Abstract
Human cyclin D1 is expressed as two isoforms derived by alternate RNA splicing, termed D1a and D1b, which differ for the inclusion of intron 4 in the D1b mRNA. Both isoforms are frequently upregulated in human cancers, but cyclin D1b displays relatively higher oncogenic potential. The splicing factors that regulate alternative splicing of cyclin D1b remain unknown despite the likelihood that they contribute to cyclin D1 oncogenicity. In this study, we report that Sam68, an RNA-binding protein frequently overexpressed in prostate cancer cells, enhances splicing of cyclin D1b and supports its expression in prostate cancer cells. Chromatin immunoprecipitation and RNA coimmunoprecipitation experiments showed that Sam68 is recruited to the human CCND1 gene encoding cyclin D1 and that it binds to cyclin D1 mRNA. Transient overexpression and RNAi knockdown experiments indicated that Sam68 acts to enhance endogenous expression of cyclin D1b. Minigene reporter assays showed that Sam68 directly affected alternative splicing of CCND1 message, with a preference for the A870 allele that is known to favor cyclin D1b splicing. Sam68 interacted with the proximal region of intron 4, and its binding correlated inversely with recruitment of the spliceosomal component U1-70K. Sam68-mediated splicing was modulated by signal transduction pathways that elicit phosphorylation of Sam68 and regulate its affinity for CCND1 intron 4. Notably, Sam68 expression positively correlates with levels of cyclin D1b, but not D1a, in human prostate carcinomas. Our results identify Sam68 as the first splicing factor to affect CCND1 alternative splicing in prostate cancer cells, and suggest that increased levels of Sam68 may stimulate cyclin D1b expression in human prostate cancers.
Introduction
Prostate cancer (PCa) cells rely on androgen-dependent activation of the androgen receptor (AR) for their growth and survival. However, although antiandrogenic therapies are initially efficacious, PCa often evolves into a hormone therapy–resistant disease in which AR activation persists in spite of the treatments (1). Thus, the identification of the molecular steps involved in acquisition of hormone therapy resistance, and/or of novel markers that can predict disease progression, represents a clinical priority. In this respect, it has been recently suggested that the analysis of mRNA variants deriving from alternative splicing (AS) of specific genes can accurately distinguish between normal and neoplastic prostate epithelial cells (2, 3). A clear example is given by regulation of AR itself. Truncated AR isoforms deriving from exon 2b AS promote androgen-independent growth and are strongly expressed in animal models of castration-refractory PCa (4). Ligand-independent splicing variants of AR were also found enriched in specimens from castration-resistant PCa patients (5), suggesting that AS can represent a primary cause of PCa progression through the production of constitutively active AR isoforms.
Another AS event with strong relevance to PCa was observed in CCND1, a proto-oncogene that is frequently deregulated in cancer cells (6). This gene encodes for two alternative transcripts: the common cyclin D1a isoform, containing all five exons, and cyclin D1b, which derives from retention of intron 4 and premature termination of the transcript (7). Cyclin D1b showed higher oncogenic potential than cyclin D1a in NIH-3T3 cells possibly due to its more pronounced nuclear localization (8, 9). Moreover, cyclin D1b is upregulated in several types of cancers, including PCa (10), and it has been correlated with higher risk of cell transformation and poor prognosis in selected tumor types (6). Interestingly, cyclin D1b favors PCa cell proliferation through an aberrant regulation of AR activity (11). In contrast with cyclin D1a, which interacts with AR and represses its transcriptional activity, cyclin D1b associates with the receptor without interfering with it (11). Thus, it has been proposed that a change in the ratio between the cyclin D1 isoforms interrupts this negative feedback and leads to unleashed growth of PCa cells. Given its implication for PCa progression, it is crucial to understand the molecular basis of cyclin D1b expression in PCa cells. A polymorphism (G870/A) at the exon 4–intron 4 boundary predisposes cells to cyclin D1b splicing, possibly by altering exon 4 definition, and might be associated with increased cancer risk in several tissues (6). However, a large study recently showed that cyclin D1b can be also expressed in tissue of patients bearing the G870 polymorphism (10), suggesting that cancer-specific changes in trans-acting factors might contribute to the regulation of this AS event.
AS is a regulatory mechanism affecting virtually all human multiexon genes (12). Through the differential assortment of exons, AS insures the production of several protein isoforms from a single gene, thereby greatly expanding the complexity and plasticity of the genome (13, 14). AS is operated by a complex ribonucleoprotein machinery, the spliceosome, which recognizes the splice site consensus sequences (15). In addition, the assembly of the spliceosome at exon-intron boundaries is modulated by several constitutive and ancillary RNA-binding proteins (RBP) that participate to the regulation of AS, and their expression levels influence the splicing decisions (16). An interesting splicing regulator is Sam68, which integrates mitogenic signaling pathways with AS of target mRNAs (17, 18).
Sam68 belongs to the STAR (signal transduction and activation of RNA metabolism) family of RBPs (18). They are characterized by a GSG (GRP33-Sam68-GLD-1) domain, which is required for RNA binding and homodimerization, flanked by regions involved in protein-protein interactions and post-translational modifications, which affect the affinity and specificity of RNA binding (18). Sam68 was shown to regulate AS of several target mRNAs, such as human CD44 (17, 19) and BCL2L1 (20) or mouse Sgce (21). Moreover, it was shown that mitogen-activated protein kinases (MAPK) and Src family kinases (SFK) phosphorylate Sam68 and modulate its AS activity (17, 20). Interestingly, Sam68 is upregulated in a subset of human PCa specimens (22, 23) and was shown to contribute to PCa cell proliferation and survival (22). Downregulation of Sam68 by RNA interference (RNAi) in PCa cells caused changes in the expression levels of several mRNAs, including cyclin D1 (22). Given its role in AS and its effects on cyclin D1 expression, herein we have investigated the implication of Sam68 in cyclin D1 AS in PCa model systems and primary tumors. Our results strongly point to Sam68 as the first splicing factor promoting cyclin D1b splicing in PCa cells.
Materials and Methods
Plasmid constructs
The pcDNA3.1-CCND1 exon 4–exon 5 minigene, G870 polymorphism, was amplified from genomic DNA purified from PC3 cells using Pwo SuperYield DNA Polymerase (Roche) with the forward primer 5′-AGGCGGCCGCATGTGAAGTTCATTTCCAATC-3′ and the reverse primer 5′-AGGATATCGCCTGCCTGGCGCCCTCAGAT-3′ and analyzed by direct sequencing (BMR Genomics). G870A pcDNA3.1-CCND1 minigenes, encoding the cyclin D1 cDNA with the intervening intron 4, and pEGFPC1-Sam68, wild-type and G178E or V229F mutants, pCMV-Fyn, and pSG-RASL61Q have been previously described (10, 17, 20).
Cell cultures, transfections, and in vivo splicing assays
PC3, LNCaP, and HEK293T cells were purchased from the American Type Culture Collection. PC3 and HEK293T cells were grown in DMEM (Life Technologies), whereas LNCaP cells were grown in RPMI 1640 (BioWhittaker Cambrex Bioscience) both supplemented with 10% fetal bovine serum (BioWhittaker Cambrex Bioscience), penicillin, and streptomycin. Transfections were performed with 0.1 to 1 µg of appropriate constructs in the presence or absence of 0.5 µg of pcDNA3.1-CCND1 minigenes together with green fluorescent protein (GFP) or GFP-Sam68 constructs using Lipofectamine 2000 (Invitrogen). For RNAi, PC3 cells were plated the day before transfection at 4 × 105 per well in six-well plates and transfected with 20 µmol/L of scramble or Sam68 small interfering RNA (siRNA) oligonucleotides (22) using Lipofectamine RNAiMAX (Invitrogen) and analyzed according to published procedures (20). Briefly, total RNA was isolated using cold Trizol reagent (Invitrogen) and resuspended in RNase-free water (Sigma-Aldrich). DNA digestion was performed using RNase-free DNase (Roche). Protein extraction for Western blot or immunoprecipitation experiments was performed as previously described (20).
Stable knockdown of sam68 in PC3 cells
PC3 cells were transfected either with pLKO.1puro or PLKO.1-KHDRBS1_527 (MISSION short hairpin RNA, Sigma-Aldrich) in a 12-multiwell plate using Lipofectamine reagent. The puromycin resistance marker was used for stable selection of PC3 colonies. Puromycin was added at a concentration of 1 µg/mL in fresh medium every 2 d. Sam68 knockdown was verified by reverse transcription-PCR (RT-PCR) and Western blot analyses.
RT-PCR analysis
DNA-free total RNA (0.2–1 µg) from transfected cells was used for RT-PCR using Moloney murine leukemia virus reverse transcriptase (Invitrogen). Five percent to 10% of the RT reaction was used as template for a PCR analysis (GoTaq, Promega) together with the following primers: forward (common for D1a and D1b), 5′-CCAGAGTGATCAAGTGTGAC-3′; reverse (D1a), 5′-CAAGGAGAATGAAGCTTTCCCTT-3′; reverse (D1b), 5′-GGGACATCACCCTCACTTAC-3′. Data are represented as the mean ± SD of at least three experiments. Statistical analysis was performed using the Student's t test.
Chromatin immunoprecipitation
LNCaP cells (~2 × 106 per sample) were incubated with 1% (v/v) formaldehyde in culture medium for the last 10 min of culture at room temperature. Cells were then washed in cold PBS, harvested, and lysed to isolate nuclei in a hypotonic buffer containing 5mmol/L PIPES (pH 8.0), 85 mmol/L KCl, and NP40 0.5%. Nuclei were then resuspended; lysed in a buffer containing 1% SDS, 10 mmol/L EDTA, and 50 mmol/L Tris-HCl (pH 8.1); and sonicated with 8 pulses (1'–90% amplitude) to yield chromatin size of 700 bp and used (100 µg of DNA/sample) for immunoprecipitation with 2 µof anti-Sam68 rabbit (Santa Cruz Biotechnology) or control rabbit IgGs (Sigma-Aldrich) as previously described (24). Coprecipitated DNA was then analyzed by quantitative real-time PCRs performed with SYBR Green mix (for LightCycler 480, Roche) according to the manufacturer's instructions. The primers used were the following: E1-I1, 5′-CCTACTTCAAATGTGTGCAGAAG-3′ (forward) and 5′-CAACAAGTTGCAGGGAAGTC-3′ (reverse); E4-I4, 5′-CCAACAACTTCCTGTCCTAC-3′ (forward) and 5′-TGGGACATCACCCTCACTTAC-3′ (reverse); E5-end, 5′-GCTTCCCAGCACCAACATGT-3′ (forward) and 5′-CTTTCATGTTTGTCTTTTTGTCTTCTG-3′ (reverse); prox-I4, 5′-CCTGTGAGGTCCGAGACACCGG-3′ (forward) and 5′-GGCCTTGGCACCAGCCTC-3′ (reverse); and dist-I4, 5′-GGTTGGGAGTTAAGTGGCAC-3′ (forward) and 5′-AGCCTGAAGTGGCTGCTTAC-3′ (reverse). The fold enrichment in Sam68 immunoprecipitates was calculated with respect to the signal detected with control IgGs.
Protein-RNA immunoprecipitation
LNCaP cells were washed with PBS and used for RNA-protein coimmunoprecipitation as previously described (20, 25) using 5 µg of anti-Sam68 (Santa Cruz Biotechnology) or 5 µg of rabbit IgGs for 3 h at 4°C under constant rotation. Immunocomplexes were incubated with RNase-free DNase (Roche) for 15 min at 37°C, washed, and incubated with 50 µg of proteinase K (Roche) for additional 15 min at 37°C. Coprecipitated RNA was then extracted by standard procedure and used for RT-PCR.
Preparation of cell extracts and Western blot analysis
Cells were lysed directly on the plate by the addition of lysis buffer as described (20). Protein concentration was determined by using Bradford reagent (Bio-Rad). Cell extracts were used for Western blot analysis as described previously (20) with the primary antibodies (1:1,000 dilution) rabbit anti-Sam68, goat anti–U1-70K, and rabbit anti–extracellular signal-regulated kinase (ERK) 2 (Santa Cruz Biotechnology); mouse anti–hnRNP A1 and mouse anti–β-tubulin (Sigma-Aldrich); and rabbit anti–cyclin D1b (11) and detected as described elsewhere (20).
In vitro synthesis of RNAs
Templates for RNA synthesis were generated using PCR products amplified with a forward primer containing the 5′ end of the T7 promoter sequence upstream of sequences in exon 4 of the CCND1 gene: 5′-ATTAATACGACTCACTATAGGGATGTGAAGTTCATTTCCA-3′. Reverse primers were complementary to sequences within intron 4: Rev1, 5′-CTCTCTGGTCTAATTTGG-3′; Rev2, 5′-CACATTGTGGCCCAAACA-3′; Rev3, 5′-GGCCTTGGCACCAGCCTC-3′. Amplified bands were purified from the agarose gel using Gel Extraction kit (Qiagen) and used as templates (1 µg) for in vitro RNA transcription using the MAXI-script RNA synthesis kit (Ambion) in the presence of labeled nucleotides (NTP mix, UTP biotinylated, Roche). After DNase digestion, RNA was purified using ProbeQuant G-50 Micro Columns (GE Healthcare) and diluted in RNase-free water (Sigma-Aldrich) for subsequent experiments.
Biotin-RNA pull-down experiments
HEK293T nuclear extracts were prepared as previously described (20). Nuclear extracts were precleared for 60 min on protein A-Sepharose beads and for 60 min on streptavidin-Sepharose beads in the presence of 0.25 mg/mL of yeast tRNA to diminish nonspecific interactions of proteins with RNAs. Precleared extracts were then incubated with streptavidin-Sepharose beads, previously preadsorbed for 1 h with 0.05% bovine serum albumin, and the specific biotinylated RNA oligonucleotides (3 µg). Incubation was carried out for 60 min at 4°C under rotation. Bound complexes were rinsed thrice with RSB100 and eluted in SDS-PAGE sample buffer and analyzed by Western blot.
Immunohistochemistry
A human prostate tissue array (Super Bio Chips Laboratories) containing 40 carcinoma specimens and 9 normal tissue specimens were used for immunostaining with anti–cyclin D1a, anti–cyclin D1b (10, 11), or anti-Sam68 (20) antibodies as previously described (10). Cytokeratin was stained with the AE1/AE3 antibody (DAKO) and visualized with an anti-mouse Alexa Fluor 488–conjugated antibody (Invitrogen). Immunohistochemical sections were quantified using the unbiased automated image acquisition system AQUA/PM2000 Imaging Platform (HistoRx) as previously described (10).
Results
Sam68 associates with cyclin D1 mRNA in PCa cells
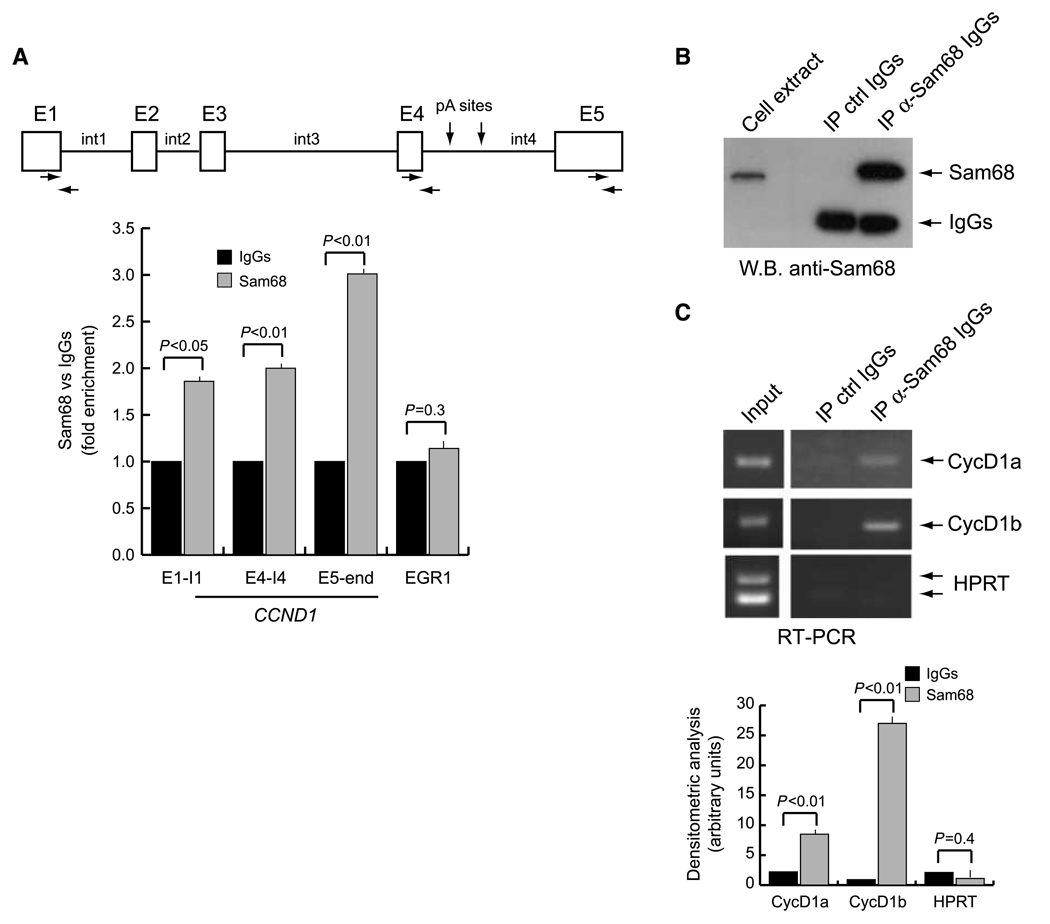
Sam68 is a multifunctional RBP that has been implicated in both transcriptional and posttranscriptional regulation of gene expression (18). Sam68 is frequently upregulated in PCa cells and it influences cyclin D1 expression in LNCaP cells (22). Because splicing regulators are often recruited onto nascent pre-mRNAs, we set out to determine whether Sam68 was associated with the CCND1 gene. Chromatin immunoprecipitation (ChIP) experiments showed that endogenous Sam68 was present along the transcription unit of this gene in LNCaP cells, where it associated with the exon 1–intron 1 and exon 4–intron 4 junctions as well as with exon 5 at the far end of the gene (Fig. 1A). Next, we asked whether Sam68 could bind cyclin D1 mRNA. CCND1 encodes for two splicing variants in PCa cells: the common cyclin D1a and the less abundant cyclin D1b (6). Interestingly, we found that endogenous Sam68 specifically coimmunoprecipitated with both mRNA isoforms, albeit with higher enrichment for the less abundant cyclin D1b mRNA, in LNCaP (Fig. 1B and C) and PC3 (data not shown) PCa cells. These observations suggest that Sam68 is directly bound to cyclin D1 mRNAs in PCa cells.
Figure 1.
Sam68 associates with CCND1 gene and with cyclin D1 mRNA in LNCaP cells. A, scheme of the CCND1 gene. Boxes identify the five exons (E1–E5), whereas the lines identify the four introns (int1–int4). Horizontal arrows, position of the primers used for the PCR of the immunoprecipitated chromatin; vertical arrows, position of the polyadenylation sites in intron 4 (28). Cross-linked chromatin derived from LNCaP cells was immunoprecipitated with anti-Sam68 antibody or control IgGs. The immunoprecipitated chromatin was analyzed by quantitative real-time PCR using the indicated primers. Early growth response 1 (EGR1) primers were used as internal control for a gene not regulated by Sam68. Results are shown as fold enrichment in the anti-Sam68 immunoprecipitates versus the control IgGs. B and C, coimmunoprecipitation of endogenous Sam68 with cyclin D1 mRNA. LNCaP cell extracts were immunoprecipitated with anti–Sam68-specific antibody; IgGs were used as mock reaction; coprecipitated RNAs were analyzed by RT-PCR with primers specific for cyclin D1a (CycD1a), cyclin D1b (CycD1b), and hypoxanthine phosphoribosyltransferase (HPRT) used as negative control (C). An aliquot of the immunoprecipitates was saved for Western blot analysis of Sam68 (B). A and C, columns, mean of three independent experiments; bars, SD. P values of Student's t test are reported.
Fluctuations in Sam68 expression affect the ratio between cyclin D1a and cyclin D1b
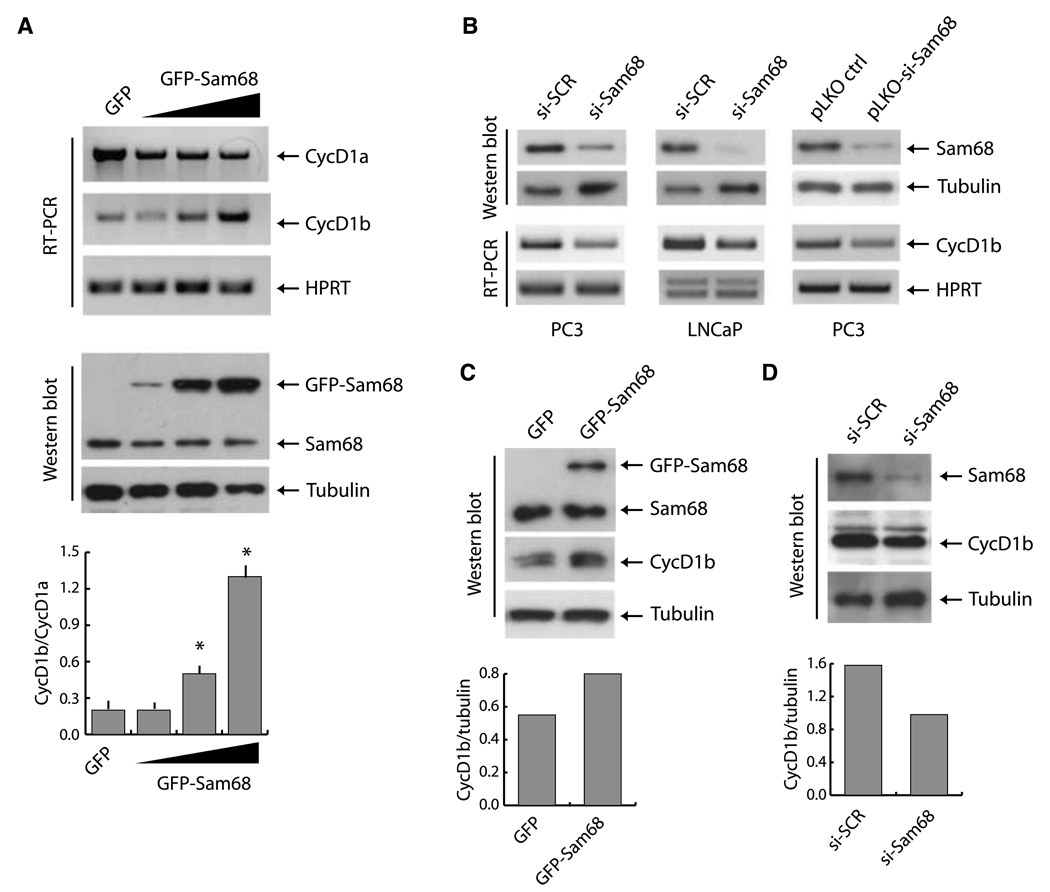
Knockdown of Sam68 in PCa cells caused accumulation of cyclin D1 mRNA and protein (22), whereas its overexpression in NIH-3T3 (26) and HeLa cells (27) inhibited cyclin D1 transcription. To further investigate the role of Sam68 in cyclin D1 expression in PCa cells, we transiently transfected increasing amounts of GFP-Sam68 in PC3 cells, which express low levels of cyclin D1b (10). Upregulation of Sam68 caused a dose-dependent decrease of the more abundant cyclin D1a mRNA, whereas it concomitantly stimulated the expression of the cyclin D1b transcript, with a consequent shift in the ratio between the levels of the two isoforms in the cell (Fig. 2A). To test the hypothesis that Sam68 supports cyclin D1b expression in PCa cells, we knocked it down by RNAi in PC3 and LNCaP cells. Remarkably, depletion of Sam68 decreased cyclin D1b expression in both PCa cell lines (Fig. 2B), indicating that it was required to maintain high levels of cyclin D1b mRNA. A similar result was obtained by downregulation of Sam68 in PC3 cells stably transfected with a plasmid (pLKO-si-Sam68) encoding a different siRNA (Fig. 2B). Moreover, transient transfection of GFP-Sam68 in PC3 cells caused an increase also in cyclin D1b protein (Fig. 2C), whereas depletion of the endogenous Sam68 by RNAi decreased cyclin D1b expression (Fig. 2D). These results strongly indicate that fluctuations in Sam68 levels modulate the expression of cyclin D1b in PCa cells.
Figure 2.
Sam68 enhances expression of cyclin D1b in PCa cells. A, PC3 cells were transfected either with GFP or with increasing amounts of GFP-Sam68 constructs. Top, cyclin D1a and cyclin D1b transcripts were analyzed by RT-PCR; middle, Western blot analysis of GFP-Sam68 and endogenous Sam68. Staining of the blot with α-tubulin was used as internal loading control. Bottom, densitometric analysis of the cyclin D1b/cyclin D1a ratio. Columns, mean of three separate experiments; bars, SD. Statistical significance was determined by Student's t test. *, P < 0.05. B, effect of Sam68 knockdown on cyclin D1b expression was detected after silencing of Sam68 mRNA in PC3 and LNCaP cells, either with siRNA oligo or with stable transfection of pLKO or pLKO-si-Sam68. Cells were harvested 48 h after transfection and analyzed by immunoblot for Sam68 and tubulin expression or by RT-PCR to determine the expression of cyclin D1b or HPRT. C and D, Sam68 overexpression (C) and downregulation (D) modulate cyclin D1b protein level. Immunoblot analysis of Sam68 and cyclin D1b was normalized for the β-tubulin abundance. Bottom, densitometric analysis of the ratio of cyclin D1b/tubulin.
Sam68 promotes AS of cyclin D1b
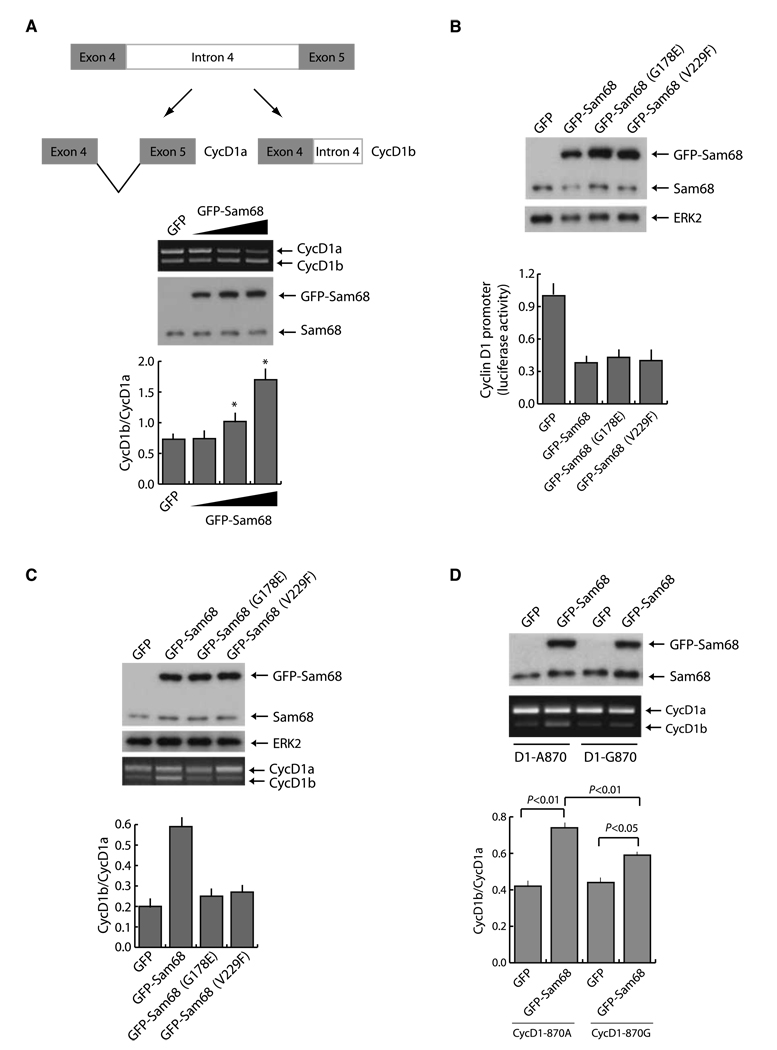
To determine whether Sam68 increased cyclin D1b expression through an effect on AS, we constructed a minigene containing only the alternatively spliced region of the gene, comprised between exon 4 and exon 5, under the control of the cytomegalovirus promoter (Fig. 3A). Expression of this minigene in PC3 cells yielded both cyclin D1 transcripts. Upregulation of wild-type Sam68 caused an increase in the D1b transcript and a concomitant decrease in the D1a transcript (Fig. 3A), indicating that Sam68 affects the AS of the cyclin D1 minigene in a dose-dependent manner.
Figure 3.
Sam68 modulates the AS of cyclin D1. A, schematic representation of the cyclin D1 minigene and of the PCR products from the in vivo splicing assay. Top, splicing assay of the cyclin D1 minigene in PC3 cells transfected with the cyclin D1 minigene together with increasing amounts of GFP-Sam68. Cells were harvested 20 h after transfection and processed for RT-PCR and protein extraction. Middle, immunoblot analysis of the same samples of the splicing assay. Bottom, columns, mean of the cyclin D1b/cyclin D1a ratio from three independent experiments; bars, SD. *, P < 0.01. B, cyclin D1 luciferase reporter assay of PC3 cells transfected with pGL3-cyclinD1-luc and GFP-Sam68wt or G178E and V229F mutants. Top, Western blot analysis of the transfected cells; bottom, cell extracts were prepared and assayed for firefly and Renilla luciferase (to normalize for transfection efficiency). Columns, mean of three experiments; bars, SD. C, splicing assay of the cyclin D1 minigene described in A in PC3 cells transfected with the indicated GFP or GFP-Sam68 constructs. Top, immunoblot analysis of the same samples of the splicing assay; bottom, PCR from the splicing assay with the cyclin D1 minigene. Columns, mean of the cyclin D1b/cyclin D1a ratio from three independent experiments; bars, SD. D, in vivo splicing assay of cyclin D1 minigenes 870G or 870A in the presence of GFP-Sam68. PC3 cells were transfected with the indicated constructs and collected 30 h after transfection for RNA and protein extraction. Top, immunoblot analysis of the transfected cells. Middle, PCR analysis for cyclin D1a and cyclin D1b. Bottom, densitometric analysis of three different experiments. Columns, mean; bars, SD. P value is indicated.
Recent evidence showed that a change in cyclin D1 AS could be achieved through a decrease in the elongation rate of transcription of the gene exerted by the transcriptional regulator EWS-FLI (28). Because Sam68 represses cyclin D1 and viral promoters independently of its RNA-binding activity (18, 26, 27, 29), we asked whether the effect on splicing was direct or a consequence of the decreased transcription. PC3 cells were transfected with a reporter constructs encoding the firefly luciferase gene under the control of the cyclin D1 promoter in the presence or absence of wild-type Sam68, Sam68G178E, or Sam68V229F. These Sam68 GSG domain mutants are defective in RNA binding and splicing (20) but retain their effect on transcription (23). As expected, we found that both alleles strongly decreased cyclin D1 transcription (Fig. 3B). However, when assayed on cyclin D1 AS in vivo, neither Sam68G178E nor Sam68V229F was able to significantly affect the cyclin D1b/cyclin D1a ratio (Fig. 3C). These experiments strongly indicate that direct binding of Sam68 to the cyclin D1 RNA is required for cyclin D1b AS.
It has been proposed that a transition from G to A (G870A) at the exon 4–intron 4 junction can favor cyclin D1b expression and predispose patients to cancer risk (6). To investigate whether this polymorphism also affected the activity of Sam68 in AS, we used two previously validated minigenes encoding the open reading frame of cyclin D1 and containing intron 4 (10). Although Sam68 stimulated cyclin D1b splicing with both alleles, its effect was significantly stronger in the presence of A at position 870 (Fig. 3D), indicating that this polymorphism in CCND1 favors Sam68 activity.
Sam68 binds cyclin D1 intron 4 RNA sequences and negatively affects U1 recruitment to the exon 4–intron 4 junction
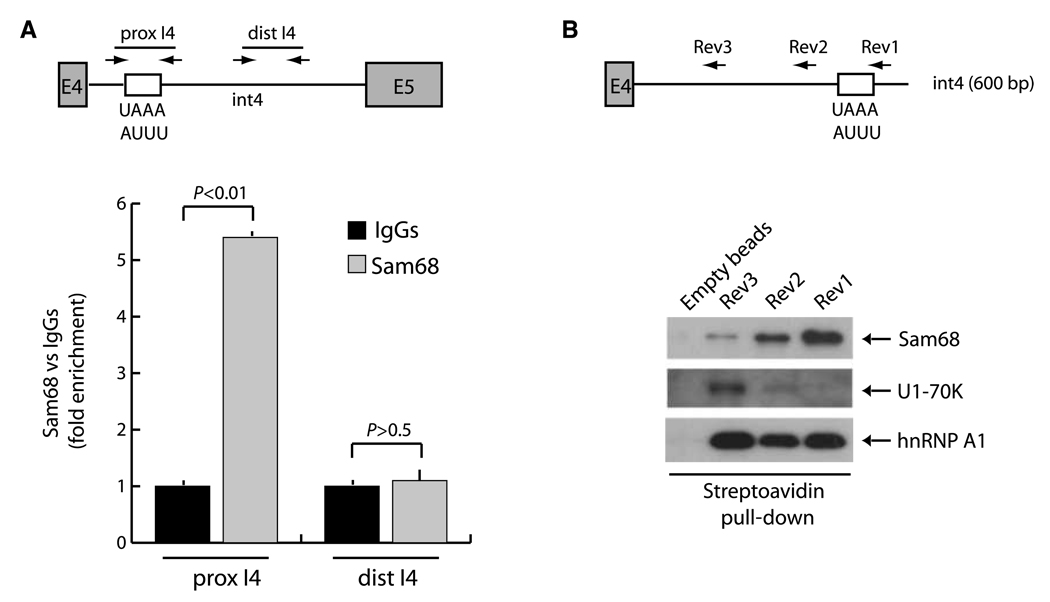
Cyclin D1b derives from retention of intron 4 in the pre-mRNA (6–8). This AS event might be due to the lack of recognition of the exon 4–intron 4 junction by the U1 small nuclear ribonucleoprotein (snRNP) that binds the 5′ splice site. Analysis of exon 4 and intron 4 sequences revealed several potential binding sites for Sam68 (UAAA or UUUA) within intron 4 (Fig. 4A), starting 390 bases downstream of the exon 4–intron 4 boundary. To determine whether endogenous Sam68 was enriched nearby these sites in vivo, we performed ChIP experiments in LNCaP cells. We observed a strong accumulation of Sam68 within intron 4 in proximity of its potential binding sites (prox-I4) but not in the distal part of the intron (dist-I4; Fig. 4A). To confirm that Sam68 bound this region in the RNA, we performed RNA oligo pull-down experiments. In vitro synthesized biotinylated RNAs containing (Rev1) or not (Rev2 and Rev3) the region in intron 4 enriched in Sam68-binding sites were incubated in vitro with nuclear extracts in the presence of streptavidin-Sepharose beads to isolate protein-RNA complexes. Immunoblot analysis showed that Sam68 was strongly associated with the Rev1 RNA, whereas its binding was substantially impaired when UAAA/UUUA sites were deleted in the Rev2 and Rev3 RNAs (Fig. 4B). Remarkably, we observed a complementary behavior for the U1-70K protein, a constitutive component of the U1 snRNP that recognizes the 5′ splice site at the end of the exon (13, 15). In this case, the U1 complex was recruited only when Sam68 binding was decreased or abolished in the Rev2 and Rev3 RNAs. By contrast, hnRNP A1, which binds heterogeneous nuclear pre-mRNAs with low specificity (16), associated equally well with all RNAs (Fig. 4B). These results suggest that Sam68 binding within intron 4 interferes with the recruitment of the U1 snRNP and the spliceosome at the intron 4–exon 4 junction.
Figure 4.
Sam68 binds to intron 4 sequences in the cyclin D1 pre-mRNA. A, scheme of the CCND1 gene comprised between exon 4 (E4) and exon 5 (E5). White box, position of the Sam68-binding sites in intron 4 (int4). Arrows, genomic regions amplified in the ChIP experiment. Cross-linked chromatin derived from LNCaP cells was immunoprecipitated and analyzed by quantitative real-time PCR using the indicated primers as described in Fig. 1. Columns, mean of three independent experiments; bars, SD. P values of Student's t test are reported. B, pull-down assays of Sam68, U1-70K, and hnRNP A1 with biotinylated RNA containing CCND1 exon 4 and different regions of intron 4 with (Rev1) or without (Rev2 and Rev3) the potential Sam68-binding sites. The proteins pulled down with the RNAs were analyzed by Western blot with antibodies for Sam68, U1-70K, or hnRNP A1.
Signal transduction pathways impinging on Sam68 activity modulate cyclin D1 AS
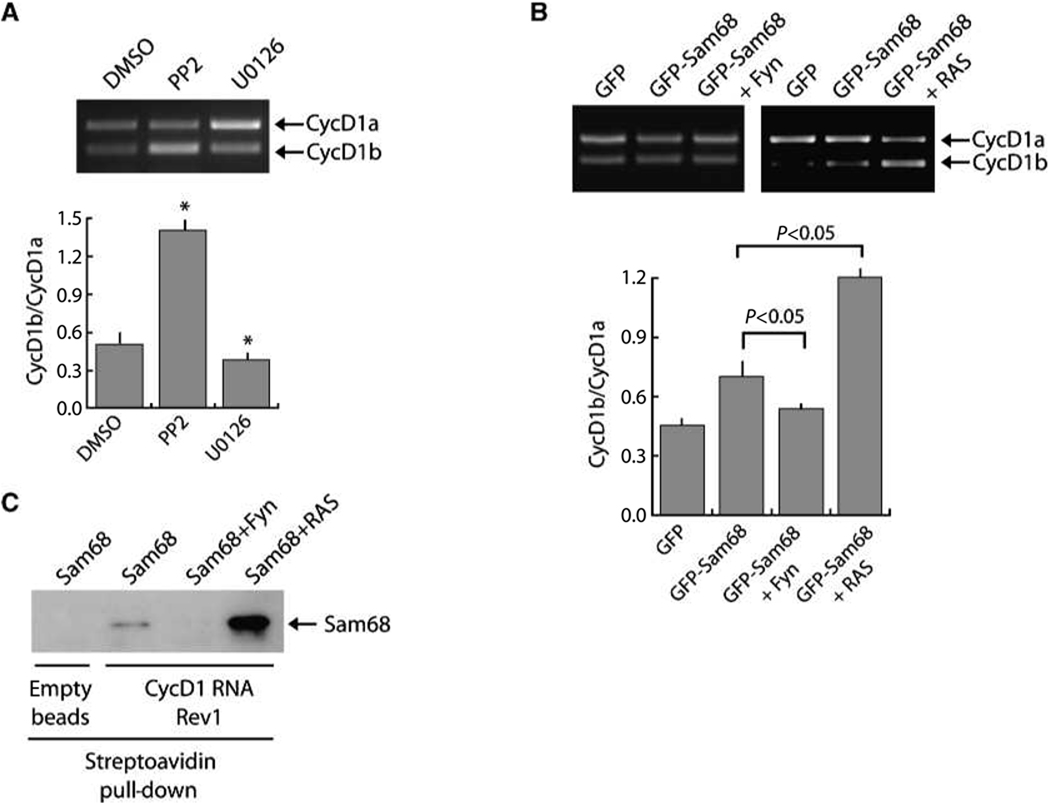
Serine-threonine phosphorylation by the RAS–MAPK/ERK kinase (MEK) 1/2–ERK1/2 pathway strongly enhances Sam68-dependent exon v5 inclusion in CD44 pre-mRNA (17). On the other hand, tyrosine phosphorylation by SFKs suppresses Sam68-mediated splicing of BCL2L1 (20). Thus, we asked whether interference with the activity of these kinases in PCa cells affected cyclin D1 splicing. To this end, PC3 cells were transfected with the CCND1 minigene (Fig. 3A) and treated for 12 hours with the Src inhibitor PP2 or the MEK1/2 inhibitor U0126 to prevent activation of ERK1/2. Remarkably, we found that inhibition of Src kinases increased the cyclin D1b/cyclin D1a ratio, whereas inhibition of the ERK1/2 pathway decreased it (Fig. 5A). These effects seemed specific because inhibition of the mammalian target of rapamycin pathway or the c-Jun NH2-terminal kinase MAPK pathway, which is not known to influence Sam68 splicing activity, exerted no effect on the cyclin D1b/cyclin D1a ratio (data not shown).
Figure 5.
RAS/ERKs and Src-related kinases modulate Sam68-dependent AS of cyclin D1. A, PC3 cells were transfected with the CCND1 minigene and treated for 12 h with the Src inhibitor PP2 or the MEK1/2 inhibitor U0126. Cells were harvested after the treatment, RNA was extracted, and RT-PCR was performed. Columns, mean of the cyclin D1b/cyclin D1a ratio from three independent experiment; bars, SD. *, P < 0.05. B, in vivo splicing assay of cyclin D1 870G minigene in the presence of GFP-Sam68 coexpressed with Fyn or with RASL61Q, which stimulates the ERK1/2 pathway. Columns, mean of the cyclin D1b/D1a ratio from three independent experiments; bars, SD. P value is indicated above. C, pull-down assay with biotinylated RNA containing CCND1 exon 4 and the proximal 600 bases of intron 4 containing the potential Sam68-binding sites. HEK293T cells were transfected with Sam68 alone or together with Fyn or RASL61Q. Nuclear extracts prepared from the transfected cells were used for pull-down assays with biotinylated cyclin D1 RNA and bound proteins were analyzed by immunoblot for Sam68.
To determine if the effects of PP2 and U0126 were due to direct action on Sam68, we coexpressed the SFK Fyn or a constitutively active form of RAS (RASL61Q), which stimulates the ERK1/2 pathway (17), with suboptimal amounts of Sam68 and determined the effect on the cyclin D1 minigene AS. We observed the opposite results of the corresponding inhibitors. Upregulation of Fyn completely suppressed Sam68-mediated cyclin D1b splicing, whereas overexpression of RASL61Q substantially increased the cyclin D1b/cyclin D1a ratio (Fig. 5B). Moreover, binding of Sam68 to the intron 4 sequences was strongly increased in the presence of RASL61Q, whereas it was abolished by coexpression of Fyn. These results show that signal transduction pathways impinging on Sam68 phosphorylation modulate cyclin D1 AS through regulation of Sam68 affinity for the cyclin D1 pre-mRNA in cell model systems.
Correlation between Sam68 and cyclin D1b expression in human PCa
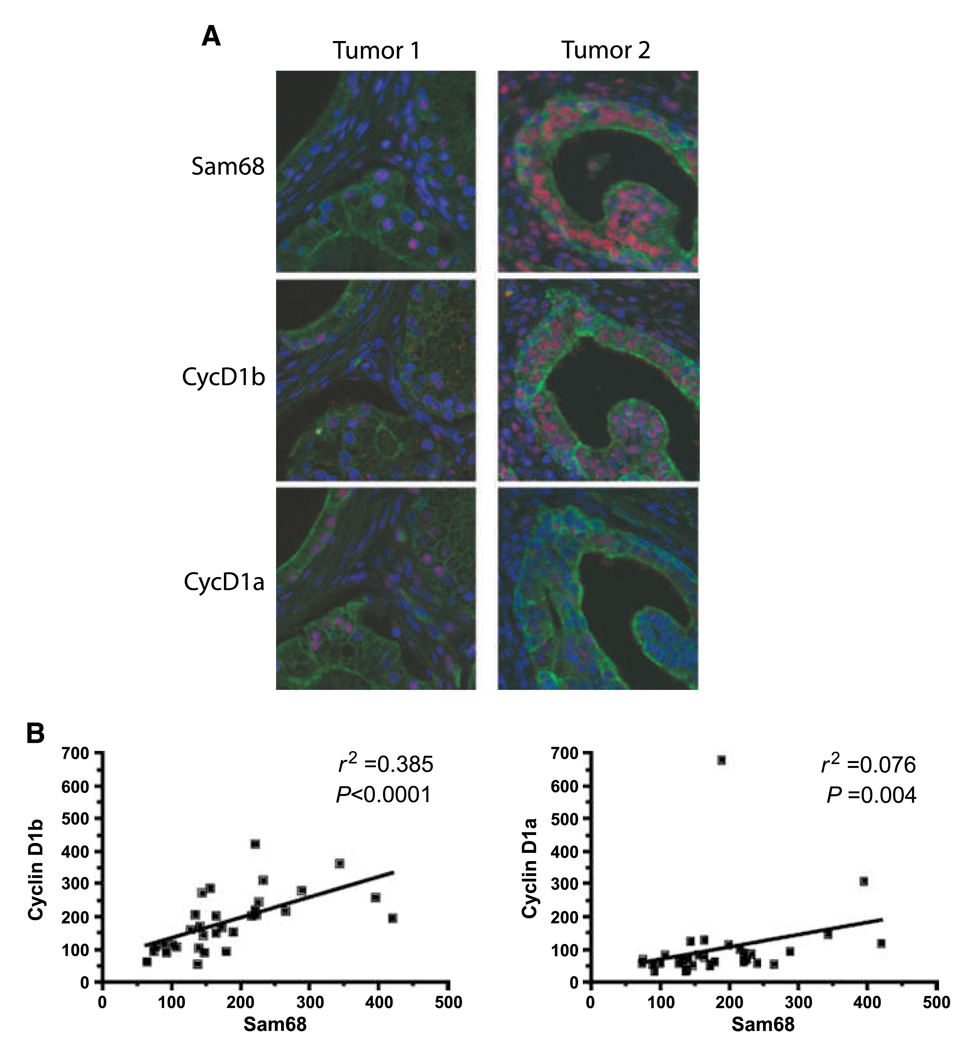
Recent evidence, obtained by using isoform- specific antisera (11), showed that cyclin D1b is elevated in a significant fraction of human PCa, and it is associated with tumorigenesis (10). Thus, we sought to determine if Sam68 and cyclin D1b expression levels correlate in PCa specimens. Fluorescent-based immunohistochemistry was used to analyze serial-sectioned human prostate tissue microarrays. Representative staining (Fig. 6A) for low and high Sam68 (top, tumor 1 and tumor 2, respectively) with corresponding cyclin D1b (middle) and cyclin D1a (bottom) suggested that individual glands with elevated Sam68 correlate with elevated cyclin D1b but not cyclin D1a. This observation was substantiated by analysis using unbiased quantification (AQUA system) of signal intensities for Sam68, cyclin D1b, and cyclin D1a. In these analyses, a positive correlation between Sam68 and cyclin D1b (Fig. 6B, left; compared with cyclin D1a, right) was observed using linear regression (r2 = 0.385) and Spearman correlations (P < 0.0001). These results suggest that Sam68 is an important component for the regulation of cyclin D1 AS in human PCa.
Figure 6.
Sam68 correlates with cyclin D1b levels in human PCa. A, representative fluorescence-based immunohistochemistry for Sam68 (top), cyclin D1b (middle), and cyclin D1a (bottom) in serial sections from two human PCa specimens with low (Tumor 1) and high Sam68 (Tumor 2). Red fluorescence, Sam68, cyclin D1b, and cyclin D1a; green fluorescence, epithelial cytokeratin; blue fluorescence, 4′,6-diamidino-2-phenylindole–stained nuclei. B, correlation analysis between Sam68 and cyclin D1b (left) or cyclin D1a (right) using nuclear intensities obtained from cytokeratin-positive epithelia using the AQUA system. Linear regression analysis is indicated by an r2 value. Spearman correlation statistics are indicated by a P value.
Discussion
The alternatively spliced cyclin D1b variant of the CCND1 gene has been proposed to have higher oncogenic potential than cyclin D1a (8, 9). In breast cancer, aberrant cyclin D1b expression confers resistance to therapeutic treatment (30) and is associated with poor prognosis in patients (31). Cyclin D1b was also recently shown to enhance cell invasiveness and anchorage-independent growth of bladder cancer cells (32), and this isoform has been detected in various other cancer types (8, 28, 33). In PCa, changes in the cyclin D1b/cyclin D1a ratio are of particular relevance. Indeed, whereas both isoforms support cell cycle progression, they behave differently in the interaction with the AR pathway. Cyclin D1a was reported to associate with AR and to negatively regulate its transcriptional activity, thereby representing a brake for uncontrolled proliferation of PCa cells (6). By contrast, this negative feedback function is lacking in cyclin D1b (11), and its expression positively correlated with PCa tumorigenesis in patients (10). However, the factors that determine the induction of cyclin D1b expression in PCa cells are still largely unknown. Herein, we show that the splicing regulator Sam68 modulates the ratio of cyclin D1 isoform expression in favor of cyclin D1b through regulation of AS. Interestingly, Sam68 is frequently upregulated in PCa (22, 23), and we show a positive correlation between Sam68 and cyclin D1b expression in PCa patients. Thus, our results identify Sam68 as the first splicing factor capable to regulate the cyclin D1b/cyclin D1a ratio in PCa cells.
We previously showed that Sam68 is upregulated in human PCa and that expression of this RBP supports LNCaP cell proliferation and their survival to genotoxic stresses (22). Microarray analyses showed that cyclin D1 was among the mRNAs regulated by Sam68 in these cells. In particular, we found that downregulation of Sam68 caused increased cyclin D1 levels (22). The microarray platform used did not distinguish between the cyclin D1 isoforms, and it was likely that the observed changes reflected the levels of the more abundant cyclin D1a. Our current experiments support this hypothesis. Transient overexpression of Sam68 led to a decrease in cyclin D1a expression (Fig. 2A). However, we found that upregulation of Sam68 caused a concomitant increase in the cyclin D1b isoform, whereas its depletion had the opposite effect. Although Sam68 is known to also regulate cyclin D1 transcription (26, 27), several observations indicate that the effects elicited are due to modulation of intron 4 AS in the cyclin D1 pre-mRNA. First, upregulation of Sam68 caused a dose-dependent increase in the cyclin D1b/cyclin D1a ratio also from a minigene driven by a viral promoter. Modulation of the cyclin D1 isoform ratio required the RNA-binding activity of Sam68, whereas the effect on transcription did not. Moreover, we found that Sam68 was particularly enriched in the proximal region of intron 4 in the CCND1 gene, and it could associate with intron 4 RNA sequences in vitro. Finally, activation of signal transduction pathways known to modulate its RNA-binding activity (17, 18, 20) regulated the association of Sam68 with intron 4 RNA sequences and resulted in modulation of cyclin D1 AS in live cells. These results strongly suggest that Sam68 is recruited to the CCND1 gene during transcription and promotes intron 4 retention and cyclin D1b expression in PCa cells.
Sam68 binds to U- and A-rich RNA sequences, with a preference for the UAAA or UUUA consensus (34), and it was recently shown that these sequences are enriched 500 bp upstream or downstream of the Sam68-regulated splice sites (21). Interestingly, we found that the proximal region of intron 4 contains several potential Sam68-binding sites and that these sequences are required for efficient recruitment of Sam68 to the CCND1 intron 4 and for binding to the RNA in vitro. Moreover, deletion of the Sam68-binding sites caused an increase in the association of the U1-70K spliceosomal component with intron 4 RNA, which mirrored the decreased association of Sam68. U1-70K is part of the U1 snRNP, which recognizes the 5′ splice site and defines the border of the upstream exon during pre-mRNA splicing (13). Thus, although further in vitro work is necessary to define the mechanism of regulation of this splicing event, our results suggest that binding of Sam68 to the proximal region of intron 4 interferes with recruitment of the U1 snRNP and with exon 4 definition, thereby causing intron retention and cyclin D1b splicing.
A polymorphism in the human CCND1 gene at the exon 4–intron 4 junction, G870/A, seems to affect cyclin D1 AS in a context-specific manner (6). In benign cultured cells and normal prostate tissue, the A870 allele favors cyclin D1b splicing (10). However, this effect of the genotype is lost in cancer and in tumor cell lines, and cyclin D1b was frequently observed also in specimens from patients bearing the G870 polymorphism (10). Moreover, analysis of >3,000 patients and controls showed that the polymorphism is not independently predictive of cancer risk. These observations collectively suggest that trans-acting factors and/or activation of signaling pathways regulating this splicing event in cells of patients carrying the G870 polymorphism compensate for the less efficient splicing of cyclin D1b. Our study indicates that Sam68 is one of such trans-acting factors affecting this AS event and identifies a signal transduction pathway that increases its splicing activity in cancer cells. We found that Sam68 was less active in enhancing cyclin D1b AS from a G870 minigene. However, its activity could be strongly increased by coexpression of a constitutively active form of RAS (17) and activation of the MEK/ERK pathway. Because this pathway is frequently upregulated in advanced PCa and correlates with the onset of refractoriness to hormonal therapy (35), it is possible that Sam68-dependent regulation of cyclin D1b splicing by the RAS pathway contributes to this step of prostate tumorigenesis. Importantly, the AS function of Sam68 likely contributes to cyclin D1b production in human disease, as Sam68 expression significantly correlated with cyclin D1b levels in clinical specimens. Collectively, the present findings obtained using in vitro models and through analyses of human tumor specimens strongly support a causative role for Sam68 in CCND1 AS.
Cyclin D1b splicing was increased by pharmacologic inhibition of SFKs in PCa cells. This effect can also be related to Sam68 function because Src kinases are known to phosphorylate this RBP in tyrosine residues at the COOH terminus and decrease its affinity for RNA (18). Moreover, we have previously shown that phosphorylation by the SFK Fyn decreased binding of Sam68 to BCL2L1 pre-mRNA and suppressed its effect on AS of this target (20). Herein, we found that tyrosine phosphorylation by Fyn also inhibits Sam68-dependent cyclin D1b splicing, suggesting that tyrosine phosphorylation of this RBP might negatively regulate this splicing event in PCa cells. These results could explain why cyclin D1b is upregulated in advanced and less differentiated PCa. Indeed, it was observed that the tyrosine kinase BRK, which also phosphorylates Sam68 and reduces its affinity for RNA like Fyn (36), is aberrantly regulated in advanced PCa. BRK is normally localized in the nucleus of prostate epithelial cells like Sam68, whereas it is less active and aberrantly relocalized in the cytoplasm in more advanced PCa patient cells (37). Thus, it is possible that reduced tyrosine phosphorylation of Sam68 by BRK in these patients favors cyclin D1b expression through the regulation of CCND1 AS.
In conclusion, our results identify Sam68 as a trans-acting factor that modulates cyclin D1 AS in PCa cells and suggest that the increased levels of Sam68 in human prostate carcinomas contribute to the elevated expression of cyclin D1b in this tumor type.
Acknowledgments
We thank Dr. Harald Konig for the gift of the RASL61Q plasmids and Dr. Maria Loiarro for assistance with luciferase reporter assays.
Grant Support
Associazione Italiana per la Ricerca sul Cancro, Association for International Cancer Research, and the Istituto Superiore della Sanità (ISS Project n.527/B/3A/5).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 2.Li HR, Wang-Rodriguez J, Nair TM, et al. Two-dimensional transcriptome profiling: identification of messenger RNA isoform signatures in prostate cancer from archived paraffin-embedded cancer specimens. Cancer Res. 2006;66:4079–4088. doi: 10.1158/0008-5472.CAN-05-4264. [DOI] [PubMed] [Google Scholar]
- 3.Zhang C, Li HR, Fan JB, et al. Profiling alternatively spliced mRNA isoforms for prostate cancer classification. BMC Bioinformatics. 2006;7:202–213. doi: 10.1186/1471-2105-7-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–5477. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu R, Dunn TA, Wei S, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knudsen KE, Diehl JA, Haiman CA, Knudsen ES. Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25:1620–1628. doi: 10.1038/sj.onc.1209371. [DOI] [PubMed] [Google Scholar]
- 7.Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WD, Heighway J. Alternate splicing produces a novel cyclin D1 transcript. Oncogene. 1995;11:1005–1011. [PubMed] [Google Scholar]
- 8.Lu F, Gladden AB, Diehl JA. An alternatively spliced cyclin D1 isoform, cyclin D1b, is a nuclear oncogene. Cancer Res. 2003;63:7056–7061. [PubMed] [Google Scholar]
- 9.Solomon DA, Wang Y, Fox SR, et al. Cyclin D1 splice variants. Differential effects on localization, RB phosphorylation, and cellular transformation. J Biol Chem. 2003;278:30339–30347. doi: 10.1074/jbc.M303969200. [DOI] [PubMed] [Google Scholar]
- 10.Comstock CES, Augello MA, Benito RP, et al. Cyclin D1 splice variants: polymorphism, risk, and isoform regulation in prostate cancer. Clin Cancer Res. 2009;15:5338–5349. doi: 10.1158/1078-0432.CCR-08-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burd CJ, Petre CE, Morey LM, et al. Cyclin D1b variant influences prostate cancer growth through aberrant androgen receptor regulation. Proc Natl Acad Sci U S A. 2006;103:2190–2195. doi: 10.1073/pnas.0506281103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartmann B, Valcárcel J. Decrypting the genome's alternative messages. Curr Opin Cell Biol. 2009;21:377–386. doi: 10.1016/j.ceb.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 14.Mendes Soares LM, Valcárcel J. The expanding transcriptome: the genome as the ‘Book of Sand’. EMBO J. 2006;25:923–931. doi: 10.1038/sj.emboj.7601023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 17.Matter N, Herrlich P, Konig H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- 18.Lukong KE, Richard S. Sam68, the KH domain-containing super-STAR. Biochim Biophys Acta. 2003;1653:73–86. doi: 10.1016/j.bbcan.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Cheng C, Sharp PA. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol Cell Biol. 2006;26:362–370. doi: 10.1128/MCB.26.1.362-370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol. 2007;176:929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chawla G, Lin CH, Han A, Ares M, Black DL. Sam68 regulates a set of alternatively spliced exons during neurogenesis. Mol Cell Biol. 2008;29:201–213. doi: 10.1128/MCB.01349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Busà R, Paronetto MP, Farini D, et al. The RNA-binding protein Sam68 contributes to proliferation and survival of human prostate cancer cells. Oncogene. 2007;26:4372–4382. doi: 10.1038/sj.onc.1210224. [DOI] [PubMed] [Google Scholar]
- 23.Rajan P, Gaughan L, Dalgliesh C, et al. The RNA-binding and adaptor protein Sam68 modulates signal-dependent splicing and transcriptional activity of the androgen receptor. J Pathol. 2008;215:67–77. doi: 10.1002/path.2324. [DOI] [PubMed] [Google Scholar]
- 24.Gurtner A, Manni I, Fuschi P, et al. Requirement for down-regulation of the CCAAT-binding activity of the NF-Y transcription factor during skeletal muscle differentiation. Mol Biol Cell. 2003;14:2706–2715. doi: 10.1091/mbc.E02-09-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paronetto MP, Messina V, Bianchi E, et al. Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J Cell Biol. 2009;185:235–249. doi: 10.1083/jcb.200811138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor SJ, Resnick RJ, Shalloway D. Sam68 exerts separable effects on cell cycle progression and apoptosis. BMC Cell Biol. 2004;5:5. doi: 10.1186/1471-2121-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Babic I, Cherry E, Fujita DJ. SUMO modification of Sam68 enhances its ability to repress cyclin D1 expression and inhibits its ability to induce apoptosis. Oncogene. 2006;25:4955–4964. doi: 10.1038/sj.onc.1209504. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez G, Bittencourt D, Laud K, et al. Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer. Proc Natl Acad Sci U S A. 2008;105:6004–6009. doi: 10.1073/pnas.0710748105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong W, Resnick RJ, Rakowski C, et al. Physical and functional interaction between the transcriptional cofactor CBP and the KH domain protein Sam68. Mol Cancer Res. 2002;1:48–55. [PubMed] [Google Scholar]
- 30.Wang Y, Dean JL, Millar EK, et al. Cyclin D1b is aberrantly regulated in response to therapeutic challenge and promotes resistance to estrogen antagonists. Cancer Res. 2008;68:5628–5638. doi: 10.1158/0008-5472.CAN-07-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Millar EK, Dean JL, McNeil CM, et al. Cyclin D1b protein expression in breast cancer is independent of cyclin D1a and associated with poor disease outcome. Oncogene. 2009;28:1812–1820. doi: 10.1038/onc.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim CJ, Nishi K, Isono T, et al. Cyclin D1b variant promotes cell invasiveness independent of binding to CDK4 in human bladder cancer cells. Mol Carcinog. 2009;48:953–964. doi: 10.1002/mc.20547. [DOI] [PubMed] [Google Scholar]
- 33.Lévêque C, Marsaud V, Renoir JM, Sola B. Alternative cyclin D1 forms a and b have different biological functions in the cell cycle of B lymphocytes. Exp Cell Res. 2007;313:2719–2729. doi: 10.1016/j.yexcr.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 34.Lin Q, Taylor SJ, Shalloway D. Specificity and determinants of Sam68 RNA binding. Implications for the biological function of K homology domains. J Biol Chem. 1997;272:27274–27280. doi: 10.1074/jbc.272.43.27274. [DOI] [PubMed] [Google Scholar]
- 35.Shen MM, Abate-Shen C. Pten inactivation and the emergence of androgen-independent prostate cancer. Cancer Res. 2007;67:6535–6538. doi: 10.1158/0008-5472.CAN-07-1271. [DOI] [PubMed] [Google Scholar]
- 36.Derry JJ, Richard S, Valderrama Carvajal H, et al. Sik (BRK) phosphorylates Sam68 in the nucleus and negatively regulates its RNA binding ability. Mol Cell Biol. 2000;20:6114–6126. doi: 10.1128/mcb.20.16.6114-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Derry JJ, Prins GS, Ray V, Tyner AL. Altered localization and activity of the intracellular tyrosine kinase BRK/Sik in prostate tumor cells. Oncogene. 2003;22:4212–4220. doi: 10.1038/sj.onc.1206465. [DOI] [PubMed] [Google Scholar]