Abstract
KChIP4a shows a high homology with other members of the family of Kv channel-interacting proteins (KChIPs) in the conserved C-terminal core region, but exhibits a unique modulation of Kv4 channel gating and surface expression. Unlike KChIP1, the KChIP4 splice variant KChIP4a has been shown to inhibit surface expression and function as a suppressor of channel inactivation of Kv4. In this study, we sought to determine whether the multitasking KChIP4a modulates Kv4 function in a clamping fashion similar to that shown by KChIP1. Injection of Kv4.3 T1 zinc mutants into Xenopus oocytes resulted in the nonfunctional expression of Kv4.3 channels. Coexpression of Kv4.3 zinc mutants with WT KChIP4a gave rise to the functional expression of Kv4.3 current. Oocyte surface labeling results confirm the correlation between functional rescue and enhanced surface expression of zinc mutant proteins. Chimeric mutations that replace the Kv4.3 N-terminus with N-terminal KChIP4a or N-terminal deletion of KChIP4a further demonstrate that the functional rescue of Kv4.3 channel tetramerization mutants depends on the KChIP4a core region, but not its N-terminus. Structure-guided mutation of two critical residues of core KChIP4a attenuated functional rescue and tetrameric assembly. Moreover, size exclusion chromatography combined with fast protein liquid chromatography showed that KChIP4a can drive zinc mutant monomers to assemble as tetramers. Taken together, our results show that KChIP4a can rescue the function of tetramerization-defective Kv4 monomers. Therefore, we propose that core KChIP4a functions to promote tetrameric assembly and enhance surface expression of Kv4 channels by a clamping action, whereas its N-terminus inhibits surface expression of Kv4 by a mechanism that remains elusive.
Introduction
Kv channel formation requires early tetramerization of the pore-forming α-subunit T1 domain at the cytoplasmic N-terminus of the protein, followed by additional trafficking steps and interactions with auxiliary proteins for completion of the final functional channel complex at the membrane surface (1–6). In Kv4 Shal channels, the T1 (tetramerization) domain features a hallmark intersubunit Zn2+ ion that is found on non-Shaker-type Kv channel T1 domains (7,8). The Zn2+-binding site at the intersubunit interface reveals the characteristic motif in which the cysteine doublet and histidine are from the same subunit and the third cysteine is from a neighboring subunit (8–13). One important function of the intersubunit Zn2+-binding sites is to mediate the T1-T1 domain interaction, which promotes and stabilizes tetrameric channel assembly. Mutations of the Zn2+-binding residues give rise to nonfunctional channel expression by disrupting subunit tetramerization (9,10). Coexpression of auxiliary Kv channel-interacting proteins (KChIPs) can rescue the functional expression of Kv4 Zn2+-binding-site mutants by promoting tetrameric channel assembly and surface expression (9,14).
Cytosolic Kv channel-interacting proteins KChIPs1–4 (216–256 amino acids) coassemble with pore-forming Kv4 subunits to form a native complex that encodes a somatodendritic A-type K+ current. This current operates in the subthreshold range of membrane potentials and plays a critical role in shaping the action potential waveforms and regulating the interspike interval and action potential duration in neurons (15–24). In hippocampal CA1 neurons, Kv4 channels are primarily expressed in distal dendrites, where they function to regulate the amplitude of back-propagating action potentials (25,26). Auxiliary KChIPs1–4 have a conserved carboxy-terminal core region that contains four EF-hand-like calcium-binding motifs, and in general they increase Kv4 current expression by promoting channel trafficking to the membrane surface. It is of interest that KChIPs exist as splice variants with alternative amino-terminal regions that have been proposed to mediate the diverse modulation of Kv4 function (13,15–19,27–29). For instance, KChIP1 increases Kv4 surface expression, speeds up and reshapes the overall development of inactivation, and accelerates recovery from inactivation (15,30). In contrast, the KChIP4 splice variant KChIP4a, which overlaps in its C-terminal core region with KChIP1, neither promotes Kv4 surface expression nor has any effect on the recovery time constant after inactivation. Instead, it suppresses inactivation by eliminating fast inactivation of Kv4 channels (6,27,31–33). The effect of KChIP4a on inactivation has been attributed to the distinct function of its K-channel inactivation suppressor (KIS) domain, which resides in the N-terminus (residues 1–34) of KChIP4a (27).
X-ray crystal structures of auxiliary KChIP1 from previous studies have provided some structural insights into how different KChIPs might modulate Kv4 channel function (8,14,34–36). The structural complex of N-terminal Kv4.3 and KChIP1 revealed a unique clamping action, in which a single KChIP1 molecule as a monomer laterally clamps two neighboring Kv4.3 N-termini in a 4:4 manner, forming an octameric channels. The proximal N-terminal peptide of Kv4.3 binds to an elongated groove on the surface of KChIP1, and the same KChIP1 molecule binds to an adjacent T1 domain of another Kv4.3 subunit to stabilize the tetrameric Kv4.3 channels (8,14,34–36). A recent structural study revealed a distinctive fork-like conformation of the KChIP4a N-terminal α-helices that distinguishes it from other KChIPs (37). Biochemical analysis of competitive binding showed that the Kv4.3 N-terminal peptide competes with the N-terminus of KChIP4a for binding to the hydrophobic groove in the core of KChIP4a, freeing the KChIP4a N-terminus so that it can suppress the inactivation of Kv4.3 channels. This gave rise to the hypothesis that the distinctive KChIP4a N-terminus and its core function independently to exert the diverse modulation characteristic of Kv4 channels (37). Given the structural similarity of the conserved core of these two molecules, the question arises as to whether KChIP4a functions in a manner generally similar to KChIP1, which is known to promote tetrameric assembly and trafficking by clamping Kv4 subunits together, and whether the core of KChIP4a can override the effect of its N-terminus, which has been proposed to function as a transmembrane domain that causes endoplasmic reticulum retention (38,39).
In this study, we demonstrate that coexpression of Kv4.3 zinc mutants with wild-type (WT) KChIP4a gives rise to functional expression of the mutant Kv4.3. An oocyte surface-labeling assay further confirmed that KChIP4a can increase surface expression of Kv4.3 C110A mutant proteins. A biochemistry assay with size exclusion chromatography (SEC) and fast protein liquid chromatography (FPLC) demonstrated that KChIP4a can drive zinc mutant monomers to assemble as tetramers. Taken together, our results indicate that core KChIP4a modulates and interacts with Kv4 by a clamping action, resulting in the functional rescue of Kv4 mutant subunits residing predominantly in a monomeric state. The N-terminus of KChIP4a, however, functions independently as an inhibitory domain to suppress surface expression of Kv4 by a mechanism that remains elusive.
Results
Functional rescue and characterizations of Kv4.3 zinc mutant channels by KChIP4a
To test whether KChIP4a can rescue the function of Kv4.3 zinc mutants that disrupt tetrameric assembly, we generated three zinc mutations: a single mutant (Kv4.3 T1 C110A), a double mutant (C131A-C132A), and a triple mutant (C110A-C131A-C132A). Each of these mutations within the Zn2+-binding sites has been reported to disrupt tetrameric assembly. Consistent with previous reports, expression of each zinc mutant alone in Xenopus oocytes resulted in nonfunctional currents, whereas WT Kv4.3 showed a typical transient outward current with fast inactivation (Fig. 1 A). By contrast, coexpression of each zinc mutant with KChIP4a gave rise to a functional current with characteristics of instantaneous activation and ablated fast inactivation (Fig. 1 A). The peak current amplitude of rescued C110A mutant channels was 9.2 ± 0.7 μA (n = 21), which is fourfold greater than the peak amplitude of WT Kv4.3 and KChIP4a channels (1.8 ± 0.2 μA, n = 13; Fig. 1 B and Table S1 in the Supporting Material). Since KChIP4a was capable of rescuing all three zinc Kv4.3 mutants with similar gating effects, we chose the less-disruptive point mutation C110A for the remainder of these studies (Fig. 1, C and D).
Figure 1.
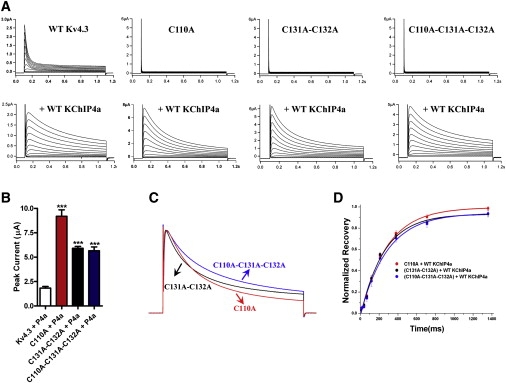
Functional rescue of Kv4.3 zinc mutants by coexpression of KChIP4a. (A) Current traces of WT Kv4.3 or zinc mutant channels with and without coexpression with WT KChIP4a were recorded in oocytes held at −80 mV in response to a family of depolarizing potentials from −60 mV to +40 mV in 10 mV increments for 1 s. Upper panel: Current traces of WT Kv4.3 or zinc mutant channels alone. Bottom panel: Current traces of coexpression of WT Kv4.3 or zinc mutant channels with WT KChIP4a. (B) Summary of peak currents recorded at +40 mV from panel A. Data are shown as the mean ± SE for 8–24 oocytes, and statistical significance (Student's t-test) is indicated as ∗∗∗P < 0.001. (C) Scaled and superimposed currents recorded at +40 mV from panel A are displayed to compare the development of inactivation kinetics among three zinc mutants with coexpression with WT KChIP4a. (D) Summary of normalized recovery from inactivation fitted with a single exponential function for varying lengths of time in steps from –80 to +40 mV, measured at +40 mV from zinc mutants with coexpression with WT KChIP4a.
The activation (τact) and fast components of inactivation time constants (τinact, fast) of rescued C110A channels were 3.0 ± 0.2 ms (n = 9) and 149.5 ± 9.1 ms (n = 13), respectively. Both are faster than those found for WT Kv4.3 coexpressed with KChIP4a (9.0 ± 0.7 ms, n = 8 for activation, and 413.4 ± 42.1 ms, n = 8 for inactivation; Fig. 2 A and Table S1). The speed of recovery from inactivation of rescued C110A channels is also faster at 205.1 ± 4.6 ms (n = 8), as compared with 328.2 ± 15.8 ms (n = 5) for the WT Kv4.3/KChIP4a channel complex (Fig. 2 B and Table S1). Analysis of tail current activation of the rescued C110A mutant showed that the half activation voltage (V1/2) calculated by Boltzmann distribution was 6.1 mV (n = 12), similar to 5.8 mV (n = 10) for WT Kv4.3/KChIP4a. In contrast, the prepulse inactivation of rescued C110A channel showed a significant rightward shift of ∼25 mV compared to the WT Kv4.3/KChIP4a complex (Fig. 2 C and Table S1).
Figure 2.

Gating properties of rescued Kv4.3 C110A zinc mutant by KChIP4a. (A) Left panel: Scaled and superimposed currents at +40 mV to compare the development of macroscopic activation (τact) and inactivation kinetics between coexpression of WT Kv4.3/KChIP4a and C110A zinc mutant/KChIP4a. Middle and right panels: Comparison of activation and fast components of inactivation time constants (τinact, fast) at +40 mV between WT Kv4.3/KChIP4a and C110A/KChIP4a (in the panels, P4a denotes KChIP4a). The activation was fitted to curve with a single exponential equation, and the inactivation was fitted to curve with a double exponential equation. (B) Left panel: Recovery from inactivation of WT or C110A mutant channels coexpressed with KChIP4a. Right panel: Comparison of time constants for recovery from inactivation between WT Kv4.3/KChIP4a and C110A/KChIP4a. (C) Left panel: Tail current activation and prepulse inactivation plots of WT and C110A mutant channels. Middle and right panels: Comparison of V1/2 values for tail current activation and prepulse inactivation. A significant difference was found between C110A and WT Kv4.3 in the V1/2 of prepulse inactivation, but not in the tail current activation. Note that there is a strong (25 mV) rightward shift in prepulse inactivation for C110A mutant channels compared to WT Kv4.3. All data are shown as the mean ± SE for 5–13 oocytes, and statistical significance is indicated as ∗∗P < 0.005 and ∗∗∗P < 0.001.
These results show that KChIP4a can drive nonfunctional mutant subunits to form functional channels, and suggest that functional rescue of the Kv4.3 zinc mutant by KChIP4a may utilize a clamping action mechanism similar to that observed for KChIP1, which clamps Kv4 together to promote and stabilize the tetrameric assembly. To further confirm that KChIP4a promotes tetrameric assembly of Kv4 by clamping, we performed mutagenesis experiments in the region that mediates Kv4 and KChIP1 interaction and clamping (14).
Attenuation of functional rescue by mutating residues critical for clamping
Our previous studies of the cocrystal structure of the Kv4.3N/KChIP1 complex showed that KChIP1 modulates Kv4.3 in a clamping mode (37). In this mode, a single KChIP1 molecule, as a monomer, laterally binds to two neighboring Kv4.3 N-termini in a 4:4 manner. The interaction involves two contact interfaces. In the first interface, the proximal N-terminal peptide of Kv4.3 is sequestered and buried deep within an elongated groove on the surface of KChIP1. In the second interface, the same KChIP1 molecule binds to another adjacent T1 domain of a second Kv4.3 subunit, resulting in a stabilized tetrameric Kv4.3 T1-KChIP1 complex. If KChIP4a modulates the Kv4.3 zinc mutant in a clamping mode similar to that observed for KChIP1, then two intersubunit interfaces must be involved in mediating the interaction between the Kv4.3 zinc mutant and KChIP4a. To prove this hypothesis, we made mutations or deletions within the two interfaces and tested whether these mutations can attenuate the functional rescue of zinc mutants by KChIP4a.
Based on their structural alignment, the two residues Y70 and K74 of N-terminal KChIP4a are identical to those of KChIP1, which were previously shown to mediate the second contact interface for the clamping action of Kv4 channels (Fig. 3 A). To test the effects of these two residues on functional rescue, we made a KChIP4a Y70A-K74A double mutant. Coexpression of this double mutant resulted in a fourfold reduction of the rescued current of the C110A mutant channel as compared to coexpression of WT KChIP4a, demonstrating that these two residues, which are critical for KChIP1 clamping of Kv4, are also critical for the functional rescue of KChIP4a (Fig. 3, B–D, and Table S1). Although the rescue current was significantly reduced by the double mutant, the inactivation kinetics of the C110A mutant remained almost unchanged (τinact, fast = 192.5 ± 8.7 ms, n = 8) in comparison with WT KChIP4a (τinact, fast = 149.5 ± 9.1 ms, n = 13; Fig. 3 E and Table S1). The speed of recovery from inactivation of C110A channels rescued by the double mutant was slower at 444.1 ± 31.2 ms (n = 8), compared to 278.0 ± 8.7 ms (n = 8) for the Kv4.3 C110A/KChIP4a channel complex (Fig. 3 F and Table S1).
Figure 3.

Double mutant Y70A-K74A of KChIP4a attenuates the functional rescue of the C110A zinc mutant. (A) Sequence alignment of the KChIP4a peptide (64-83) with the corresponding sequence of KChIP1 (residues 51–70). Residues shadowed in yellow denote amino acids that are conserved in KChIP1 and KChIP4a. Asterisks denote the two residues that when mutated in KChIP1 disrupt the second interface interaction with Kv4.3, as reported in our previous study. (B) Coexpression of Kv4.3 C110A and KChIP4a (Y70A-K74A); current traces were recorded with the same protocol as in Fig. 1. (C) Representative current traces of Kv4.3 C110A alone (black line), C110A/WT KChIP4a (red), and C110A/KChIP4a Y70A-K74A (blue) recorded at +40 mV from Xenopus oocytes. (D) Summary of normalized peak current amplitudes measured at +40 mV from Kv4.3 C110A alone and coexpression with WT or double mutant KChIP4a. Data are shown as mean ± SE for 8–25 oocytes. ∗∗∗P < 0.001 (Student's t-test), comparison between C110A as control and C110A/WT KChIP4a or C110A/KChIP4a Y70A-K74A double mutant; ##P < 0.001 (Student's t-test), comparison between C110A/WT KChIP4a and C110A/KChIP4a Y70A-K74A double mutant. (E) Fast component of the time constant of inactivation (τinact, fast) measured at +40 mV from C110A/WT KChIP4a and C110A/KChIP4a Y70A-K74A. Data are shown as mean ± SE for 8–13 oocytes. (F) Summary of time constants of recovery (τrecovery) from inactivation for varying lengths of time in steps from −80 to +40 mV, measured at +40 mV for coexpression of Kv4.3-C110A and different KChIPs. Data are shown as mean ± SE for 8–10 oocytes. ∗∗∗P < 0.001, comparison between controls (either C110A/WT KChIP1 or C110A/WT KChIP4a) and C110A/ KChIPs mutants.
Previous studies have shown that the proximal N-terminus of Kv4 is a primary binding site in the first interface for KChIP interaction, as N-terminal deletions in Kv4 resulted in the loss of KChIP modulation (8,14). Therefore, we constructed a C110A zinc mutant with N-terminal deletion of residues 2–24 to test whether N-terminal Kv4.3 is critical for mediating the rescue effect of KChIP4a. To our surprise, this N-terminal deletion of the C110A mutant (Kv4.3Δ24-C110A) resulted in a functional current when expressed in oocytes (Fig. 4 A), whereas the C110A point mutation alone was nonfunctional (Fig. 1 A). A similar deletion with a triple mutation, Kv4.3Δ24-C110A-C131A-C132A, also gave rise to functional expression (data not shown). These results suggest that N-terminal deletion of Kv4.3 has some rescuing effect on the C110A zinc mutant, consistent with the report that deletion of the N-terminal retention signal of Kv4 channels resulted in increased current expression (40).
Figure 4.

The proximal N-terminus of Kv4.3 is necessary for interaction of the C110A mutant and KChIP4a. (A and B) Current traces of the deletion mutant Kv4.3Δ24-C110A with and without WT KChIP4a were recorded in oocytes held at −80 mV in response to a family of depolarizing potentials from −60 mV to +40 mV in 10 mV increments for 1 s. (C) The scaled and superimposed currents at +40 mV (from panels A and B) are shown to compare the development of inactivation kinetics among Kv4.3Δ24-C110A alone and coexpression of Kv4.3Δ24-C110A and WT KChIP4a. (D) Summary of peak current amplitudes measured at +40 mV from C110A alone, coexpression of C110A and WT KChIP4a, Kv4.3Δ24-C110A alone, and coexpression of Kv4.3Δ24-C110A and WT KChIP4a. All data are shown as mean ± SE for 20–25 oocytes. Statistical significance (∗∗∗P < 0.001) was compared between groups as indicated; ns: no statistical significance between Kv4.3Δ24-C110A alone and coexpression of Kv4.3Δ24-C110A with WT KChIP4a.
Coexpression of KChIP4a with the Kv4.3Δ24-C110A mutant failed to further increase the functional current, in contrast to coexpression of C110A and KChIP4a, demonstrating that the proximal N-terminus of Kv4.3 is primarily required for KChIP4a modulation of Kv4 channels (Fig. 4, B–D, and Table S1). Furthermore, when the residue Y147 of KChIP4a, the counterpart within the first interface, was mutated (Y147E), the effect on functional rescue of Kv4.3C110A mutant was also abolished, further confirming the clamping effect of KChIP4a on Kv4.3 function (Fig. S1).
Enhanced surface expression of Kv4.3 zinc mutants by KChIP4a
To test whether functional rescue is correlated with surface expression, we engineered an extracellularly exposed c-myc tag between S1 and S2 of Kv4.3 for detection of surface expression. We first tested the current expression of myc-Kv4.3 channels in oocytes expressing myc-Kv4.3 alone. As shown in Fig. S2, the voltage-dependent activation and recovery from inactivation of myc-Kv4.3 are identical to those of WT untagged Kv4.3 channels, showing that myc-tag does not affect Kv4.3 channel expression or function. Protein surface labeling, as measured by the chemiluminescence signal in each individual oocyte, was then carried out. As a positive control, the effect of KChIP1 on surface expression was also measured. KChIP1 enhanced surface expression threefold, but KChIP4a did not increase the surface expression of WT myc-Kv4.3 (Fig. 5 A, left panel), consistent with previous reports that KChIP4a does not enhance current expression (27,32,37,39). In contrast, KChIP4a increased the surface expression of the zinc mutant C110A by eightfold compared to the myc-C110A mutant alone (Fig. 5 A, right panel), consistent with the results of our functional assay. When the KChIP4a double mutant Y70A-K74A was used, the increased surface expression was significantly attenuated (Fig. 5 A, right panel). This result indicates that functional rescue correlates well with increased surface expression as evaluated by the surface-labeling assay.
Figure 5.

Enhanced surface expression of the myc-tagged C110A mutant and tetrameric assembly by KChIP4a. (A) In oocyte labeling, normalized protein expression by surface labeling in oocytes expressing the myc-Kv4.3 channel alone and in the presence of WT KChIP4a, compared with uninjected oocytes as a negative control or WT KChIP1 coinjected with Kv4.3 as a positive control. Right panel: Normalized protein expression by surface labeling in oocytes expressing myc-Kv4.3 C110A mutant alone and in the presence of either WT KChIP4a or KChIP4a double mutant (Y70A-K74A), which attenuated the surface expression increased by WT KChIP4a. For statistical comparison, WT Kv4.3 or C110A alone was compared with that of coexpressed KChIPs, ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 (Student's t-test); #P < 0.05 is only denoted for comparison between Kv4.3 C110A/WT KChIP4a and Kv4.3 C110A/KChIP4a Y70A-K74A double mutant. (B) Top panels: SEC-FPLC profiles of Kv4 channel proteins expressed with or without KChIPs and Western-blotted to identify fractions containing Kv4.3 subunits. WT Kv4.3 channels ran in the tetrameric fractions; however, Kv4.3 C110A channels ran as monomers. Bottom panels: Channels expressed with KChIPs Western-blotted for Kv4.3 C110A protein. Kv4.3 C110A channels showed a significant shift to tetrameric fractions with KChIP1 or KChIP4a proteins, but ran mainly as monomers with KChIP4a Y70A-K74A mutant. (C) Semiquantitative analysis of the ratio of tetramer/total protein based on panels in B.
Enhanced tetrameric assembly of Kv4.3 zinc mutant proteins by KChIP4a
It has been reported that monomeric Kv4 zinc mutant proteins can be driven to assemble as tetramers by association with KChIP1 or KChIP3 (9,14). To test the hypothesis that KChIP4a can rescue the function of Kv4.3 zinc mutants by driving tetrameric assembly, we transiently expressed KChIP4a and WT Kv4.3 or zinc mutant proteins in HEK 293 cells. The proteins were then solubilized and characterized by SEC-FPLC analysis to evaluate the state of tetrameric assembly. As shown in Fig. 5 B, WT Kv4.3 proteins alone were found in fractions consistent with self-assembly into tetramers as determined by Western blot in sodium dodecyl sulfate polyacrylamide gel electrophoresis gels, whereas the Kv4.3 C110A zinc mutant was found in fractions of lower molecular size, reflecting a monomeric status. In contrast, coexpression of the C110A mutant with KChIP4a resulted in a significant shift of C110A mutant proteins to fractions of greater molecular size (Fig. 5 B), consistent with the positive control of coexpressed KChIP1 proteins found in tetrameric fractions. However, the KChIP4a double mutant Y70A-K74A proteins, which attenuated the surface expression of zinc mutant channels in oocyte current recording and labeling assays, resulted in Kv4.3 C110A mutant proteins remaining as monomers. These results further confirm that KChIP4a can rescue the function and increase surface expression of the Kv4.3 C110A zinc mutant by promoting tetrameric assembly of the mutant channels (Fig. 5, B and C).
KIS and core KChIP4a function independently to modulate Kv4.3
Since KChIP4a shares a conserved C-terminal core domain with other members of the KChIP family and exerts a distinct modulation of Kv4 function, it is of interest to dissect whether distinctive KIS and core KChIP4a can function independently to exert a diverse modulation of Kv4.3 channels. To address the role of KIS and core domains in modulating surface expression and inactivation of Kv4.3 channels, we constructed a KIS-Kv4.3Δ24 chimera in which N-terminal residues 2–24 of Kv4.3 (Kv4.3Δ24) were replaced with the KIS domain of KChIP4a, consisting of residues 2–34, and then tested whether the KIS affects surface expression and gating of Kv4.3. Fig. 6 A shows that the deletion mutant Kv4.3Δ24 yielded functional currents with a 1.9-fold greater peak amplitude and a moderately slow inactivation compared to WT Kv4.3 (Fig. 6 A), consistent with previous observations (8,40,41). In contrast, the chimera KIS-Kv4.3Δ24 resulted in much smaller currents (0.3-fold compared to WT Kv4.3 alone) and slower inactivation, compared to either N-terminal deletion mutant of Kv4.3Δ24 or coexpression of Kv4.3/WT KChIP4a (0.7-fold compared to WT Kv4.3 alone). Inactivation in the chimera was even slower than with either the N-terminal deletion mutant of Kv4.3Δ24 alone, or coexpression of Kv4.3/WT KChIP4a. These results demonstrate that KIS (N-terminal KChIP4a) suppresses both surface expression and inactivation of Kv4.3 channels (Fig. 6, B–D).
Figure 6.

KIS suppresses surface expression of Kv4.3 channels and causes slow inactivation. (A) Kv4.3Δ24 current traces were recorded in oocytes held at −80 mV in response to a family of depolarizing potentials from −60 mV to +40 mV with a 10 mV increments for 1 s. The inset compares the inactivation-scaled Kv4.3Δ24 and WT Kv4.3 currents at +40 mV. (B) Chimera (KIS-Kv4.3Δ24) current traces were recorded with the same protocol as in panel A. (C) The scaled and superimposed currents at +40 mV (from panels A and B) are shown to compare the development of inactivation kinetics among Kv4.3Δ24 alone, WT Kv4.3/WT KChIP4a, and chimera (KIS-Kv4.3Δ24). (D) Summary of normalized peak current amplitudes measured at +40 mV from WT Kv4.3 alone, Kv4.3/WT KChIP4a, Kv4.3Δ24 alone, and chimera (KIS-Kv4.3Δ24). All data are shown as the mean ± SE for 20–25 oocytes. Statistical significance (Student's t-test) was compared between groups as indicated; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
To test the effect of core KChIP4a on WT Kv4.3 or C110A zinc mutant channels, we constructed a KChIP4aΔ34 in which the KIS domain was deleted, and then investigated whether core KChIP4a can function in a manner similar to that of KChIP1, which clamps Kv4 for tetrameric assembly. Since several studies have reported that truncating the proximal N-terminus of KChIP4a can convert KChIP4a into KChIP1 when coexpressed with WT Kv4.3, we started to replicate the effect of core KChIP4a on WT Kv4.3 function (27,39). Coexpression of core KChIP4a (KChIP4aΔ34) with WT Kv4.3 resulted in an increased peak current amplitude (5.2 ± 0.3-fold, n = 8), accelerated inactivation (τinact, fast = 75.5 ± 3.3 ms, n = 8), and accelerated recovery from inactivation (100.6 ± 10.6 ms, n = 5) compared to WT KChIP4a/WT Kv4.3, resembling the effects of KChIP1 on WT Kv4 channels (Fig. 7 and Table S1). We then sought to determine whether core KChIP4a could behave similarly when coexpressed with C110A zinc mutant channels. Coexpression of core KChIP4a with Kv4.3 C110A mutant led to an increased peak current (5.9 ± 1.0-fold, n = 10) compared to C110A/WT KChIP4a (3.7 ± 0.3-fold, n = 13; Fig. 7). The fast component of time constant of inactivation (τinact, fast) for core KChIP4a/C110A was significantly accelerated at 40.3 ± 1.5 ms (n = 6) compared to WT KChIP4a/C110A (149.5 ± 9.1 ms, n = 13), but similar to WT KChIP1/C110A (29.2 ± 2.2 ms, n = 6; Fig. 7). Moreover, the time constant of recovery from inactivation for core KChIP4a/C110A was significantly increased at 50.2 ± 1.6 ms (n = 5) compared to WT KChIP4a/C110A (205.1 ± 4.6 ms, n = 8) or WT KChIP1/C110A (105.4 ± 8.4 ms, n = 5; Fig. 7). These results further show that core KChIP4a behaves like KChIP1 in functioning in a clamping manner to modulate Kv4 function, and that functional rescue of Kv4.3 channel tetramerization-defective mutants by KChIP4a does not involve the KIS domain.
Figure 7.

Core KChIP4a-dependent rescue of Kv4.3 channel tetramerization mutants. (A) Upper panel: WT Kv4.3 current traces in the absence or presence of WT KChIP1, WT KChIP4a, and KChIP4a Δ34 were recorded in oocytes held at −80 mV in response to a family of depolarizing potentials from −60 mV to +40 mV with a 10 mV increments for 1 s, respectively. The normalized peak current amplitude at +40 mV is onefold (n = 21) for WT Kv4.3 alone, compared with 3.5 ± 0.2-fold (n = 19) for WT Kv4.3/WT KChIP1, 0.7 ± 0.1-fold (n = 13) for WT Kv4.3/WT KChIP4a, and 5.2 ± 0.3-fold (n = 8) for WT Kv4.3/ KChIP4a Δ34, respectively. Lower panel: Kv4.3-C110A current traces in the absence or presence of WT KChIP1, WT KChIP4a, and KChIP4a Δ34 were recorded using the same protocol as in the upper panel. The C110A mutant shows nonfunctional current. (B) The scaled and superimposed currents at +40 mV (from upper panel A) are shown to compare the development of the inactivation kinetics of WT Kv4.3 alone (black line) or WT Kv4.3/WT KChIP1 (red line) as controls with WT Kv4.3/WT KChIP4a (green line) and WT Kv4.3/KChIP4a Δ34 (blue line). The fast component of time constant of inactivation (τinact, fast) is 32.6 ± 1.7 (n = 12) for WT Kv4.3 alone, compared with 74.9 ± 1.1 (n = 8) for WT Kv4.3/WT KChIP1, 413.4 ± 42.1 (n = 12) for WT Kv4.3/WT KChIP4a, and 75.5 ± 3.3 (n = 5) for WT Kv4.3/ KChIP4a Δ34, respectively. Note the crossover of the inactivation kinetics of Kv4.3 current and coexpression of WT KChIP1 or KChIP4a Δ34 at +40 mV. (C) The scaled and superimposed currents at +40 mV (from lower side panel A) are shown to compare the development of inactivation kinetics among coexpression of C110A and WT KChIP1 (red line), coexpression of C110A and WT KChIP4a (green line), and coexpression of C110A and KChIP4a Δ34 (blue line). The fast component of the time constant of inactivation (τinact, fast) is 40.3 ± 1.5 (n = 6) for C110A/KChIP4a Δ34, as compared with 29.2 ± 2.2 (n = 6) for C110A/WT KChIP1 and 149.5 ± 9.1 (n = 13) for C110A/WT KChIP4a, respectively. (D) The normalized peak current amplitude at +40 mV is 3.9 ± 0.2-fold (n = 21) for C110A/WT KChIP1, 3.7 ± 0.3-fold (n = 13) for C110A/WT KChIP4a and 5.9 ± 1.0-fold (n = 10) for C110A/KChIP4a Δ34, as compared with WT Kv4.3 alone. (E) Summary of the time constants of recovery (τrecovery) from inactivation for varying lengths of time at steps from −80 to +40 mV, measured at +40 mV from WT Kv4.3 or Kv4.3-C110A and different KChIPs. All data are shown as mean ± SE; the statistical comparison (Student's t-test) for ∗P < 0.05, ∗P < 0.01, and ∗P < 0.001 was made with regard to WT Kv4.3 alone; and the groups denoted with #P < 0.05, ##P < 0.01, and ###P < 0.001 were compared with either WT Kv4.3/WT KChIP4a or Kv4.3 C110A/ WT KChIP4a complexes.
Discussion
We were intrigued by the fact that KChIP4a, which shares a conserved C-terminal core domain with other members of the KChIP family, exerts a distinct modulation of Kv4 function. Based on their structural similarity, we hypothesized that core KChIP4a predominantly functions to promote and stabilize the tetrameric assembly of Kv4.3 subunits by means of a clamping mechanism like that utilized by other KChIPs, whereas KChIP4a N-terminus functions independently to affect surface expression and gating of Kv4.3. To test this hypothesis, we took advantage of unique mutations in the T1 domain that have been shown to disrupt tetrameric assembly of Kv4 channels (7–10). We found that the auxiliary KChIP4a subunit proteins can indeed rescue the function of tetramerization-defective Kv4.3 channel mutants, further confirming the clamping action of KChIP4a on Kv4 channels.
Our previous studies of the cocrystal structure of the Kv4.3N/KChIP1 complex showed that this complex has a clamping mode in which a single KChIP1 monomer laterally clamps two neighboring Kv4.3 N-termini in a 4:4 manner, and this interaction involves two contact interfaces (14,36). At the first interface, the proximal N-terminal peptide of Kv4.3 is sequestered and deeply buried within an elongated groove on the surface of KChIP1. The second interface is formed by the binding of KChIP1 to another adjacent T1 domain, resulting in a stabilized tetrameric Kv4.3 T1-KChIP1 complex. It is known that both KChIP4a and KChIP1 show 79% amino acid homology in their C-terminal core region (27). The recently demonstrated crystal structure of KChIP4a reveals distinctive N-terminal α-helices that distinguish this molecule from other known KChIPs. Moreover, the core of KChIP4a shows significant alignment with bound KChIP1, which laterally clamps Kv4 channels (37), suggesting that core KChIP4a regulates Kv4 in a clamping mechanism similar to that used by KChIP1. The results of this study show that KChIP4a is capable of binding to unassembled monomeric subunits and driving them to form functional channels. Since disruption of residues Y70–K74, which are critical for clamping, resulted in attenuation of functional rescue, KChIP4a probably uses the same clamping mechanism employed by KChIP1 to promote and stabilize tetrameric assembly of Kv4 channels.
The mechanism that underlies the functional rescue and gating of KChIP4a on Kv4 channels remains elusive. It was previously shown that increasing the ratio of KChIP3 to Kv4.2 can result in a complete rescue of current in a Kv4.2 zinc mutant to the level of WT Kv4.2 channels, indicating that KChIP3 binding to zinc-site mutants promotes functional expression of mutant channels (9). In this study, we observed that KChIP4a enhances surface expression of a Kv4.3 monomeric zinc-site mutant that normally cannot form tetramers that will reach the surface, but does not enhance surface expression of tetrameric WT Kv4.3. In fact, surface expression of WT Kv4.3 is to some degree inhibited by KChIP4a. This unexpected functional phenomenon and discrepancy suggest that KChIP4a possesses a distinct dual function in which core KChIP4a plays a dominant role in promoting tetrameric assembly of monomeric subunits, whereas its N-terminus functions in an inhibitory manner to affect channel trafficking and override the effect of the core domain on assembled Kv4 subunits.
The proximal N-terminus of Kv4 has been shown to be the primary binding site (as the first interface) for KChIP interaction (8,14). In this study, we generated a C110A zinc mutant with Kv4.3 N-terminal deletion of residues 2–24 to better understand the role of N-terminal Kv4.3 in the functional rescue of zinc mutant. The N-terminal deletion of Kv4.3 in the C110A background unexpectedly revealed functional expression in the absence of KChIP4a. Although the signal for trafficking of channel proteins has been shown to be the RXR endoplasmic reticulum retention motif (42), such a signal has not been specifically identified in Kv4.2 (32) or, in particular for Kv4.3, where the RXR-like motif is located in the distal N-terminal residues (after residue 24) (43). This suggests that the proximal N-terminal hydrophobic residues of Kv4.3 may influence the stability of the tetramer or the tetrameric assembly when the zinc site is mutated; however, the mechanism underlying the negative regulation of tetrameric stability requires further investigation.
It is noteworthy that inactivation was affected by KChIP4a in both zinc-site mutants and WT Kv4.3 channels compared with KChIP1 coexpressed channels, indicating that surface expression and inactivation are tightly regulated by the concerted action of a structurally distinct hydrophobic N-terminus and core region of KChIP4a (37). KChIP4a can be structurally divided into two distinct parts: the core region and the KIS domain, which exerts a unique modulation of Kv4 channels. To further dissect the role of KIS and core KChIP4a in modulating Kv4.3 function, we generated a chimeric mutant. We found that the KIS domain suppresses surface expression of Kv4.3 channels, and that core KChIP4a behaves like KChIP1 in modulating either WT Kv4.3 or C110A zinc mutant channels in a clamping manner. This indicates that the functional rescue of Kv4.3 channel tetramerization mutants by KChIP4a is a core-dependent process. Our findings suggest that the core of KChIP4a, like other KChIPs, functions primarily as a clamp to enhance Kv4.3 surface expression by promoting and stabilizing tetrameric assembly. The KIS domain, however, serves as an autoinhibitory domain that probably interacts with its intracellular receptors to regulate inactivation and recovery from the inactivation properties of Kv4 channels. All of these results support the concept that KChIP4a regulates Kv4 channels by a clamping action, whereas the existing KIS can override some effects of core KChIP4a. Since the KIS domain and core KChIP4a can exhibit some opposite effects on modulation of Kv4.3 channels, the functional phenotype of Kv4 modulation is likely to depend on their net effect between the KIS and the core (Fig. S3). Therefore, it is possible that the ultimate phenotype of functional modulation of Kv4 channels by multitasking KChIPs is activity-dependent and involves different distinct parts of the KChIP4a structure in native cells.
Supporting Material
Experimental procedures, one table, and three figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(10)00409-1.
Supporting Material
Acknowledgments
We thank Drs. M.A. McNutt and S. Lin for helpful comments on this manuscript, and laboratory members J. Su, S. Liu, Y.Q Tang, and X. Bian for discussions and valuable help with the molecular and cell biology. K.W.W. thanks J.M. Wang for her consistent support during this research.
This work was supported by research grants to K.W.W. from the National Science Foundation of China (30630017), the Ministry of Science Technology of China (2006AA02Z183 and 973 Program 2007CB512100) and the Ministry of Education of China (706002).
References
- 1.Nagaya N., Papazian D.M. Potassium channel α and β subunits assemble in the endoplasmic reticulum. J. Biol. Chem. 1997;272:3022–3027. doi: 10.1074/jbc.272.5.3022. [DOI] [PubMed] [Google Scholar]
- 2.Kreusch A., Pfaffinger P.J., Choe S. Crystal structure of the tetramerization domain of the Shaker potassium channel. Nature. 1998;392:945–948. doi: 10.1038/31978. [DOI] [PubMed] [Google Scholar]
- 3.Yi B.A., Minor D.L., Jan L.Y. Controlling potassium channel activities: interplay between the membrane and intracellular factors. Proc. Natl. Acad. Sci. USA. 2001;98:11016–11023. doi: 10.1073/pnas.191351798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu J., Yu W., Li M. Assembly of voltage-gated potassium channels. Conserved hydrophilic motifs determine subfamily-specific interactions between the α-subunits. J. Biol. Chem. 1995;270:24761–24768. doi: 10.1074/jbc.270.42.24761. [DOI] [PubMed] [Google Scholar]
- 5.Li M., Isacoff E., Jan L.Y. Assembly of potassium channels. Ann. N. Y. Acad. Sci. 1993;707:51–59. doi: 10.1111/j.1749-6632.1993.tb38041.x. [DOI] [PubMed] [Google Scholar]
- 6.Cui Y.Y., Liang P., Wang K.W. Enhanced trafficking of tetrameric Kv4.3 channels by KChIP1 clamping. Neurochem. Res. 2008;33:2078–2084. doi: 10.1007/s11064-008-9688-7. [DOI] [PubMed] [Google Scholar]
- 7.Bixby K.A., Nanao M.H., Choe S. Zn2+-binding and molecular determinants of tetramerization in voltage-gated K+ channels. Nat. Struct. Biol. 1999;6:38–43. doi: 10.1038/4911. [DOI] [PubMed] [Google Scholar]
- 8.Scannevin R.H., Wang K., Rhodes K.J. Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1. Neuron. 2004;41:587–598. doi: 10.1016/s0896-6273(04)00049-2. [DOI] [PubMed] [Google Scholar]
- 9.Kunjilwar K., Strang C., Pfaffinger P.J. KChIP3 rescues the functional expression of Shal channel tetramerization mutants. J. Biol. Chem. 2004;279:54542–54551. doi: 10.1074/jbc.M409721200. [DOI] [PubMed] [Google Scholar]
- 10.Strang C., Kunjilwar K., Pfaffinger P.J. The role of Zn2+ in Shal voltage-gated potassium channel formation. J. Biol. Chem. 2003;278:31361–31371. doi: 10.1074/jbc.M304268200. [DOI] [PubMed] [Google Scholar]
- 11.Wang G., Shahidullah M., Covarrubias M. Functionally active t1-t1 interfaces revealed by the accessibility of intracellular thiolate groups in kv4 channels. J. Gen. Physiol. 2005;126:55–69. doi: 10.1085/jgp.200509288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang G., Strang C., Covarrubias M. Zn2+-dependent redox switch in the intracellular T1-T1 interface of a Kv channel. J. Biol. Chem. 2007;282:13637–13647. doi: 10.1074/jbc.M609182200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G., Covarrubias M. Voltage-dependent gating rearrangements in the intracellular T1-T1 interface of a K+ channel. J. Gen. Physiol. 2006;127:391–400. doi: 10.1085/jgp.200509442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H., Yan Y., Chai J. Structural basis for modulation of Kv4 K+ channels by auxiliary KChIP subunits. Nat. Neurosci. 2007;10:32–39. doi: 10.1038/nn1822. [DOI] [PubMed] [Google Scholar]
- 15.An W.F., Bowlby M.R., Rhodes K.J. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403:553–556. doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- 16.Kuo H.C., Cheng C.F., Chien K.R. A defect in the Kv channel-interacting protein 2 (KChIP2) gene leads to a complete loss of I(to) and confers susceptibility to ventricular tachycardia. Cell. 2001;107:801–813. doi: 10.1016/s0092-8674(01)00588-8. [DOI] [PubMed] [Google Scholar]
- 17.Holmqvist M.H., Cao J., An W.F. Kinetic modulation of Kv4-mediated A-current by arachidonic acid is dependent on potassium channel interacting proteins. J. Neurosci. 2001;21:4154–4161. doi: 10.1523/JNEUROSCI.21-12-04154.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nadal M.S., Amarillo Y., Rudy B. Evidence for the presence of a novel Kv4-mediated A-type K(+) channel-modifying factor. J. Physiol. 2001;537:801–809. doi: 10.1111/j.1469-7793.2001.00801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata R., Nakahira K., Ikenaka K. A-type K+ current mediated by the Kv4 channel regulates the generation of action potential in developing cerebellar granule cells. J. Neurosci. 2000;20:4145–4155. doi: 10.1523/JNEUROSCI.20-11-04145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffman D.A., Magee J.C., Johnston D. K+ channel regulation of signal propagation in dendrites of hippocampal pyramidal neurons. Nature. 1997;387:869–875. doi: 10.1038/43119. [DOI] [PubMed] [Google Scholar]
- 21.Kim J., Wei D.S., Hoffman D.A. Kv4 potassium channel subunits control action potential repolarization and frequency-dependent broadening in rat hippocampal CA1 pyramidal neurones. J. Physiol. 2005;569:41–57. doi: 10.1113/jphysiol.2005.095042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song W.J., Tkatch T., Surmeier D.J. Somatodendritic depolarization-activated potassium currents in rat neostriatal cholinergic interneurons are predominantly of the A type and attributable to coexpression of Kv4.2 and Kv4.1 subunits. J. Neurosci. 1998;18:3124–3137. doi: 10.1523/JNEUROSCI.18-09-03124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheng M., Tsaur M.L., Jan L.Y. Contrasting subcellular localization of the Kv1.2 K+ channel subunit in different neurons of rat brain. J. Neurosci. 1994;14:2408–2417. doi: 10.1523/JNEUROSCI.14-04-02408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheng M., Tsaur M.L., Jan L.Y. Subcellular segregation of two A-type K+ channel proteins in rat central neurons. Neuron. 1992;9:271–284. doi: 10.1016/0896-6273(92)90166-b. [DOI] [PubMed] [Google Scholar]
- 25.Johnston D., Christie B.R., Yuan L.L. Active dendrites, potassium channels and synaptic plasticity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003;358:667–674. doi: 10.1098/rstb.2002.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai X., Liang C.W., Thompson S.M. Unique roles of SK and Kv4.2 potassium channels in dendritic integration. Neuron. 2004;44:351–364. doi: 10.1016/j.neuron.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 27.Holmqvist M.H., Cao J., An W.F. Elimination of fast inactivation in Kv4 A-type potassium channels by an auxiliary subunit domain. Proc. Natl. Acad. Sci. USA. 2002;99:1035–1040. doi: 10.1073/pnas.022509299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng X.Y., Cai F., Xia J.H. Identification of the alternative promoters of the KChIP4 subfamily. Acta Biochim. Biophys. Sin. (Shanghai) 2005;37:241–247. doi: 10.1111/j.1745-7270.2005.00034.x. [DOI] [PubMed] [Google Scholar]
- 29.Guo W., Li H., Nerbonne J.M. Role of heteromultimers in the generation of myocardial transient outward K+ currents. Circ. Res. 2002;90:586–593. doi: 10.1161/01.res.0000012664.05949.e0. [DOI] [PubMed] [Google Scholar]
- 30.Beck E.J., Bowlby M., Covarrubias M. Remodelling inactivation gating of Kv4 channels by KChIP1, a small-molecular-weight calcium-binding protein. J. Physiol. 2002;538:691–706. doi: 10.1113/jphysiol.2001.013127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morohashi Y., Hatano N., Iwatsubo T. Molecular cloning and characterization of CALP/KChIP4, a novel EF-hand protein interacting with presenilin 2 and voltage-gated potassium channel subunit Kv4. J. Biol. Chem. 2002;277:14965–14975. doi: 10.1074/jbc.M200897200. [DOI] [PubMed] [Google Scholar]
- 32.Shibata R., Misonou H., Trimmer J.S. A fundamental role for KChIPs in determining the molecular properties and trafficking of Kv4.2 potassium channels. J. Biol. Chem. 2003;278:36445–36454. doi: 10.1074/jbc.M306142200. [DOI] [PubMed] [Google Scholar]
- 33.Xiong H., Kovacs I., Zhang Z. Differential distribution of KChIPs mRNAs in adult mouse brain. Brain Res. Mol. Brain Res. 2004;128:103–111. doi: 10.1016/j.molbrainres.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 34.Zhou W., Qian Y., Choe S. Structural insights into the functional interaction of KChIP1 with Shal-type K(+) channels. Neuron. 2004;41:573–586. doi: 10.1016/s0896-6273(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 35.Kim L.A., Furst J., Grigorieff N. Three-dimensional structure of I(to); Kv4.2-KChIP2 ion channels by electron microscopy at 21 Angstrom resolution. Neuron. 2004;41:513–519. doi: 10.1016/s0896-6273(04)00050-9. [DOI] [PubMed] [Google Scholar]
- 36.Pioletti M., Findeisen F., Minor D.L. Three-dimensional structure of the KChIP1-Kv4.3 T1 complex reveals a cross-shaped octamer. Nat. Struct. Mol. Biol. 2006;13:987–995. doi: 10.1038/nsmb1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang P., Wang H., Wang K. Structural insights into KChIP4a modulation of Kv4.3 inactivation. J. Biol. Chem. 2009;284:4960–4967. doi: 10.1074/jbc.M807704200. [DOI] [PubMed] [Google Scholar]
- 38.Jerng H.H., Pfaffinger P.J. Multiple Kv channel-interacting proteins contain an N-terminal transmembrane domain that regulates Kv4 channel trafficking and gating. J. Biol. Chem. 2008;283:36046–36059. doi: 10.1074/jbc.M806852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwenk J., Zolles G., Bentrop D. NMR analysis of KChIP4a reveals structural basis for control of surface expression of Kv4 channel complexes. J. Biol. Chem. 2008;283:18937–18946. doi: 10.1074/jbc.M800976200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bähring R., Dannenberg J., Isbrandt D. Conserved Kv4 N-terminal domain critical for effects of Kv channel-interacting protein 2.2 on channel expression and gating. J. Biol. Chem. 2001;276:23888–23894. doi: 10.1074/jbc.M101320200. [DOI] [PubMed] [Google Scholar]
- 41.Gebauer M., Isbrandt D., Bähring R. N-type inactivation features of Kv4.2 channel gating. Biophys. J. 2004;86:210–223. doi: 10.1016/S0006-3495(04)74097-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zerangue N., Schwappach B., Jan L.Y. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron. 1999;22:537–548. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 43.Dilks D., Ling H.P., Numann R. Cloning and expression of the human kv4.3 potassium channel. J. Neurophysiol. 1999;81:1974–1977. doi: 10.1152/jn.1999.81.4.1974. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


