Abstract
Lateral diffusion of cell membrane constituents is a prerequisite for many biological functions. However, the diffusivity (or mobility) of a membrane-bound species can be influenced by many factors. To provide a better understanding of how the conformation and location of a membrane-bound biological molecule affect its mobility, herein we study the diffusion properties of a pH low insertion peptide (pHLIP) in model membranes using fluorescence correlation spectroscopy. It is found that when the pHLIP peptide is located on the membrane surface, its lateral diffusion is characterized by a distribution of diffusion times, the characteristic of which depends on the peptide/lipid ratio. Whereas, under conditions where pHLIP adopts a well-defined transmembrane α-helical conformation the peptide still exhibits heterogeneous diffusion, the distribution of diffusion times is found to be independent of the peptide/lipid ratio. Taken together, these results indicate that the mobility of a membrane-bound species is sensitive to its conformation and location and that diffusion measurement could provide useful information regarding the conformational distribution of membrane-bound peptides. Furthermore, the observation that the mobility of a membrane-bound species depends on its concentration may have important implications for diffusion-controlled reactions taking place in membranes.
Introduction
The lateral diffusion of lipids and absorbed biomolecules (e.g., proteins and peptides) in cell membranes is thought to play an important role in many biological processes as it may control the timescale within which key molecular events take place (1–6). For example, to form the functional oligomeric structures, membrane-bound antimicrobial peptides have to diffuse laterally to find each other. Thus, in this case the diffusion rate of the membrane-bound peptides may limit the rate of oligomer formation. Similarly, the lateral diffusion rate of membrane-bound α-helices may play an important role in determining the folding time of membrane proteins, as the two-state model of membrane protein folding (7) suggests that secondary structure formation precedes the formation of tertiary structures. Because of its importance, many studies have been performed to examine the diffusion properties of membrane-bound biological molecules (8–41).
Although these studies have demonstrated that the charge, size, and molecular shape of the diffusing species are important determinants of its diffusivity, many factors still remain unexplored. For example, the diffusion of a membrane-bound species could be significantly influenced by its conformation, location, and concentration. Among these potential influences, the effect of concentration is particularly worthwhile to investigate, as such effects would have important implications for interpreting the kinetics of diffusion-controlled reactions. Herein, in an attempt to provide further insight into factors that affect the diffusion of membrane-bound peptides, we measure the diffusivity of a pH (low) inserting peptide (pHLIP) in model membranes under different conditions using fluorescence correlation spectroscopy (FCS).
The pHLIP peptide used in this study is based on that designed by Engelman and co-workers (42–47). Our motives for choosing this peptide include the following:
-
1.
The interaction of pHLIP with model membranes is well studied and understood (46–48).
-
2.
The conformation and location of membrane-bound pHLIP molecules can be easily controlled by changing the solution pH: near neutral pH, the pHLIP binds to membrane surface as an extended chain, whereas at low pH it forms a transmembrane (TM) α-helix due to protonation of the aspartic acid residues.
-
3.
Perhaps most importantly, unlike antimicrobial peptides, the membrane-bound pHLIP molecules remain monomeric and do not induce membrane fusion or damage even at high peptide/lipid ratios.
Taken together, these unique membrane-binding properties of pHLIP make it an ideal model system to probe how those aforementioned factors affect the diffusion of membrane-bound peptides.
Fluorescence correlation spectroscopy (FCS) is based on correlating fluorescence intensity fluctuations arising from fluorescent molecules diffusing in and out of a small confocal volume, thus providing a convenient means to measure the diffusion time and hence the diffusion constant of the diffusing species (49,50). In addition, FCS allows observation of single molecules, and thus can be used to probe diffusion heterogeneity (51–53). Indeed, our results show that the diffusion of pHLIP in two model membranes, namely, supported lipid bilayers and giant unilamellar vesicles (GUVs), is heterogeneous and that the extent of the heterogeneity depends on pH and peptide concentration. Interestingly, the diffusion time distribution of the surface-bound pHLIP consists of two distinct peaks at high peptide/lipid ratios, indicating the feasibility of using FCS to probe the conformational distribution of membrane-bound peptides and proteins.
Materials and Methods
Materials
All materials were used as received. Fmoc-protected amino acids were purchased from Advanced Chem Tech (Louisville, KY). Rink amide PEGA resin was purchased from Novabiochem (San Diego, CA). Tetramethylrhodamine-5-maleimide (TMR-maleimide) was purchased from AnaSpec (San Jose, CA). Phospholipid 1-palmitoyl-2-oleoyl phosphatidylcholine (POPC) and 1-palmitoyl-2-oleoyl-phosphaticylglycerol (POPG) was purchased from Avanti Polar Lipids (Alabaster, AL). 2-[methoxy(polyethyleneoxy)propyl] trimethoxysilane (PEG) was purchased from Gelest (Morrisville, PA). Texas Red 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine, triethylammonium salt (Texas-Red-DHPE) was purchased from Molecular Probes (Eugene, OR).
Peptide synthesis and labeling
The pHLIP peptide (sequence: ACEQNPIYWARYADWLFTTPLLLLDLALLVDADEGTG) was synthesized using standard Fmoc-based solid-phase synthesis protocols employing a double-coupling strategy on a PS3 peptide synthesizer (Protein Technologies, Boston, MA) and purified by reverse-phase HPLC (1100 Series; Agilent Technologies, Quantum Analytics, Foster City, CA). The identity of the peptide was verified by MALDI-TOF mass spectrometry. The fluorescence probe, TMR-maleimide (AnaSpec), was attached to the peptide via the cysteine residue and the resultant TMR-labeled peptide product was further purified using a G-10 size-exclusion column.
Preparation of supported lipid bilayers and giant unilamellar vesicles
All membranes were made from POPC. Specifically, supported lipid bilayers were prepared from a pre-prepared POPC vesicle solution (2 mg/mL lipid and 10 mM phosphate buffer, pH 8.0 or 4.0). First, an aliquot (80 μL) of this vesicle solution was pipetted into a preheated (40°C) petri dish, and then a dry, PEG-covered glass-slip was placed on top of the vesicle solution. This assembly was further incubated at 40°C for ∼30 min, allowing for the formation of the lipid bilayer on top of the PEG surface of the glass-slip. Then the glass-slip was carefully removed from the bottom of the petri dish and rinsed out by copious phosphate buffer to remove any nonfused vesicles.
Giant unilamellar vesicles (GUVs) were prepared by the standard method of electroswelling (54) using a custom-made closed perfusion chamber and indium-tin-oxide (ITO) coated coverslips (Delta Technologies, Stillwater, MN) as electrodes. Briefly, 100 μL of 1 μM/mL POPC or POPC/POPG (3/1, mol/mol) lipid mixture solution in chloroform was first deposited on an ITO coverslip. After evaporation of the solvent, the chamber was assembled from two lipid-coated ITO coverslips separated by a rubber spacer and filled with 100 mM sucrose solution. A voltage of 1.2 V/mm at a frequency of 5 Hz was applied to the system for 2 h while incubating the chamber at 60°C. The final pH of the GUV solution (50 mM phosphate buffer) was adjusted to either 4.0 or 8.0. Same procedures were used to prepare POPC GUVs containing Texas Red-DHPE. The only difference is that the 1 μM/mL POPC chloroform solution also contains 0.002 mol % Texas Red-DHPE.
FCS sample preparation
For experiments conducted with supported lipid bilayers, a 30 μL peptide stock solution at the desired concentration and pH (4 or 8, 50 mM phosphate buffer) was added on top of the supported lipid bilayer and allowed to equilibrate for 1 h before FCS measurements. In all cases, the concentration of the TMR-labeled pHLIP in the peptide stock solution is 1 nM.
For experiments involving GUVs, the sample solution was prepared by mixing equal volumes of a peptide solution at the desired concentration and pH (50 mM phosphate buffer) and a GUV suspension in 100 mM sucrose solution. This mixed solution was then introduced into a custom-made, sealed FCS sample chamber and was allowed to equilibrate for 2 h, during which the GUVs settled to the bottom of the coverslip and remained stationary over the course of the experiment. For measurements involving 0.1 and 1 nM peptide, only TMR-labeled pHLIP was used. In all other cases, both labeled and unlabeled peptides were used, and the concentration of the labeled peptide was maintained at 1 nM. Finally, for diffusion measurements of the lipids (i.e., Texas Red-DHPE), only unlabeled peptides were used.
FCS setup and data analysis
The detail of the FCS apparatus has been described elsewhere (55). In this study, each FCS curve (1 μs–10 s) measuring the diffusion of the membrane-bound TMR-labeled pHLIP peptides was obtained by correlating the fluorescence signals for a duration of 40 s and further fit to the equation
| (1) |
where represents the characteristic two-dimensional diffusion time constant of membrane-bound species, and represents the three-dimensional diffusion time constant of unbound species in solution, which was determined from the diffusion of TMR-labeled pHLIP in buffer solution (see below). In all cases, we found that fb was <10%. Thus, only the two-dimensional diffusion time constants were considered and discussed in the Result and Discussion sections. Specifically, they were presented in the format of probability distribution, wherein the data were binned every 200 μs, and also the format of cumulative distribution.
On the other hand, those FCS curves obtained with Texas Red-DHPE in membranes were fit to the following two-dimensional diffusion model:
| (2) |
We found that for Texas Red-DHPE diffusion in membrane in the absence of pHLIP, all FCS curves can be adequately fit by a single diffusion component (i.e., n = 1). For Texas Red-DHPE diffusion in membrane in the presence of pHLIP, a small number of FCS curves required a second component (i.e., n = 2) to yield a good fit.
For those FCS curves obtained with TMR-labeled pHLIP in buffer solution, the following three-dimensional diffusion model was used to fit the data:
| (3) |
In all the equations above, ω refers to the axial/lateral dimension ratio of the confocal volume element, N represents the number of fluorescent molecules in the confocal volume, fi represents the fraction of the diffusion component i, τtriplet is the triplet lifetime of the fluorophore, and T represents the corresponding triplet amplitude. The value of ω was determined using the diffusion coefficient and the measured diffusion time of the fluorescent dye, R6G, in water. A nonlinear least-squares curve-fitting algorithm, which uses Gauss-Newton approach for the initial search of the fitting parameters and then the method of gradient descent for a finer search, was used to fit all the FCS data without weighting. For each case, representative FCS curves and the corresponding fits and residuals were given in Fig. S1, Fig. S2, Fig. S3, Fig. S4, Fig. S5, Fig. S6, Fig. S7, Fig. S8, and Fig. S9, which can be found in the Supporting Material.
Result
Diffusion behavior of pHLIP bound to a supported lipid bilayer
Supported lipid bilayers provide a convenient two-dimensional membrane environment for diffusion measurements. Thus, we first investigated the diffusion properties of pHLIP molecules that are bound to a supported lipid bilayer as a function of peptide concentration. To reduce any potential interactions between the peptide and the microscope coverslip on which the lipid bilayer was prepared, a layer of PEG polymer was added between the glass surface and the bilayer. As shown (Fig. 1 A and Fig. S10 A), at pH 8 and relatively low peptide concentration, wherein the peptide remains unfolded and is located at the membrane surface (42–47), repeating FCS measurements, each with an acquisition time of 40 s, indicate that the diffusion of pHLIP is very heterogeneous. Interestingly, as indicated (Fig. 1 B and Fig. S10 B), increasing the peptide concentration appears to decrease the heterogeneity of the diffusion time (τD) and also leads to a decrease in the average diffusion time of the peptide. However, FCS measurements using a fluorescent-tracer-labeled lipid (i.e., Texas-Red-DHPE) in the absence of pHLIP showed that the diffusion of the lipid is heterogeneous (inset of Fig. 2 B). Although this result is in agreement with that of Burns et al. (51), it also suggests that the heterogeneous diffusion of pHLIP observed in the current case could arise from interactions between peptide and the substrate or between lipid and the substrate. Nevertheless, control experiments showed that the characteristic diffusion times of pHLIP in buffer, obtained from multiple repeated measurements, are only distributed within a very narrow time range (Fig. 2). In addition, the diffusion constant of pHLIP in aqueous solution is determined to be (1.3 ± 0.1) × 10−6 cm2/s, which is consistent with those obtained on other peptides of similar size (56,57), whereas the modest decrease in the diffusion time at pH 4 compared to that at pH 8 is likely due to the increased hydrophobicity of the peptide upon protonation of the Asp residues. Thus, these control experiments suggest that a better model membrane is required to determine whether pHLIP exhibits heterogeneous diffusion when bound to membranes.
Figure 1.
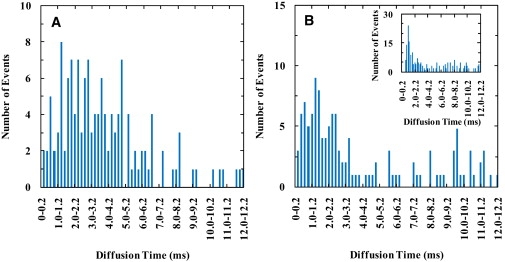
Diffusion time (τD) distributions of TMR-labeled pHLIP bound to a supported lipid bilayer of POPC at pH 8 and different total peptide concentrations: (A) 1 nM and (B) 1 μM. (Inset) Diffusion time (τD) distribution of Texas Red-DHPE (0.002 mol %) in supported lipid bilayer of POPC at pH 8.
Figure 2.
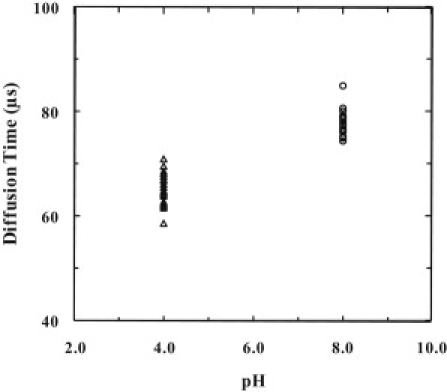
Diffusion times of TMR-labeled pHLIP in water at pH 4 and 8, respectively.
Diffusion behavior of pHLIP bound to GUVs
To eliminate any potential effects of the coverslip on the diffusion of the membrane-bound peptide, we carried out similar FCS measurements using GUVs consisting of either POPC or POPC/POPG mixture (3:1).
As shown (Fig. 3), the GUVs used in this study have a diameter of 5–50 μm and remain static on the timescale of the FCS experiments. Thus, by placing the focus of the excitation laser beam near the center of the upper membrane of the GUV, any interference arising from the coverslip can be eliminated (58). Because membrane undulations/drifting movements are known to introduce additional (or nondiffusion) components in the FCS curves (59), cares have been taken to exclude those FCS curves that contain such interferences. This was achieved by rejecting those fluorescence traces that showed an apparent decrease in the counting rate (during the FCS measurement) that was reversible upon refocusing (58,60) and those FCS curves that exhibit a very long (i.e., >1 s) component (61). As shown (Fig. 4, A–C), for POPC membranes the diffusion times of pHLIP at pH 8 clearly show a distribution (hereafter referred to as τD-distribution) whose position and width depend on the peptide concentration or the peptide/lipid ratio. It is apparent that at relatively high concentration, the distribution becomes bimodal, and the average diffusion time becomes shorter. As indicated (Fig. S11 and Fig. S12), a similar concentration dependence is also observed for POPC/POPG membranes at pH 8 although the overall change is less pronounced. In stark contrast, at pH 4 the τD-distribution of pHLIP shows little or no dependence on peptide concentration (Fig. 4, D–F). Taken together, these results indicate that the location and conformation of a membrane-bound peptide are important determinants of its diffusibility.
Figure 3.
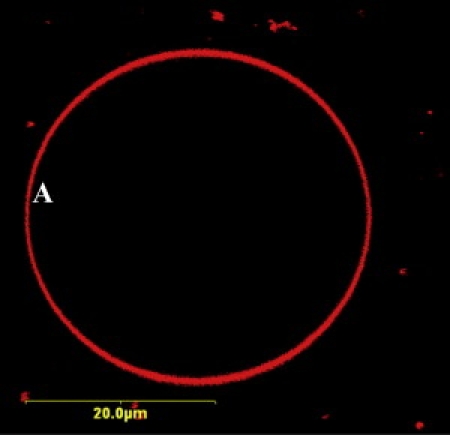
Confocal image of a representative POPC GUV. The fluorescence was derived from a small number of dye-labeled lipids (0.1 mol %, Texas Red-DHPE).
Figure 4.
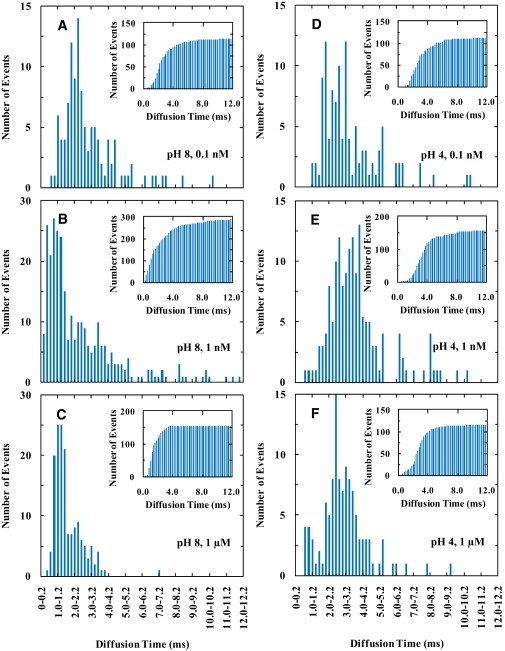
Diffusion time (τD) distributions of TMR-labeled pHLIP bound to the lipid bilayer of GUVs measured at different total peptide concentrations and pH values, as indicated. (Insets) Corresponding cumulative distribution of the diffusion times.
To better understand these data and to serve as a control, we also measured the lipid diffusion times in the POPC GUV membrane using a fluorescent-tracer-labeled lipid (i.e., Texas-Red-DHPE) in the presence and absence of pHLIP. As shown (Fig. 5 and Fig. S13), the measured lipid diffusion times in the absence of the peptide are narrowly distributed at ∼1.0 ms, indicating that the intrinsic lipid diffusion is more or less homogeneous. However, addition of the peptide affects the lipid diffusion in such a way that the mobility of the lipids depends on the concentration and location of the peptide (Figs. 6 and 7, and Fig. S14 and Fig. S15). For example, at pH 8 and relatively high peptide concentration (Fig. 6 and Fig. S14), the τD-distribution clearly shows a longer component, as compared to that obtained in the absence of pHLIP.
Figure 5.
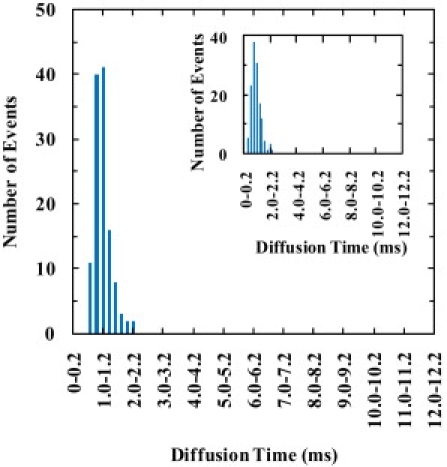
Diffusion time (τD) distribution of Texas Red-DHPE (0.002 mol %) in GUV membranes at pH 8. (Inset) Diffusion time (τD) distribution of Texas Red-DHPE (0.002 mol %) in GUV membranes at pH 4.
Figure 6.
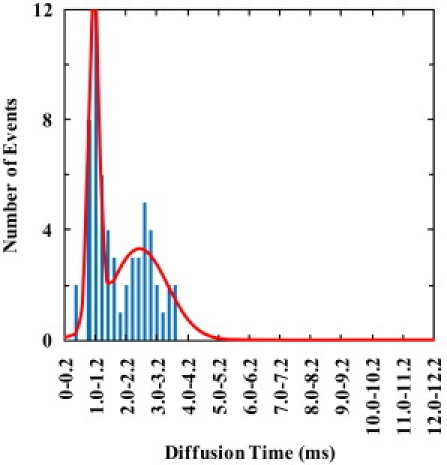
Diffusion time (τD) distribution of Texas Red-DHPE (0.002 mol %) in GUV membranes with the presence of 1 μM pHLIP (total concentration) at pH 8.
Figure 7.
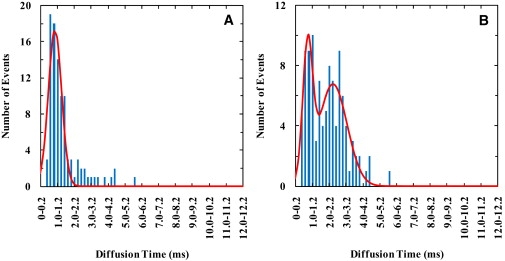
Diffusion time (τD) distributions of Texas Red-DHPE (0.002 mol %) in GUV membranes with the presence of (A) 1 nM and (B) 1 μM pHLIP (total concentration) at pH 4.
Discussion
While the diffusion of chemical constituents of cell membranes is of fundamental importance for many biological events and functions, very little is known about the timescale during which such diffusions occur and what affects their rate. Herein, we use a model peptide system (pHLIP) and FCS technique to investigate how the location, conformation, and concentration of the peptide affect its mobility in supported lipid bilayers and the membrane of GUVs. We find that even for such a relatively simple system, its diffusion is heterogeneous and shows complex dependence on those aforementioned factors.
Diffusion of pHLIP bound to membrane surface
At pH 8 and relatively low peptide concentrations (e.g., 0.1 nM), the lateral diffusion of pHLIP molecules on the membrane surface shows a broad τD-distribution (Fig. 4 A and Fig. S11 D), indicating the heterogeneous nature of the peptide's mobility. Previously it has been shown that FCS is able to follow protein unfolding transition (62) and that repeat FCS measurements are even capable of unmasking the conformational heterogeneity of unfolded proteins, provided that the unfolded conformers have distinctly different hydrodynamic radii and they interconvert slowly in comparison to their respective transit times (i.e., τD) through the confocal volume (63). The observation that the diffusion of pHLIP is characterized by a broad τD distribution indicates that the membrane-bound peptide samples an ensemble of slowly interconverting conformations. This picture is entirely consistent with the study of Engelman and co-workers (45), which showed that under similar conditions the surface-bound pHLIP adopts an ensemble of extended and unstructured conformations (45). In addition, a correlation existing between peptide conformation and diffusivity is readily explicable by the fact that the lateral mobility of a surfaced-bound molecule depends on the strength of its interactions with the membrane, a quantity directly related to the molecular conformation.
Of particular interest is that the (mean) diffusion time of the surface-bound pHLIP molecules depends on peptide concentration (or more precisely the peptide/lipid ratio), which could have important implications for non-unimolecular reactions. As shown (Fig. 4, A–C, and Fig. S11), increasing the peptide/lipid ratio results in a narrower τD-distribution and also a shift of the peak position of the distribution toward a shorter time for both POPC and POPC/POPG membranes. In addition, the τD-distribution obtained at high peptide concentrations can be described reasonably well by two components (Fig. 4 C), centered at ∼1.0 and 2.5 ms for POPC membrane, respectively, indicating that the pHLIP molecules can sample (at least) two distinguishable conformational ensembles that interconvert slowly.
The diffusion behavior of surface-bound pHLIP at pH 8 can be understood in terms of peptide surface coverage. At low concentration (e.g., 0.1 nM), where the peptide surface coverage is low, peptide molecules rarely encounter each other. Thus, each can freely explore the entire conformational space determined by the underlying pHLIP-membrane interactions, generating a broad distribution of conformations (45), which in turn gives rise to a broad τD-distribution as different conformations show different diffusion rates. Furthermore, at low surface coverage, the average number of lipids that interact with one peptide molecule is expected to be maximized, causing the peptide to be less mobile and hence a slower diffusion time. On the other hand, at high surface coverage (e.g., at 1 μM peptide concentration), the surface-bound peptides are more densely packed and thus experience self-crowding. As a result, each peptide molecule interacts, on average, with a smaller number of lipid molecules, making the molecules in the ensemble look alike in terms of their interactions with the membrane. Therefore, the τD-distribution becomes narrower and the peptides diffuse, on average, faster. In fact, the major component of the τD-distribution obtained at 1 μM pHLIP (Fig. 4 C) almost coincides with that of the lipids in the membrane (Fig. 5), which further corroborates the idea that at high surface coverage each peptide is interacting with fewer lipids. The above assessments are also consistent with a previous study by Engelman and co-workers, albeit in a qualitative manner, which showed that at pH 8 the interactions between pHLIP and POPC membranes depend on the peptide/lipid ratio (47). By analyzing the free energy change associated with binding of pHLIP to POPC bilayers at different peptide/lipid ratios, they concluded that there are two types of pHLIP-membrane interactions. Specifically, their results suggest that when the peptide/lipid ratio is low enough so that the entire peptide chain can interact with the membrane, type II interaction dominates and one peptide interacts (on average) with 124 lipid molecules. On the other hand, at high peptide/lipid ratios wherein the total accessible membrane surface area is not large enough to prevent peptide self-crowding, the membrane-bound peptides adopt a bent conformation wherein only part of the peptide chain is interacting with the membrane. In this partial, or type I interaction, one peptide interacts with 57 lipids, on average (47). In addition, our results are in agreement with those of Liu et al. (64), who have shown that the diffusivity of a series of homologous amphiphilic molecules on model membranes depends on their size and position. In particular, their results show that the diffusion constant of those amphiphiles that reside near the membrane surface decreases with increasing the molecular surface area. In other words, the diffusivity of an adsorbate molecule depends on the number of lipids it is interacting with or its interfacial area.
Because the present pHLIP peptide carries a net charge of −6 at pH 8, its diffusion behavior is expected to depend on the electrostatics of the membrane. Indeed, addition of negatively charged lipids (i.e., POPG) to the POPC membrane affects the τD-distribution of pHLIP. As shown (Fig. S12), the τD-distribution exhibits a weaker dependence on (bulk) peptide concentration, as compared to that obtained with zwitterionic POPC membranes. Because at pH 8 pHLIP is expected to have a weaker affinity toward negatively charged membranes and hence a lower membrane surface coverage for a given peptide concentration, this result thus corroborates the aforementioned notion that the peptide density on the membrane surface is one of the key determinants of its diffusion heterogeneity.
Peptide adsorption is expected to also affect the mobility of the membrane lipids, especially those that are directly interacting with the adsorbate molecules. Indeed, the τD-distribution of the lipids, which is determined by measuring the diffusion time of a very small number of fluorescently-labeled lipids that are added into the membrane of GUVs, depends on whether pHLIP is absent (Fig. 5) or present (Fig. 6). As shown (Fig. 6), in the presence of 1 μM pHLIP the τD-distribution of the lipids consists of two distinct components, centered at ∼1.0 and 2.6 ms, respectively. It is apparent that the fast component is similar to that observed when pHLIP is absent and that the entire distribution is similar to the τD-distribution of the peptide (i.e., Fig. 4 C) obtained under the same conditions. Thus, these results suggest that the surface-bound pHLIP molecules are moving together, as a loosely packed cluster, with the lipids with which they interact. Similar to what is observed here, a previous study by Zhang and Granick (52) has shown that when the number of lipids that interact with one surface-bound polymer molecule is >80, the diffusion of the lipids is slaved to the diffusion of the absorbed substrates and is slow, whereas when the number of lipids that interact with one polymer molecule is <80, the lipids diffuse faster with a rate similar to that of unperturbed lipids.
Diffusion of transmembrane-bound pHLIP molecules
The diffusion times of the TM-bound pHLIP, obtained at pH 4, show a single distribution which is insensitive to the peptide concentration (Fig. 4, D–F). This result suggests that the TM-bound pHLIP samples one conformational ensemble, a picture that is consistent with the study of Engelman and co-workers (45–47). However, the τD-distribution is still considerably broader than that (i.e., Fig. 5) obtained with unperturbed lipids, indicative of heterogeneity in pHLIP diffusion. Because, under the current experimental conditions, pHLIP does not form aggregates or oligomers (45), this broad τD-distribution thus manifests the heterogeneity in the solvation of the TM-bound pHLIP by lipid molecules. To provide further insights into this point, we measured the mobility of the lipids in the presence of pHLIP at pH 4. As shown (Fig. 7), the τD-distributions of the lipids obtained at different peptide/lipid ratios show that the mobility of certain lipid molecules is affected by the presence of pHLIP. At 1 μM peptide concentration, it is apparent that a slow diffusion component emerges as a result of peptide-membrane interactions. Based on the Saffman and Delbruck model (64–66), a diffusing object that shows a diffusion time comparable to the slow diffusion component would have a radius in the range of 1.0–3.0 nm, which was estimated to be equivalent to a diffusing species consisting of one pHLIP molecule and 6–26 lipids (assuming that the radius of the α-helix and that of the lipid headgroup are 0.6 and 0.5 nm, respectively, and that the lipids are tightly packed). Remarkably, this simple estimate is in excellent agreement with the result of Engelman and co-workers (47) that a single TM-bound pHLIP is solvated by 9–16 lipid molecules, distributed in two solvation layers surrounding the α-helix, thus further demonstrating the sensitivity, as well as the uniqueness, of the FCS method in studying peptide-membrane interactions. In addition, we believe that this technique is potentially applicable to probe the transiently populated local lipid clusters in membranes (67).
Conclusions
In summary, we have measured the diffusion times (or rates) of a membrane-bound pHLIP peptide through a well-defined confocal volume under different conditions. Our results show that the lateral mobility of this peptide in membranes depends on its conformation as well as its location. When the peptide is adsorbed to the membrane surface where it is unstructured and thus can sample a large number of conformations, individual molecules can show very different diffusion times although the shape and position of the diffusion time distribution depends on the peptide/lipid ratio or surface coverage of the peptide. At sufficiently low peptide/lipid ratios where individual peptide molecules do not experience self-crowding, the diffusion time distribution is composed of one component. Interestingly, increasing the peptide/lipid ratio causes an increase in the average diffusion rate of the surface-bound peptides but a decrease in the width of the distribution. Moreover, at high peptide/lipid ratios, the diffusion time distribution is composed of two distinct components, indicating that under such conditions two distinguishable (by the current method) conformational ensembles are populated. In particular, at sufficiently high peptide/lipid ratios, the diffusion time of the fast diffusion component of the peptide approaches that of the lipid in the membrane. On the other hand, under conditions where the peptide adopts a TM-oriented α-helical conformation, the diffusion time distribution is independent of the peptide/lipid ratio, indicating that the binding of the peptide perturbs fewer lipid molecules. Taken together, these results show that FCS is a useful technique for revealing the conformational heterogeneity of membrane-bound peptides or proteins and that the characteristic FCS diffusion time is a good indicator of how many lipid molecules are interacting with the diffusing species.
Supporting Material
Fifteen figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(10)00418-2.
Supporting Material
Acknowledgments
We thank Hector J. Melendez Gonzalez for labeling the pHLIP peptide with TMR and Dr. Tobias Baumgart and his group for assisting with the preparation of the GUVs. We also thank Dr. Pramit Chowdhury for his critical reading of the manuscript and helpful comments.
We gratefully acknowledge financial support from the National Science Foundation (grant No. DMR05-20020).
References
- 1.Liebman P.A., Pugh E.N. The control of phosphodiesterase in rod disk membranes: kinetics, possible mechanisms, and significance for vision. Vision Res. 1979;19:375–380. doi: 10.1016/0042-6989(79)90097-x. [DOI] [PubMed] [Google Scholar]
- 2.Pastan I.H., Willingham M.C. Journey to the center of the cell: role of the receptosome. Science. 1981;214:504–509. doi: 10.1126/science.6170111. [DOI] [PubMed] [Google Scholar]
- 3.Hackenbrock C.R. Lateral diffusion and electron transfer in the mitochondrial inner membrane. Trends Biochem. Sci. 1981;6:151–154. [Google Scholar]
- 4.Koppel D.E. Measurement of membrane protein lateral mobility. In: Hesketh T.R., Kornberg H.L., Metcalfe J.C., Northcote D.H., Pogson C.I., Tipton K.F., editors. Techniques in the Life Sciences, B4/II. Lipid and Membrane Biochemistry. Elsevier Biomedical Press; County Clare, Ireland: 1982. [Google Scholar]
- 5.Gross D., Webb W.W. Molecular counting of low-density lipoprotein particles as individuals and small clusters on cell surfaces. Biophys. J. 1986;49:901–911. doi: 10.1016/S0006-3495(86)83718-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters R. Lateral mobility of proteins and lipids in the red cell membrane and the activation of adenylate cyclase by β-adrenergic receptors. FEBS Lett. 1988;234:1–7. doi: 10.1016/0014-5793(88)81290-0. [DOI] [PubMed] [Google Scholar]
- 7.Popot J.-L., Engelman D.M. Helical membrane protein folding, stability, and evolution. Annu. Rev. Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- 8.Koppel D.E., Sheetz M.P., Schindler M. Matrix control of protein diffusion in biological membranes. Proc. Natl. Acad. Sci. USA. 1981;78:3576–3580. doi: 10.1073/pnas.78.6.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters R., Cherry R.J. Lateral and rotational diffusion of bacteriorhodopsin in lipid bilayers: experimental test of the Saffman-Delbrück equations. Proc. Natl. Acad. Sci. USA. 1982;79:4317–4321. doi: 10.1073/pnas.79.14.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frey S., Tamm L.K. Membrane insertion and lateral diffusion of fluorescence-labeled cytochrome c oxidase subunit IV signal peptide in charged and uncharged phospholipid bilayers. Biochem. J. 1990;272:713–719. doi: 10.1042/bj2720713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bussell S.J., Koch D.L., Hammer D.A. Effect of hydrodynamic interactions on the diffusion of integral membrane proteins: diffusion in plasma membranes. Biophys. J. 1995;68:1836–1849. doi: 10.1016/S0006-3495(95)80360-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pralle A., Keller P., Hörber J.K. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J. Cell Biol. 2000;148:997–1008. doi: 10.1083/jcb.148.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwille P. Fluorescence correlation spectroscopy and its potential for intracellular applications. Cell Biochem. Biophys. 2001;34:383–408. doi: 10.1385/CBB:34:3:383. [DOI] [PubMed] [Google Scholar]
- 14.Fujiwara T., Ritchie K., Kusumi A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 2002;157:1071–1081. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niv H., Gutman O., Henis Y.I. Activated K-Ras and H-Ras display different interactions with saturable nonraft sites at the surface of live cells. J. Cell Biol. 2002;157:865–872. doi: 10.1083/jcb.200202009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pampel A., Kärger J., Michel D. Lateral diffusion of a transmembrane peptide in lipid bilayers studied by pulsed field gradient NMR in combination with magic angle sample spinning. Chem. Phys. Lett. 2003;379:555–561. [Google Scholar]
- 17.Shvartsman D.E., Kotler M., Henis Y.I. Differently anchored influenza hemagglutinin mutants display distinct interaction dynamics with mutual rafts. J. Cell Biol. 2003;163:879–888. doi: 10.1083/jcb.200308142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cézanne L., Lecat S., Lopez A. Dynamic confinement of NK2 receptors in the plasma membrane. Improved FRAP analysis and biological relevance. J. Biol. Chem. 2004;279:45057–45067. doi: 10.1074/jbc.M404811200. [DOI] [PubMed] [Google Scholar]
- 19.Kenworthy A.K., Nichols B.J., Lippincott-Schwartz J. Dynamics of putative raft-associated proteins at the cell surface. J. Cell Biol. 2004;165:735–746. doi: 10.1083/jcb.200312170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusumi A., Ike H., Fujiwara T. Single-molecule tracking of membrane molecules: plasma membrane compartmentalization and dynamic assembly of raft-philic signaling molecules. Semin. Immunol. 2005;17:3–21. doi: 10.1016/j.smim.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 21.Scheidt H.A., Huster D., Gawrisch K. Diffusion of cholesterol and its precursors in lipid membranes studied by 1H pulsed field gradient magic angle spinning NMR. Biophys. J. 2005;89:2504–2512. doi: 10.1529/biophysj.105.062018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vrljic M., Nishimura S.Y., McConnell H.M. Cholesterol depletion suppresses the translational diffusion of class II major histocompatibility complex proteins in the plasma membrane. Biophys. J. 2005;88:334–347. doi: 10.1529/biophysj.104.045989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gambin Y., Lopez-Esparza R., Urbach W. Lateral mobility of proteins in liquid membranes revisited. Proc. Natl. Acad. Sci. USA. 2006;103:2098–2102. doi: 10.1073/pnas.0511026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golebiewska U., Gambhir A., McLaughlin S. Membrane-bound basic peptides sequester multivalent (PIP2), but not monovalent (PS), acidic lipids. Biophys. J. 2006;91:588–599. doi: 10.1529/biophysj.106.081562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ries J., Schwille P. Studying slow membrane dynamics with continuous wave scanning fluorescence correlation spectroscopy. Biophys. J. 2006;91:1915–1924. doi: 10.1529/biophysj.106.082297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conchonaud F., Nicolas S., Matarazzo V. Polysialylation increases lateral diffusion of neural cell adhesion molecule in the cell membrane. J. Biol. Chem. 2007;282:26266–26274. doi: 10.1074/jbc.M608590200. [DOI] [PubMed] [Google Scholar]
- 27.Jin S., Haggie P.M., Verkman A.S. Single-particle tracking of membrane protein diffusion in a potential: simulation, detection, and application to confined diffusion of CFTR Cl− channels. Biophys. J. 2007;93:1079–1088. doi: 10.1529/biophysj.106.102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lajoie P., Partridge E.A., Nabi I.R. Plasma membrane domain organization regulates EGFR signaling in tumor cells. J. Cell Biol. 2007;179:341–356. doi: 10.1083/jcb.200611106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mashanov G.I., Molloy J.E. Automatic detection of single fluorophores in live cells. Biophys. J. 2007;92:2199–2211. doi: 10.1529/biophysj.106.081117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pooler A.M., McIlhinney R.A.J. Lateral diffusion of the GABAB receptor is regulated by the GABAB2 C terminus. J. Biol. Chem. 2007;282:25349–25356. doi: 10.1074/jbc.M702358200. [DOI] [PubMed] [Google Scholar]
- 31.Gaborski T.R., Clark A., McGrath J.L. Membrane mobility of β2 integrins and rolling associated adhesion molecules in resting neutrophils. Biophys. J. 2008;95:4934–4947. doi: 10.1529/biophysj.108.132886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guigas G., Weiss M. Influence of hydrophobic mismatching on membrane protein diffusion. Biophys. J. 2008;95:L25–L27. doi: 10.1529/biophysj.108.136069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malengo G., Andolfo A., Caiolfa V.R. Fluorescence correlation spectroscopy and photon counting histogram on membrane proteins: functional dynamics of the glycosylphosphatidylinositol-anchored urokinase plasminogen activator receptor. J. Biomed. Opt. 2008;13:031215. doi: 10.1117/1.2940570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sung B.J., Yethiraj A. Lateral diffusion of proteins in the plasma membrane: spatial tessellation and percolation theory. J. Phys. Chem. B. 2008;112:143–149. doi: 10.1021/jp0772068. [DOI] [PubMed] [Google Scholar]
- 35.Wieser S., Schütz G.J. Tracking single molecules in the live cell plasma membrane—Do's and Don't's. Methods. 2008;46:131–140. doi: 10.1016/j.ymeth.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 36.Alcor D., Gouzer G., Triller A. Single-particle tracking methods for the study of membrane receptors dynamics. Eur. J. Neurosci. 2009;30:987–997. doi: 10.1111/j.1460-9568.2009.06927.x. [DOI] [PubMed] [Google Scholar]
- 37.Casuso I., Kodera N., Scheuring S. Contact-mode high-resolution high-speed atomic force microscopy movies of the purple membrane. Biophys. J. 2009;97:1354–1361. doi: 10.1016/j.bpj.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciobanasu C., Harms E., Kubitscheck U. Cell-penetrating HIV1 TAT peptides float on model lipid bilayers. Biochemistry. 2009;48:4728–4737. doi: 10.1021/bi900365s. [DOI] [PubMed] [Google Scholar]
- 39.Hammond G.R.V., Sim Y., Irvine R.F. Reversible binding and rapid diffusion of proteins in complex with inositol lipids serves to coordinate free movement with spatial information. J. Cell Biol. 2009;184:297–308. doi: 10.1083/jcb.200809073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knight J.D., Falke J.J. Single-molecule fluorescence studies of a PH domain: new insights into the membrane docking reaction. Biophys. J. 2009;96:566–582. doi: 10.1016/j.bpj.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tannert A., Tannert S., Schaefer M. Convolution-based one- and two-component FRAP analysis: theory and application. Eur. Biophys. J. 2009;38:649–661. doi: 10.1007/s00249-009-0422-4. [DOI] [PubMed] [Google Scholar]
- 42.Hunt J.F., Rath P., Engelman D.M. Spontaneous, pH-dependent membrane insertion of a transbilayer α-helix. Biochemistry. 1997;36:15177–15192. doi: 10.1021/bi970147b. [DOI] [PubMed] [Google Scholar]
- 43.Reshetnyak Y.K., Andreev O.A., Engelman D.M. Translocation of molecules into cells by pH-dependent insertion of a transmembrane helix. Proc. Natl. Acad. Sci. USA. 2006;103:6460–6465. doi: 10.1073/pnas.0601463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andreev O.A., Dupuy A.D., Reshetnyak Y.K. Mechanism and uses of a membrane peptide that targets tumors and other acidic tissues in vivo. Proc. Natl. Acad. Sci. USA. 2007;104:7893–7898. doi: 10.1073/pnas.0702439104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reshetnyak Y.K., Segala M., Engelman D.M. A monomeric membrane peptide that lives in three worlds: in solution, attached to, and inserted across lipid bilayers. Biophys. J. 2007;93:2363–2372. doi: 10.1529/biophysj.107.109967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zoonens M., Reshetnyak Y.K., Engelman D.M. Bilayer interactions of pHLIP, a peptide that can deliver drugs and target tumors. Biophys. J. 2008;95:225–235. doi: 10.1529/biophysj.107.124156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reshetnyak Y.K., Andreev O.A., Engelman D.M. Energetics of peptide (pHLIP) binding to and folding across a lipid bilayer membrane. Proc. Natl. Acad. Sci. USA. 2008;105:15340–15345. doi: 10.1073/pnas.0804746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang J., Gai F. Dissecting the membrane binding and insertion kinetics of a pHLIP peptide. Biochemistry. 2008;47:8250–8252. doi: 10.1021/bi801103x. [DOI] [PubMed] [Google Scholar]
- 49.Magde D., Elson E.L., Webb W.W. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers. 1974;13:29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 50.Haustein E., Schwille P. Fluorescence correlation spectroscopy: novel variations of an established technique. Annu. Rev. Biophys. Biomol. Struct. 2007;36:151–169. doi: 10.1146/annurev.biophys.36.040306.132612. [DOI] [PubMed] [Google Scholar]
- 51.Burns A.R., Frankel D.J., Buranda T. Local mobility in lipid domains of supported bilayers characterized by atomic force microscopy and fluorescence correlation spectroscopy. Biophys. J. 2005;89:1081–1093. doi: 10.1529/biophysj.105.060327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang L., Granick S. Slaved diffusion in phospholipid bilayers. Proc. Natl. Acad. Sci. USA. 2005;102:9118–9121. doi: 10.1073/pnas.0502723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo L., Chowdhury P., Gai F. Heterogeneous and anomalous diffusion inside lipid tubules. J. Phys. Chem. B. 2007;111:14244–14249. doi: 10.1021/jp076562n. [DOI] [PubMed] [Google Scholar]
- 54.Mathivet L., Cribier S., Devaux P.F. Shape change and physical properties of giant phospholipid vesicles prepared in the presence of an AC electric field. Biophys. J. 1996;70:1112–1121. doi: 10.1016/S0006-3495(96)79693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chowdhury P., Wang W., Gai F. Fluorescence correlation spectroscopic study of serpin depolymerization by computationally designed peptides. J. Mol. Biol. 2007;369:462–473. doi: 10.1016/j.jmb.2007.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tjernberg L.O., Pramanik A., Rigler R. Amyloid β-peptide polymerization studied using fluorescence correlation spectroscopy. Chem. Biol. 1999;6:53–62. doi: 10.1016/S1074-5521(99)80020-9. [DOI] [PubMed] [Google Scholar]
- 57.Sengupta P., Garai K., Maiti S. The amyloid β peptide (Aβ1–40) is thermodynamically soluble at physiological concentrations. Biochemistry. 2003;42:10506–10513. doi: 10.1021/bi0341410. [DOI] [PubMed] [Google Scholar]
- 58.Kahya N., Schwille P. Fluorescence correlation studies of lipid domains in model membranes. Mol. Membr. Biol. 2006;23:29–39. doi: 10.1080/09687860500489099. [DOI] [PubMed] [Google Scholar]
- 59.Milon S., Hovius R., Wohland T. Factors influencing fluorescence correlation spectroscopy measurements on membranes: simulations and experiments. Chem. Phys. 2003;288:171–186. [Google Scholar]
- 60.Bacia K., Scherfeld D., Schwille P. Fluorescence correlation spectroscopy relates rafts in model and native membranes. Biophys. J. 2004;87:1034–1043. doi: 10.1529/biophysj.104.040519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fradin C., Abu-Arish A., Elbaum M. Fluorescence correlation spectroscopy close to a fluctuating membrane. Biophys. J. 2003;84:2005–2020. doi: 10.1016/S0006-3495(03)75009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sherman E., Itkin A., Haran G. Using fluorescence correlation spectroscopy to study conformational changes in denatured proteins. Biophys. J. 2008;94:4819–4827. doi: 10.1529/biophysj.107.120220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo L., Chowdhury P., Gai F. Denaturant-induced expansion and compaction of a multi-domain protein: IgG. J. Mol. Biol. 2008;384:1029–1036. doi: 10.1016/j.jmb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu C., Paprica A., Petersen N.O. Effects of size of macrocyclic polyamides on their rate of diffusion in model membranes. Biophys. J. 1997;73:2580–2587. doi: 10.1016/S0006-3495(97)78287-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saffman P.G., Delbrück M. Brownian motion in biological membranes. Proc. Natl. Acad. Sci. USA. 1975;72:3111–3113. doi: 10.1073/pnas.72.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramadurai S., Holt A., Poolman B. Lateral diffusion of membrane proteins. J. Am. Chem. Soc. 2009;131:12650–12656. doi: 10.1021/ja902853g. [DOI] [PubMed] [Google Scholar]
- 67.Falck E., Róg T., Vattulainen I. Lateral diffusion in lipid membranes through collective flows. J. Am. Chem. Soc. 2008;130:44–45. doi: 10.1021/ja7103558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


