Abstract
Primary cells, such as HUVEC, are notoriously difficult to transfect and are susceptible to the toxic effects of transfection reagents. A transfection reagent with a high transfection efficiency and low cytotoxicity was sought to retain sufficient viability of transfected HUVEC for subsequent assays. Nine chemical transfection reagents, currently commercially available, were compared for their ability to transfect HUVEC in vitro. A plasmid expressing the enhanced GFP (EGFP) was used for transfection, followed by flow cytometry of transfected HUVEC to determine the proportion of EGFP-expressing cells as a measure of transfection efficiency. Lipofectamine 2000 and Lipofectamine LTX (Invitrogen, Carlsbad, CA, USA) gave the highest transfection efficiencies of the reagents tested. Lipofectamine LTX was identified as the optimal transfection reagent as a result of its higher transfection efficiency at shorter periods of time following transfection when cytotoxicity was limited, allowing sufficient yield of transfected HUVEC for use in subsequent assays.
Keywords: HUVEC, cytotoxicity
INTRODUCTION
Primary cells are considered more difficult to transfect than immortalized cell lines, as they are more susceptible to toxic agents and may degrade exogenous nucleic acids in the cytoplasm.1,2 In vitro genetic modification of primary endothelial cells, such as HUVEC, is used in the study of gene function,3 angiogenesis,4 and for applications in gene therapy,5 among others. HUVEC have a limited lifespan and a relatively low proliferation rate.2,6 These characteristics provide further challenges for transient transfection but also mean that HUVEC may better reflect the in vivo situation than immortalized cell lines and subsequently, are a widely used cell type as in vitro models for endothelial cells lining the vasculature7 or as gene-delivery vehicles.8
Cells can be gene-modified in vitro and in vivo using physical, viral, or chemical methods.9–11 Physical methods, including electroporation, biolistics, and injection, are used with varying success and are cell cycle-independent but may be more toxic for some cell types and usually require cell suspensions in vitro and specialized equipment (reviewed by Villemejane and Mir12). Viral transduction is particularly efficient for gene transfer and is favored for in vivo use.13 However, depending on the viral vector used, viral transduction can potentially cause insertional mutagenesis, immunogenicity, or replication-induced infection in vivo, and the size of the delivered gene is often limited.14,15 In addition, higher safety measures are generally required for laboratory production and use of viral vectors.16,17 Chemical methods of transfection are widely used, as they are relatively simple, cheap, and safe.18 They include calcium phosphate, liposomes, cationic lipids [e.g., dioleoyl trimethylammonium propane (DOTAP)], cationic and biodegradable polymers, peptides [e.g., polyethylenimine (PEI), dendrimers], and cationic polysaccharides.19
Here, we describe the comparison of transfection of HUVEC using nine chemical transfection reagents, currently commercially available, to identify the reagent that elicits the highest transfection efficiency without compromising cell viability.
MATERIALS AND METHODS
Isolation and Culture of HUVEC
HUVEC were extracted from umbilical cords, according to the method described by Jaffe et al.20 Written, informed consent was obtained from each woman who donated an umbilical cord (approved by Canterbury Ethics Committee R, New Zealand). Isolated HUVEC were grown in M199 medium (Invitrogen, Carlsbad, CA, USA) with 10% cosmic calf serum (HyClone, Thermo Scientific, Tauranga, New Zealand) and 1 ng/mL basic fibroblast growth factor (Invitrogen; complete medium) in 75 cm2 flasks precoated with 0.1% w/v gelatin (Calbiochem, La Jolla, CA, USA) in water. Cells were grown in 100 μg/mL streptomycin, 60 μg/mL benzylpenicillin, and 1 μg/mg amphotericin B (Invitrogen) for the first passage only and frozen at Passage 3 in 10% DMSO (Sigma-Aldrich, St. Louis, MO, USA) for storage in liquid nitrogen. HUVEC were cultured routinely at 37°C with 5% CO2 in a water-jacketed incubator (Forma Scientific Inc., Marietta, OH, USA), and medium was replaced every 2 days. At Passage 4, HUVEC were dissociated from plasticware using 0.05% trypsin-EDTA (Invitrogen) in PBS (Invitrogen) and seeded into gelatin-coated six-well plates for transfection.
Plasmid Preparation and Transfection
Stocks of enhanced green fluorescent protein (EGPF)-encoding plasmid (pEGFP-N1) (ClonTech, Mountain View, CA, USA; GenBank Accession #U55762) were prepared in Top10 Escherichia coli (Invitrogen) using the endotoxin-free Maxiprep plasmid kit (Qiagen GmbH, Hilden, Germany). DNA concentration was adjusted to 1 mg/mL in Tris-EDTA, and plasmids were stored at −20°C.
For transfections, HUVEC were seeded in six-well plates at a density of 1–2 × 105 cells/well (1–2×104 cells/cm2) in 2 mL complete medium and grown for 1–3 days until 70–80% confluent. Transfection complexes were formed at room temperature in serum-free medium prior to drop-wise addition to HUVEC, followed by incubation for various periods and replacement with complete medium for 24 or 48 h. Antibiotics and antifungal agents were not used during transfection procedures.
Nine commercially available transfection reagents were tested using a range of DNA:reagent ratios (Table 1), according to the manufacturers' recommendations, described below briefly. All protocols are per-well of a six-well plate.
TABLE 1.
Commercial Transfection Reagents Tested in HUVEC
| Transfection reagent | Formulation | Reagent (μL):DNA (μg)/well (9.4 cm2) | Manufacturer |
|---|---|---|---|
| Effectene | Nonliposomal lipid | 20:2 | Qiagen GmbH (Hilden, Germany) |
| 8:0.8 | |||
| 20:0.8 | |||
| 40:0.8 | |||
| SuperFect | Activated dendrimer | 10:5 | Qiagen GmbH |
| 4:2 | |||
| 10:2 | |||
| 20:2 | |||
| Escort IV | Polycationic lipid and neutral nontransfecting lipid | 20:5 | Sigma-Aldrich (St. Louis, MO, USA) |
| 8:2 | |||
| 4:1 | |||
| ExGen 500 | Cationic polymer of linear 22 kDa PEI | 16.5:5 | Fermentas International Inc. (Burlington, Ontario, Canada) |
| 9.87:3 | |||
| 8.23:3 | |||
| 11.52:3 | |||
| FuGene 6 | Lipids and other components | 3:1 | Roche (Basel, Switzerland) |
| 3:2 | |||
| 6:1 | |||
| FuGene HD | Lipids and other components | 15:5 | Roche |
| 6:2 | |||
| GeneJammer | Polyamine and other components | 6:2 | Stratagene (La Jolla, CA, USA) |
| 9:3 | |||
| Lipofectamine 2000 | Cationic lipid | 2:4 | Invitrogen (Carlsbad, CA, USA) |
| 8:4 | |||
| 10:4 | |||
| 10:5 | |||
| Lipofectamine LTX | Cationic lipid | 7.5:2.5 | Invitrogen |
| 3.75:2.5 | |||
| 6.25:2.5 | |||
| 3:1 |
Effectene
DNA was diluted in Buffer EC, Enhancer was added to a total volume of 100 μL, and the mixture was incubated for 5 min. Effectene was added and complexes incubated for 10 min, followed by addition of 600 μL complete medium. Cell medium was replaced with 1600 μL fresh complete medium prior to addition of complexes and incubation.
SuperFect
DNA was diluted in 100 μL M199 and incubated for 5 min. SuperFect was added, and complexes incubated for 10 min, 600 μL complete medium was added to the complexes, and this was added drop-wise to HUVEC without other medium present. Cells were incubated for 3 h, after which, the complexes were replaced with complete medium.
Escort IV
DNA was diluted in 400 μL M199. Escort IV was diluted in 600 μL M199. The DNA mixture was added to the Escort IV mixture and incubated for 15 min. Complete medium (1 mL; containing 20% serum) was added to the complexes. Medium was removed from the cells, replaced with complexes, and incubated.
ExGen 500
DNA was diluted in 200 μL 150 mM NaCl, followed by addition of ExGen 500. Complexes were incubated for 10 min. The complexes were added to cells in 2 mL complete medium and incubated.
FuGene 6
FuGene 6 was diluted in M199 to give a total volume of 100 μL and incubated for 5 min. DNA was added to the mixture and the complexes incubated for 15 min. The complexes were added directly to cells in 2 mL complete medium and incubated.
FuGene HD
DNA was diluted in 100 μL M199. FuGene HD was added to the DNA mixture and incubated for 15 min. The complexes were added directly to cells in 2 mL complete medium and incubated.
GeneJammer
GeneJammer was added to 100 μL M199 and incubated for 10 min. DNA was added to the mixture and incubated for a further 10 min. The medium was removed from the cells and replaced with 900 μL complete medium. The complexes were incubated on the cells for 3 h before 1 mL complete medium was added.
Lipofectamine 2000
DNA was diluted in 250 μL Opti-MEM I (Reduced Serum Medium, Invitrogen). Lipofectamine 2000 was diluted in 250 μL Opti-MEM I. Mixtures were incubated for 5 min and then combined together for a further 20 min. Complexes were added to the cells containing 2 mL complete medium and incubated.
Lipofectamine LTX
DNA was diluted into 500 μL Opti-MEM I. An equal volume of PLUS reagent was added. The mixture was incubated for 5 min. Lipofectamine LTX was added, and the complexes were allowed to form by incubation for 25 min. Cell medium was replaced with 2 mL Opti-MEM I, to which the mixture was added, and incubated for 4 h, after which the complexes were replaced with complete medium.
Quantification of EGFP
Transiently transfected HUVEC (adherent cells only) were trypsinised at 24 h and 48 h, centrifuged, and resuspended in Opti-MEM I. Samples were analyzed using a Cytomics FC 500 MPL flow cytometer (Beckman Coulter Inc., Brea, CA, USA) and MXP Version 2.2 software. Untransfected HUVEC and an EGFP-expressing, stable T24 cell line (G. U. Dachs, unpublished) were used as negative and positive controls, respectively. EGFP fluorescence was detected using the fluorescence 1 (FL1) detector (525 nm band-pass filter). The percentage of HUVEC that expressed EGFP was measured by creating a gated region with <1% of untransfected HUVEC in the EGFP gate.
Quantification of Cytotoxicity
Forward-scatter (cell size) versus log side-scatter (granularity; FSLin/SSLog) analysis was performed using standard flow cytometry settings. Transfected and control HUVEC (adherent cells only) were stained using SYTOX AADvanced dead-cell stain (AAD, Invitrogen) 24 h or 48 h after transfection. AAD was diluted in 200 μL buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2, pH 7.4) containing approximately 2 × 105 HUVEC cells to a final concentration of 5 μM for at least 5 min prior to analysis. FL1 and FL4 (675 nm band-pass filter) detectors were used to simultaneously detect EGFP and AAD fluorescence, respectively, by flow cytometry under a compensation protocol.
RESULTS
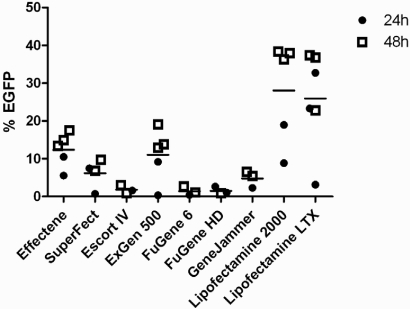
Transfection efficiencies, according to the proportion of EGFP-expressing cells measured by flow cytometry (Fig. 1), varied by over 50-fold (range: 0.15–49%) among the transfection reagents tested (Fig. 2). Differences in EGFP expression were dependent mainly on transfection reagent. The length of incubation prior to analysis made a difference in four of the nine reagents tested, and higher expression levels were detected after 48 h compared with 24 h (Fig. 2). Different reagent:DNA ratios were tested, but no clear association with transfection efficiency was detected (data not shown). For three of the reagents (Effectene, Escort IV, and ExGen 500), the final amount of DNA played a role in transfection efficiency, with more DNA in the mixture associated with increased transfection efficiency (results not shown). Of the nine reagents tested, Lipofectamine 2000 and Lipofectamine LTX demonstrated the highest transfection efficiency in HUVEC (Fig. 2). Lipofectamine 2000 (reagent:DNA ratio 2:4) resulted in 19 ± 9% and 38 ± 2% transfected cells at 24 h and 48 h, respectively (n≥3; mean±sem), and Lipofectamine LTX resulted in 33 ± 8% transfected cells at 24 h (reagent:DNA 6.25:2.5 n=4) and 23 ± 2% transfected cells at 48 h (reagent:DNA 3:1; n=5; mean±sem). At 24 h after transfection, the most efficient reagents were Lipofectamine LTX > Lipofectamine 2000 > Effectene, whereas at 48 h, Lipofectamine 2000 > Lipofectamine LTX > ExGen 500.
FIGURE 1.
EGFP expression detection by flow cytometry. Representative traces of HUVEC, incubated for 48 h after transfection before analysis by flow cytometry. For each experiment, the EGFP gate was set so that untransfected HUVEC had <1% EGFP. The proportion of EGFP-positive cells is presented on the graphs. (A) Mock-transfected HUVEC (DNA only). (B) Lipofectamine 2000 (2:4). (C) Lipofectamine LTX (3:1). (D) Lipofectamine LTX (6.25:2.5).
FIGURE 2.
Comparison of nine transfection reagents. The percentage of EGFP-expressing HUVEC is presented 24 h or 48 h following transfection. Each point represents a different ratio of reagent:DNA tested. Mean transfection efficiency for each reagent is shown (horizontal bars). Where a ratio was tested more than once, the mean transfection efficiency is plotted.
Mock transfection controls using DNA only or using Lipofectamine 2000 or Lipofectamine LTX only showed no background autofluorescence in the FL1 or FL4 channels of the flow cytometer. Mock transfection controls had similar profiles to untransfected HUVEC for FSLin/SSLog plots, AAD staining (data not shown), and EGFP expression (Fig. 1A).
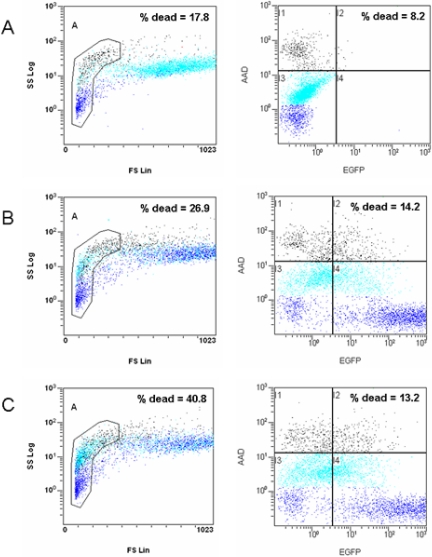
The cytotoxic effects of the transfection reagents were similar between Lipofectamine 2000 and Lipofectamine LTX (Fig. 3, right columns). A previous study had demonstrated that cellular debris and necrotic cells could be excluded from analysis using FSLin/SSLog analysis by flow cytometry.21 However, our study demonstrated this technique to be ambiguous for transfected HUVEC, as flow cytometry analysis of specific dead-cell staining (AAD) did not agree with results obtained from the FSLin/SSLog analysis (Fig. 3, left columns). In particular, AAD-positive cells tended to be spread equally between the regions identified as viable or necrotic by FSLin/SSLog in transfected HUVEC (black, Fig 3B and C), and AAD-negative, viable cells accumulated in the necrotic region according to FSLin/SSLog (light blue, Fig 3B and C). Flow cytometry (black, AAD-positive cells) indicated that cells analyzed after 48 h tended to have more dead cells compared with cells analyzed after 24 h, e.g., Lipofectamine 2000 2:4, 10% versus 15% cell death, and Lipofectamine LTX 6.25:2.5, 9% versus 14%, 24 h versus 48 h, respectively. Mock transfection data showed no difference in cell death at 24 h versus 48 h, i.e., 8% versus 8% (DNA only, 4 μL), 11% versus 8% (DNA only, 2.5 μL), 10% versus 7% (Lipofectamine 2000, 2 μL), or 12% versus 9% (Lipofectamine LTX, 6.25 μL).
FIGURE 3.
Cell viability after transfection. Representative example of HUVEC incubated for 48 h after transfection before staining with AAD and analysis by flow cytometry. Five thousand cells/experiment are displayed, and the percentage of dead cells according to each method is shown. (A) Mock-transfected HUVEC (DNA only). (B) Lipofectamine 2000 (2:4). (C) Lipofectamine LTX (6.25:2.5). FSLin/SSLog plots: The gated region for cell debris or necrosis is indicated, as well as the percentage of dead cells according to this method. AAD versus EGFP plots: For each experiment, the EGFP gate was set so that untransfected HUVEC had <1% EGFP. The percentage of dead cells according to AAD staining is indicated (gates I1 and I2). Dark blue, Low AAD staining; light blue, medium AAD staining; black, high AAD staining.
DISCUSSION
Our study demonstrated that a small selection of commercially available chemical transfection reagents was able to transfer exogenous genes efficiently to primary human cells. The most efficient reagents were the cationic lipid reagents, Lipofectamine 2000 and LTX, from Invitrogen, which were able to gene-modify up to 38% of HUVEC.
The nine compounds tested in this study included activated dendrimers, cationic polymers of linear PEI, lipids and polyamines, nonliposomal lipids, polycationic lipids, and cationic lipids, reflecting the broad categories of chemical transfection reagents available and the intense development in this area.22 Chemical transfection reagents act as a packaging mechanism to condense and deliver DNA to the nucleus of cells, usually by endocytosis.23 Liposomal transfection reagents have a lipid bilayer that encloses the DNA, allowing it to fuse with the cell membrane to deliver the DNA to the cells. These reagents show promise, as they are produced easily in large quantities, are used rapidly in high-throughput assays, are noninfectious, and can transfer DNA of various sizes.24–26 However, they can be susceptible to nuclease degradation, are potentially harmful,1 and are usually, but not always, cell cycle-dependent.27 Liposomal transfection reagents may be more effective in dividing cells, as the nuclear membrane becomes fragmented during replication (prometaphase),28 potentially enabling access of foreign DNA to the nucleus. Hence, this study used semi-confluent HUVEC throughout.
Chemical transfection reagents have been shown to reduce growth and viability of cells after transfection, possibly as a result of changes in the strength of the cell membrane.29 Specifically, cationic lipids have been shown to cause apoptosis by the generation of reactive oxygen species.30
Previously reported chemical transfection efficiencies in HUVEC vary, ranging from a maximum of 0.45% using Lipofection,31 10.9% using SuperFect,32 50% using the cationic liposome DOTAP,1 and up to 77% using Effectene, although 22% of cells were nonviable.33 This study demonstrated reproducible transfection efficiencies of 19 ± 9% and 33 ± 8% using Lipofectamine 2000 and Lipofectamine LTX, respectively, which is within the range of reported data but lower than those advertised.
A number of reasons have been considered to explain these differences. Optimal expression of transfected genes in vitro is influenced by many factors, including cell type, passage history, confluence, vector structure, size and purity, promoters, a DNA:transfection reagent complex ratio, incubation time, and presence of serum,34 but these were constant across the nine reagents, with the exception of complex ratios and incubation times that did not affect efficiencies consistently. However, it was observed in this study that transfection efficiencies appear to vary with some batches of HUVEC, and cells extracted from some cords are able to be transfected more efficiently than those from others. Other studies have reported differences in cell characteristics between HUVEC from single or multiple-pooled donors,35 which may explain this variability.
Increasing the time to analysis from 24 h to 48 h resulted in an increased proportion of EGFP-positive cells for some reagents but also caused a reduction in cell viability. Cell death analysis, using a simple assay (FSLin/SSLog)21 and specific dead-cell staining (AAD), indicated that significant cell death occurred as a result of the transfection reagents but also showed that the simple FSLin/SSLog method may not always be relied on. It is of note that some dead cells lose their EGFP expression (Fig. 3 and own unpublished data).36 Necrotic cells reportedly lose all of their EGFP signal, whereas apoptotic cells lose less EGFP signal.36 However, in Figure 3, AAD-positive cells lost only some EGFP intensity, which may reflect the presence of late apoptotic cells in that population. Therefore, measurement of EGFP expression by flow cytometry may underestimate the total number of cells that was gene-modified initially. Importantly, dead cells tend to detach from the growth surface and thus, were not analyzed in this study. This led to an underestimation of the total amount of cell death caused by transfection but a more accurate representation of the number of live cells (and proportion of EGFP-expressing cells) left to plate out for subsequent assays.
A range of reporter genes is in use to determine transfection efficiencies, including luciferase, β-galactosidase, and EGFP. EGFP gene expression allows easy determination of the proportion of cells that is gene-modified on a single-cell basis, detecting the number of cells expressing EGFP and their level of EGFP expression via flow cytometry. On the other hand, luciferase activity, detected via conversion of a substrate, resulting in amplified signal, determines the behavior of the entire population, thereby losing information about single cells.37
In this study, a stringent cut-off was used to determine EGFP expression by setting the flow cytometry region to include <1% of control cells, whereas other studies may have set other criteria for quantification of transfection efficiencies. It has been reported that some cationic liposome transfection reagents could lead to autofluorescence in fluorescent microscopy and flow cytometry analysis,38 but our results for mock transfection using Lipofectamine 2000 and Lipofectamine LTX showed no autofluorescence.
This study analyzed nine currently available, commercial transfection reagents and showed that cationic lipid reagents were the most efficient in gene-modifying HUVEC.
ACKNOWLEDGMENTS
This project was funded by the Cancer Society of New Zealand. M. A. H. was a recipient of a University of Otago Postgraduate Scholarship, a Postgraduate Tassell Scholarship in Cancer Research, and a South Canterbury Finance Research Scholarship. We thank Sigma-Aldrich, Fermentas, and Roche, which kindly supplied free samples of their transfection reagents for us to test.
Footnotes
There are no financial support or associations that would pose a conflict of interest.
University of Otago Institutional Biological Safety Committee (GMO06/UO012) approval for transient transfections.
Written, informed consent was obtained from each woman who donated an umbilical cord (approved by Canterbury Ethics Committee R).
REFERENCES
- 1.Colombo MG, Citti L, Basta G, De Caterina R, Biagini A, Rainaldi G. Differential ability of human endothelial cells to internalize and express exogenous DNA. Cardiovasc Drugs Ther 2001;15:25–29 [DOI] [PubMed] [Google Scholar]
- 2.van Beijnum JR, van der Linden E, Griffioen AW. Angiogenic profiling and comparison of immortalized endothelial cells for functional genomics. Exp Cell Res 2008;314:264–272 [DOI] [PubMed] [Google Scholar]
- 3.Nathwani AC, Gale KM, Pemberton KD, Crossman DC, Tuddenham EG, McVey JH. Efficient gene transfer into human umbilical vein endothelial cells allows functional analysis of the human tissue factor gene promoter. Br J Haematol 1994;88:122–128 [DOI] [PubMed] [Google Scholar]
- 4.Jin Y, An X, Ye Z, Cully B, Wu J, Li J. RGS5, a hypoxia-inducible apoptotic stimulator in endothelial cells. J Biol Chem 2009;284:23436–23443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang WY, Huang ZH, Lin LJ, et al. Kinase domain insert containing receptor promoter controlled suicide gene system selectively kills human umbilical vein endothelial cells. World J Gastroenterol 2006;12:5331–5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim TW, Kim HJ, Lee C, et al. Identification of replicative senescence-associated genes in human umbilical vein endothelial cells by an annealing control primer system. Exp Gerontol 2008;43:286–295 [DOI] [PubMed] [Google Scholar]
- 7.Fife K, Bower M, Cooper RG, et al. Endothelial cell transfection with cationic liposomes and herpes simplex-thymidine kinase mediated killing. Gene Ther 1998;5:614–620 [DOI] [PubMed] [Google Scholar]
- 8.Ojeifo JO, Su N, Ryan US, Verma UN, Mazumder A, Zwiebel JA. Towards endothelial-cell-directed cancer immunotherapy: in vitro expression of human recombinant cytokine genes by human and mouse primary endothelial cells. Cytokines Mol Ther 1996;2:89–101 [PubMed] [Google Scholar]
- 9.Azzam T, Domb AJ. Current developments in gene transfection agents. Curr Drug Deliv 2004;1:165–193 [DOI] [PubMed] [Google Scholar]
- 10.Greco O, Scott SD, Marples B, Dachs GU. Cancer gene therapy: “delivery, delivery, delivery”. Front Biosci 2002;7:d1516–d1524 [DOI] [PubMed] [Google Scholar]
- 11.Kawakami S, Higuchi Y, Hashida M. Nonviral approaches for targeted delivery of plasmid DNA and oligonucleotide. J Pharm Sci 2008;97:726–745 [DOI] [PubMed] [Google Scholar]
- 12.Villemejane J, Mir LM. Physical methods of nucleic acid transfer: general concepts and applications. Br J Pharmacol 2009;157:207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Work LM, Ritchie N, Nicklin SA, Reynolds PN, Baker AH. Dual targeting of gene delivery by genetic modification of adenovirus serotype 5 fibers and cell-selective transcriptional control. Gene Ther 2004;11:1296–1300 [DOI] [PubMed] [Google Scholar]
- 14.Glasgow JN, Bauerschmitz GJ, Curiel DT, Hemminki A. Transductional and transcriptional targeting of adenovirus for clinical applications. Curr Gene Ther 2004;4:1–14 [DOI] [PubMed] [Google Scholar]
- 15.Kreppel F, Kochanek S. Modification of adenovirus gene transfer vectors with synthetic polymers: a scientific review and technical guide. Mol Ther 2008;16:16–29 [DOI] [PubMed] [Google Scholar]
- 16.Manilla P, Rebello T, Afable C, et al. Regulatory considerations for novel gene therapy products: a review of the process leading to the first clinical lentiviral vector. Hum Gene Ther 2005;16:17–25 [DOI] [PubMed] [Google Scholar]
- 17.Raty JK, Lesch HP, Wirth T, Yla-Herttuala S. Improving safety of gene therapy. Curr Drug Saf 2008;3:46–53 [DOI] [PubMed] [Google Scholar]
- 18.Douglas KL. Toward development of artificial viruses for gene therapy: a comparative evaluation of viral and non-viral transfection. Biotechnol Prog 2008;24:871–883 [DOI] [PubMed] [Google Scholar]
- 19.Eliyahu H, Barenholz Y, Domb AJ. Polymers for DNA delivery. Molecules 2005;10:34–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaffe EA, Nachman RL, Becker CG, Minick CR. Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J Clin Invest 1973;52:2745–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovala AT, Harvey KA, McGlynn P, Boguslawski G, Garcia JG, English D. High-efficiency transient transfection of endothelial cells for functional analysis. FASEB J 2000;14:2486–2494 [DOI] [PubMed] [Google Scholar]
- 22.Morille M, Passirani C, Vonarbourg A, Clavreul A, Benoit JP. Progress in developing cationic vectors for non-viral systemic gene therapy against cancer. Biomaterials 2008;29:3477–3496 [DOI] [PubMed] [Google Scholar]
- 23.Vijayanathan V, Thomas T, Thomas TJ. DNA nanoparticles and development of DNA delivery vehicles for gene therapy. Biochemistry 2002;41:14085–14094 [DOI] [PubMed] [Google Scholar]
- 24.Ewert KK, Ahmad A, Bouxsein NF, Evans HM, Safinya CR. Non-viral gene delivery with cationic liposome-DNA complexes. Methods Mol Biol 2008;433:159–175 [DOI] [PubMed] [Google Scholar]
- 25.Li S, Tan Y, Viroonchatapan E, Pitt BR, Huang L. Targeted gene delivery to pulmonary endothelium by anti-PECAM antibody. Am J Physiol Lung Cell Mol Physiol 2000;278:L504–L511 [DOI] [PubMed] [Google Scholar]
- 26.Muller K, Nahde T, Fahr A, Muller R, Brusselbach S. Highly efficient transduction of endothelial cells by targeted artificial virus-like particles. Cancer Gene Ther 2001;8:107–117 [DOI] [PubMed] [Google Scholar]
- 27.Brunner S, Furtbauer E, Sauer T, Kursa M, Wagner E. Overcoming the nuclear barrier: cell cycle independent nonviral gene transfer with linear polyethylenimine or electroporation. Mol Ther 2002;5:80–86 [DOI] [PubMed] [Google Scholar]
- 28.Georgatos SD, Pyrpasopoulou A, Theodoropoulos PA. Nuclear envelope breakdown in mammalian cells involves stepwise lamina disassembly and microtubule-drive deformation of the nuclear membrane. J Cell Sci 1997;110:2129–2140 [DOI] [PubMed] [Google Scholar]
- 29.Zhi D, Zhang S, Wang B, Zhao Y, Yang B, Yu S. Transfection efficiency of cationic lipids with different hydrophobic domains in gene delivery. Bioconjug Chem 2010;21:563–577 [DOI] [PubMed] [Google Scholar]
- 30.Aramaki Y, Takano S, Tsuchiya S. Cationic liposomes induce macrophage apoptosis through mitochondrial pathway. Arch Biochem Biophys 2001;392:245–250 [DOI] [PubMed] [Google Scholar]
- 31.Teifel M, Heine LT, Milbredt S, Friedl P. Optimization of transfection of human endothelial cells. Endothelium 1997;5: 21–35 [DOI] [PubMed] [Google Scholar]
- 32.Uchida E, Mizuguchi H, Ishii-Watabe A, Hayakawa T. Comparison of the efficiency and safety of non-viral vector-mediated gene transfer into a wide range of human cells. Biol Pharm Bull 2002;25:891–897 [DOI] [PubMed] [Google Scholar]
- 33.Cook-Johnson RJ, Demasi M, Cleland LG, Gamble JR, Saint DA, James MJ. Endothelial cell COX-2 expression and activity in hypoxia. Biochim Biophys Acta 2006;1761:1443–1449 [DOI] [PubMed] [Google Scholar]
- 34.Colosimo A, Goncz KK, Holmes AR, et al. Transfer and expression of foreign genes in mammalian cells. Biotechniques 2000;29:314–318, 320,–312, 324 [DOI] [PubMed] [Google Scholar]
- 35.Glee PM, Cutler JE, Benson EE, Bargatze RF, Hazen KC. Inhibition of hydrophobic protein-mediated Candida albicans attachment to endothelial cells during physiologic shear flow. Infect Immun 2001;69:2815–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strebel A, Harr T, Bachmann F, Wernli M, Erb P. Green fluorescent protein as a novel tool to measure apoptosis and necrosis. Cytometry 2001;43:126–133 [DOI] [PubMed] [Google Scholar]
- 37.Welsh S, Kay SA. Reporter gene expression for monitoring gene transfer. Curr Opin Biotechnol 1997;8:617–622 [DOI] [PubMed] [Google Scholar]
- 38.Guo B, Pearce AG, Traulsen KE, Rintala AC, Lee H. Fluorescence produced by transfection reagents can be confused with green fluorescent proteins in mammalian cells. Biotechniques 2001;31:314–316, 318,, 320–311 [DOI] [PubMed] [Google Scholar]