Summary
The tyrosine kinase receptor KIT and the transcription factor MITF, each required for melanocyte development, have been shown to interact functionally both in vitro and in vivo. In vitro, KIT signaling leads to MITF phosphorylation, affecting MITF activity and stability. In vivo, the presence of the MitfMi-wh allele exacerbates the spotting phenotype associated with heterozygosity for Kit mutations. Here we show that among a series of other Mitf alleles, only the recessive Mitfmi-bws mimics the effect of MitfMi-wh on Kit. Intriguingly, Mitfmi-bws is characterized by a splice defect that leads to a reduction of RNAs containing MITF exon 2B which encodes serine-73, a serine phosphorylated upon KIT signaling. Nevertheless, other Mitf alleles that generally affect Mitf RNA levels, or carry a serine-73-to-alanine mutation that specifically reduces exon 2B-containing RNAs, do not show interactions with Kit in vivo. We conclude that the recessive Mitfmi-bws is a complex allele that can display a semi-dominant effect when present in a Kit-sensitized background. We suggest that human disease variability may equally be due to complex, allele-specific interactions between different genes.
Keywords: transcription factor, signaling, gene interactions, pigmentation, mouse
Introduction
Interactions between different genes are often difficult to assess because they may be subtle and the corresponding phenotypes not easily visible to the naked eye. Such interactions can be probed readily, however, for genes affecting pigmentation because pigmentary alterations can serve as a highly sensitive read-out of the modification of the action of one gene by another (Quevedo and Holstein, 1992, Barsh, 1996, Spritz, 1997, Baxter et al., 2009). In fact, given that over 200 loci are known to affect pigmentation in mice alone, pigmentation may be among the first phenotypes for which an extensive if not complete genetic network can be established (Hearing and Jimenez, 1989, Bennett and Lamoreux, 2003, Baxter et al., 2004, Hou and Pavan, 2008).
Genetic interactions often reflect functional interactions where the gene products in question feed into common molecular pathways (Drees et al., 2005). This is perhaps best illustrated by interactions between transcription factor genes on the one hand and genes controlling signaling pathways that modify the activities of these transcription factor genes or their products on the other. For instance, mice that are heterozygous for a mutation in the gene encoding the signaling receptor Kit and also heterozygous for a mutation in the gene encoding the microphthalmia-associated transcription factor Mitf (a basic-helix-loop-helix-leucine zipper protein which binds DNA as homo- or heterodimers) can show much more extensive white spotting than would be expected from heterozygosity for mutations in either gene alone (Beechey and Harrison, 1994, Hou et al., 2000, Diwakar et al., 2008). This phenomenon may indeed reflect functional interactions as it is well documented in vitro that Mitf is needed for the maintenance of KIT expression in melanoblasts and that Kit signaling affects Mitf both at the transcriptional and post-translational levels (Opdecamp et al., 1997, Hemesath et al., 1998, Price et al., 1998). Interactions have also been observed between genes encoding signaling components, such as endothelin receptor B (Ednrb) and Kit ligand (Kitl), or transcription factors, such as Sox10 and Mitf (Potterf et al., 2000, Rhim et al., 2000, Hou et al., 2006). Moreover, an interaction has been observed between Mitf and Bcl2, a gene that is involved in the cell death pathway and is regulated by Mitf (McGill et al., 2002).
The fact that pigmentation defects caused by mutations in one gene can be affected by mutations in a second gene have also made it possible to search for mutant pigmentation genes that by themselves would hardly produce a visible phenotype in heterozygotes. For example, a number of novel mutations affecting the pigment cell lineage have recently been identified in mice by combining a null mutation for Sox10 with germline mutations obtained after treatment with N-ethyl-N-nitrosourea (Matera et al., 2008). When designing such sensitized genetic screens, however, one has to take into account that gene interactions may be allele-specific. The above mentioned interaction between Kit and Mitf, for instance, although seen with several Kit alleles (Beechey and Harrison, 1994, Hou et al., 2000, Diwakar et al., 2008), has so far been tested only for the semi-dominant MitfMi-wh (Microphthalmia-white) allele. MitfMi-wh is characterized by a codon change in exon 7 and produces at least one protein isoform that is unable to bind DNA but still able to form dimers with wild-type dimerization partners (Hemesath et al., 1994). The enhanced spotting phenotype of MitfMi-wh/Kit mutant mice could be explained, therefore, by reduced phosphorylation, and hence increased stability and dominant-negative action, of this isoform (Arnheiter et al., 2006). If so, Mitf alleles lacking dominant-negative characteristics might not show similar interactions with Kit. In fact, as demonstrated here, many other Mitf alleles showed no interactions with the Kit null allele Kittm1Alf (Bernex et al., 1996, Hou et al., 2000), with the intriguing exception of a recessive allele, Mitfmi-bws (microphthalmia-black and white spots), prompting us to analyze this allele and its interactions with Kittm1Alf in more detail.
Mitfmi-bws, when homozygous, produces extensive white spotting but when heterozygous, there is no visible phenotype. The allele is characterized by a point mutation in the Mitf intron preceding exon 2 but otherwise the coding region remains wild-type (Hallsson et al., 2000). Mitfmi-bws is associated with a severe reduction of Mitf RNA levels when measured in an organ that shows no obvious phenotype, the heart (Bauer et al., 2009). It is also associated with a skewed splicing pattern that enriches RNAs excluding a part of exon 2, called exon 2B, at the expense of RNAs including this particular subexon (Hallsson et al., 2000). Intriguingly, exon 2B contains the codon for a serine, serine-73, that is phosphorylated in response to Kit signaling and whose phosphorylation affects the transcriptional activity and stability of MITF protein (Hemesath et al., 1998). Therefore, we used additional Mitf alleles that separately probe changes in Mitf RNA levels and changes in splicing patterns and mutations in serine-73, and analyzed the effects of these alleles in mice heterozygous for Kit. The results imply that Mitfmi-bws is a complex allele that may act in a semi-dominant fashion similar to MitfMi-wh, but that is dissimilar from MitfMi-wh in that its semi-dominant activity may only be revealed when placed on a Kit-sensitized background.
Results and discussion
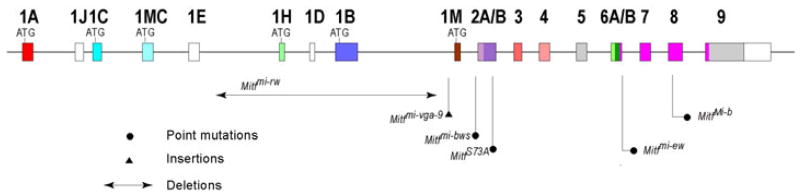
The Kit allele used in this study, Kittm1Alf, is characterized by an in-frame insertion of the bacterial LacZ gene, encoding nuclear LACZ, in the first exon of Kit in a way that the Kit gene becomes a functional null allele (Bernex et al., 1996, Hou et al., 2000). Homozygotes usually die in utero but heterozygotes display a belly spot, white feet and a white tail tip. Such mice were crossed with mice carrying either one of the following Mitf alleles: microphthalmia-brownish (MitfMi-b), microphthalmia-vga9 (Mitfmi-vga9), microphthalmia-eyeless-white Mitfmi-ew, microphthalmia red-eyed white (Mitfmi-rw), Mitftm1.1Arnh (here called MitfS73A), and microphthalmia-black and white spots (Mitfmi-bws). Their molecular characteristics are schematically shown in Fig. 1 and their phenotypes described in Table 1. MitfMi-b is inherited semi-dominantly and contains a point mutation in exon 8 that affects DNA binding. Homozygotes have a white coat and red eyes while heterozygotes have a brownish coat color, and pale ears and tails (Steingrimsson et al., 1996). The recessive allele Mitfmi-vga9 is a transgenic insertional null allele and its homozygotes are white with small eyes (Hodgkinson et al., 1993). The recessive allele Mitfmi-ew has a point mutation close to the exon 6B/intron 6 splice junction, and its mRNA is characterized by the absence of exon 6A/B and retention of the open reading frame between exons 5 and 7. The resulting protein affects DNA binding, with homozygotes showing a similar phenotype as Mitfmi-vga9 homozygotes (Opdecamp et al., 1997, Nakayama et al., 1998). The recessive allele Mitfmi-rw contains a genomic deletion that encompasses the exons 1H, 1D, and 1B1a/1B1b and their flanking sequences (Bharti et al., 2008). Mitfmi-rw homozygotes have eyes of variable sizes and a coat that is white except for a black spot on the head and/or belly or the base of the tail. The allele MitfS73A has been generated by gene targeting and encodes a non-phosphorylatable alanine instead of the phosphorylatable serine at position 73 (Bismuth et al., 2008). Because the codon change affects an exonic splice enhancer sequence (Wang et al., 2009), it leads to efficient skipping of exon 2B. Nevertheless, the corresponding mice, heterozygous or homozygous, have no visible phenotype. Mitfmi-bws is characterized by a point mutation in intron 1 that results in partial skipping of exon 2B (Hallsson et al., 2000). Homozygotes have widespread white spotting but normal eyes.
Fig. 1.
Schematic diagram of the mouse Mitf gene and the mutations used in this study. Filled boxes represent coding exons, and open boxes non-coding exons or non-coding parts of exons.
Table 1.
Allele-specific interactions between Mitf alleles and the Kittm1Alf allele visualized by white spotting of the coat
| Genotype 1 | Inheritance | Genotype 2 | Inheritance | Spotting phenotype | Interaction | Reference |
|---|---|---|---|---|---|---|
| Mitf+/+ | – | Kit+/+ | – | None | – | |
| Mitf+/+ | – | Kittm1Alf/+ | Sd | Belly spot, white feet and tail | – | Bernex et al., 1996 |
| MitfMi-b/+ | Sd | Kit+/+ | – | None | – | Steingrimsson et al., 1996 |
| Mitfmi-ew/+ | Re | Kit+/+ | – | None | – | Nakayama et al., 1998 |
| Mitfmi-rw/+ | Re | Kit+/+ | – | None | – | Bharti et al., 2008 |
| Mitfmi-vga-9/+ | Re | Kit+/+ | – | None | – | Hodgkinson et al., 1993 |
| MitfS73A/+ | Re | Kit+/+ | – | None | – | Bismuth et al., 2008 |
| Mitf mi-bws/+ | Re | Kit+/+ | – | None | Hallsson et al., 2000 | |
| Mitf Mi-b/+ | Sd | Kittm1Alf/+ | Sd | Belly spot, white feet and tail | NI | This paper |
| Mitf mi-ew/+ | Re | Kittm1Alf/+ | Sd | Belly spot, white feet and tail | NI | This paper |
| Mitf mi-rw/+ | Re | Kittm1Alf/+ | Sd | Belly spot, white feet and tail | NI | This paper |
| Mitf mi-vga-9/+ | Re | Kittm1Alf/+ | Sd | Belly spot, white feet and tail | NI | This paper |
| MitfS73A/+ | Re | Kittm1Alf/+ | Sd | Belly spot, white feet and tail | NI | This paper |
| Mitf mi-bws/+ | Re | Kittm1Alf/+ | Sd | Extensive spotting around trunk, white feet, white tail | IN | This paper |
| Ednrbtm1Myks/+ | Re | Kit+/+ | – | Occasional belly spot | NI | This paper |
| Ednrbtm1Myks/+ | Re | Kittm1Alf/+ | Sd | Occasional belly spot | NI | This paper |
Mice containing the indicated alleles were obtained by appropriate crosses to yield 14 to 18 doubly heterozygous mice. Note that while many mutant Mitf alleles, when homozygous, cause small eyes (microphthalmia), there were no obvious eye phenotypes in Mitf heterozygotes alone or in combination with Kittm1Alf/+.
Backgrounds of parental strains: Mitf +/+; Kit +/+ (C57BL/6), Mitf+/+; Kittm1Alf (mixed C57BL/6; C3H/He), Mitf Mi-b (C57BL/6J; C3H/RI), Mitf mi-ew (C57BL/6Bn), Mitf mi-rw (C57BL/6J), Mitf mi-vga-9 (mixed C57BL/6J; C3H/He), MitfS73A (129S1/Sv; C57BL/6), Mitf mi-bws (C57BL/10).
Abbreviations: “–“: not applicable; NI: no gene interaction; IN: gene interaction; Sd: semi-dominant; Re: recessive.
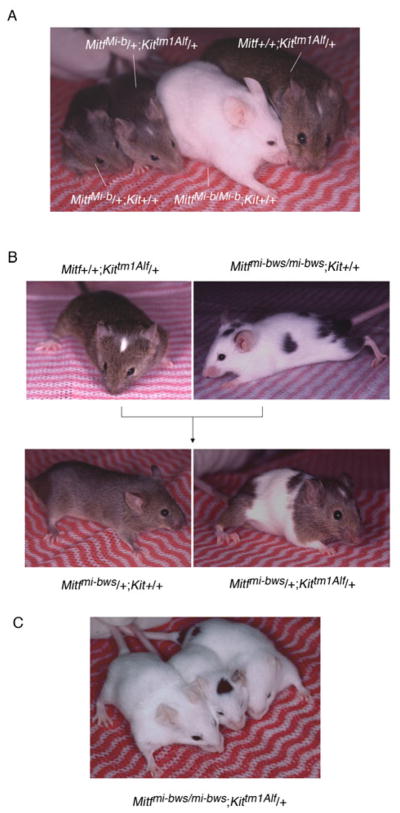
It was possible that in contrast to the previously observed strong MitfMi-wh/Kit interactions, the recessive Mitf alleles may show no or only mild (additive) interactions with Kit. Indeed, the recessive Mitfmi-ew or Mitfmi-rw, for instance, were unable to enhance the extent of the white spotting of Kittm1Alf/+, and even the mildy semi-dominant MitfMi-b showed no interactions with Kittm1Alf (Fig. 2A). In contrast, Mitfmi-bws/+; Kittm1Alf/+ mice were extensively spotted (Fig. 2B), and Mitfmi-bws/mi-bws; Kittm1Alf/+ mice were either completely white or occasionally retained just small pigmented spots on the head or the rump (Fig. 2C, compare with Mitfmi-bws/mi-bws; Kit+/+ mouse in Fig. 2B). To assay whether this interaction is Kit-specific or can also be seen with alterations in a different signaling pathway critical for melanocyte development, we generated double heterozygous combinations of Mitfmi-bws and Ednrbtm1Myks in which the receptor for endothelin-3, a G protein-coupled receptor, is non-functional (Lee et al., 2003, Hou et al., 2004, Saldana-Caboverde and Kos, in press). These mice, however, did not show the exacerbation of white spotting seen in Mitfmi-bws/+; Kittm1Alf/+ mice (Supplementary Fig. 1A).
Fig. 2.
Genetic interactions between Mitf and Kit. A. Lack of genetic interactions between MitfMi-b and Kittm1Alf. The corresponding genotypes are indicated in the figure. Note that MitfMi-b/+; Kittm1Alf/+ mice show a phenotype indistinguishable from that of Mitf+/+; Kittm1Alf/+ mice. B. Interaction between Mitfmi-bws and Kittm1Alf. Mice of the indicated genotypes were crossed and their offspring genotyped. Note that Mitfmi-bws/+; Kit+/+ mice are fully pigmented whereas Mitfmi-bws/+; Kittm1Alf/+ offspring show extensive white spotting in the trunk area, different from Mitf+/+; Kittm1Alf/+ mice. C. Mitfmi-bws/mi-bws; Kittm1Alf/+ mice are either totally white or retain just small pigmented spots.
Because Mitfmi-bws reduces both the overall Mitf RNA levels (as measured in the heart) and the relative amounts of Mitf RNA that contains exon 2B (for short, 2B+) versus Mitf RNA that lacks exon 2B (for short, 2B-), it was not a priori clear whether the above mentioned interaction with Kit was brought about by the general or the exon-specific reduction of Mitf RNA levels. Hence, we used additional alleles allowing us to test overall RNA levels separately from changes in exon 2B splicing. The allele Mitfmi-vga9 eliminates the production of Mitf RNA entirely so that in Mitfmi-vga-9/+ heterozygotes, Mitf RNA, all of which contributed from the remaining wild-type allele, accumulates to approximately 50% of what is seen in a normal animal, measured either in heart or skin (Bauer et al., 2009). However, Mitfmi-vga-9/+; Kittm1Alf/+ mice showed a spotting phenotype just as Mitf+/+; Kittm1Alf/+ mice, suggesting that the exacerbated phenotype in Mitfmi-bws/+; KittmAlf/+ mice is not simply due to a reduction in total Mitf RNA.
Because the above mentioned exon 2B splicing alteration in Mitfmi-bws mice has not so far been demonstrated specifically for melanocytes, we analyzed by standard RT-PCR reactions the exon distribution in the melanocyte-specific M-Mitf RNA. For these tests, we used dorsal skin from C57BL/6 control mice, and dorsal black skin from Mitfmi-bws/mi-bws and Kittm1Alf/+ mice (Supplementary Fig. 2). The results showed exon 2B splicing patterns as expected from the analysis of heart RNA from the respective mice. There was, however, no obvious reduction in M-Mitf RNA levels in black Mitfmi-bws/mi-bws skin. Conceivably, the corresponding melanocytes represent a pool of cells with higher Mitf levels which have allowed them to escape the developmental demise of Mitfmi-bws melanoblasts which on average may express lower levels of Mitf. In Kittm1Alf/+ skin, however, M-Mitf and total Mitf RNA were indeed reduced, consistent with the pigment dilution characteristics of these mice.
To test whether a skewed exon 2B splicing might contribute to the Mitf/Kit interaction independently of overall M-Mitf RNA level changes, we used the MitfS73A allele. This allele produces normal levels of total Mitf RNA in heart or skin, normal levels of M-Mitf in skin (not shown), but a 2B+/2B− ratio of approximately 0.1, as compared to wild type, where this ratio is approximately 0.9 (Bismuth et al., 2008). The phenotype of MitfS73A/+; Kittm1Alf/+ and MitfS73A/S73A; Kittm1Alf/+ mice (Supplementary Fig. 1B), suggests, however, that the exon 2B splice change can also not account separately for the Mitfmi-bws/Kit interaction, with the only caveat that the remaining 10% of 2B+ RNA derived from the MitfS73A allele contains the serine-to-alanine mutation while in Mitfmi-bws, the serine residue is unchanged. Given the above results, we also tested whether a combination of splice changes and a reduction in overall RNA levels would mimic the Mitfmi-bws/Kit gene interactions. To this end, we generated compound heterozygotes between Mitfmi-vga9 and MitfS73A and combined them with KittmAlf/+, but even this allelic combination (Mitfmi-vga9/S73A; Kittm1Alf/+) yielded mice with just the Kit phenotype (not shown). Quantitations of Mitf RNA (total, 2B+ and 2B-) for heart and skin of mice carrying the various alleles alone and in some combinations are shown in Supplementary Fig. 3.
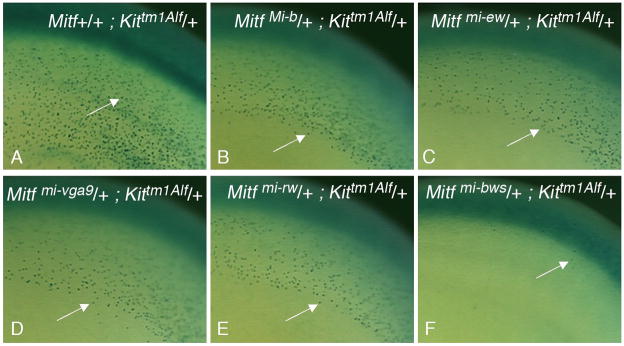
Finally, to determine whether the extensive white spotting in postnatal Kittm1Alf/+; Mitfmi-bws/+ mice already originates during development, at the melanoblast stage, or, conversely, whether the absence of enhanced white spotting in the other crosses might be the result of a postnatal compensation of an early developmental alteration, we made use of the fact that the Kittm1Alf allele produces melanoblasts that can be labeled by the X-GAL reaction. Fig. 3 shows examples of labeled embryos at day 12.5 of gestation from groups that contained 9–15 embryos and that showed little embryo-to-embryo variation within one genotypic cohort. As shown in Fig. 3A, in Kittm1Alf/+ embryos, individual β-Gal-positive cells in the area of the trunk are normally distributed in the dorsolateral migration pathway underneath the surface ectoderm where earlier studies have identified MITF-positive cells (Nakayama et al., 1998) that depend on functional MITF (Hou et al., 2000). Double heterozygous embryos for Kittm1Alf and either MitfMi-b, Mitfmi-vga-9, Mitfmi-ew, Mitfmi-rw or MitfS73A displayed a distribution pattern of β-Gal-positive melanoblasts that was similar to that observed in Mitf wild-type embryos heterozygous for Kittm1Alf (Fig. 3B-E, and data not shown). In contrast, the numbers of labeled cells were severely reduced in the middle trunk region of Kittm1Alf/+; Mitfmi-bws/+ embryos (Fig. 3F). Hence, these latter embryos, but not the former ones, have a severe defect in melanoblasts, suggesting that the adult white spotting of Kittm1Alf/+; Mitfmi-bws/+ mice is set at an early developmental stage.
Fig. 3.
Tracking of β-Gal-positive melanoblasts in embryogenesis. Embryos of the indicated genotypes were harvested at 12.5 days of gestation and processed for β-Gal labeling. Arrows point to individual β-Gal-positive melanoblasts in the dorsolateral migration pathway underneath the surface ectoderm in the trunk area. (A-E) Note similar distribution but slightly reduced numbers and densities of β-Gal-positive melanoblasts in embryos carrying a mutant Mitf allele together with the Kittm1Alf allele as opposed to a wild-type Mitf allele along with the Kittm1Alf allele, consistent with earlier findings that Mitf gene dosage affects melanoblast numbers early in development (Hornyak et al., 2001). (F) Melanoblasts in embryos double heterozygous for Mitfmi-bws and Kittm1Alf are very sparse and largely restricted to the area over the neural tube.
Earlier results have clearly indicated that Kit-mediated phosphorylation of serine-73 regulates MITF protein activity and stability in vitro (Hemesath et al., 1998). Nevertheless, neither direct targeting of the corresponding codon in the endogenous gene or in transgenic, bacterial artificial chromosome rescue constructs have been able to demonstrate a clear role for this serine in vivo (Bismuth et al., 2008, Bauer et al., 2009). An isolated serine-to-alanine change, with no effect on exon 2B splicing, however, has not so far been obtained, and so the exact role of serine-73 in an otherwise entirely normal gene has not been formally addressed. Nevertheless, as here demonstrated genetically, neither RNA level changes, nor exon 2B splice changes associated with the serine73-to-alanine mutation, alone or in combinations, can account for the Mitfmi-bws/Kit interaction phenotype. Hence, Mitfmi-bws is an allele that is likely altered in one or several more ways than would be suggested solely from its point mutation in intron 1. In fact, an explanation for the Mitfmi-bws phenotype may be found only when additional promoter- and splice-isoforms have been analyzed, an undertaking that may require the sequencing of the entire 200 kb Mitfmi-bws gene.
Supplementary Material
A. The Mitfmi-bws/Kit interaction is Kit-specific as no interaction is seen between Mitfmi-bws and Ednrbtm1Myks, an Ednrb null allele. B. No interaction between Kittm1Alf and MitfS73A. Note white feet characteristic of the Kittm1Alf/+ genotype but no extensive spotting, unlike what is seen in Mitfmi-bws/+; Kittm1Alf/+ mice.
RT-PCR using RNA from black dorsal skin of C57BL/6 wild type (wt), Mitfmi-bws/mi-bws (mi-bws/mi-bws), and Kittm1Alf/+ mice. Primers mapped to Mitf or Usf1 (upstream stimulatory factor-1) exons as indicated. Note that wt contains more M-Mitf exon 2B+ compared to M-Mitf exon 2B- cDNA while the M-Mitf exon 2B+/2B- cDNA ratio is reversed in mi-bws/mi-bws (primers 1M-3). The total levels of M-Mitf cDNA is, however, similar for wt and mi-bws/mi-bws skin and only lower for Kittm1Alf/+ skin for which, because of the lower levels, it becomes difficult to assess the relative 2B+/2B- ratio. Total Mitf cDNA levels (measured using primers 6B-8, amplifying Mitf from melanocytes and the remainder of skin tissue), are only slightly lower in mi-bws/mi-bws or Kittm1Alf/+ skin compared to wt. Note similar levels of control Usf1 cDNA for all samples.
Real time RT-PCR for the tissue samples and genotypes indicated. Separate quantitation was performed for exon 2B+ and exon 2B- cDNA, using primers giving equal length products as described in Bharti et al., in press. A. Wild type, and heterozygous and homozygous single-gene mutant genotypes. B. Wild type and Mitfmi-vga9/S73A compound mutants either lacking or carrying Kittm1Alf.
Acknowledgments
We would like to thank Dr. Jean-Jacques Panthier for providing Kittm1Alf mice and Dr. Myung K. Shin for providing Ednrbtm1Myks mice. All animals were handled according to the regulations of the Institutional Animal Care and Use Committee. This research was supported in part by the National Basic Research Program (973 Program) of China (2009CB526502), the National Natural Science Foundation of China (30771149), the Research Development Grant of Wenzhou Medical College (to L. H.), and the Intramural Research Program of NINDS, NIH.
References
- Arnheiter H, Hou L, Nguyen MTT, Bismuth K, Csermely T, Murakami H, Skuntz S, Liu W, Bharti K. MITF-A matter of life and death for developing melanocytes. In: Hearing VJ, Leong SPL, editors. From melanocytes to melanoma: The progression to malignancy. Totowa, N. J: Human Press, Inc; 2006. [Google Scholar]
- Barsh GS. The genetics of pigmentation: from fancy genes to complex traits. Trends Genet. 1996;12:299–305. doi: 10.1016/0168-9525(96)10031-7. [DOI] [PubMed] [Google Scholar]
- Bauer GL, Praetorius C, Bergsteinsdottir K, et al. The role of MITF phosphorylation sites during coat color and eye development in mice analyzed by bacterial artificial chromosome transgene rescue. Genetics. 2009;183:581–94. doi: 10.1534/genetics.109.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter LL, Hou L, Loftus SK, Pavan WJ. Spotlight on spotted mice: a review of white spotting mouse mutants and associated human pigmentation disorders. Pigment Cell Res. 2004;17:215–24. doi: 10.1111/j.1600-0749.2004.00147.x. [DOI] [PubMed] [Google Scholar]
- Baxter LL, Loftus SK, Pavan WJ. Networks and pathways in pigmentation, health, and disease. Wiley Interdiscip Rev Syst Biol Med. 2009;1:359–371. doi: 10.1002/wsbm.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beechey CV, Harrison MA. A new spontaneous W allele, W36H. Mouse Genome. 1994;92:502–502. [Google Scholar]
- Bennett DC, Lamoreux ML. The color loci of mice--a genetic century. Pigment Cell Res. 2003;16:333–44. doi: 10.1034/j.1600-0749.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- Bernex F, De Sepulveda P, Kress C, Elbaz C, Delouis C, Panthier JJ. Spatial and temporal patterns of c-kit-expressing cells in WlacZ/+ and WlacZ/WlacZ mouse embryos. Development. 1996;122:3023–33. doi: 10.1242/dev.122.10.3023. [DOI] [PubMed] [Google Scholar]
- Bharti K, Liu W, Csermely T, Bertuzzi S, Arnheiter H. Alternative promoter use in eye development: the complex role and regulation of the transcription factor MITF. Development. 2008;135:1169–78. doi: 10.1242/dev.014142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bismuth K, Skuntz S, Hallsson JH, Pak E, Dutra AS, Steingrimsson E, Arnheiter H. An unstable targeted allele of the mouse Mitf gene with a high somatic and germline reversion rate. Genetics. 2008;178:259–72. doi: 10.1534/genetics.107.081893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwakar G, Zhang D, Jiang S, Hornyak TJ. Neurofibromin as a regulator of melanocyte development and differentiation. J Cell Sci. 2008;121:167–77. doi: 10.1242/jcs.013912. [DOI] [PubMed] [Google Scholar]
- Drees BL, Thorsson V, Carter GW, Rives AW, Raymond MZ, Avila-Campillo I, Shannon P, Galitski T. Derivation of genetic interaction networks from quantitative phenotype data. Genome Biol. 2005;6:R38. doi: 10.1186/gb-2005-6-4-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallsson JH, Favor J, Hodgkinson C, et al. Genomic, transcriptional and mutational analysis of the mouse microphthalmia locus. Genetics. 2000;155:291–300. doi: 10.1093/genetics/155.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing VJ, Jimenez M. Analysis of mammalian pigmentation at the molecular level. Pigment Cell Res. 1989;2:75–85. doi: 10.1111/j.1600-0749.1989.tb00166.x. [DOI] [PubMed] [Google Scholar]
- Hemesath TJ, Price ER, Takemoto C, Badalian T, Fisher DE. MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature. 1998;391:298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–80. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- Hornyak TJ, Hayes DJ, Chiu LY, Ziff EB. Transcription factors in melanocyte development: distinct roles for Pax-3 and Mitf. Mech Dev. 2001;101:47–59. doi: 10.1016/s0925-4773(00)00569-4. [DOI] [PubMed] [Google Scholar]
- Hou L, Arnheiter H, Pavan WJ. Interspecies difference in the regulation of melanocyte development by SOX10 and MITF. Proc Natl Acad Sci U S A. 2006;103:9081–5. doi: 10.1073/pnas.0603114103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Panthier JJ, Arnheiter H. Signaling and transcriptional regulation in the neural crest-derived melanocyte lineage: interactions between KIT and MITF. Development. 2000;127:5379–89. doi: 10.1242/dev.127.24.5379. [DOI] [PubMed] [Google Scholar]
- Hou L, Pavan WJ. Transcriptional and signaling regulation in neural crest stem cell-derived melanocyte development: do all roads lead to Mitf? Cell Res. 2008;18:1163–76. doi: 10.1038/cr.2008.303. [DOI] [PubMed] [Google Scholar]
- Hou L, Pavan WJ, Shin MK, Arnheiter H. Cell-autonomous and cell non-autonomous signaling through endothelin receptor B during melanocyte development. Development. 2004;131:3239–47. doi: 10.1242/dev.01193. [DOI] [PubMed] [Google Scholar]
- Lee HO, Levorse JM, Shin MK. The endothelin receptor-B is required for the migration of neural crest-derived melanocyte and enteric neuron precursors. Dev Biol. 2003;259:162–75. doi: 10.1016/s0012-1606(03)00160-x. [DOI] [PubMed] [Google Scholar]
- Matera I, Watkins-Chow DE, Loftus SK, Hou L, Incao A, Silver DL, Rivas C, Elliott EC, Baxter LL, Pavan WJ. A sensitized mutagenesis screen identifies Gli3 as a modifier of Sox10 neurocristopathy. Hum Mol Genet. 2008;17:2118–31. doi: 10.1093/hmg/ddn110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill GG, Horstmann M, Widlund HR, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–18. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- Nakayama A, Nguyen MT, Chen CC, Opdecamp K, Hodgkinson CA, Arnheiter H. Mutations in microphthalmia, the mouse homolog of the human deafness gene MITF, affect neuroepithelial and neural crest-derived melanocytes differently. Mech Dev. 1998;70:155–66. doi: 10.1016/s0925-4773(97)00188-3. [DOI] [PubMed] [Google Scholar]
- Opdecamp K, Nakayama A, Nguyen MT, Hodgkinson CA, Pavan WJ, Arnheiter H. Melanocyte development in vivo and in neural crest cell cultures: crucial dependence on the Mitf basic-helix-loop-helix-zipper transcription factor. Development. 1997;124:2377–86. doi: 10.1242/dev.124.12.2377. [DOI] [PubMed] [Google Scholar]
- Potterf SB, Furumura M, Dunn KJ, Arnheiter H, Pavan WJ. Transcription factor hierarchy in Waardenburg syndrome: regulation of MITF expression by SOX10 and PAX3. Hum Genet. 2000;107:1–6. doi: 10.1007/s004390000328. [DOI] [PubMed] [Google Scholar]
- Price ER, Ding HF, Badalian T, Bhattacharya S, Takemoto C, Yao TP, Hemesath TJ, Fisher DE. Lineage-specific signaling in melanocytes. C-kit stimulation recruits p300/CBP to microphthalmia. J Biol Chem. 1998;273:17983–6. doi: 10.1074/jbc.273.29.17983. [DOI] [PubMed] [Google Scholar]
- Quevedo WC, Jr, Holstein TJ. Molecular genetics and the ontogeny of pigment patterns in mammals. Pigment Cell Res. 1992;5:328–34. doi: 10.1111/j.1600-0749.1992.tb00557.x. [DOI] [PubMed] [Google Scholar]
- Rhim H, Dunn KJ, Aronzon A, Mac S, Cheng M, Lamoreux ML, Tilghman SM, Pavan WJ. Spatially restricted hypopigmentation associated with an Ednrbs-modifying locus on mouse chromosome 10. Genome Res. 2000;10:17–29. [PubMed] [Google Scholar]
- Saldana-Caboverde A, Kos L. Roles of endothelin signaling in melanocyte development and melanoma. Pigment Cell Melanoma Res. doi: 10.1111/j.1755-148X.2010.00678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spritz RA. Piebaldism, Waardenburg syndrome, and related disorders of melanocyte development. Semin Cutan Med Surg. 1997;16:15–23. doi: 10.1016/s1085-5629(97)80031-4. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Nii A, Fisher DE, Ferre-D’Amare AR, McCormick RJ, Russell LB, Burley SK, Ward JM, Jenkins NA, Copeland NG. The semidominant Mi(b) mutation identifies a role for the HLH domain in DNA binding in addition to its role in protein dimerization. Embo J. 1996;15:6280–9. [PMC free article] [PubMed] [Google Scholar]
- Wang X, Debbache J, Arnheiter H. Alternative splicing and cell regulation in vertebrate pigment cells. Dev Biol. 2009;331:A108. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. The Mitfmi-bws/Kit interaction is Kit-specific as no interaction is seen between Mitfmi-bws and Ednrbtm1Myks, an Ednrb null allele. B. No interaction between Kittm1Alf and MitfS73A. Note white feet characteristic of the Kittm1Alf/+ genotype but no extensive spotting, unlike what is seen in Mitfmi-bws/+; Kittm1Alf/+ mice.
RT-PCR using RNA from black dorsal skin of C57BL/6 wild type (wt), Mitfmi-bws/mi-bws (mi-bws/mi-bws), and Kittm1Alf/+ mice. Primers mapped to Mitf or Usf1 (upstream stimulatory factor-1) exons as indicated. Note that wt contains more M-Mitf exon 2B+ compared to M-Mitf exon 2B- cDNA while the M-Mitf exon 2B+/2B- cDNA ratio is reversed in mi-bws/mi-bws (primers 1M-3). The total levels of M-Mitf cDNA is, however, similar for wt and mi-bws/mi-bws skin and only lower for Kittm1Alf/+ skin for which, because of the lower levels, it becomes difficult to assess the relative 2B+/2B- ratio. Total Mitf cDNA levels (measured using primers 6B-8, amplifying Mitf from melanocytes and the remainder of skin tissue), are only slightly lower in mi-bws/mi-bws or Kittm1Alf/+ skin compared to wt. Note similar levels of control Usf1 cDNA for all samples.
Real time RT-PCR for the tissue samples and genotypes indicated. Separate quantitation was performed for exon 2B+ and exon 2B- cDNA, using primers giving equal length products as described in Bharti et al., in press. A. Wild type, and heterozygous and homozygous single-gene mutant genotypes. B. Wild type and Mitfmi-vga9/S73A compound mutants either lacking or carrying Kittm1Alf.