiTrap QFF) column (GE Healthcare) equilibrated with 20mM Tris-HCl (pH 7.4), 100mM NaCl, 4mM EDTA, and 5mM benzamidine-HCl (Sigma-Aldrich). The column was washed with 20mM Tris-HCl (pH 7.4), 100mM NaCl, and 5mM benzamidine-HCl. Protein S was eluted from the column with 20mM Tris-HCl (pH 7.4), 0.5M NaCl, and 5mM benzamidine-HCl.
Two volumes of 7.5mM CaCl2 were added to the elution fraction from the anionic QFF column to lower the ionic strength, before injecting the fraction onto an affinity column prepared by coupling 2.5 mg of monoclonal antibody against protein S, MK21, to a 1-mL HiTrap NHS HP column (GE Healthcare), according to the manufacturer's instructions. All monoclonal antibodies against protein S used in this study are described in the paper of Dahlbäck et al.23 The protein S–containing fraction was applied to the equilibrated column (equilibration buffer: 50mM Tris-HCl [pH 7.5], 150mM NaCl, 2mM CaCl2). After washing with 1M NaCl, the ionic strength was lowered by passing additional 3 column volumes of equilibration buffer over the column prior to elution of protein S with 50mM Tris-HCl (pH 7.5), 150mM NaCl, and 10mM EDTA. An excess of CaCl2 was present in the tubes collecting the eluted protein S to neutralize the EDTA. The column was regenerated by stripping with 0.1M glycine-HCl (pH 2.7). Purified protein S was dialyzed in TBS and concentrated, as required.
Determination of protein S concentration by ELISA
Protein S concentrations in media were determined by an in-house ELISA. Polyclonal rabbit anti–protein S antibody (1 μg/mL; DAKO) was immobilized in a 96-well Nunc Maxisorp microplate in 50mM sodium carbonate buffer (pH 9.6) at 4°C overnight. Washing steps were performed in triplicate with 250 μL of phosphate-buffered saline, 0.1% Tween between each step. Wells were blocked with phosphate-buffered saline containing 3% bovine serum albumin (BSA; Sigma-Aldrich) for 2 hours. Bound protein S was detected by a mouse monoclonal antibody directed against the EGF3-4 domain of protein S, MK55, followed by a horseradish peroxidase (HRP)–conjugated goat anti–mouse antibody (DAKO). The plate was developed with 100 μL/well chromogenic substrate O-phenylene diamide dihydrochloride; OPD (Sigma-Aldrich), the enzymatic reaction was stopped with 50 μL/well 2M H2SO4, and the absorbance was read at 492 nm.
SDS-PAGE and Western blotting
SDS-PAGE and Western blotting were performed using standard techniques. Briefly, 4% to 12% or 10% precast NuPAGE Novex Bis-Tris gels (Invitrogen) were used. For silver staining and Western blotting, standard techniques were used. The SeeBlue-prestained marker from Invitrogen was used. Protein S was detected either by a polyclonal rabbit anti–protein S antibody followed by an HRP-conjugated goat anti–rabbit antibody or by a monoclonal mouse antibody recognizing only γ-carboxylated Glu residues (American Diagnostica Inc), followed by a goat anti–mouse HRP-conjugated antibody (DAKO).
Phospholipid vesicle preparation
Phospholipids mixtures (Avanti Polar Lipids Inc) in chloroform were prepared and the chloroform was evaporated under a nitrogen stream. The phospholipids were resuspended in TBS or 25mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (pH 7.7), 150mM NaCl (HN) for the plasma and purified assays, respectively. Unilamellar phospholipids vesicles were obtained by sonicating the phospholipids in an ice container at amplitude 22% for 7 minutes. Extruded phospholipids were prepared as previously described33 and used in the FVa inactivation assay. Synthetic phospholipids 1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-Dioleoyl-sn-glycero-3-phosphoserine (DOPS), and 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) were used in the plasma assay and the plate-binding assay. Natural phospholipids l-α-phosphatidylserine (PS; brain extract), l-α-phosphatidylethanolamine (PE; egg extract), and l-α-phosphatidylcholine (PC; egg extract) were used in the FVa inactivation and prothrombinase assays.
Binding of protein S to phospholipids
Phospholipids vesicles (DOPS/DOPC/DOPE, 20:60:20), 25 μg/mL, were coated as described for the ELISA. Washing steps were performed in triplicate with 250 μL/well TBS, 5mM CaCl2, 0.3% BSA between each step and incubations were carried out in a plate shaker at 37°C. Wells were blocked with TBS, 5mM CaCl2, 3% BSA for 2 hours. Protein S partially purified by barium citrate precipitation, 0 to 120nM, was incubated in the wells in duplicate for 2 hours and was detected with an HRP-conjugated anti–protein S antibody for 45 minutes (Affinity Biologicals). Data were analyzed with GraphPad Prism 4.03 and curves were fitted to a one-site binding hyperbola.
Binding of protein S to domain-specific monoclonal antibodies
Domain-specific monoclonal antibodies MK21 (Gla domain), MK54 (EGF1 domain), and MK61 (SHBG domain), 1 μg/mL, were coated as described for the ELISA. Washing steps were performed in triplicate with 250 μL of TBS, 3mM CaCl2, 0.1% Tween between each step. Incubations were carried out in a plate shaker at 37°C for 1 hour unless stated otherwise. Wells were blocked with 200 μL of TBS, 3mM CaCl2, 3% BSA for 2 hours. Protein S, 0 to 160nM was incubated in the plate for 1 hour and was detected by a rabbit polyclonal antibody against protein S followed by a goat anti–rabbit HRP-conjugated antibody.
Thrombin generation assay by calibrated automated thrombography
Thrombin generation was assessed using a Fluoroscan Ascent FL Plate Reader (Thermo Lab System) in combination with Thrombinoscope software (SYNAPSE BV). Protein S–deficient plasma (Affinity Biologicals), 80 μL, was incubated with 65 μg of corn trypsin inhibitor (Haematologic Technologies Inc) per milliliter of plasma to inhibit contact activation, 50μM phospholipid vesicles (DOPS/DOPC/DOPE, 20:60:20), 1pM tissue factor (Dade Innovin; Dade Behring), 4 to 16nM APC (Enzyme Research Laboratories Ltd) with 0 to 120nM protein S, in a final volume of 100 μL (all concentrations are final). A polyclonal antibody against TFPI (Haematologic Technologies Inc), 100nM, was used to inhibit any protein S cofactor activity toward TFPI. Although protein S cofactor activity toward TFPI was not observed at low concentrations of APC, it was observed to a small extent when lower amounts of thrombin were generated (eg, at higher concentrations of APC in the presence of protein S). Polyclonal antibodies against protein S and protein C were used to show the specificity of the system and were from, respectively, DAKO and Sigma-Aldrich. Thrombin generation was initiated by automatic dispensation of 20 μL of 2.5mM Z-GlyArg-AMC-HCl (Bachem), 2.5% Me2SO, 20mM Tris-HCl (pH 7.5), 60 mg/mL BSA, 100mM CaCl2 into each well. The reaction was performed at 37°C and measurements were performed with an excitation and emission wavelength of 390 nm and 460 nm, respectively.
Protein S–dependent APC-mediated factor FVa inactivation assay
A previously prepared and reported34 double mutant of FV, FV R506Q/R679Q, was activated with 0.5 U/mL human thrombin at 37°C for 10 minutes. The reaction was stopped by the addition of hirudin (5 U/mL, final concentration). The activated FVa variant was used as a substrate for APC to determine the ability of protein S to enhance cleavage of Arg306 of FVa by APC. The highly purified protein S preparations, 0 to 100nM, were incubated with 0.5nM APC, 25μM phospholipids vesicles (PS/PC/PE, 10:70:20), and 0.8nM FVa R506Q/R679Q in HN, 5mM CaCl2, 5 mg/mL BSA (HNBSACa2+) in a total volume of 50 μL (all concentrations are final). The solution was incubated at 37°C for 10 minutes, and the reaction was stopped by performing a 1:25-fold dilution in ice-cold HNBSACa2+. The remaining FVa activity was measured in a prothrombinase assay.
Time course experiments were also performed to derive the rate constant of FVa cleavage. APC, 0.5 and 3nM, and 50nM protein S were then used and aliquots were quenched at intervals between 0 and 20 minutes. The remaining FVa activity was again measured in a prothrombinase assay. To calculate the pseudo–first-order rate constant for APC-mediated cleavage at Arg306, the previously reported equation33 was used: Vat = Va0 × e−k306 × t + C × Va0 × (1 − e−k306 × t).
Vat represents the cofactor activity determined at time t, Va0 is the cofactor activity determined at time point 0, C is the remaining procoagulant cofactor activity of FVa after cleavage at position 306, and k306 is the rate constant of cleavage at position 306.
Prothrombinase assay
An aliquot, 25 μL, of the FVa inactivation reaction was incubated with phospholipid vesicles (PS/PC 10:90) and FXa in the presence of CaCl2. The reaction was initiated by the addition of prothrombin, in a final volume of 125 μL. The buffer contained HN and 0.5 mg/mL ovalbumin. Final concentrations were 50μM PS/PC, 5nM FXa, and 0.5μM prothrombin. The solution was incubated at 37°C for 2 minutes and the reaction was terminated by an 8-fold dilution in 50mM Tris-HCl (pH 7.9), 100mM NaCl, 20mM EDTA, 1% PEG 6000. The amount of thrombin generated was measured by cleavage of its chromogenic substrate S-2238 (Chromogenix) at 405 nm for 15 minutes at 30-second intervals.
Results
Screening of protein S mutants for APC cofactor activity in plasma
Concentrated and dialyzed conditioned media samples containing WT protein S and the 27 protein S variants were normalized with respect to protein S concentration, using the protein S–specific ELISA. Western blotting revealed single-band protein S in all samples (nonreducing conditions).
To assess the APC cofactor activity of protein S, thrombin generation was assessed by calibrated automated thrombography (CAT) using protein S–deficient plasma supplemented with recombinant WT protein S and its variants in conditioned media. Under the conditions of the assay, APC (0-10nM) did not affect peak, endogenous thrombin potential (ETP), lag time, or time to peak, when coagulation was initiated in protein S–deficient plasma (Figure 1A). Concentrated conditioned media from cells not expressing protein S had no influence on thrombin generation (data not shown). When 120nM WT protein S was progressively introduced into the assay, dose-dependent effects were observed, primarily on reduction of the peak and of ETP values (Figure 1B). The extent of inhibition of peak and ETP was dependent both on the concentration of APC and protein S. The higher the APC concentration, the less protein S was necessary to completely inhibit thrombin generation. In these experimental conditions, 10nM APC and 120nM protein S reduced both the peak and ETP by approximately 84%. To verify that those effects were a direct consequence of APC cofactor activity of protein S, polyclonal antibodies against either protein S (Figure 1C) or protein C (Figure 1D) were used. These inhibited the anticoagulant APC–protein S effect.
Figure 1.
Effect of APC and protein S on thrombin generation. Thrombin generation was performed in protein S–deficient plasma with 100nM inhibitory antibodies against TFPI. Up to 10nM APC had no effect on thrombin generation in the absence of protein S. All concentrations generate lines that are superimposable (A). After addition of 120nM protein S (at 0-10nM APC), an APC dose-dependent effect was observed (B). The top single line represents 0 to 10nM APC in the absence of protein S. Protein S in the presence of no or 2.5nM APC generated lines that were superimposable. Conditions used are noted adjacent to the peaks to which they refer. The anticoagulant effect of 10nM APC and 120nM protein S was inhibited by polyclonal antibodies against protein S (C) or against protein C (D). PS indicates protein S; PC, protein C. Representative experiments are shown (n = 3).
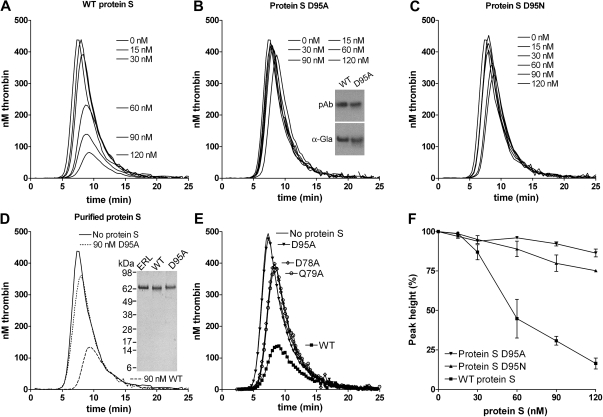
The screening of the protein S variants for their APC cofactor activity was performed at high concentration of APC (16nM) to achieve almost complete inhibition of thrombin generation with 100nM WT protein S (Figure 2). These screening experiments identified a single point mutant in EGF1 of protein S, D95A, which had a severely impaired APC cofactor activity. Under all conditions used, the reduction in cofactor activity was greater than that observed for the previously reported variant with 7 amino acids exchanged (prothrombin residue swap), GLA2/Face2.
Figure 2.
Screening of protein S variants for APC cofactor activity. The APC cofactor activity of protein S was evaluated at 16nM APC and 100nM protein S by CAT. The peak height in the absence of protein S was set to 100%. A high concentration of APC, leading to almost complete inhibition of thrombin generation with 100nM WT protein S, was chosen specifically for screening purposes as this allows widening of the assay window at which mutants with reduced APC cofactor activity are visualized. Results were confirmed by evaluating protein S (at 60 and 90nM) cofactor activity toward 4 or 9nM APC.
Evaluation of the importance of Asp95 for APC cofactor activity of protein S in plasma
To further evaluate the APC cofactor activity of protein S D95A, a titration (0-120nM) of the mutant (Figure 3B) was performed alongside WT protein S (Figure 3A), using 9nM APC. The protein S D95A had a severely impaired APC cofactor activity, a finding replicated when different concentrations of APC were used (results not shown). In these experimental conditions, whereas 120nM WT protein S reduced peak and ETP by approximately 80%, 120nM protein S D95A reduced peak and ETP by only approximately 10%. To confirm the importance of Asp95 in protein S APC cofactor function, Asp95 was also substituted with Asn, rather than Ala. Asn is structurally more similar to Asp than Ala and it can also be β-hydroxylated. When the protein S D95N mutant was titrated, it was observed that it also had severely reduced APC cofactor activity (Figure 3C) with 120nM D95N inhibiting peak and ETP by approximately 20%. All results in Figure 3A through C were conducted with protein S and its variants contained in concentrated conditioned media. To confirm that they were not influenced by the media, WT protein S and the protein S D95A variant were purified to homogeneity using anion exchange and affinity chromatography (see Figure 3D inset for SDS-PAGE gel visualized with silver staining). Addition of 90nM purified WT protein S to protein S–deficient plasma in the presence of 9nM APC appreciably attenuated thrombin generation (Figure 3D), replicating the findings with WT protein S in conditioned media. Moreover, addition of 90nM purified protein S D95A to APC produced only a minimal effect on thrombin generation (Figure 3D).
Figure 3.
Effect of WT protein S, D95A, D95N, D78A, and Q79A variants on thrombin generation. Thrombin generation was measured in protein S–deficient plasma supplemented with 9nM APC, 100nM antibodies against TFPI, and 0 to 120nM WT protein S (A), protein S D95A (B), protein S D95N (C), or 90nM purified WT (dashed line) or purified protein S D95A (dotted line; D). Protein S concentrations are positioned adjacent to the peaks to which they refer. The cofactor activity of 60nM WT protein S and protein S variants D95A, D78A, and Q79A was compared at 9nM APC (E). Typical experiments are shown (n = 3). Whereas the cofactor activity of WT protein S is highly dependent on the APC concentration used (Figure 1B), that of protein S D95A is not, explaining the difference in fold activity between WT protein S and protein S D95A in Figures 2 and 3. Dose-response data from titrations with WT protein S, protein S D95A, and protein S D95N in the presence of 9nM APC are shown in panel F (data are expressed as mean ± SD of 2 independent experiments performed in duplicate). Inset in panel B shows recognition of WT protein S and protein S D95A in media by polyclonal antibodies and a monoclonal antibody recognizing only γ-carboxylated Gla domains. Inset in panel D shows the SeeBlue-prestained marker, plasma-purified protein S from Enzyme Research Laboratories Ltd (lane 1), purified recombinant WT protein S (lane 2), and purified protein S D95A (lane 3) visualized with silver staining.
When analyzing a model35 of Gla-TSR-EGF1 domains of protein S, 2 residues with a solvent-exposed R residue in close proximity to that of Asp95 were identified, Asp78 and Gln79. These were also mutated to alanine and expressed. When protein S D78A and Q79A were analyzed in the thrombin generation assay alongside WT protein S, a severely reduced APC cofactor activity was also observed (Figure 3E). This further confirms the importance of this region of EGF1 in APC cofactor activity of protein S. Dose-response experiments of WT protein S, protein S D95A, and protein S D95N, shown in Figure 3F, confirm reduced APC cofactor function of these variants.
Phospholipid binding of WT protein S and protein S D95A
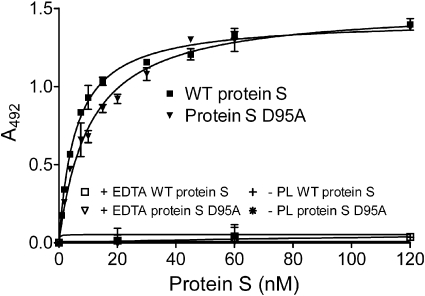
As phospholipid binding is a prerequisite for protein S function, the binding of WT protein S and of protein S D95A to phospholipids was evaluated. Partially purified (using barium citrate precipitation) protein S, 0 to 120nM, was incubated for 2 hours on a plate coated with 25 μg/mL phospholipids and was detected by an HRP-conjugated antibody against protein S. As shown in Figure 4, both WT protein S and protein S D95A were able to bind phospholipids, with apparent dissociation constant (Kdapp) of 5.69 plus or minus 1.24 and 9.54 plus or minus 2.26nM, respectively (Table 2). Kdapp values were analyzed by the Mann-Whitney test and were found not to be statistically different (P > .05). Binding was observed in the presence of Ca2+, but not in the presence of EDTA or in the absence of phospholipids, as expected. These findings are broadly consistent with previously reported Kd values for protein S binding to phospholipid surfaces.17–19,36,37 Our results suggest the loss of APC cofactor activity observed for protein S D95A is not due to loss of binding to phospholipid surfaces.
Figure 4.
Binding of protein S to phospholipid surfaces. Protein S (0-120nM) was incubated in a plate coated with 25 μg/mL phospholipids. Bound protein S was detected with an HRP-conjugated polyclonal antibody against protein S. A representative experiment is shown. The apparent Kd values, 5.69 ± 1.24 and 9.54 ± 2.26nM for WT protein S and protein S D95A, respectively, were obtained by calculating the mean ± SD of 3 independent experiments performed in duplicate. PL indicates phospholipids.
Table 2.
Binding of protein S to phospholipids and domain-specific monoclonal antibodies
| Phospholipid vesicles | MK21 (Gla) | MK54 (EGF1) | MK61 (SHBG) | |
|---|---|---|---|---|
| WT protein S | 5.69 ± 1.24 | 2.18 ± 0.97 | 0.81 ± 0.11 | 5.31 ± 1.11 |
| Asp95 protein S mutant | 9.54 ± 2.26 | 2.53 ± 1.06 | 0.83 ± 0.03 | 5.39 ± 0.54 |
Kdapp values (nM) of WT protein S and the Asp95 protein S mutant for phospholipid vesicles and conformational domain-specific monoclonal antibodies are expressed as mean plus or minus SD of 3 independent experiments performed in duplicate. Binding of WT protein S and the Asp95 mutant to phospholipids and domain-specific monoclonal antibodies was analyzed by Mann-Whitney test; results were not statistically significant (P > .05).
Binding to domain-specific monoclonal antibodies
Binding of protein S to conformational domain-specific antibodies was evaluated to assess the integrity of the domain structure and folding. Protein S in concentrated conditioned media was incubated in a plate coated with monoclonal antibodies recognizing either the Gla domain (MK21), the EGF1 domain (MK54), or the C-terminal SHBG domain (MK61). Bound protein S was detected with polyclonal antibodies. Binding curves were fitted with a one-site binding equation, and the Kdapp values were obtained. Kdapp values were analyzed by the Mann-Whitney test and were found not to be statistically different (P > .05). Table 2 represents the mean plus or minus SD Kdapp values of 3 independent experiments performed in duplicate. The results suggest that mutation of Asp95 does not result in a significant change in the domain structure of protein S.
Protein S enhancement of APC-mediated cleavage of FVa at Arg306
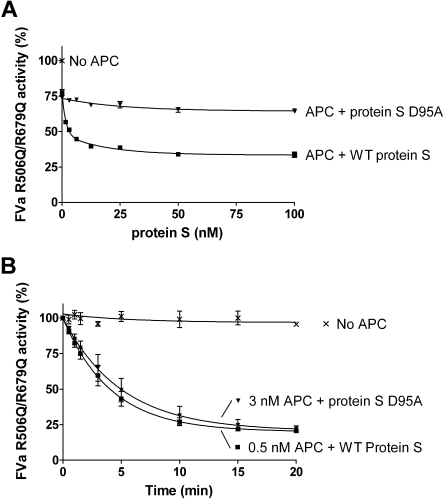
The FVa variant, R506Q/R679Q, was used to evaluate protein S–enhanced APC-mediated cleavage at Arg306 in FVa. FVa inactivation was performed in the presence or absence of 0.5nM APC and a titration of purified protein S (0-100nM) was performed. After 10 minutes, the remaining FVa activity was measured in the prothrombinase assay. WT protein S efficiently enhanced APC-mediated cleavage at Arg306 of FVa, in contrast to the D95A mutant (Figure 5A).
Figure 5.
Protein S enhancement of APC-mediated cleavage of FVa in Arg306. Protein S (0-120nM) in the presence of 0.5nM APC was incubated with 0.8nM FVa R506Q/R679Q in the presence of phospholipids for 10 minutes. The remaining FVa actvity was measured with a prothrombinase assay. Results are plotted as mean ± SD from 3 independent experiments performed in duplicate (A). A time course experiment was performed to calculate the apparent pseudo–first-order rate constants of WT protein S and protein S D95A. It is observed that approximately 6-fold more APC is needed in the presence of protein S D95A to obtain a similar amount of APC-mediated FVa R506Q/R679Q inactivation as with WT protein S (B).
To quantify the rate of cleavage at FVa Arg306 in the presence and absence of protein S, time course experiments were performed (Figure 5B). In the absence of protein S, or in the presence of protein S D95A, 3nM APC was used and aliquots were quenched at different time points (0-20 minutes). In the presence of WT protein S, the APC concentration was lowered to 0.5nM, as APC is efficiently enhanced by protein S. The remaining FVa activity at all time points was measured in the prothrombinase assay. Using the FVa inactivation curves obtained, the apparent pseudo–first-order rate constants were calculated and corrected for the APC concentrations used. Under these experimental conditions, protein S had no effect on FVa activity in the absence of APC (data not shown). APC-mediated cleavage at FVa Arg306 was enhanced by WT protein S by 13.9-fold plus or minus 3.6-fold, whereas protein S D95A was only able to enhance APC by 1.8-fold plus or minus 0.4-fold.
Discussion
We initially constructed and expressed 27 protein S variants with mutations in the Gla, TSR, EGF1, and EGF2 domains and evaluated their ability to enhance APC-mediated anticoagulant activity in plasma. The anticoagulant function of these 27 variants, and the subsequently expressed variants D78A, D79A, and D95N, were assessed by a thrombin generation assay, the specificity of which was demonstrated using polyclonal antibodies against either protein S or protein C that completely inhibited the anticoagulant response observed when adding protein S in the presence of APC. The advantage of the thrombin generation assay conducted in plasma is that it shows very clearly how APC is heavily dependent on protein S.38 In the absence of protein S, 10nM APC has no effect on thrombin generation. In contrast, in the presence of protein S, maximal anticoagulant effects with near ablation of thrombin generation are obtainable with APC concentrations less than 10nM.
Our initial screening results identified 2 protein S variants with appreciable reduction in APC cofactor function. These were the already reported GLA2/Face2 variant (a 7-residue composite mutant in the Gla domain) and the novel D95A point variant in the EGF1 domain. The former variant, reported by Saller et al, retains binding to phospholipids and yet has appreciably reduced APC cofactor activity.19 It is suggested that the 7 residues substituted in this variant collectively present a face of the Gla domain to APC and form a potential contact region. It was selected for investigation here because it is a well-characterized protein S variant with substantially reduced APC cofactor activity and could therefore act as a representative control for a dysfunctional protein S. Saller et al,19 however, did not assess GLA2/Face2 APC cofactor activity in the CAT assay. We can confirm a reduction in APC cofactor activity in plasma, assessed by CAT. The important novel results here, however, concerned the protein S D95A variant, which was almost devoid of APC cofactor activity in plasma. The natural protein S variants T103N39 and K155E40,41 have been reported to have reduced APC cofactor activity. We could not observe any significant reduction in APC cofactor activity of protein S T103A in the CAT assay. It is, however, important to point out that defective APC cofactor activity of protein S T103N was not observed in the APC-mediated inactivation of WT-FVa.39 In addition, at higher concentrations of APC (8nM), no difference was observed between WT protein S and protein S T103N in the FVIIIa degradation assay.39 We did, however, observe a decrease in K155A cofactor activity toward APC, although this was quite moderate under conditions used in the screening assay.
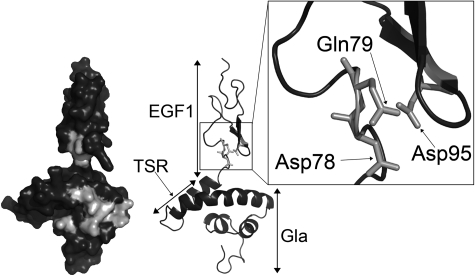
Analysis of protein S function is complicated by its propensity to multimerize upon purification.42,43 Accordingly, we have performed APC cofactor assays with unpurified protein S in concentrated conditioned media as well as with fully purified preparations. In the event, our results were all consistent. Both forms of the protein S D95A variant, purified or in concentrated conditioned media, had an appreciable reduction in APC cofactor activity compared with the respective WT protein S preparation. Although it has not been formally assessed, in all comparison experiments carried out, the protein S D95A variant had less APC cofactor than the GLA2/Face2 variant. This suggests that Asp95 in protein S may occupy a pivotal position with respect to its interaction with APC. The importance of Asp95 in APC cofactor activity was further confirmed by the severely reduced activity observed when mutating to alanine the 2 residues in close proximity to Asp95, Asp78, and Gln79. Asp78, Gln79, and Asp95 are conserved across species with the exception of Asp78, which is replaced by the structurally similar Asn78 in birds. Using a working model of the Gla-TSR-EGF1 domains of protein S, we demonstrate the likely proximal spatial location of these 3 residues (Figure 6) and their relationship to GLA2/Face2 residues.
Figure 6.
Location of Asp78, Gln79, and Asp95 within the protein S Gla-TSR-EGF1 model. Domains are labeled on the right side of the model. Residues mutated in the GLA2 variant, Asp78, Gln79, and Asp95, are in light gray on the left side surface model. Residues Asp78, Gln79, and Asp95 are highlighted by the box to show their proximal spatial location. The model is taken from Villoutreix et al.35
We have investigated the mechanism of reduction of activity of protein S Asp95 variants. Whereas EGF1 of protein S has not been shown to contain a calcium-binding site (in contrast to EGF2, EGF3, and EGF4),44 Asp95 could be part of a partially conserved calcium-binding motif. We therefore mutated Asp95 to Asn rather than Ala. Asn is structurally similar to Asp, can be β-hydroxylated, and is able to coordinate a calcium ion (as has been observed in EGF2, EGF3, and EGF4). As the protein S D95N variant also had reduced activity, however, it is unlikely that a calcium-binding site is disrupted. Furthermore, whereas Asp95 is known to be β-hydroxylated, it has previously been shown that β-hydroxylation itself is not a requirement for anticoagulant activity of protein S.45 Our evidence favors a direct effect of Asp95 in APC cofactor activity. Alternatively, the dramatic effect on protein S activity of the Asp95 residue substitution could be through a conformational repositioning of other functional domains. To assess this, we performed binding to phospholipids, as this property underpins all protein S function. Plate binding assays, however, indicated no major functional defect on phospholipid binding. Furthermore, domain-specific monoclonal antibody binding to WT and mutant protein S (against Gla domain, EGF1 domain, and the C-terminal SHBG domain) was also normal, suggesting that substitution of Asp95 does not reduce APC cofactor function by disrupting adjacent domain structure.
A current view of protein S–enhanced APC cofactor activity suggests a functional repositioning of the APC cleavage site away from FVa Arg506 toward FVa Arg306.9 We therefore performed a FVa inactivation assay using a FVa variant, FVa R506Q/R679Q, that cannot be cleaved at position 506 and 679. This allowed us to analyze directly cleavage at Arg306 by APC, which is at the site enhanced mainly by protein S. Using both concentration-dependent and time course assays, we were able to confirm reduced cleavage of this variant by APC in the presence of protein S D95A.
The available results therefore suggest that Asp95 of protein S may play an important and a direct role in APC recognition, resulting in enhanced APC function. Our results are compatible with the study performed by Hackeng et al,27 who used the isolated EGF1 domain of protein S to functionally disrupt the protein S and APC interaction. They suggested EGF1 as a potentially important APC contact site on protein S. Our results suggest that Asp95 constitutes a critical residue within EGF1, mediating the APC cofactor function. Together with Asp78 and Gln79, Asp95 could form the principal functional interaction site for APC.
Acknowledgments
The authors are grateful to Dr José Renan da Cunha Melo, Dr José Nélio Januário, and Dr Marcos Vinícius Andrade for their kind support on the sharing of cloning and tissue culture room. We are also grateful to Jamil Silvano de Oliveira for his helpful advice on purification techniques.
This work was supported by the International Research Development Award from Wellcome Trust, British Heart Foundation (PG/07/005), an unrestricted educational grant from Novartis, a Swedish council grant (07143), and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (PPM 2007).
Footnotes
Presented in part in abstract form at the International Society on Thrombosis and Haemostasis, Boston, MA, July 14, 2009; at the British Society for Haemostasis and Thrombosis, Newcastle, United Kingdom, October 7, 2009; and at the Congresso Brasileiro de Hematologia e Hemoterapia, Florianópolis, Brazil, November 11, 2009.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: H.M.A., D.A.L., S.M.R., B.D., and J.T.B.C. designed the research, analyzed the results, and wrote the paper; and H.M.A., M.J.A., S.M.R., B.M.L., and S.T. performed the experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David Lane, Department of Haematology, Faculty of Medicine, Imperial College London, Hammersmith Hospital Campus, Du Cane Rd, London W12 0NN, United Kingdom; e-mail: d.lane@imperial.ac.uk.
References
- 1.Dahlbäck B, Villoutreix BO. The anticoagulant protein C pathway. FEBS Lett. 2005;579(15):3310–3316. doi: 10.1016/j.febslet.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Hackeng TM, Sere KM, Tans G, Rosing J. Protein S stimulates inhibition of the tissue factor pathway by tissue factor pathway inhibitor. Proc Natl Acad Sci U S A. 2006;103(9):3106–3111. doi: 10.1073/pnas.0504240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ndonwi M, Broze G., Jr Protein S enhances the tissue factor pathway inhibitor inhibition of factor Xa but not its inhibition of factor VIIa-tissue factor. J Thromb Haemost. 2008;6(6):1044–1046. doi: 10.1111/j.1538-7836.2008.02980.x. [DOI] [PubMed] [Google Scholar]
- 4.Gandrille S, Borgel D, Sala N, et al. Protein S deficiency: a database of mutations: summary of the first update. Thromb Haemost. 2000;84(5):918. [PubMed] [Google Scholar]
- 5.Makris M, Leach M, Beauchamp NJ, et al. Genetic analysis, phenotypic diagnosis, and risk of venous thrombosis in families with inherited deficiencies of protein S. Blood. 2000;95(6):1935–1941. [PubMed] [Google Scholar]
- 6.Burstyn-Cohen T, Heeb MJ, Lemke G. Lack of Protein S in mice causes embryonic lethal coagulopathy and vascular dysgenesis. J Clin Invest. 2009;119(10):2942–2953. doi: 10.1172/JCI39325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saller F, Brisset AC, Tchaikovski SN, et al. Generation and phenotypic analysis of protein S-deficient mice. Blood. 2009;114(11):2307–2314. doi: 10.1182/blood-2009-03-209031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosing J, Hoekema L, Nicolaes GA, et al. Effects of protein S and factor Xa on peptide bond cleavages during inactivation of factor Va and factor VaR506Q by activated protein C. J Biol Chem. 1995;270(46):27852–27858. doi: 10.1074/jbc.270.46.27852. [DOI] [PubMed] [Google Scholar]
- 9.Yegneswaran S, Smirnov MD, Safa O, et al. Relocating the active site of activated protein C eliminates the need for its protein S cofactor: a fluorescence resonance energy transfer study. J Biol Chem. 1999;274(9):5462–5468. doi: 10.1074/jbc.274.9.5462. [DOI] [PubMed] [Google Scholar]
- 10.Tran S, Dahlback B. Novel APC-cleavage sites in FVa providing insights into mechanisms of action of APC and its cofactor protein S. J Thromb Haemost. 2010;8(1):129–136. doi: 10.1111/j.1538-7836.2009.03657.x. [DOI] [PubMed] [Google Scholar]
- 11.Mann KG, Hockin MF, Begin KJ, Kalafatis M. Activated protein C cleavage of factor Va leads to dissociation of the A2 domain. J Biol Chem. 1997;272(33):20678–20683. doi: 10.1074/jbc.272.33.20678. [DOI] [PubMed] [Google Scholar]
- 12.O'Brien LM, Mastri M, Fay PJ. Regulation of factor VIIIa by human activated protein C and protein S: inactivation of cofactor in the intrinsic factor Xase. Blood. 2000;95(5):1714–1720. [PubMed] [Google Scholar]
- 13.Gershagen S, Fernlund P, Lundwall A. A cDNA coding for human sex hormone binding globulin. Homology to vitamin K-dependent protein S. FEBS Lett. 1987;220(1):129–135. doi: 10.1016/0014-5793(87)80890-6. [DOI] [PubMed] [Google Scholar]
- 14.Lundwall A, Dackowski W, Cohen E, et al. Isolation and sequence of the cDNA for human protein S, a regulator of blood coagulation. Proc Natl Acad Sci U S A. 1986;83(18):6716–6720. doi: 10.1073/pnas.83.18.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rezende SM, Simmonds RE, Lane DA. Coagulation, inflammation, and apoptosis: different roles for protein S and the protein S-C4b binding protein complex. Blood. 2004;103(4):1192–1201. doi: 10.1182/blood-2003-05-1551. [DOI] [PubMed] [Google Scholar]
- 16.Maurissen LF, Thomassen MC, Nicolaes GA, et al. Re-evaluation of the role of the protein S-C4b binding protein complex in activated protein C-catalyzed factor Va-inactivation. Blood. 2008;111(6):3034–3041. doi: 10.1182/blood-2007-06-089987. [DOI] [PubMed] [Google Scholar]
- 17.Rezende SM, Lane DA, Mille-Baker B, et al. Protein S Gla-domain mutations causing impaired Ca(2+)-induced phospholipid binding and severe functional protein S deficiency. Blood. 2002;100(8):2812–2819. doi: 10.1182/blood-2002-03-0909. [DOI] [PubMed] [Google Scholar]
- 18.Baroni M, Mazzola G, Kaabache T, et al. Molecular bases of type II protein S deficiency: the I203-D204 deletion in the EGF4 domain alters GLA domain function. J Thromb Haemost. 2006;4(1):186–191. doi: 10.1111/j.1538-7836.2005.01682.x. [DOI] [PubMed] [Google Scholar]
- 19.Saller F, Villoutreix BO, Amelot A, et al. The gamma-carboxyglutamic acid domain of anticoagulant protein S is involved in activated protein C cofactor activity, independently of phospholipid binding. Blood. 2005;105(1):122–130. doi: 10.1182/blood-2004-06-2176. [DOI] [PubMed] [Google Scholar]
- 20.Chang GT, Aaldering L, Hackeng TM, et al. Construction and characterization of thrombin-resistant variants of recombinant human protein S. Thromb Haemost. 1994;72(5):693–697. [PubMed] [Google Scholar]
- 21.Heeb MJ, Griffin JH. Activated protein C-dependent and -independent anticoagulant activities of protein S have different structural requirements. Blood Cells Mol Dis. 2002;29(2):190–199. doi: 10.1006/bcmd.2002.0558. [DOI] [PubMed] [Google Scholar]
- 22.Long GL, Lu D, Xie RL, Kalafatis M. Human protein S cleavage and inactivation by coagulation factor Xa. J Biol Chem. 1998;273(19):11521–11526. doi: 10.1074/jbc.273.19.11521. [DOI] [PubMed] [Google Scholar]
- 23.Dahlbäck B, Hildebrand B, Malm J. Characterization of functionally important domains in human vitamin K-dependent protein S using monoclonal antibodies. J Biol Chem. 1990;265(14):8127–8135. [PubMed] [Google Scholar]
- 24.Giri TK, Villoutreix BO, Wallqvist A, Dahlback B, de Frutos PG. Topological studies of the amino terminal modules of vitamin K-dependent protein S using monoclonal antibody epitope mapping and molecular modeling. Thromb Haemost. 1998;80(5):798–804. [PubMed] [Google Scholar]
- 25.He X, Shen L, Villoutreix BO, Dahlback B. Amino acid residues in thrombin-sensitive region and first epidermal growth factor domain of vitamin K-dependent protein S determining specificity of the activated protein C cofactor function. J Biol Chem. 1998;273(42):27449–27458. doi: 10.1074/jbc.273.42.27449. [DOI] [PubMed] [Google Scholar]
- 26.Stenberg Y, Drakenberg T, Dahlback B, Stenflo J. Characterization of recombinant epidermal growth factor (EGF)-like modules from vitamin-K-dependent protein S expressed in Spodoptera cells: the cofactor activity depends on the N-terminal EGF module in human protein S. Eur J Biochem. 1998;251(3):558–564. doi: 10.1046/j.1432-1327.1998.2510558.x. [DOI] [PubMed] [Google Scholar]
- 27.Hackeng TM, Yegneswaran S, Johnson AE, Griffin JH. Conformational changes in activated protein C caused by binding of the first epidermal growth factor-like module of protein S. Biochem J. 2000;349(pt 3):757–764. doi: 10.1042/bj3490757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mille-Baker B, Rezende SM, Simmonds RE, et al. Deletion or replacement of the second EGF-like domain of protein S results in loss of APC cofactor activity. Blood. 2003;101(4):1416–1418. doi: 10.1182/blood-2002-08-2353. [DOI] [PubMed] [Google Scholar]
- 29.Rezende SM, Razzari C, Simmonds RE. In vitro high level protein S expression after modification of protein S cDNA. Thromb Haemost. 2003;90(6):1214–1215. [PubMed] [Google Scholar]
- 30.Komoriya A, Hortsch M, Meyers C, et al. Biologically active synthetic fragments of epidermal growth factor: localization of a major receptor-binding region. Proc Natl Acad Sci U S A. 1984;81(5):1351–1355. doi: 10.1073/pnas.81.5.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Persson KE, Stenflo J, Linse S, et al. Binding of calcium to anticoagulant protein S: role of the fourth EGF module. Biochemistry. 2006;45(35):10682–10689. doi: 10.1021/bi0601151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorand L, Mann KG. San Diego, CA: Academic Press; 1993. Proteolytic Enzymes in Coagulation, Fibrinolysis, and Complement Activation, Part A: Mammalian Blood Coagulation Factors and Inhibitors. [PubMed] [Google Scholar]
- 33.Tran S, Norstrom E, Dahlback B. Effects of prothrombin on the individual activated protein C-mediated cleavages of coagulation factor Va. J Biol Chem. 2008;283(11):6648–6655. doi: 10.1074/jbc.M708036200. [DOI] [PubMed] [Google Scholar]
- 34.Norstrøm EA, Steen M, Tran S, Dahlback B. Importance of protein S and phospholipid for activated protein C-mediated cleavages in factor Va. J Biol Chem. 2003;278(27):24904–24911. doi: 10.1074/jbc.M303829200. [DOI] [PubMed] [Google Scholar]
- 35.Villoutreix BO, Teleman O, Dahlback B. A theoretical model for the Gla-TSR-EGF-1 region of the anticoagulant cofactor protein S: from biostructural pathology to species-specific cofactor activity. J Comput Aided Mol Des. 1997;11(3):293–304. doi: 10.1023/a:1007912929828. [DOI] [PubMed] [Google Scholar]
- 36.Saller F, Kaabache T, Aiach M, Gandrille S, Borgel D. The protein S thrombin-sensitive region modulates phospholipid binding and the gamma-carboxyglutamic acid-rich (Gla) domain conformation in a non-specific manner. J Thromb Haemost. 2006;4(3):704–706. doi: 10.1111/j.1538-7836.2006.01827.x. [DOI] [PubMed] [Google Scholar]
- 37.Heeb MJ, Schuck P, Xu X. Protein S multimers and monomers each have direct anticoagulant activity. J Thromb Haemost. 2006;4(2):385–391. doi: 10.1111/j.1538-7836.2006.01743.x. [DOI] [PubMed] [Google Scholar]
- 38.Seré KM, Rosing J, Hackeng TM. Inhibition of thrombin generation by protein S at low procoagulant stimuli: implications for maintenance of the hemostatic balance. Blood. 2004;104(12):3624–3630. doi: 10.1182/blood-2004-03-1146. [DOI] [PubMed] [Google Scholar]
- 39.Giri TK, Garcia de Frutos P, Dahlback B. Protein S Thr103Asn mutation associated with type II deficiency reproduced in vitro and functionally characterised. Thromb Haemost. 2000;84(3):413–419. [PubMed] [Google Scholar]
- 40.Hayashi T, Nishioka J, Shigekiyo T, Saito S, Suzuki K. Protein S Tokushima: abnormal molecule with a substitution of Glu for Lys-155 in the second epidermal growth factor-like domain of protein S. Blood. 1994;83(3):683–690. [PubMed] [Google Scholar]
- 41.Tsuda H, Urata M, Tsuda T, et al. Four missense mutations identified in the protein S gene of thrombosis patients with protein S deficiency: effects on secretion and anticoagulant activity of protein S. Thromb Res. 2002;105(3):233–239. doi: 10.1016/s0049-3848(02)00015-4. [DOI] [PubMed] [Google Scholar]
- 42.Seré KM, Janssen MP, Willems GM, et al. Purified protein S contains multimeric forms with increased APC-independent anticoagulant activity. Biochemistry. 2001;40(30):8852–8860. doi: 10.1021/bi002500a. [DOI] [PubMed] [Google Scholar]
- 43.Seré KM, Willems GM, Rosing J, Hackeng TM. Protein S multimers are generated in vitro and affect protein S structure-function analyses. Semin Hematol. 2006;43(1 suppl 1):S111–S120. doi: 10.1053/j.seminhematol.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 44.Stenflo J, Stenberg Y, Muranyi A. Calcium-binding EGF-like modules in coagulation proteinases: function of the calcium ion in module interactions. Biochim Biophys Acta. 2000;1477(1-2):51–63. doi: 10.1016/s0167-4838(99)00262-9. [DOI] [PubMed] [Google Scholar]
- 45.Nelson RM, VanDusen WJ, Friedman PA, Long GL. Beta-Hydroxyaspartic acid and beta-hydroxyasparagine residues in recombinant human protein S are not required for anticoagulant cofactor activity or for binding to C4b-binding protein. J Biol Chem. 1991;266(31):20586–20589. [PubMed] [Google Scholar]