Abstract
Despite recent advances in measuring cellular immune responses, the quantitation of antigen-specific T cell clones in infections or diseases remains challenging. Here, we employed combined megaplex TCR isolation and SMART-based real-time quantitation methods to quantitate numerous antigen-specific T cell clones using limited amounts of specimens. The megaplex TCR isolation covered the repertoire comprised of recombinants from 24 Vβ families and 13 Jβ segments, and allowed us to isolate TCR VDJ clonotypic sequences from one or many PPD-specific IFNγ-producing T cells that were purified by flow cytometry sorting. The SMART amplification technique was then validated for its capacity to proportionally enrich cellular TCR mRNA/cDNA for real-time quantitation of large numbers of T cell clones. SMART amplified cDNA was shown to maintain relative expression levels of TCR genes when compared to unamplified cDNA. While the SMART-based real-time quantitative PCR conferred a detection limit of 10−5 to 10−6 antigen-specific T cells, the clonotypic primers specifically amplified and quantitated the target clone TCR but discriminated other clones that differed by ≥2 bases in the DJ regions. Furthermore, the combined megaplex TCR isolation and SMART-based real-time quantiation methods allowed us to quantitate large numbers of PPD-specific IFNγ-producing T cell clones using as few as 2×106 PBMC collected weekly after mycobacterial infection. This assay system may be useful for studies of antigen-specific T cell clones in tumors, autoimmune and infectious diseases.
Keywords: T cell receptor repertoires, Clonal responses of Ag-specific T cells, SMART cDNA, Gene expression
1. Introduction
T cells play an important role in immune regulation as well as control of tumors, autoimmune and infectious diseases. Characterizing antigen-specific T cells is therefore of central importance for developing immune-based therapeutic modalities including T cell-targeted vaccines. Recent advances in immunoassays such as MHC/peptide tetramers and antigen-specific intracellular cytokine capture procedures provide powerful tools for tracking antigen-specific T cells (Altman et al., 1996; Kuroda et al., 1998; Larsson et al., 1999; Pitcher et al., 1999; Klenerman et al., 2002). Such advances should facilitate the assessment of immune responses of antigen-specific T cell clones during infections. Elucidating clonal T cell responses may provide important information concerning dominant T cell clones and their respective epitopes that are relevant to vaccine development efforts.
Despite recent progress in measuring cellular immune responses, the quantitation of antigen-specific T cell clones during infections or diseases remains challenging. While MHC/peptide tetramers and intracellular cytokine staining/ELISPOT have proven to be powerful tools for measuring the development of peptide-specific T cell immune responses, these assays do not provide information concerning the frequency and dominance of individual antigen-specific T cell clones. Although peptide-specific T cells can be purified and cloned for clonotypic TCR sequence analyses, such analyses may not be quantitative due to the cloning bias (Chen et al., 2001; Meyer-Olson et al., 2004). Recently, real-time quantitative PCR has been used to assess clonal representation in MHC/tetramer-bound T cells (Cohen et al., 2002; Douek et al., 2002; Gallard et al., 2002; Lim et al., 2002). However, this application is limited to analyses of small numbers of T cell clones specific for few well-defined dominant epitopes (Douek et al., 2002; Lim et al., 2002; Meyer-Olson et al., 2004). Moreover, such analyses of limited numbers of T cell clones require large initial samples of PBMC or tissue cells. Since clinical sample sizes of blood and biopsies available for analyses of clonal immune responses are usually small or limited, it is important to develop an assay system that allows extensive quantitation of large numbers of T cell clones using a limited amount of specimen.
Over the past decade, we have characterized molecular aspects of MHC-restricted cytotoxic T lymphocytes, superantigen-reactive Vβ+ T cell subpopulations, mycobacterium-selected CDR3-bearing T cells, and MHC/viral peptide tetramer-bound T cells in monkey models of infections (Chen et al., 1995, 1996, 2001; Kou et al., 1998; Zhou et al., 1999). We also utilized the Altas Switch Mechanism At the 5′-end of Reverse Transcript (SMART)-derived cDNA to clone and sequence TCR in a MHC/peptide tetramer-reactive CD8 T cell population (Chen et al., 2001). SMART-based amplification of cDNA has proven to be a useful technique to enrich total cellular mRNA/cDNA from a low level of <50 ng to a large quantity (Herrler, 2000; Zhumabayeva et al., 2001, 2004; Seth et al., 2003; Wang et al., 2003; Gustincich et al., 2004; Rihl et al., 2004; Wellenreuther et al., 2004). This novel technique has made it possible to perform a large-scale quantitation of gene expression and arrays using a limited amount of clinical specimens (Zhumabayeva et al., 2001, 2004; Gustincich et al., 2004; Rihl et al., 2004). However, the SMART-based technique has not been validated for the quantitation of T cell clones in small amounts of specimen. In the present study, we combined our experience in SMART–cDNA and TCR-based molecular work to develop an assay system for quantitating T cell clonal responses during infections of monkeys. First, we successfully optimized a megaplex PCR reaction that covered the whole TCR repertoire comprised of recombinants from 24 Vβ families and 13 Jβ segments. This allowed us to isolate TCR VDJ clonotypic sequences in an antigen-specific IFNγ-producing T cell population that was fixed by formaldehyde and purified by flow sorting. Then, we validated SMART-amplification of cDNA derived from PBMC comprised of small numbers of cells collected over time after infections. Finally, we performed real-time quantitative PCR using clonotypic primers and SMART-amplified cDNA to measure the frequencies of antigen-specific T cell clones using small quantities of PBMC. The sensitivity, specificity and utility of this assay system were assessed in the present studies.
2. Materials and methods
2.1. Outline of approaches
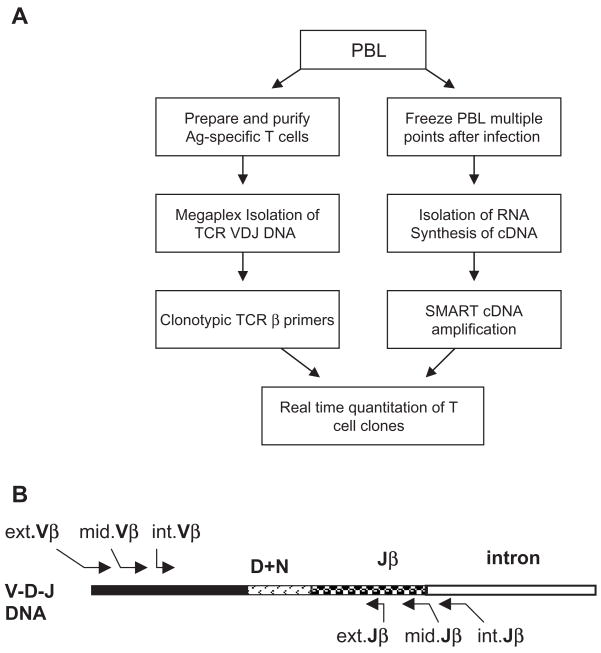
Two parallel approaches were employed to quantitate antigen-specific T cell clones in infection (Fig. 1A). To isolate clonotypic TCR and design clonotypic primers for real-time quantitation, peripheral blood mononuclear cells (PBMC) from Mycobacterium bovis BCG-infected monkeys were used to generate antigen-specific T cells by means of peptide stimulation and intracellular IFNγ staining. The antigen-specific IFNγ-producing T cells were purified by flow sorting; TCR VDJ DNA was isolated by megaplex PCR and sequenced for designing clonotypic primers. In parallels, 2×106 PBMC were collected and frozen down weekly after M. bovis BCG infection. RNA isolated from the frozen cells was used to synthesize SMART cDNA. SMART-amplified cDNA and clonotypic primers were used to quantitate T cell clones during BCG infection.
Fig. 1.
Combined megaplex TCR isolation and SMART-based real-time quantitative PCR methods for quantitating antigen-specific T cell clones. (A) Megaplex TCR isolation and SMART-based real-time quantitative methods. (B) Complementary regions of each of the 25 Vβ family-specific primers and 13 Jβ-specific primers. Three sets of nested Vβ family-specific primers were designated as external (ext.Vβ), middle (mid.Vβ), and internal (int.Vβ) Vβ family-specific primers. Likewise, three sets of Jβ family-specific primers were designated as ext.Jβ, mid.Jβ and int.Jβ. Note that ext.Jβ-specific primers were designed based on intron sequences close to Jβ segments.
2.2. Collection of PBMC and antigen-specific T cells
2.2.1. Separation of PBMC
PBMC were isolated from whole blood using ficoll-hypaque density gradient centrifugation. Briefly, blood was loaded with Ficoll Hypaque-diatrizoate and centrifuged at 500×g for 25 min at 20 °C. Isolated mononuclear cells (primarily lymphocytes and monocytes) were washed with Hanks buffer containing 2% fetal calf serum.
2.2.2. Intracellular IFNγ staining
PBMC were cultured at 37 °C in a 5% CO2 incubator for 1 h in the presence of RPMI medium alone, RPMI medium with the addition of 25 μg/ml purified protein derivative (PPD) or RPMI medium with 100 ng/ml PMA +1 μg/ml lonomycin. The cultures were incubated for an additional 5 h in the presence of Brefeldin A (10 μg/ml; GolgiPlug, BD Pharmingen) to disrupt golgi apparatus transport, as well as 2 μg/ml anti-CD28 and anti-CD49d. The cultured cells were then stained with Abs specific for CD3, CD4 and CD8 to determine their collective responses to the PPD stimulation. The PBMC were washed once with PBS/2% FCS, and then permeabilized with Cell Fix/Perm solution (BD Pharmingen). The cytofix/cytoperm was washed out by permeabilization buffer and the cells were maintained in the permeabilization buffer throughout staining to keep cells permeably. The samples were stained by anti-IFNγAPC (1 μg/106 cells) for 45 min at room temperature. Stained cells were washed with permeabilization buffer, and fixed in 2% formaldehyde. IFNγ+ CD4+CD3+ cells were then purified and enumerated by flow cytometry sorting, as previously described (Chen et al., 2001). The formaldehyde-fixed PPD-specific CD4 T cells were subjected to TCR VDJ DNA isolation using the megaplex PCR.
2.3. Isolation of TCR VDJ DNA from small numbers of formaldehyde-fixed T cells
2.3.1. Designing of PCR primers
To isolate and amplify TCR β VDJ DNA from formaldehyde-fixed cells, three sets of 24 forward Vβ family-specific primers for nested PCR were designed based on the published human and monkey sequences in GenBank database (Table 1A). The positions of these external, middle and internal Vβ primers (ext.Vβ, mid.Vβ, and int.Vβ) are outlined in Fig. 1. Each of Vβ primers was derived from highly variant regions and shown to be Vβ family-specific. Thermokinetic principles such as GC content and certain nucleotide repeats were also considered as factors in determining the position and length of a primer. Similarly, three sets of 13 reverse Jβ primers were designed based on genomic DNA sequences of corresponding monkey Jβ genes (Cheynier et al., 1998). Thirteen external JR primers (ext.Jβ) were designed based on intron DNA sequences flanked at 3′to each of TCR Jβ regions (11), whereas middle and internal Jβ primers (mid.Jβ and int.Jβ) were complementary in sequences to the regions within the Jβ segments. There were five bases overlapping between each set of mid.Jβ and in.Jβ primers (Table 1B).
Table 1A.
External, middle and internal sets of Vβ family-specific primers for isolating clonotypic VDJ DNA from a single cell or small numbers of Ag-specific T cells
| 5′Vβ family-specific primers |
GenBank accession number |
||||
|---|---|---|---|---|---|
| Ext.Vβ primers | Mid.Vβ primers | Int.Vβ primers | Monkey | Human | |
| Vβ1 | TAATGGGGAAGAGAGAGC | AAAGATTCTCAGCACAAC | GCACAACAGTTCCCTGACTTGCA | M60537 (PanAY168688) | NG001333, AF430690, AF395525, AF387759, AF387744, AJ549953 |
| Vβ2 | GAAGATCGAGTGCCGTTC | GGACTTTCAGGTCACAAC | CAGTTCCAGACACAGAGC | M60541 | NG001333, AY232285, AF043182, X72719, X57604, D13088 |
| Vβ3 | TTTCTGGAATGTGTCCAG | TCCAGGATATGGACCATG | TGAGGGGTACAGTGTCTC | AF143618 (PanAY168706) | AF043183, X63456, X57610, Z23040, X74846, AJ301512 |
| Vβ4 | CAAGCAGGGATGTCTGTC | AGTCGATAGCCAAGTCAC | TCAGGGCTCTGAGGCCAC | L27622, U04581, U04579, U04568 | Z29582, Z29580, L27623, X04926, X04921, U51445 |
| Vβ5 | ACACGACACTGTCTACTG | CAGCAGGGTCCAGGTCAG | TCCCTGGTCGATTCTCAG | U04576, M60530, M60545, M60535 | X57613, X74849, X58799, AJ405811 |
| Vβ6 | ACAGGATGTAGCTCTCAG | AATTTCGGGTCATGTAGC | CTTCTCTGCAGAGAGGAC | M60538, U04601 | X588061, X61441, X04931 |
| Vβ7 | AGAAGTCTTTGAAATGTG | ACATATGGGACACAATGC | CCAGTCGCTTCTCACCTG | U04565, M60551, M60550, M60543 | X57617, X57728, X74842, AF316870, X58813 |
| Vβ8 | AAGTGACTCTGAGATGCG | CCAATTTCAGGCCACAGC | CAGACAGACCATGATGCG | U04573, U4569, M60548, M60547 | X61439, AF430681, AF430678, AF430676, X57615 |
| Vβ9 | GACGGGAAAAAACGAGTC | AAATGTGAACAAAATCTG | TTGGTATAAGCAGGACTC | M60549, M60546, AY168669 | AF020637, NG001333, U66059, L36092, L06889, X57535 |
| Vβ10 | AGATGGATTGTGTTCCTG | TAAAAGGACATAGTTATG | AAGCTGGACGAAGAGCTC | L27617, U04575 | NG001333, AJ549946, L27608 |
| Vβ11 | GATCACTCTGGAATGTTC | CAAACCATGGGCCATGAC | GAATTACACCTCATCCAC | L27618, U04572 | L27610, L05152, M13861, M14266 |
| Vβ12 | ACTCTGAGATGTCACCAG | CAGACTGAGAACCACCGC | AATGGGCTGAGGCTGATC | U04578, M60531, U04564 | M14268, AF009663, U03115, U17047, L33100, XM353798 |
| Vβ13.1 | GTCCTGAAAACAGGACAG | ACACTGCAGTGTGCCCAG | ACGTGTCACCAGACTTGGA | M60531, M60533, M60529 | AF043179, X04932, Z26594, M64353 |
| Vβ13.2 | ACAAGGTCACAGAGACAG | GCAGGTGACCTCGACGTG | ACGTGTCACCAGACTTGGA | M60539, M60540, (PanAY168690) | L43588, U50073, U66059, M64352 |
| Vβ14 | GAAGTTGACAGTCACTTG | CAGAATATGAACCATGAG | GGCCTGAGGCAGATCTAC | M60542, M60532 | M13865, M14267, AF272745, AF272742, U66061, M17200 |
| Vβ15 | CCCAAGGAATAAGATCGCAAAGA | AGGAAAGAGGATTATGCTGGAATG | TGTTCTCAGACTAAGGGTCATGATCA | AF143623 | M14269, M11950, AF321238, M11951 |
| Vβ16 | GTAACTCTGAGATGTGAC | ACCCAATTTCTGGACATG | CTGAGAGCGTCTATGCAG | U04592, U04570 | X57723, X06154, X57722, X04933, AJ301406, AJ403805, AJ403762 |
| Vβ17 | GCAAGACTGAGATGCAGC | AGCCCAATGAAAGGACAC | TATCGGCAGCTCCCAGAG | AF143624 | AJ007777, M97711, M27389, M14270, AJ007773 |
| Vβ18 | ACCCTGGTGCAGCCTGTG | CACTGTGGAGGGAACATC | TGTTGACCAGATCAGCTC | AF143625 | M13554, Z13967, D13086, L06893 |
| Vβ19 | TGACCCTGAGTTGTGAAC | AACAGAATTTGAACCACG | ACCGACAGGACCCAGGTC | U04591 | AF043180, Y10201, Y10199, AJ405848 |
| Vβ20 | GAAAACAAAGATGGATTG | CCCCGAAAAAGGACATAC | CAAGAAACGGAGCTGCAC | U04586, U04583, L03738, L07373 | L27614, M27390, L48730 |
| Vβ21 | GAAAAGCCAGGCTGTGAC | TCCTATATCTGGCCATGC | TACTGGTACCAGCAGATC | AF143626 | Z23042, X57536, X57724, X58796, M33235 |
| Vβ22 | TCAGTGGCTGCTGGAGTC | CTGCAATGCTATCCTCC | GAACACGACACTGTCTAC | U04576, M60545, M60535 | M62378, U96844, X57613, X58799, X74849 |
| Vβ23 | GGAAGTGATCTTGCGGTG | GTCCCCATCCCTAATCAC | ACTTCTATTGGTACAGAC | U04582, U04574, U04571, M87476 | AC0737576, X57727, X58798, AJ549945, AJ007770 |
| Vβ24 | AAAGCCAGTGACCTTGAG | AGTTGTTCTCAGAATCTG | ACTGGTACCAGCAGAAGC | AF143627 | X57725, X58800, X74854, AJ007779 |
Table 1B.
External, middle and internal sets of Jβ-specific primers for isolating clonotypic VDJ DNA from a single cell or small numbers of Ag-specific T cells
| 3′ Jβ-specific primers |
GenBank accession number | |||
|---|---|---|---|---|
| Ext.Jβ primers | Mid.Jβ primers | Int.Jβ primers | ||
| Jβ1.1 | TGGACCCACTTTTCCCTATGACGG | AAAAAGTTTACCTAAAACTG | AAAACTGTGAGTCTGGTGCC | L43137 |
| Jβ1.2 | TCTTTGACATTTCCCAGGACAGAG | CCCAGCCTTACCTACAAC | ACAACAGTTAACTTGGTC | |
| Jβ1.3 | TTAACCTAAGCACTGGAGACAAGG | TGACTACTCACCTACAAC | TACAACAGTGAGCCGAGTTC | |
| Jβ1.4 | CAGTGTCTACCCTCCCGCAAGGGG | CCTTTTACATACCCAAGAC | AAGACAGAGAGCTGGGTTC | |
| Jβ1.5 | ATCCTCCTCACCAGTGCTGGAAGG | TTCTGCAACTTACCTAGGAC | TAGGACGGAGAGTCGAGTG | |
| Jβ1.6 | CTTCCCAGCAACAGATAATTGCGG | GAGCCCCCATACCCGTCAC | GTCACAGTGAGCCTGGTCC | |
| Jβ2.1 | GGTTCTGGGGTCGTCCAGGCTGGG | CCTGGAGCCCCCTTCTTAC | GCTCACCGTGCTAGGTAAG | |
| Jβ2.2 | AGCACAGCCCCTCTCGGAGTGGGG | CCGCCTCCTTACCCAGCAC | AGCACGGTCAGCCTAGAGC | |
| Jβ2.3 | AGCACAAAAACTGGCCCTGCTATG | AGCCCCCGCTTACCGAGCAC | GAGCACTGTCAGCCGGGTG | |
| Jβ2.4 | AACCCGACGCCGTCTCTCCCCAGG | CCCCCAGCTTACCCAGCAC | AGCACTGAGAGCCGGGTCC | |
| Jβ2.5 | TCCCCACAAAAACCGGACCCAAG | GGCCCGCGCTCACCGAGCAC | AGCACCAGGAGCCGCGTGC | |
| Jβ2.6 | CCTAGCACAGGGCGGGACCAGGTG | GCGGCAACTCACCCAGCAC | AGCACGGTCAGCCGGCTGC | |
| Jβ2.7 | GAGCGGGGAGGTGACGCTGCAGAG | GCCCGAATCTCACCTATGAC | TATGACTGTGAGCCTGGTG | |
ext.Jβ primers correspond to the sequences downstream to Jβ.
2.3.2. Megaplex PCR for isolation of TCR βDNA (megaplex TCR isolation)
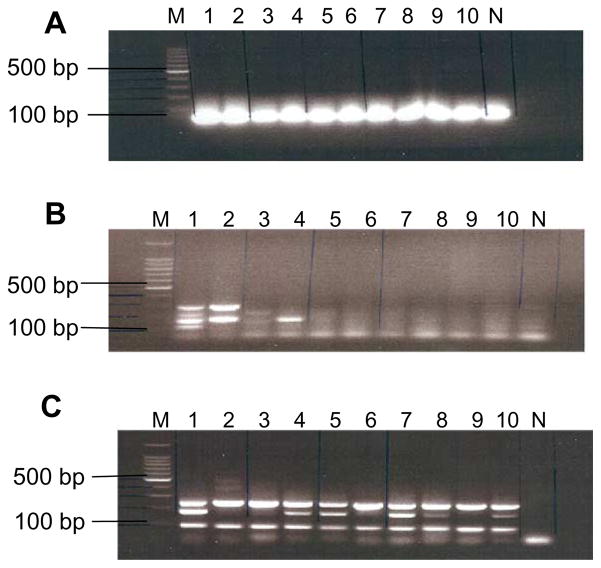
Initial experiments were performed to determine the sensitivity of three consecutive rounds of nested megaplex PCR for isolation of TCRβ DNA from defined numbers of formaldehyde-fixed cells. The 1st, 2nd, and 3rd rounds of nested megaplex PCR were performed in a single PCR tube using external, middle and internal sets of mixed 25 forward Vβ family-specific primers and mixed 13 reverse Jβ primers, respectively (Tables 1A and 1B). These experiments showed that the 3rd round of PCR could amplify TCR β VDJ DNA from one cell (Fig. 2). Therefore, subsequent experiments were designed to amplify and display each TCR Vβ–Jβ clonotype for direct sequencing. The methods for isolation of TCR DNA from limited numbers of antigen-specific T cells are described below:
Fig. 2.
Three-round nested megaplex PCR were able to amplify TCR VDJ DNA from small numbers of formaldehyde-fixed T cells. Lane M on the 2% agarose gel was DNA size markers. Lanes 1 and 2 were PCR products amplified from sample DNA of 200 T cells; Lanes 3 and 4 were PCR products each starting from 20 T cells. Lanes 5 and 6 were PCR products each starting from 4 cells; Lanes 7, 8, 9 and 10 were PCR products each amplified from single cell DNA. (A) The 2% agarose gel showed TCR VDJ DNA amplified from primary megaplex PCR using 25 ext.Vβ and 13 ext.Jβ primers (Tables 1A–1C). The expected size ranged from 280 to 350 bp. Only primer dimers/multimers were seen. (B) VDJ DNA amplified from the 2nd megaplex PCR using 1 μl of the 1st round PCR product as template, 25 mid.Vβ and 13 mid.Jβ primers. The expected VDJ DNA sizes ranged from 150 to 280 bp. (C) VDJ DNA amplified from the 3rd megaplex PCR using 1 μl of 500-fold diluted 2nd PCR product as template, 25 int.Vβ, and 13 int.Jβ primers. The expected sizes ranged from 120 to 250 bp. The VDJ DNA were confirmed by direct sequencing.
2.3.2.1. Preparation of DNA
All cell samples fixed with 2% formaldehyde were purified by flow cytometry sorting. The sorted cells were placed in ependorf tubes containing 12–20 μl of lysis buffer (50 mM Tris–HCl pH 8.0, 1 mM EDTA, 5% Tween 20, 200 μg/ml proteinase K). The lysate was incubated at 55 °C for 1.5 h followed by heat inactivation of the proteinase K at 99 °C for 10 min. DNA samples were then kept at −20 °C until use for PCR reactions.
2.3.2.2. The 1st megaplex PCR
Primary PCR amplification was performed in a single tube with 50 μl volume containing mixtures of ext.Vβ family-specific primers (24 families, 0.25 μM each), mixtures of ext.Jβ-specific primers (13 segments, 0.25 μM each), 1 × Titanium DNA Polymerase (Clontech) and 12–20 μl of DNA template sample. PCR conditions were as follows: 95 °C for 12 min, 35 cycles at 95 °C for 30 s, 60 °C for 30 s, 68 °C 1 for min, and then 68 °C for 1 min. The PCR products were separated and visualized with ethidium bromide on a 2% agarose gel. The sizes of the primary PCR products ranged from 250 to 300 bp.
2.3.2.3. The 2nd megaplex PCR
The 2nd megaplex PCR amplified and displayed Vβ–Jβ clonotypes in the amplified DNA at the level of Vβ families. This PCR reaction narrowed down the complex DNA to individual Vβ families employed by T cell clones. In these experiments, the 1st round PCR product was added to 25 PCR tubes (1 μl for each tube), and each tube contained one of 25 mid.Vβ primers and the mixture of 13 mid.Jβ primers. A 35-cycle PCR was performed under the condition as described above. The 2nd round PCR product was run and visualized with ethidium bromide on a 2% agarose gel. At this time, visible DNA with estimated VDJ sizes was seen on the gel for some but not other sample tubes. The sample tubes with or without visible DNA were treated differently for the 3rd round megaplex PCR.
2.3.2.4. The 3rd megaplex PCR
The 3rd megaplex PCR amplified and displayed Vβ–Jβ clonotypes at the level of individual Vβ–Jβ recombinations for direct sequencing. The 2nd PCR samples containing presumptive VDJ DNA were used for the 3rd PCR amplification and display. For each of these samples, one int.Vβ family-specific primer and one of 13 int.Jβ specific primers were used in 13 PCR tubes. In contrast, the 2nd PCR sample tubes without visible Vβ-bearing DNA individually underwent the 3rd round Vβ family-leveled PCR using one int.Vβ primer and the mixture of 13 int.Jβ primers. The 3rd round PCR reaction was done using 0.1 U/μl of TaqGolden DNA polymerase (ABI) and the following PCR conditions: hot start 95 °C for 12 min; 35 cycles at 95 °C for 30 s, 65 °C for 30 s, 72 °C for 1.5 min; last extension 72 °C for 7 min.
2.3.3. Direct sequencing of TCR VDJ DNA
The predicted single Vβ–Jβ DNA was recovered using a gel extraction kit from QIAGEN (Valencia, CA), and then directly sequenced using third round PCR primers. Since DNA from <50 cells was only visible after the 3rd round PCR amplification (one Vβ with thirteen Jβs), the PCR product was initially sequenced by the direct sequencing approach, if the direct sequencing showed multiple templates, the DNA was then inserting to the plasmid SP65 (Promega, Madison, WI) for cloning and sequencing, as previously described (Chen et al., 2001).
2.4. Isolation of RNA from frozen cells
Total RNA was isolated from PBMC using the TRIzol® isolation method (Shen et al., 2002). PBMC were mixed with 800 μl of TRIzol® reagent, and homogenized gently by repeated pipepting and by vortexing for 10 s. 160 μl of chloroform was added to each sample and mixed by vortexing for 30 s. Samples were spun at 14000 rpm on a desktop centrifuge for 5 min to separate phases. The upper aqueous phase was collected, and 10 μl of glycogen was added to it. It was mixed vigorously by vortex and brief centrifugation. 400 μl of ice-cold isopropanol was added to the sample and precipitated at −20 °C for 1 h. The RNA was precipitated by centrifuging at 14000×g for 15 min at room temperature. The RNA pellet was washed in 200 μl of 70% ethanol and centrifuged at 14000×g for 10 min, and was air-dried for 8 min after removing the supernatant. The RNA pellet was resuspended in RNase-free deionized water, and then used immediately for synthesis of cDNA.
2.5. SMART-based cDNA synthesis and amplification
cDNA was synthesized using a method based on the Altas Switch Mechanism At the 5′-end of Reverse Transcript (SMART™). The synthesis was done using the protocol provided in the SMART cDNA Synthesis Kit from CLONTECH Laboratories (Palo Alto, CA). Briefly, the cDNA reaction was catalyzed using PowerScript Reverse Transcriptase (Gibco BRL, Rockville, MD), the 3′SMART™ CDS primer and the 5′SMART primer provided in the CLONTECH Kit. The double strand cDNA was made and enriched by an 18-cycle PCR using the primers and reagents provided in the SMART cDNA synthesis Kit from CLONTECH. The cycling conditions were as follow: 95 °C for 1 min, 18 cycles at 95 °C for 5 s, 65 °C for 5 s and 68 °C for 6 min.
2.6. Real-time quantitative PCR for quantitating antigen-specific T cell clones
2.6.1. Designing of TCR clonotypic primers and probes
The TCR β clonotypic primers and TaqMan probes were designed based on the recommendation from Primer Express™ computer software (PE Applied Bio-systems). Forward Vβ family-specific primers for real-time quantitative PCR (Table 1C) were designed at the position close to the 3′end of Vβ regions. Jβ clonotypic reverse primers were designed based on the sequences in D-J regions. 24 Vβ family-specific TaqMan probes were synthesized to cover all VDJ clonotypes of the whole TCR β repertoire in clonotypic primer-based real-time quantitative PCR (Fig. 1B and Table 1C). Vβ-specific probes were complementary to the Vβ regions downstream of the sequences corresponding to forward Vβ family-specific primers. The probes were labeled at the 5′-end with the reporter dye 6-carboxyfluorescein (FA) and with the quencher dye 6-carboxytetramethyl-rodamine (TAMRA).
Table 1C.
Clonotypic primers and TaqMan probes for real-time quantitation of antigen-specific T cell clones
| 5′Vβ family primers | Vβ family probes | 3′clonotypic primers | |
|---|---|---|---|
| Vβ1 | TGCACTCTGAACTAAACCTGAGCT | CCAGCAGCGAATATGTCAGAGTATCAGCAAC | Designed based on the unique CDR3 sequences of individual TCR VDJ DNA clones. |
| Vβ2 | GGACAAGTTTCCCATCAACCA | CCAAACCTGACCCCCATCCTGAAGA | |
| Vβ3 | TGAACGAAAAAGGAGATATTCCTGA | AGAAGGAGCGCTTCTCCCTGATTCTGG | |
| Vβ4 | AAACCTAACATTCTCAACTCTAACTGTGA | CAACACGAGCCCTGAAGACAGCAGC | |
| Vβ5 | ACACAGAGACAAAAAGGAAACTTCTCT | CAACACGAGCCCTGAAGACAGCAGC | |
| Vβ6 | GAGGACTGAGGGATCCGTCTC | ACTCTGAAGATCCAGCGCACAGAGCA | |
| Vβ7 | TCTCACTTACACCTTCACCTACGC | TGCAGCCAGGAGACTCAGCCCTG | |
| Vβ8 | ACGATGCCTGACGCATCA | TCTCCACTCTGAAGATCCAGCCCTCAGA | |
| Vβ9 | TGAATCTCCAGACAAAGCTCATTTA | CACATCAAGTCTGTGGAGCTTGGTGACTCT | |
| Vβ10 | AATGAGAGATTTTCAGCCCAATGT | CCCAAAACTCATCCTGTACCTTGGAGATCC | |
| Vβ11 | AGAGGATTTTCCCCTGACCCT | AGTCTGCCAGCCCCTCACACACCT | |
| Vβ12 | TCTCTAGATCAAAGACAGAGGATTTCC | CTCTGGAGTCCGCTACCCGCTCC | |
| Vβ13.1 | TCCCCGATGGCTACAATGTC | CCAGATCAAACACAGAGGATTTCCCACTCA | |
| Vβ13.2 | CCCAGATGGCTACAGTGTCTCTAG | TCAAATAAAGAGGATTTCCCCCTCACTCTGG | |
| Vβ14 | TTGAGATGGTTGATAAGGGAGATATTC | TGAAGGGTACAACGTCTCTCGAAAAGAGAAGA | |
| Vβ15 | TCGACAGGAACAGGCTAAATTCT | TGTCCCTAGAGTCTGCCA | |
| Vβ16 | AAGGACTGGAGGGACGTATTCTACT | TGAAGGTGCAGCCTGCAGAACTGG | |
| Vβ17 | GCTGAATTTCCCAAAGAAGGC | TGAAGGTGCAGCCTGCAGAACTGG | |
| Vβ18 | GTGCCCCAGAATCTCTCAGC | AGGCGGTTCATCCTGAGTTCTAAGAAGCTC | |
| Vβ19 | AATCCTTTCCTCTCACTGTGACATC | CCCAAAGGAACCCAACAGCTTTCTATCTCTG | |
| Vβ20 | CCTACCTTGCAGCCTGGAGAT | TCGGAACCAGGAGACACCGCACTGTAT | |
| Vβ21 | TCTGCAGAGAGGCTCAAAGGA | AACTCCACTCTCAAGATCCAGCCTGCAG | |
| Vβ22 | GGAAGCATCCCTGATCGATTC | ACTCAGCCGTGTACCTCTGTGCCAGC | |
| Vβ23 | GATGGATCAAATTTCACTCTGAAGATC | CCACAAAGCTGGAGGACTCAGCCATG | |
| Vβ24 | GAGGCCAAACACTTCTTTCTGC | TCTTGACATCCGCTCACCAGGCCT |
2.6.2. Real-time quantitative PCR
The real-time quantitative PCR was performed using PE Applied Biosystems 7700 single reporter Sequence Detection Systems 1.6.3. The total reaction volume was 12.5 μl. It contained 6.25 μl of master mix (PE Applied Biosystems), 0.5 μl of 12.5 pmol/μl forward primer, 0.5 μl of 5 pmol/μl FAM-probe (Applied Biosystems), 2.75 μl of H2O, 2 μl of 1:100 diluted SMART-amplified cDNA. Reaction wells for each sample were done in triplicate. The real-time PCR conditions were: 50 °C 2 min, 95 °C for 10 min, 40 cycles at 95 °C 15 s, 60 °C 1 min. All amplifications were carried out in a MicroAmp optical 96-well reaction plate with optical membrane cover (PE Applied Biosystems). All data were analyzed using the GeneAmp 7700 SDS software.
2.6.3. Calculation and expression of frequencies of antigen-specific Vβ+ T cell clones
To quantitate clonotypic T cell clones in a T cell pool, individual clonotypic TCR transcripts were measured for their frequencies in total TCR β transcripts (Cβ), as previously described (Chen et al., 1995, 2001). To this end, the threshold cycle (CT) values for individual Vβ-bearing clones (CT Vβ) and Cβ (CT Cβ) were calculated in each of cDNA samples. Each of CT Vβ represented the level of the particular clonotypic TCR transcript as derived from real-time quantitative PCR using a forward Vβ family-specific primer, a Vβ family-specific Taq-Man probe and a reverse clonotypic primer. CT Cβ represented the level of a total of TCR β transcripts, which was derived by real-time quantitation using a pair of forward and reverse primers and a TaqMan probe designed according to sequences of the TCR β chain constant region. To standardize expression levels of Vβ and Cβ transcripts, CT Vβ and CT Cβ were verified by CT values for a house-keeping gene, β actin (CT β actin). The expression levels of individual clonotypic TCR transcripts among a total of TCR β transcripts within the sample were then calculated using the formula, 2−[(CTVβ −CTβactin) −(CTCβ−CTβactin)], as previously described (Livak and Schmittgen, 2001; Semighini et al., 2002; Gallagher et al., 2003). The calculation gave rise to the ratio of clonotypic Vβ-bearing transcripts versus Cβ transcripts. The data were then expressed as a relative expression of antigen-specific IFNγ-producing T cell clones, (Vβ transcripts/Cβ transcripts)×105. Such an expression of data was based on the consideration that the expression levels of clonotypic Vβ–Jβ-bearing transcripts were always less than those of Cβ transcripts, and that the detection limit for Ag-specific T cell clones in our assay system was 10−5 cells.
3. Results
3.1. Sensitivity and utility of megaplex TCR isolation
To test the sensitivity and utility of the nested megaplex PCR for TCR isolation, we first amplified complex TCR VDJ DNA in defined numbers of T cells from normal rhesus monkeys, and then applied this assay system to isolate TCR VDJ clonotypic sequences from small numbers of antigen-specific T cells generated from PBMC of mycobacterium-infected monkeys. DNA was extracted from 2% formaldehyde-fixed T cells, 1, 4, 20, or 200 in numbers, and used in sequential three-round megaplex PCR reactions to amply TCR VDJ DNA. While the first- and second-round megaplex PCR generated visible TCR β DNA bands on the gel only from DNA samples in the 20- and 200-cell tubes analyzed, the third-round megaplex PCR was able to yield TCR β DNA from the 1- and 4-cell tubes as well (Fig. 2A, B, C). We then sought to isolate TCR β VDJ sequences from antigen-specific T cells purified from PBMC of mycobacterium-infected monkeys using this sensitive megaplex PCR amplification. The 1st round megaplex PCR amplified complex TCR VDJ DNA in a single sample PCR tube; the 2nd and 3rd round megaplex PCR re-amplified and displayed VDJ clonotypes at the levels of Vβ families and individual Vβ–Jβ recombinations, respectively. The three-round megaplex PCR followed by direct sequencing allowed us to readily isolate ninety-one TCR clonotypic sequences from 50 to 500 PPD-specific IFNγ-producing CD4 T cells purified from PBMC of three monkeys 3 and 4 weeks after BCG infection. These clonotypic VDJ sequences therefore allowed us to design individual reverse clonotypic primers and TaqMan probes for real-time quantitation of the frequencies of PPD-specific CD4 T cell clones in limited numbers of PBMC collected over time after mycobacterial infections (see real-time quantitative data in Section 3.5).
3.2. Validation of SMART-based real-time quantitative PCR for quantitating clonotypic T cell clones in limited amounts of samples
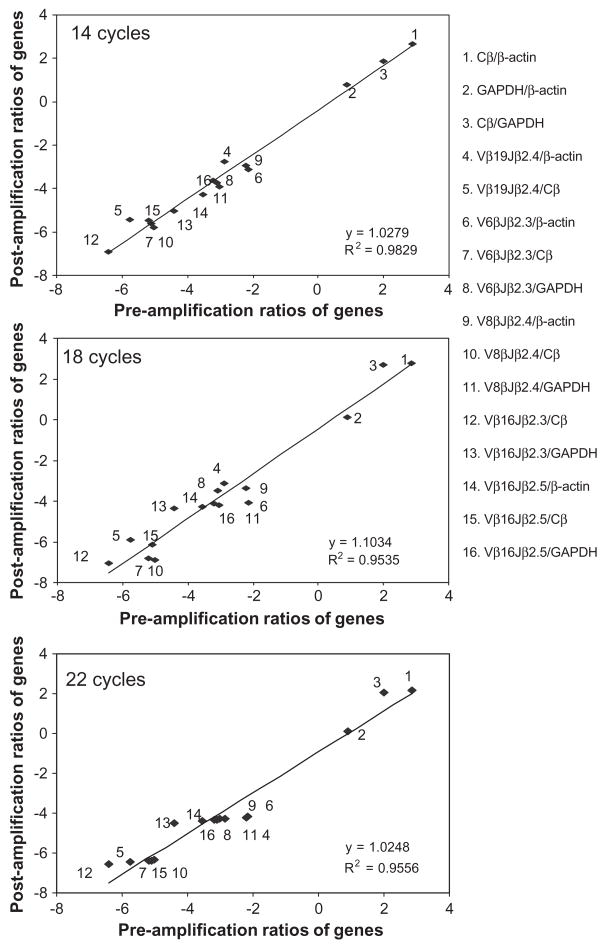
Since experimental or clinical samples are usually used for many different analyses, quantities of specimens available for assessing clonal immune responses of antigen-specific T cells are often limited. It would therefore be useful to be able to enrich limited amounts of mRNA/cDNA and employ the enriched cDNA for real-time quantitation of antigen-specific T cell clones in the limited amounts of clinical specimens. The SMART-based amplification technique has proven to be a useful method to enrich a limited amount of mRNA/cDNA for a large-scale quantitation of gene expression or arrays (Herrler, 2000; Chen et al., 2001; Zhumabayeva et al., 2001, 2004; Seth et al., 2003; Wang et al., 2003; Gustincich et al., 2004; Rihl et al., 2004; Wellenreuther et al., 2004). However, SMART-based real-time quantitation technology has not been validated for its utility in quantitating or measuring frequencies of antigen-specific T cell clones in small amounts of PBMC or tissue. We therefore sought to determine whether SMART-amplified cDNA could maintain consistent relative expression levels of TCR genes in real-time quantitative PCR when compared to mRNA/cDNA templates that did not undergo SMART amplification. To this end, RNA from monkey PBMC was used to synthesize SMART cDNA. The cDNA were equally divided into 4 tubes, and underwent SMART amplification for 0, 14, 18, or 22 PCR cycles. The cDNA and SMART-amplified cDNA were then compared side by side in real-time quantitative PCR for the frequency or representation of TCR genes bearing Cβ, Vβ19Jβ2.4, Vβ6Jβ2.3, Vβ8Jβ2.4, Vβ16Jβ2.3, or Vβ16β2.4 relative to the two house-keeping genes β-actin and GAPDH. When the 16 individual ratios of genes (Cβ/β-actin, GAPDH/β-actin, Cβ/GAPDH, Vβ19Jβ2.4/Cβ or/β-actin, Vβ6Jβ2.3/Cβ or/β-actin or/GAPDH, Vβ8Jβ2.4/Cβ or/β-actin or/GAPDH, Vβ16Jβ2.3/β-actin or/GAPDH, Vβ16Jβ2.4/Cβ or/β-actin or/GAPDH) were calculated in five repeated experiments, the profiles of the 16 expression ratios of these genes in cDNA and SMART-amplified cDNA samples were comparable regardless of the number of SMART amplification cycles employed (Fig. 3). Scattergrams and regression plots of gene-ratios indicated positive correlations between gene-ratios of 16-paired genes in pre-amplified cDNA and those in SMART-amplified cDNA (p <0.01, Fig. 3). These results therefore suggested that SMART amplification did not significantly change TCR gene profiles and that SMART-amplified cDNA could be used in real-time quantitative PCR to quantitate expression levels of clonotypic TCR genes using small amounts of PBMC or tissues.
Fig. 3.
SMART amplification of cDNA enriched cellular mRNA/cDNA without altering TCR β gene profiles. Shown are scattergrams and regression plots comparing sixteen gene expression ratios of individual VβJβ-bearing genes versus Cβ genes or the house-keeping β-actin or GAPDH gene between cDNA (pre-amplification) and SMART-amplified cDNA (post-amplification) derived after different PCR cycles. Data are shown as log10 scales, and presented as mean ratios of genes (transcripts of a VβJβ gene/transcripts of Cβ or β-actin or GAPDH gene), which were calculated from the results in three different assays. SMART-amplified cDNA was generated from the same cDNA derived from a BCG-infected monkey.
3.3. Specificity of the real-time quantitation of antigen-specific clonotypic T cells
TCR VDJ recombination plus N region addition determines the diversity of TCR repertoires. Although clonotypic primers derived from CDR3 sequences have been employed to detect clonotypic T cells, it is not known whether a pair of clonotypic primers used in real-time PCR can be employed for highly specific quantitation of the target clonotypic TCR gene without non-specific amplification of other TCR genes that differ by only few bases in CDR3. To address this important technical issue, we used multiple clonotypic primers in real-time PCR to amplify multiple clonotypic TCR β DNA clones that shared Vβ8 and Jβ1.2 regions but differed by one to eight bases in D–J regions of the beta chain. The forward Vβ8 primer and Vβ8 TaqMan probe were 100% matched with each of Vβ8Jβ1.2 clones; Jβ1.2-based reverse clonotypic primers were variously mismatched with TCR DNA templates (Tables 2A–2C).
Table 2A.
No unspecific real time amplification was seen when a reverse clonotypic primer had ≥4 bases mismatched at the DJ region with the target DNA template
| Reverse clonotypic primer (underlined bases were mismatched at the DJ region with the TCR DNA template) | TCR DNA template | Mean CT values obtained in PCR using 10–106 copies of targeted DNA templates |
|||||
|---|---|---|---|---|---|---|---|
| 10 | 102 | 103 | 104 | 105 | 106 | ||
| 227Vβ8Jβ1.2R93 5′ACCGAAGGTGTAGTCATCGTCC3′ 100% matched |
227Vββ8Jβ1.2 | 32.67 | 30.32 | 29.25 | 27.02 | 23.41 | 19.76 |
| 3050d28Vβ8Jβ1.2 5′GTGTAGTCATAGCACCCTGTC3′ 4 bases mismatched |
227Vβ8Jβ1.2 | 40a | 40 | 40 | 40 | 40 | 40 |
| 329Vβ8Jβ1.2R107 5′ACCGAAGGTGTAGTCATAGTTTGGAG3′ 4 bases mismatched |
2995Vβ8Jβ1.2 | 40 | 40 | 40 | 40 | 40 | 40 |
| 2995Vβ8Jβ1.2R109 5′GAACCGAAGGTGTAGTCATAGTCTGA3′ 5 bases mismatched |
3050d28Vβ8Jβ1.2 | 40 | 40 | 40 | 40 | 40 | 40 |
| 329Vβ8Jβ1.2R107 5′ACCGAAGGTGTAGTCATAGTTTGGAG3′ 6 bases mismatched |
227Vβ8Jβ1.2 | 40 | 40 | 40 | 40 | 40 | 40 |
| 329Vβ8Jβ1.2R107 5′ACCGAAGGTGTAGTCATAGTTTGGAG3′ 6 bases mismatched |
3050d28Vβ8Jβ1.2 | 40 | 40 | 40 | 40 | 40 | 40 |
| 3050d28Vββ8Jβ1.2 5′GACAGGGTGCTATGACTACAC3′ 8 bases mismatched |
329Vβ8Jβ1.2 | 40 | 40 | 40 | 40 | 40 | 40 |
Mismatched bases were underlined.
Ct value of 40 indicated that no signal was detected after 40 cycles of real-time PCR. Each PCR well contained only one single TCR DNA template. Real-time quantitation was done using a forward Vβ8 primer, Vβ8 probe (Table 1C) and individual reverse clonotypic primers. Mismatched bases were underlined.
Table 2C.
TCR clonotypic primers specifically amplified the target DNA template but discriminated, even at high concentrations, the mismatched DNA template mixtures with 2, 4 or 5 substitutions at DJ regions
| Target and mismatched templates (their DJ region sequences complementary to the reverse clonotypic primer 2995Vβ8Jβ1.2R109) | Addition of mismatched template mixtures | Mean CT values obtained in PCR using 10–106 copies of the target DNA template |
|||||
|---|---|---|---|---|---|---|---|
| 10 | 102 | 103 | 104 | 105 | 106 | ||
| Target DNA template (2995Vβ8Jβ1.2– TCAGACTATGACTACACCTTCGGTTC–) | None | 40a (0) | 39.21 (0) | 36.72 (0) | 35.87 (0) | 33.21 (0) | 29.98 (0) |
| Mismatched template mixtures (329Vβ8Jβ1.2– CCAAACTATGACTACACCTTCGGTTC–) (227Vβ8Jβ1.2– AGGGACGATGACTACACCTTCGGTTC–) |
Same amounts as the target template | 40 (10)b | 38.78 (102) | 36.89 (103) | 35.46 (104) | 33.08 (105) | 29.71 (106) |
| (3050Vβ8Jβ1.2– AGGGTGCTATGACTACACCTTCGGTC–) | 10-time larger amounts than the target template | 40 (102) | 38.46 (103) | 36.13 (104) | 35.62 (105) | 32.74 (106) | 29.32 (107) |
Mismatched bases were underlined.
Ct value of 40 indicated that no signal was detected after 40 cycles of real time PCR. Real-time quantitation was done using a forward Vβ8 primer, Vβ8 probe (Table 1C) and a reverse clonotypic primer 2995Vβ8Jβ1.2R109 (5′–GAACCGAAGGTGTAGTCATAGTCTGA–3′).
Numbers in parentheses are copies of each of mismatched templates in mixtures added to individual PCR wells. Note that the addition of the 100% matched target TCR DNA template 2995Vβ8Jβ1.2 to the mismatched template mixtures eliminated the high concentration-driven nonspecific amplification of a single 2-base mismatched DNA template as seen in the setting of the clone 328Vβ8Jβ1.2 (Table 2B). Sequencing of PCR products indicated that only the targeted DNA template was amplified.
When these Vβ8Jβ1.2 DNA clones were tested individually using variant clonotypic primers in different reaction wells under different concentrations, no non-specific real-time quantitation was seen when a reverse clonotypic primer had ≥4 bases mismatched at the DJ region with the target DNA template (Table 2A). A low level of non-specific amplification of a mismatched DNA template was detected under two conditions: (1) a ≤3 base-mismatch between the DNA template and a reverse clonotypic primer; (2) a template concentration ≥103 copies (Table 2B). Since naturally derived cDNA samples from PBMC contain low copy numbers of heterogeneous TCR genes, we set up the real-time quantitation experiments using mixed DNA templates of multiple TCR clones. Surprisingly, when multiple Vβ8Jβ1.2-bearing DNA templates were mixed in the same reaction well, only the target template 2995 (100% matched with the reverse clonotypic primer) was specifically amplified and quantitated in the mixed clones (Table 2C). The specific amplification and quantitation of clone 2995 was not even interfered with by a series of 10-fold higher concentrations of the mismatched DNA template mixtures with 2, 4, or 5 substitutions in the DJ regions (Table 2C). A clone with only a 1-base mismatch at the DJ region was not tested in these mixed-template experiments because it could not be identified from >500 naturally occurring clones with the same antigen specificity. In a separate experiment, a TCR DNA clone with only a 1-base mismatch in the DJ region was created and tested for cross-reactivity in the mixed templates. This 1-base mismatch could not be discriminated by the clonotypic primer in the DNA template mixtures (data not shown). These results provided experimental evidence that clonotypic TCR primers used for real-time quantitation could specifically amplify the target clonotypic TCR DNA clone in heterogeneous TCR DNA mixtures and discriminate clones that had ≥2 bases mismatched in DJ regions.
Table 2B.
The high concentration-driven unspecific real time amplification occurred when a reverse clonotypic number had ≤3 bases mismatched at the DJ region with the TCR DNA template
| Reverse clonotypic primer (underlined bases were mismatched at the DJ region with the TCR DNA template) | TCR DNA template | Mean CT values obtained in PCR using 10–106 copies of targeted DNA templates |
|||||
|---|---|---|---|---|---|---|---|
| 10 | 102 | 103 | 104 | 105 | 106 | ||
| 227Vβ8Jβ1.2R93 5′ACCGAAGGTGTAGTCATCGTCC3′ 100% matched |
227Vβ8Jβ1.2 | 32.67 | 30.32 | 29.25 | 27.02 | 23.41 | 19.76 |
| 2995Vβ8Jβ1.2R111 5′CTGAACCGAAGGTGTAGTCATAGTCT3′ 100% matched |
2995Vβ8Jβ1.2 | 38.42 | 37.04 | 35.61 | 32.63 | 29.16 | 25.94 |
| 2995Vβ8Jβ1.2R112 5′CCTGAACCGAAGGTGTAGTCATAGTC3′ 100% matched |
2995Vβ8Jβ1.2 | 34.19 | 31.47 | 30.71 | 27.86 | 24.25 | 20.88 |
| 2995Vβ8Jβ1.2R112 5′CCTGAACCGAAGGTGTAGTCATAGTC3′ 1 base mismatched |
227Vβ8Jβ1.2 | 40a | 40 | 40 | 37.1 | 33.71 | 31.19 |
| 2995Vβ8Jβ1.2R112 5′CCTGAACCGAAGGTGTAGTCATAGTC3′ 1 base mismatched |
329Vβ8Jβ1.2 | 40 | 40 | 37.59 | 35.53 | 32.31 | 29 |
| 2995Vβ8Jβ1.2R112 5′CCTGAACCGAAGGTGTAGTCATAGTC3′ 2 bases mismatched |
3050Vβ8Jβ1.2 | 40 | 40 | 40 | 37.17 | 34.56 | 31.14 |
| 2995Vβ8Jβ1.2R109 5′GAACCGAAGGTGTAGTCATAGTCTGA3′ 2 bases mismatched |
329Vβ8Jβ1.2 | 40 | 40 | 40 | 40 | 39.52 | 38.19 |
| 2995Vβ8Jβ1.2R111 5′CTGAACCGAAGGTGTAGTCATAGTCT3′ 2 bases mismatched |
227Vβ8Jβ1.2 | 40 | 40 | 38.83 | 35.99 | 32.97 | 29.40 |
| 2995Vβ8Jβ1.2R111 5′CTGAACCGAAGGTGTAGTCATAGTCT3′ 3 bases mismatched |
3050Vβ8Jβ1.2 | 40 | 40 | 40 | 35.84 | 33.11 | 29.75 |
| 227Vβ8Jβ1.2R93 5′ACCGAAGGTGTAGTCATCGTCC3′ 3 bases mismatched |
329Vβ8Jβ1.2 | 40 | 40 | 40 | 40 | 36.75 | 37.7 |
Mismatched bases were underlined.
Ct value of 40 indicated that no signal was detected after 40 cycles of real-time PCR. Each PCR well contained only one single TCR DNA template Real-time quantitation was done using a forward Vβ8 primer, Vβ8 probe (Table 1C) and individual reverse clonotypic primers. Mismatched bases were underlined.
3.4. Detection limits of SMART-based real-time quantitation of antigen-specific T cell clones in PBMC
To determine a detection limit of SMART-based real-time quantitation of antigen-specific T cells in 1–2×106 PBMC or tissue cells, we measured detectable frequencies of the SEB-specific Vβ19Jβ2.4+ CTL clone (2H6, Chen et al., 1996) using different numbers of 2H6 cells and PBMC. We added different numbers of cloned Vβ19Jβ2.4+ CTL to monkey PBMC at the ratios of 1 in 107, 1 in 106, 10 in 106, 1000 in 106, 10000 in 106 and 10000 in 105. The mixed Vβ19 CTL and PBMC were then subjected to RNA isolation, cDNA synthesis, and an 18-cycle SMART® LD amplification. SMART-amplified cDNA was then used to determine measurable levels of clonotypic Vβ19Jβ2.4 TCR in PBMC using a forward Vβ19 primer, a Vβ19 TaqMan probe and a reverse 2H6 clonotypic primer (5′–TGGCTTACCCCCCAGATGAT–3′) in the real-time quantitative assay. The results showed that SMART-based real-time quantitative PCR was able detect one antigen-specific T cell clone in 105 or 106 PBMC (Table 3). It should be pointed out that this detection limit was based on routinely diluted amounts of our SMART-amplified cDNA from 1 to 3×106 PBMC or tissue cells. We used only 1% of SMART-amplified cDNA in the real-time quantitative PCR.
Table 3.
SMART-based real-time quantitative PCR conferred a detection limit of one Ag-specific T cell clone in 105 or 106 PBMC
| Numbers of mixed cells, 2H6 clone/PBMC |
||||||
|---|---|---|---|---|---|---|
| 1/107 | 1/106 | 10/106 | 1000/106 | 10000/106 | 10000/105 | |
| CT value for 2H6Vβ19Jβ2.4 transcripts | 40a | 36.24 | 37.24 | 35.19 | 35.47 | 35.7 |
| CT value for Cβ transcripts | 16.6 | 19.66 | 20.31 | 24.65 | 27.31 | 31.81 |
| Transcript ratio,b Vβ19Jβ2.4/Cβ (×105) | 1 | 8 | 67 | 349 | 6742 | |
CT 40=no signal detected.
Transcripts of 2H6Vβ19Jβ2.4 and Cβ were calculated based on CT values using the formula as described in Section 2.6.3.
3.5. Quantitation of PPD-specific IFNγ-producing CD4 T cell clones during mycobacterial infections using the combined megaplex TCR isolation and SMART-based read-time quantitative PCR methods
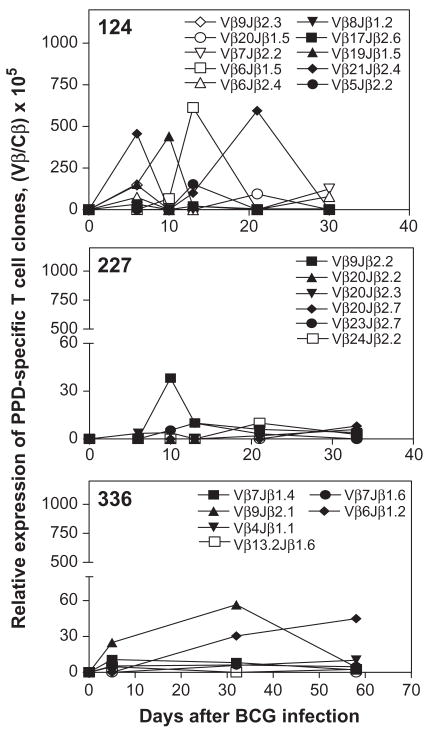
Finally, we applied the combined megaplex TCR isolation and SMART-based real-time quantitative methods to quantitate immune responses of antigen-specific T cell clones during mycobacterial infection. Ninety-one clonotypic primers for real-time quantitation were designed based on the sequences of 91 PPD-specific clonotypic TCR clones derived from PBMC of three BCG-infected rhesus monkeys (see Section 3.1). The frequencies of each of these ninety-one PPD-specific IFNγ-producing clones was then quantitated using the SMART-amplified cDNA (18 SMART cycles) generated from PBMC collected weekly after BCG infection of three monkeys. Extensive SMART-based real-time quantitative PCR analyses indicated that twenty-two of these ninety-one PPD-specific T cell clones expanded to the detectable level in PBMC over time after BCG infection of the three monkeys (Fig. 4). These twenty-two dominant PPD-specific T cell clones emerged at frequencies of 3 to 600 transcripts per 105 Cβ transcripts in PBMC during primary BCG infection (Fig. 4). Similar expression levels for dominant clones 124Vβ21Jβ2.4, 124Vβ20Jβ1.5, 336Vβ7Jβ1.6 and 336Vβ9Jβ2.1 were also confirmed by real-time PCR using conventional cDNA derived from pre- and week 4 PBMC (data not shown). Other clones could not be quantitated because of their low frequencies in PBMC (Fig. 4). Nevertheless, these results indicated that combined megaplex TCR isolation and SMART-based real-time quantitative PCR methods could be used to quantitate large numbers of antigen-specific T cell clones during mycobacterial infection using limited numbers of PBMC.
Fig. 4.
Immune responses of PPD-specific IFNβ-producing CD4 T cell clones after BCG infection of monkeys. Clonotypic TCR sequences were derived from PPD-specific IFNβ-producing CD4 T cells. These antigen-specific T cells were selected by intracellular cytokine staining of PPD-stimulated PBMC obtained 3 and 4 weeks after BCG infection, fixed by 2% formaldehyde and purified by flow cytometry sorting. Real time quantitation of PPD-specific IFNβ-producing CD4 T cell clones was done using SMART cDNA (18 cycles) derived from frozen PBMC collected before and after BCG infection. Shown are only the clones that were detectable in real-time PCR. Twenty-four, thirty and fifteen VβJβ clones isolated from monkeys 124, 227 and 336, respectively, were undetectable due to their low frequencies (<1×105) in PBMC. Similar expression levels for dominant clones 124Vβ21Jβ2.4, 124Vβ20Jβ1.5, 336Vβ7Jβ1.6 and 336Vβ9Jβ2.1 were also confirmed by real-time PCR using conventional cDNA derived from pre- and week 4 PBMC.
4. Discussion
Our combined megaplex TCR β isolation and SMART-based real-time quantitation methods represent the first effort to quantitate a large number of antigen-specific T cell clones using small quantities of clinical specimens. In fact, the megaplex TCR β isolation allowed us to co-amplify and isolate the entire macaque TCR repertoire comprised of recombinants from 24 Vβ families and 13 Jβ segments in a single sample well. This single sample PCR well underwent nested amplifications and displays of complex TCR DNA at the levels of Vβ family usage and individual Vβ–Jβ recombinations. This method allowed us to isolate clonotypic TCR VDJ elements from one or many antigen-specific T cells purified by flow cytometry sorting. The ability of megaplex TCR isolation to isolate TCR VDJ DNA from formalin-fixed PPD-specific T cells bypasses special requirements for purifying fresh antigen-specific T cells for mRNA preparation. These characteristics have advantages over previously reported multiplex or individual Vβ-based PCR approaches in human systems, in which groups of PCR tubes are set up to amplify TCR cDNA containing clustered or individual Vβ families (Maslanka et al., 1995; Akatsuka et al., 1999; van Dongen et al., 2003). While these multiplex PCR approaches employ larger sizes of samples for multiple Vβs, the use of mRNA/cDNA for isolating clonotypic TCR from highly infectious materials requires specialized flow cytometry-sorting at a BSL3 level for purifying antigen-specific T cells.
Our results suggest that SMART-amplified cDNA is useful for real-time quantitation of antigen-specific T cell clones using limited amounts of mRNA or PBMC samples. This is consistent with accumulating reports indicating that SMART-amplification can maintain relative expression levels of low and abundant cellular mRNA/cDNA (Herrler, 2000; Chen et al., 2001; Zhumabayeva et al., 2001, 2004; Seth et al., 2003; Wang et al., 2003; Gustincich et al., 2004; Rihl et al., 2004; Wellenreuther et al., 2004). In fact, our previous studies demonstrated that frequencies of Vβ families in SMART-derived cDNA were similar to those in conventional cDNA or anchored PCR-enriched cDNA when using a semi-quantitative method (Chen et al., 2001). Recent experiments have also demonstrated that conventional cDNA and SMART-amplified cDNA are comparable for quantitating expression levels of cytokines in real-time quantitative PCR (data not shown). Since most samples from experimental or preclinical studies are used for a variety of assays, available quantities of such clinical specimens shared for assessing immune responses of antigen-specific T cell clones are usually limited. It is important to note that using conventional RT–PCR quantitation, cDNA sample sizes derived from 1 to 2×106 PBMC are only sufficient to detect or quantitate a few T cell clones but not multiple clones. Since infections drive expansions of many antigen-specific T cell clones, it is important to employ versatile assay systems to quantitate large numbers of antigen-specific T cell clones in limited amounts of blood or tissue. The SMART-based real-time quantitative PCR therefore may provide a useful approach for measuring frequencies of antigen-specific T cell clones using limited quantities of specimens.
Since TCR clonotypes specific for an epitope may share some degree of sequence similarity in VDJ junctional regions, it is important to determine the extent to which a pair of clonotypic primers can specifically amplify the 100%-matched TCR clone and discriminate others that differ by only a few bases in CDR3. Surprisingly, the specificity of real-time quantitative PCR for measuring T cell clonal responses remains poorly characterized (Douek et al., 2002). Our studies indicate that a reverse clonotypic primer can ensure specific amplification of a target TCR clone while discriminating clones with ≥2 bases mismatched in DJ regions. Interestingly, the discriminatory capability of reverse clonotypic primers was more evident in the single PCR well containing template mixtures of multiple TCR clones, a condition that was similar in heterogeneity to naturally derived mRNA/cDNA. The mechanism by which mixed templates but not a single template in a PCR reaction well results in a higher level of specific amplification is currently not known. It is likely that the target template in the mixture may outperform non-targeted templates in competing for favorable PCR thermokinetics that confer dominant amplification. Given that cDNA from PBMC is comprised of low copies of heterogeneous mixtures of TCR genes, reverse clonotypic primers designed based on unique sequences at DJ regions are likely to confer specific quantitation of clonotypic TCR clones.
Our combined megaplex TCR isolation and SMART-based real-time quantitative PCR methods were useful for measuring frequencies of PPD-specific T cell clones during M. bovis BCG infection of monkeys. Using this assay system, we were able to quantitate clonal expansions of dominant PPD-specific T cell clones in PBMC obtained from monkeys after BCG infection. It should be pointed out that a number of clones were undetectable in PBMC during BCG infection because only 1% of SMART-amplified cDNA was used in the real-time quantitative PCR. We expect that an increase in the amount of SMART–cDNA would make those clones measurable using the real-time quantitation.
Thus, we have validated the utility of combined megaplex TCR isolation and SMART-based real-time quantitation methods for measuring frequencies and dynamics of antigen-specific T cell clones using limited quantities of clinical specimens. The highly sensitive and specific characteristics of this assay system make it possible to study clonal dominance, as well as tissue or mucosal migration of antigen-specific T cell clones in the setting of vaccinations, infections or diseases. Therefore, this assay system may have a broad application to studies of T cell clonal responses in the setting of tumors, autoimmunity, or infection.
Acknowledgments
Authors would like to thank other members at the Chen lab for technical assistance. This work was supported by National Institutes of Health R01 grants: HL64560 (to ZWC) and RR13601 (to ZWC).
Abbreviations
- SMART
Altas Switch Mechanism At, the 5′-end of Reverse Transcript
- TCR
T cell receptors
- BCG
Mycobacterium bovis bacille Calmette–Guérin
- PPD
purified protein derivatives
References
- Akatsuka Y, Martin EG, Madonik A, Barsoukov AA, Hansen JA. Rapid screening of T-cell receptor (TCR) variable gene usage by multiplex PCR: application for assessment of clonal composition. Tissue Antigens. 1999;53:122. doi: 10.1034/j.1399-0039.1999.530202.x. [DOI] [PubMed] [Google Scholar]
- Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- Chen ZW, Kou ZC, Lekutis C, Shen L, Zhou D, Halloran M, Li J, Sodroski J, Lee-Parritz D, Letvin NL. T cell receptor V beta repertoire in an acute infection of rhesus monkeys with simian immunodeficiency viruses and a chimeric simian–human immunodeficiency virus. J Exp Med. 1995;182:21. doi: 10.1084/jem.182.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZW, Shen L, Regan JD, Kou Z, Ghim SH, Letvin NL. The T cell receptor gene usage by simian immunodeficiency virus gag-specific cytotoxic T lymphocytes in rhesus monkeys. J Immunol. 1996;156:1469. [PubMed] [Google Scholar]
- Chen ZW, Li Y, Zeng X, Kuroda MJ, Schmitz JE, Shen Y, Lai X, Shen L, Letvin NL. The TCR repertoire of an immunodominant CD8+ T lymphocyte population. J Immunol. 2001;166:4525. doi: 10.4049/jimmunol.166.7.4525. [DOI] [PubMed] [Google Scholar]
- Cheynier R, Gratton S, Halloran M, Stahmer I, Letvin NL, Wain-Hobson S. Antigenic stimulation by BCG vaccine as an in vivo driving force for SIV replication and dissemination. Nat Med. 1998;4:421. doi: 10.1038/nm0498-421. [DOI] [PubMed] [Google Scholar]
- Cohen GB, Islam SA, Noble MS, Lau C, Brander C, Altfeld MA, Rosenberg ES, Schmitz JE, Cameron TO, Kalams SA. Clonotype tracking of TCR repertoires during chronic virus infections. Virology. 2002;304:474. doi: 10.1006/viro.2002.1743. [DOI] [PubMed] [Google Scholar]
- Douek DC, Betts MR, Brenchley JM, Hill BJ, Ambrozak DR, Ngai KL, Karandikar NJ, Casazza JP, Koup RA. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J Immunol. 2002;168:3099. doi: 10.4049/jimmunol.168.6.3099. [DOI] [PubMed] [Google Scholar]
- Gallagher RB, Yeap BY, Bi W, Livak KJ, Beaubier N, Rao S, Bloomfield CD, Appelbaum FR, Tallman MS, Slack JL, Willman CL. Quantitative real-time RT–PCR analysis of PML–RAR alpha mRNA in acute promyelocytic leukemia: assessment of prognostic significance in adult patients from intergroup protocol 0129. Blood. 2003;101:2521. doi: 10.1182/blood-2002-05-1357. [DOI] [PubMed] [Google Scholar]
- Gallard A, Foucras G, Coureau C, Guery JC. Tracking T cell clonotypes in complex T lymphocyte populations by real-time quantitative PCR using fluorogenic complementarity-determining region-3-specific probes. J Immunol Methods. 2002;270:269. doi: 10.1016/s0022-1759(02)00336-8. [DOI] [PubMed] [Google Scholar]
- Gustincich S, Contini M, Gariboldi M, Puopolo M, Kadota K, Bono H, LeMieux J, Walsh P, Carninci P, Hayashizaki Y, Okazaki Y, Raviola E. Gene discovery in genetically labeled single dopaminergic neurons of the retina. Proc Natl Acad Sci U S A. 2004;101:5069. doi: 10.1073/pnas.0400913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrler M. Use of SMART-generated cDNA for differential gene expression studies. J Mol Med. 2000;78:B23. [PubMed] [Google Scholar]
- Klenerman P, Cerundolo V, Dunbar PR. Tracking T cells with tetramers: new tales from new tools. Nat Rev, Immunol. 2002;2:263. doi: 10.1038/nri777. [DOI] [PubMed] [Google Scholar]
- Kou ZC, Halloran M, Lee-Parritz D, Shen L, Simon M, Sehgal PK, Shen Y, Chen ZW. In vivo effects of a bacterial superantigen on macaque TCR repertoires. J Immunol. 1998;160:5170. [PubMed] [Google Scholar]
- Kuroda MJ, Schmitz JE, Barouch DH, Craiu A, Allen TM, Sette A, Watkins DI, Forman MA, Letvin NL. Analysis of gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus-infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I-peptide complex. J Exp Med. 1998;187:1373. doi: 10.1084/jem.187.9.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson M, Jin X, Ramratnam B, Ogg GS, Engelmayer J, Demoitie MA, McMichael AJ, Cox WI, Steinman RM, Nixon D, Bhardwaj N. A recombinant vaccinia virus based ELISPOT assay detects high frequencies of Pol-specific CD8 T cells in HIV-1-positive individuals. Aids. 1999;13:767. doi: 10.1097/00002030-199905070-00005. [DOI] [PubMed] [Google Scholar]
- Lim A, Baron V, Ferradini L, Bonneville M, Kourilsky P, Pannetier C. Combination of MHC-peptide multimer-based T cell sorting with the Immunoscope permits sensitive ex vivo quantitation and follow-up of human CD8+ T cell immune responses. J Immunol Methods. 2002;261:177. doi: 10.1016/s0022-1759(02)00004-2. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maslanka K, Piatek T, Gorski J, Yassai M. Molecular analysis of T cell repertoires. Spectratypes generated by multiplex polymerase chain reaction and evaluated by radioactivity or fluorescence. Hum Immunol. 1995;44:28. doi: 10.1016/0198-8859(95)00056-a. [DOI] [PubMed] [Google Scholar]
- Meyer-Olson D, Shoukry NH, Brady KW, Kim H, Olson DP, Hartman K, Shintani AK, Walker CM, Kalams SA. Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape. J Exp Med. 2004;200:307. doi: 10.1084/jem.20040638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher CJ, Quittner C, Peterson DM, Connors M, Koup RA, Maino VC, Picker LJ. HIV-1-specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat Med. 1999;5:518. doi: 10.1038/8400. [DOI] [PubMed] [Google Scholar]
- Rihl M, Baeten D, Seta N, Gu J, De Keyser F, Veys EM, Kuipers JG, Zeidler H, Yu DT. Technical validation of cDNA based microarray as screening technique to identify candidate genes in synovial tissue biopsy specimens from patients with spondyloarthropathy. Ann Rheum Dis. 2004;63:498. doi: 10.1136/ard.2003.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semighini CP, Marins M, Goldman MH, Goldman GH. Quantitative analysis of the relative transcript levels of ABC transporter Atr genes in Aspergillus nidulans by real-time reverse transcription–PCR assay. Appl Environ Microbiol. 2002;68:1351. doi: 10.1128/AEM.68.3.1351-1357.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth D, Gorrell MD, McGuinness PH, Leo MA, Lieber CS, McCaughan GW, Haber PS. SMART amplification maintains representation of relative gene expression: quantitative validation by real time PCR and application to studies of alcoholic liver disease in primates. J Biochem Biophys Methods. 2003;55:53. doi: 10.1016/s0165-022x(02)00177-x. [DOI] [PubMed] [Google Scholar]
- Shen Y, Zhou D, Qiu L, Lai X, Simon M, Shen L, Kou Z, Wang Q, Jiang L, Estep J, Hunt R, Clagett M, Sehgal PK, Li Y, Zeng X, Morita CT, Brenner MB, Letvin NL, Chen ZW. Adaptive immune response of V gamma2Vdelta2+ T cells during mycobacterial infections. Science. 2002;295:2255. doi: 10.1126/science.1068819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R, van Krieken JH, Droese J, Gonzalez D, Bastard C, White HE, Spaargaren M, Gonzalez M, Parreira A, Smith JL, Morgan GJ, Kneba M, Macintyre EA. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- Wang J, Hu L, Hamilton SR, Coombes KR, Zhang W. RNA amplification strategies for cDNA microarray experiments. Biotechniques. 2003;34:394. doi: 10.2144/03342mt04. [DOI] [PubMed] [Google Scholar]
- Wellenreuther R, Schupp I, Poustka A, Wiemann S. SMART amplification combined with cDNA size fractionation in order to obtain large full-length clones. BMC Genomics. 2004;5:36. doi: 10.1186/1471-2164-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Shen Y, Chalifoux L, Lee-Parritz D, Simon M, Sehgal PK, Zheng L, Halloran M, Chen ZW. Mycobacterium bovis bacille Calmette–Guérin enhances pathogenicity of simian immunodeficiency virus infection and accelerates progression to AIDS in macaques: a role of persistent T cell activation in AIDS pathogenesis. J Immunol. 1999;162:2204. [PubMed] [Google Scholar]
- Zhumabayeva B, Diatchenko L, Chenchik A, Siebert PD. Use of SMART-generated cDNA for gene expression studies in multiple human tumors. Biotechniques. 2001;30:158. doi: 10.2144/01301pf01. [DOI] [PubMed] [Google Scholar]
- Zhumabayeva B, Chenchik A, Siebert PD, Herrler M. Disease profiling arrays: reverse format cDNA arrays complimentary to microarrays. Adv Biochem Eng Biotechnol. 2004;86:191. doi: 10.1007/b12443. [DOI] [PubMed] [Google Scholar]