Abstract
The automated volumetric output of FreeSurfer and Individual Brain Atlases using Statistical Parametric Mapping (IBASPM), two widely used and well published software packages, was examined for accuracy and consistency relative to auto-assisted manual (AAM) tracings (i.e., manual correction of automated output) when measuring the caudate, putamen, amygdala, and hippocampus in the baseline scans of 120 HIV-infected patients (86.7% male, 47.3±6.3 y.o., mean HIV duration 12.0±6.3 years) from the NIH-funded HIV Neuroimaging Consortium (HIVNC) cohort. The data was examined for accuracy and consistency relative to auto-assisted manual tracing, and construct validity was assessed by correlating automated and AAM volumetric measures with relevant clinical measures of HIV progression. When results were averaged across all patients in the eight structures examined, FreeSurfer achieved lower absolute volume difference in five, higher sensitivity in seven, and higher spatial overlap in all eight structures. Additionally, FreeSurfer results exhibited less variability in all measures. Output from both methods identified discrepant correlations with clinical measures of HIV progression relative to AAM segmented data. Overall, FreeSurfer proved more effective in the context of subcortical volumetry in HIV-patients, particularly in a multi-site cohort study such as this. These findings emphasize that regardless of the automated method used, visual inspection of segmentation output, along with manual correction if necessary, remains critical to ensuring the validity of reported results.
Introduction
Magnetic resonance imaging (MRI) based brain volumetry is a valuable technique for identifying subcortical morphometric changes in vivo and determining the regional neurological impact of psychopathology, disease progression, and advancing therapeutic regimens. This approach has been useful for characterizing the effects of dementia (Carmichael et al. 2005, Teipel et al. 2008, Thompson et al. 2001), psychiatric disorders (Csernansky et al. 1998, Hickie et al. 2005, Konarski et al. 2008, Styner et al. 2004), and normal aging (Brickman et al. 2008, Elderkin-Thompson et al. 2008, Walhovd et al. 2005), as well as uncovering regional and global neurological consequences of systemic diseases such as the Human Immunodeficiency Virus (HIV) (Carmichael et al. 2007, Sporer et al. 2005, Stout et al. 1998, Thompson et al. 2005, Thompson et al. 2006), diabetes (Jongen and Biessels 2008, Perantie et al. 2007, Tiehuis et al. 2008, Wessels et al. 2007), and scoliosis (Liu et al. 2008). As techniques in MRI continue to advance, in vivo volumetric measurement will become increasingly valuable in the drive to understand the evolution and progression of injury for CNS disorders as well as typical aging.
The range of clinical applications for MRI volumetry has generated intense interest in maximizing the accuracy and efficiency of automated segmentation techniques. For years, manual delineation by trained experts has remained the “gold standard” of accuracy in volumetric analyses. Yet while it remains the current reference standard for segmentation, the accuracy of manual volumetry relative to true structure volume is still widely debated, as results can be influenced by factors such as anatomical protocols, tracer experience, scan acquisition parameters, image quality, and even the computer hardware employed in the tracing procedure (Jack et al. 1990, Jack et al. 1995, Warfield et al. 2004). Moreover, manual tracings are time consuming, taking up to two hours per structure (though this time may vary depending on structure complexity, slice thickness, and rater experience). Thus, the required time, financial and personnel resources render manual volumetry in large cohort studies impractical.
Multiple automated methods have been developed to reduce tracing time while ensuring excellent reliability (Andersen et al. 2002, Heckemann et al. 2006, Powell et al. 2008). In particular, the FreeSurfer software package (Martinos Center, Boston, MA) and Individual Brain Atlases toolbox (IBASPM; Cuban Neuroscience Center, Havana, Cuba) of the popular Statistical Parametric Mapping package (SPM; Wellcome Trust Centre for Neuroimaging, UK) are widely used and have well-published methods. Both packages are fully automated, employing an atlas-based segmentation approach to generate an individualized anatomical label map for a spatially normalized patient image, based on an atlas composed of manually traced reference scans (Aleman-Gomez et al. 2006, Ashburner and Friston 1997, Ashburner et al. 1999, Ashburner and Friston 2005, Fischl et al. 2002, Han and Fischl 2007, Tzourio-Mazoyer et al. 2002).
While both of these packages have been validated by their creators, their accuracy and/or consistency may vary depending on image quality, scan parameters, and scanning hardware (Jovicich et al. 2009, Han and Fischl 2007, Tae et al. 2008). Additionally, previous comparisons of competing automated methods have shown notable differences in their performance relative to manual segmentation, despite examining only a limited number of structures (Cherbuin et al. 2009, Klauschen et al. 2009, Morey et al. 2009, Shen et al. 2009, Tae et al. 2008). Some have suggested the patient composition of the source atlas, particularly the inclusion of healthy or diseased subjects, may in fact influence how robust each software package will be with diseased patients or otherwise morphologically different brains (Csapo et al. 2009, Tae et al. 2008, Zhang 1996). Differences in FreeSurfer and IBASPM processing pipelines in addition to atlas composition, such as the algorithms for registration and statistical application of the information contained in the atlases, underscore the importance of re-validating these packages prior to analyzing data obtained with scan parameters or patient populations that are distinct from those of previous validation studies, especially in the case of a large sample size or multi-site study.
The purpose of this study was to address previously described inconsistencies in FreeSurfer and IBASPM subcortical segmentation results by examining the automated volumetric measurement of several clinically relevant subcortical structures from a large multisite consortium study of HIV infection. We compared the accuracy and consistency of volumetric results for the caudate, putamen, hippocampus, and amygdala obtained using three methods: AAM segmentation, FreeSurfer (Martinos Center for Biomedical Imaging, Boston, MA), and IBASPM (Cuban Neuroscience Center, Havana, Cuba). Cognitive decline is a well-described feature of HIV progression, and a small number of studies have linked this to atrophy of subcortical structures (Gonzalez-Scarano and Martin-Garcia 2005, Hall et al. 1996, Paul et al. 2002, Ragin et al. 2005, Robertson et al. 2007, Stout et al. 1998). Future investigations of this relationship will call for large-scale studies that will rely on automated volumetric procedures to efficiently obtain data. To ensure the data is interpreted correctly, it will be crucial to anticipate and thereby minimize the possible shortcomings of these automated methods. To this end, we will attempt to characterize the accuracy and variability of these methods, as well as examine the ability of each to uncover significant, valid relationships when correlated with clinical measures of HIV progression.
Materials and Methods
Subjects
One hundred twenty HIV infected patients were examined in this study (86.7% male; mean age 47.3 ± 7.2 years). Patients were recruited as part of the ongoing multisite NIH-funded MRS (magnetic resonance spectroscopy) HIV Neuroimaging Consortium (HIVNC) study based on the following inclusion criteria: HIV-positive, age ≥ 18 years, duration of HAART > 12 weeks, nadir CD4 count < 100 cells/ml during HIV history. Patients were considered to be on stable treatment (highly active antiretroviral therapies (HAART); median CD4 365.6 ± 224.7 cells/mm3; 64.2% undetectable plasma viral load (< 75 copies/mL); median HIV duration 12.0 ± 6.3 years). Patients with history of major psychiatric illness, confounding neurological disorders, hepatic or renal dysfunction, diabetes mellitus, or chronic/active alcohol or substance abuse were excluded from participation. In addition to neuroimaging, patients were also given baseline cognitive and psychiatric assessments, as well as assays for relevant clinical variables. The study was approved by the institutional review board at each data collection site. All patients provided informed consent and received approved compensation. The subjects used in this study were selected at random from the larger HIVNC cohort, and a comparison of demographic data ensured that the subset was representative.
Image acquisition
Imaging of each participant followed a strict standardized protocol established at the beginning of the study and monitored using automated and manual checking routines. Scans at all sites were performed using 1.5T GE Signa scanners with a minimum operating system requirement. T1-weighted SPGR (Spoiled Gradient Recalled) MR images were acquired for each participant using the following sequence parameters: TR=20 (minor variation allowed), TE=5 (minor variation allowed), 1×1×1.3 mm, flip angle=30, axial slices. Images were visually inspected for quality control purposes and re-acquired if necessary (e.g. presence of motion artifacts, etc.).
Automated Volumetry
FreeSurfer
FreeSurfer’s built-in mri_convert function was used to convert scans to a format compatible with FreeSurfer (.mgz). Resulting 8-bit images were processed using FreeSurfer version dev4 (released September 4th, 2008; Martinos Center, Harvard University, Boston, MA) on a local computing cluster running the Red Hat Enterprise Linux (ES 4). As implemented here, the workflow for creating subcortical segmentations consists of the following steps, which have been previously described in detail by FreeSurfer's creators (Fischl et al. 2002, FreeSurfer Wiki):
Pre-processing: Non-parametric, non-uniform intensity normalization is performed on the MRI image.
Registration: A transform matrix to Talairach space is calculated for later steps using a twelve degrees of freedom affine transformation.
Intensity normalization: Fluctuations in scan intensity are corrected and scan intensities are collectively adjusted to achieve a mean white matter intensity of 110.
Skull Stripping: The skull and meningeal surfaces are removed from the scan, leaving only the brain and overlying pial surface.
Registration: A transform matrix to align the patient volume with the FreeSurfer atlas is calculated for use when applying segmentation labels.
Labeling: Final volume labels are applied to subcortical structures based on the prior probabilities of voxel identity assigned by the atlas in addition to the probability of voxel identity based on the tissue class assignment of surrounding voxels, and volumetric statistics are computed.
In order to make spatial comparisons with IBASPM output, FreeSurfer images were resampled to the axial space with a voxel size of 0.977 × 0.977 × 1.300 mm using the FreeSurfer tkregister2 and mri_label2vol functions, as recommended by the program creators and utilized in previous studies (Klauschen et al. 2009, Morey et al. 2009). Volume statistics were obtained using the FreeSurfer mri_segstats function.
FreeSurfer and its documentation can be freely downloaded at http://surfer.nmr.mgh.harvard.edu.
IBASPM
Mri_convert (see above) was used to convert scans into the ANALYZE format (.img) to ensure compatibility with IBASPM. The resulting images were visually inspected to confirm proper orientation and the absence of warping. Images were then processed using the IBASPM toolbox (Cuban Neuroscience Center, Havana, Cuba) functions of SPM5b (Wellcome Department of Cognitive Neurology, University College, London, UK), implemented in MATLAB 7.5 (Mathworks, Natick, MA) on Mac Pro machines running OS 10.5. Using the parameters of this study, IBASPM (Individual Brain Atlases using Statistical Parametric Mapping) carries out the following five processes in order, the last three of which operate outside SPM5:
Segmentation: The images are segmented into broad tissue types: CSF, gray matter, and white matter.
Normalization: The T1 image is mapped to stereotaxic MNI space using the ICBM 152 T1 template, minimizing the sum of square differences between images in order to correct for global brain differences while maintaining feature-specific variations in images that are crucial to detecting difference. This produces a spatial transformation matrix that is used in later processing steps.
Labeling: Gray matter voxels identified in the segmentation step are mapped to the inverse of the transformation matrix produced during normalization and aligned to the 116 automatic anatomical label (AAL) structures (Tzourio-Mazoyer et al. 2002). This step ensures that each voxel is labeled exclusively as one structure and that each structure is in the space of the atlas Final volume labels are applied to subcortical structures based on the prior probabilities of voxel identity assigned by the atlas.
Atlasing: The deformation fields calculated during the normalization step are inverted and the Matlab "imfill" function is used to fill holes and isolated points in the final structure volumes.
Volume Statistic: The volume of each identified structure is calculated and descriptive statistics are compiled.
For the purpose of statistical analyses, the output of IBASPM was flipped such that RAS orientation agreed with that of FreeSurfer output and AAM tracings. The IBASPM and SPM5 software packages and their documentation are freely Available at http://www.thomaskoenig.ch/Lester/ibaspm.htm and http://www.fil.ion.ucl.ac.uk/spm/, respectively.
Auto-assisted manual segmentation
AAM segmentation was performed on Mac Pro machines (OS 10.5) using a "semi-automated" method to reduce tracing time. Rather than trace all structures from raw data, we chose to make corrections to the output of a customized version of the FreeSurfer processing pipeline. Modifications were made as follows: after intensity normalization of raw imaging data, FreeSurfer uses a set of predefined pixel intensity ranges that are considered acceptable values for gray matter regions of interest (aseg.mgz file output). Visual inspection of aseg.mgz output resulted in the modification of pixel intensity ranges to be more inclusive of additional pixel intensity values due to the consistent gross underestimation of ROIs examined in this study. These values were incrementally adjusted until visual inspection revealed the fewest amount of errors in segmentation labels.
We chose this output as a starting point in order to minimize the time required to produce a reliable and accurate segmentation while ensuring that the method would not bias the results toward default (i.e. unoptimized) FreeSurfer output and taking advantage of the FreeSurfer visualization and segmentation tools with which our delineators had already developed expertise during previous studies.
From this starting point, manual corrections were made to produce segmentations of the caudate, amygdala, putamen, and hippocampus using anatomical landmarks consistent with previously published protocols for each of these nuclei (Westmoreland and Cretsinger (a, b), Pantel et al. 2000, Pulsipher et al. 2007). To confirm that using optimized FreeSurfer output would not bias manual corrections in favor of default output, corrections were also performed on a subset of ten segmentations from IBASPM as well as from the raw data. Excellent intra-rater, intra-subject reliability was achieved between the fully manual and semi-automated segmentations in all structures (Cronbach’s α > 0.90), as well as between IBASPM- and FreeSurfer-initiated semi-automated segmentations (Cronbach’s α > 0.95), confirming the reliability and validity of the manual corrections regardless of the starting point. Additionally, high segmentation overlap was achieved between manual and AAM tracings, with dice coefficients greater than 0.9 in all structures except the right amygdala (0.86) and sensitivity measures greater than 0.85 in all structures except the left amygdala (0.823) and right amygdala (0.76). To minimize inter-subject variability, one technician oversaw quality control on every tracing for a particular structure (caudate: J.D.; putamen: J.P.; amygdala & hippocampus: T.R.) for which high intra-rater reliability had been achieved (Cronbach's α > 0.92). Raters were blinded to the clinical variables for each subject.
Statistics
While no standard metric for comparing segmentation methods has yet been established, measures of spatial overlap are common in the literature to date. Moreover, our investigation lends itself well to measures of overlap, as we are making comparisons within the feature space of each subject rather than across subjects. In the present study overlap was assessed using the following two metrics:
Dice coefficient, also called kappa index (Archibald et al. 2003, Fischl et al. 2002, Shattuck et al. 2001, Van Leemput et al. 1999, Zaidi et al. 2006), with A representing the AAM segmentation, B representing the automated segmentation, and v(A) or v(B) representing the voxel count, i.e. volume, of a structure from its respective segmentation:
Sensitivity, also called true positive volume fraction (Udupa et al. 2006):
Fully overlapping regions of interest will yield a value of 1.0 for both metrics, with lower values describing less optimal overlap. Overlap measures with AAM segmentations for both automated methods were obtained using Matlab v7.7.0 for the Mac OS.
The absolute volume difference between the output of each method was also calculated. While this measure lacks the ability to characterize positional similarity, it has been shown to be the most sensitive for detecting variances in segmentation results (Zhang 1996). Furthermore, because most studies are based solely on the volume of structures irrespective of their positions, it is important to quantify the variance that may be observed in the results of a study depending on which segmentation procedure is used. While varying forms of this metric have appeared throughout the literature (Fischl et al. 2002, Iosifescu et al. 1997, Morey et al. 2009), for ease of interpretation it was calculated here using the form proposed by Zhang (1996):
The resulting percentage represents the difference between the volumes obtained by AAM tracing and each automated method expressed as a percentage of the AAM obtained volume. Paired Student’s t-tests to assess the significance of differences between automated and AAM segmentations across all of the above metrics were performed using SPSS v16 (SPSS, Inc., Chicago, IL).
Additionally, after checking the normality of the distribution using the Kolmogorov-Smirnov test, Pearson correlations between FreeSurfer/SPM and AAM segmentations were also calculated to assess the degree of association between these measures using SPSS and R v2.9.1 (R Foundation for Statistical Computing, Vienna, Austria). As an adjunct to traditional correlation methods, we used R to construct Bland-Altman plots, which are commonly used in the methodological scientific literature to examine the association between two methods without the underlying assumption that one method is superior to the other. This is accomplished by plotting the difference between the two measures of each case against the mean of these measures. The distribution of the differences between each method and AAM tracing was also plotted in order to reveal any systematic error present in the automated measurements.
As a secondary set of analyses, volumetric data from both automated methods, as well as AAM tracing, was examined in conjunction with several commonly measured clinical markers of HIV disease severity: nadir CD4 count, age, duration of infection, CD4 count at time of image acquisition, AIDS dementia complex (ADC) stage, and plasma viral load (PVL) (Table 1). The rationale for this approach was that an effective automated segmentation method will return results accurate enough to detect relationships with clinical data similar to those that can be detected with AAM tracing. The correlations between continuous variables were estimated using Pearson’s correlation coefficient, with the following adjustments: 1) in order to account for skewed distributions, nadir CD4 and CD4 counts were natural log transformed prior to analyses; 2) ADC stage was treated as ordinal data and a polyserial correlation was used; 3) plasma viral load was dichotomized to account for different assay sensitivity limits across sites and a skewed distribution, and a biserial correlation was used. Results from these correlational analyses were directly compared to one another for each volume/clinical variable comparison using the paired.r function of the R psych package, because it can account for the inherent correlation between the volumetric measurements of the methods being compared.
Table 1.
Common clinical measures of HIV progression.
| Nadir CD4 | Lowest CD4 count since HIV diagnosis (cells/ml) |
| Age | Patient age in years |
| HIV duration | Years since HIV diagnosis |
| CD4 count | CD4 count at time of scan acquisition (cells/ml) |
| ADC stage | AIDS Dementia Complex state (0: normal; 0.5: subclinical; 1–4: mild to end stage) |
| Plasma viral load |
Level of HIV RNA present in plasma (copies/ml) |
Results
Spatial overlap with AAM segmentation
As measured by the dice coefficient, FreeSurfer (FS) segmentations exhibited significantly higher (paired t-test, p < 0.001) mean spatial overlap in all structures (Figure 1). This difference was most pronounced in the right amygdala (FS 0.740 ± 0.071; IBASPM 0.259 ± 0.114) and right hippocampus (FS 0.749 ± 0.069; IBASPM 0.374 ± 0.112). While the difference in dice coefficients was smallest in the right caudate (FS 0.813 ± 0.065; IBASPM 0.721 ± 0.128), the difference was nonetheless significant (p < 0.001).
Figure 1.
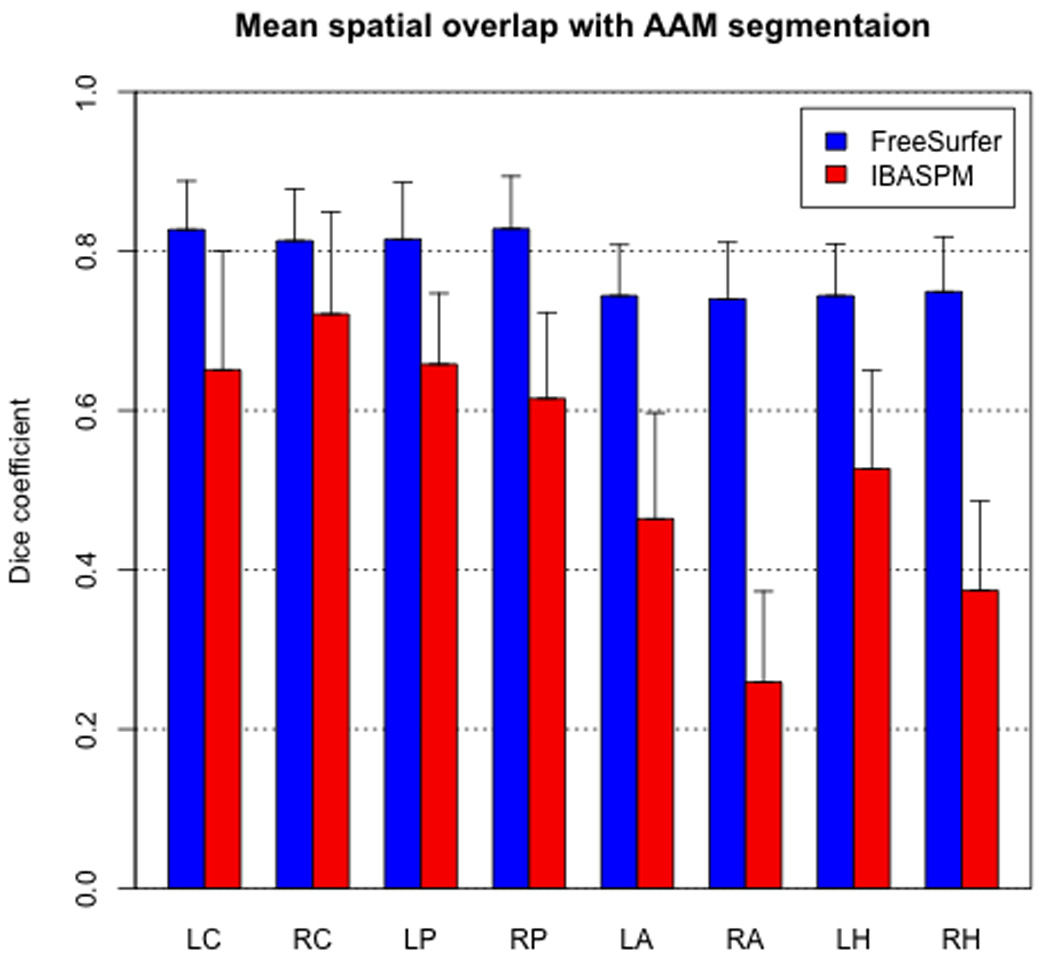
FreeSurfer achieved significantly higher spatial overlap with AAM segmentation in every structure. Error bars indicate one standard deviation.
The mean sensitivity of IBASPM did surpass that of FreeSurfer in the right caudate (FS 0.666 ± 0.060; IBASPM 0.714 ± 0.150; p < 0.001). However, FreeSurfer sensitivity was significantly higher (p < 0.001) in all other structures examined (Figure 2).
Figure 2.
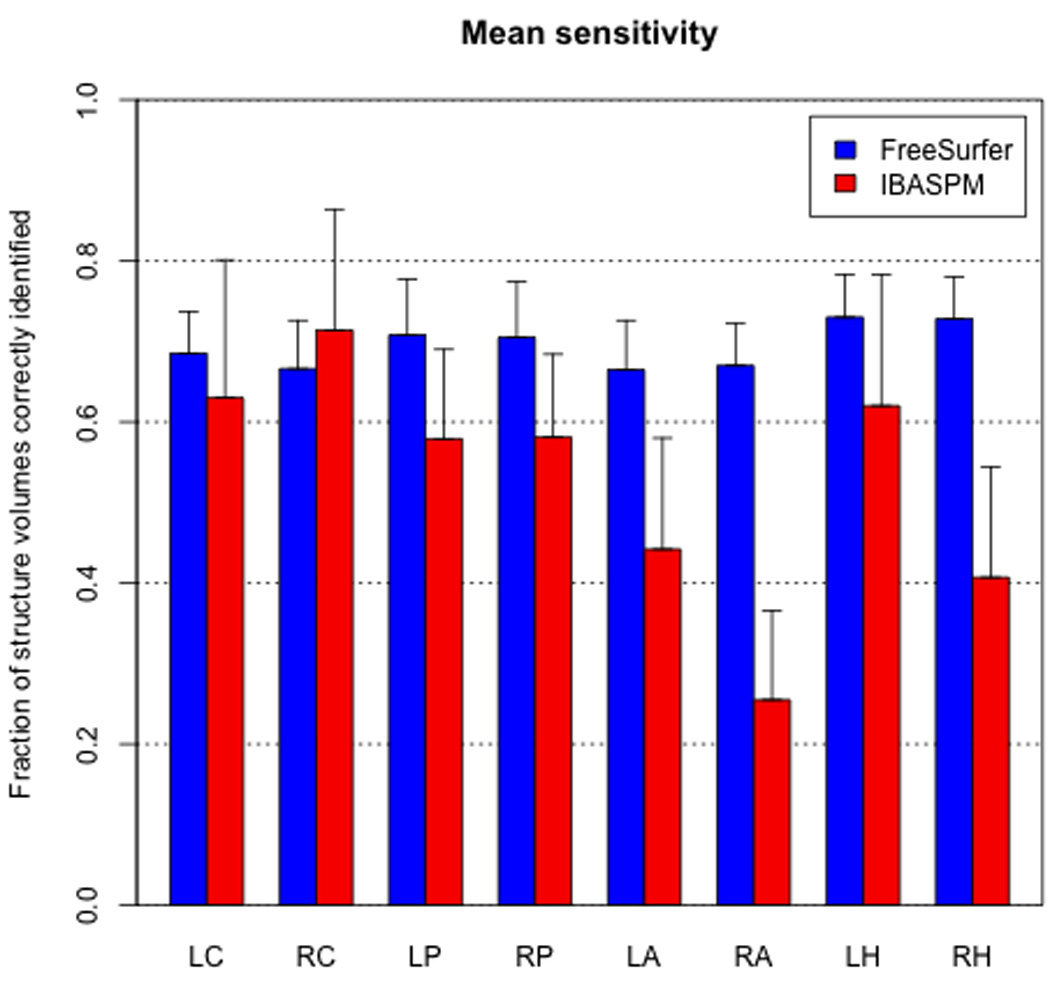
FreeSurfer achieved significantly higher mean sensitivity (i.e., fraction of structure voxels correctly identified) in all structures except the right caudate, in which IBASPM sensitivity was significantly higher. Error bars indicate one standard deviation.
Absolute volume difference relative to AAM measurement
FreeSurfer volumes were significantly closer (paired t-test, p < 0.01) to the AAM segmented results in five of the eight structures examined (Figure 3), with the difference being most prominent in the left caudate (FS 6.76% ± 5.71%; IBASPM 26.05% ± 47.76%) and left putamen (FS 13.99% ± 5.44%; IBASPM 40.48% ± 28.38%). IBASPM volumes were more accurate in the right amygdala and the hippocampus in both hemispheres (p < 0.01), with the largest discrepancy in the right hippocampus (FS 28.64% ± 7.13%; IBASPM 15.43% ± 10.62%). In all cases, these differences were found to be significant (p < 0.01). IBASPM exhibited higher variability in measurement accuracy across all structures.
Figure 3.
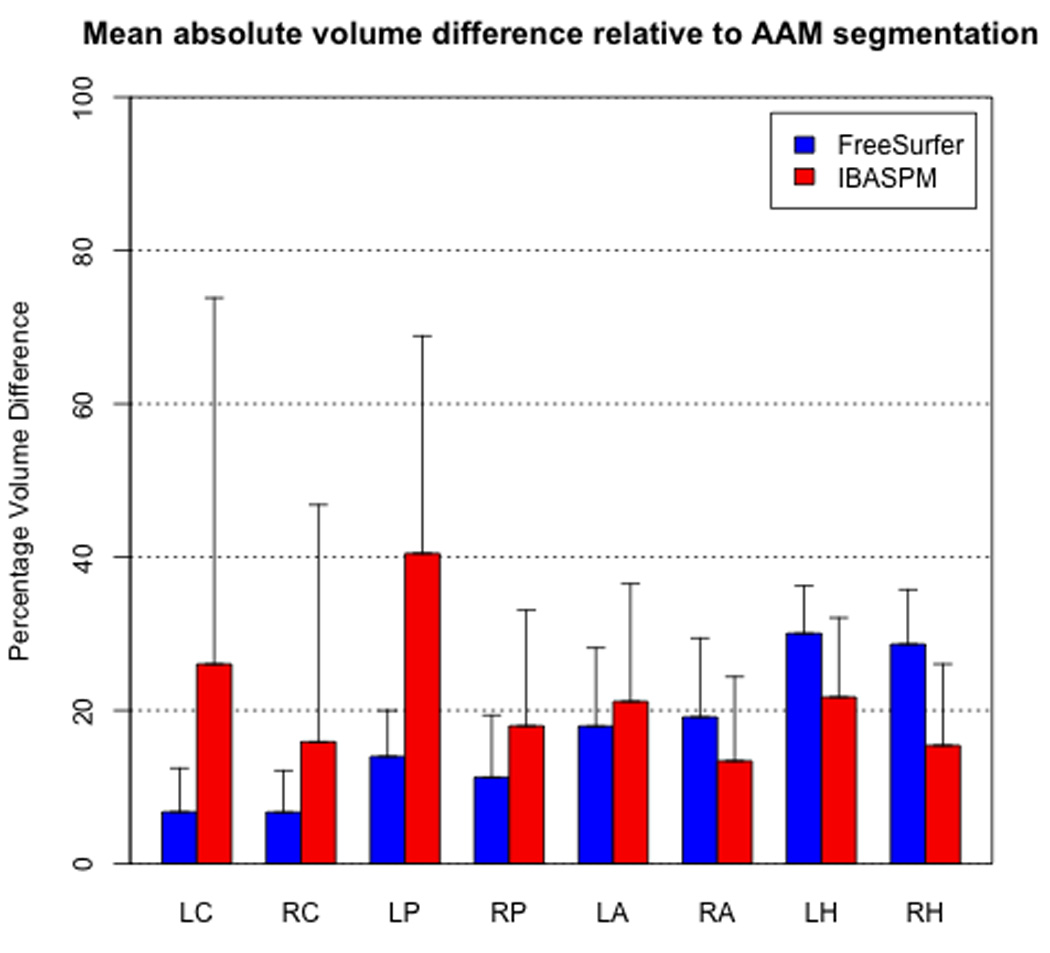
FreeSurfer achieved significantly lower mean absolute volume difference relative to AAM segmentation in five of eight structures, while IBASPM differences were significantly lower in the right amygdala and left/right hippocampus. Error bars indicate one standard deviation.
Consistency of automated methods relative to AAM segmentation
Consistency between FreeSurfer/IBASPM and AAM tracing was measured using Pearson’s rho. The coefficients of FreeSurfer volumes to those of AAM tracing were significantly higher (p<0.003) than those of IBASPM to AAM results across all structures (Figure 4). This difference was particularly apparent in the putamen in both left (FS: 0.874; IBASPM: 0.171) and right (FS: 0.771; IBASPM: 0.364) hemispheres, as well as the right hippocampus (FS: 0.754; IBASPM: 0.345).
Figure 4.
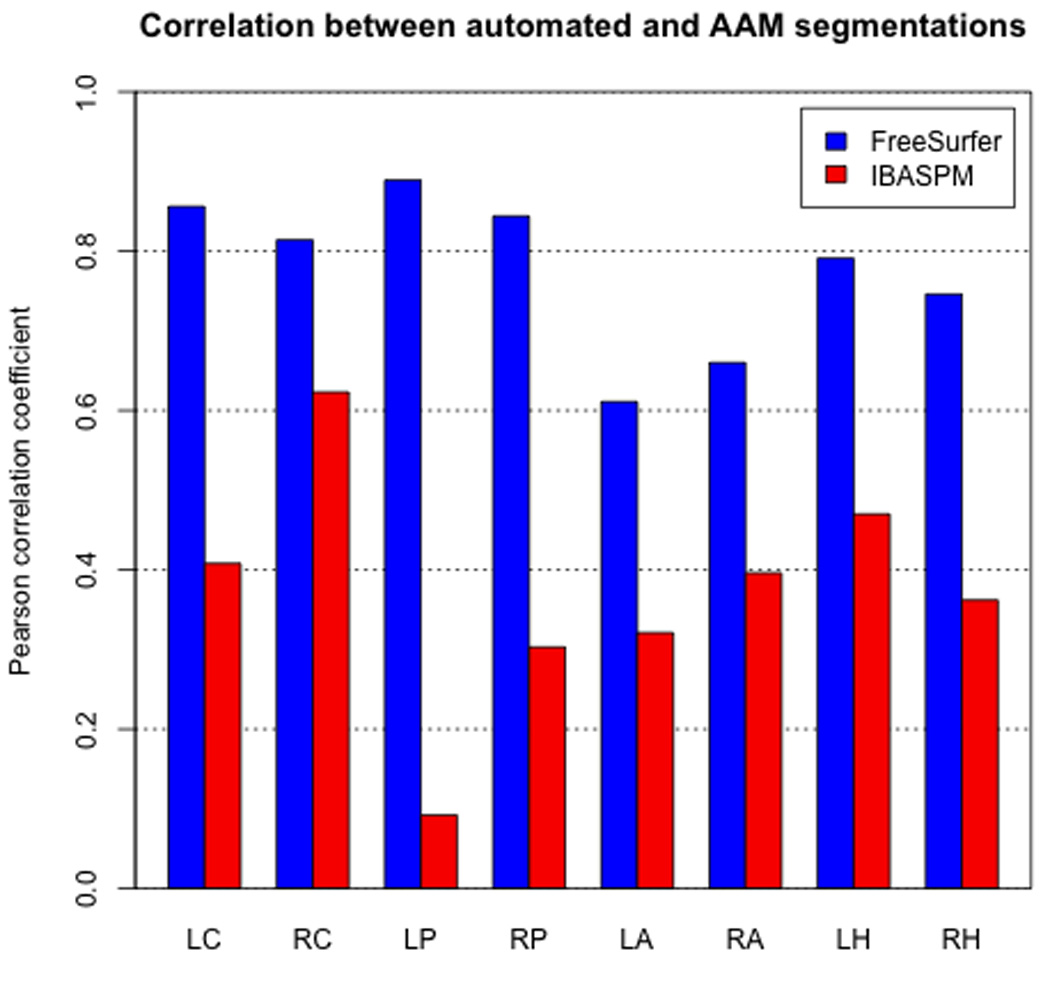
Correlations between FreeSurfer and AAM obtained volumes were significantly higher in all eight structures.
A distribution of volumetric differences, i.e. (automated volume)-(AAM volume), across structures demonstrated that FreeSurfer systematically overestimated the volume of the hippocampus, amygdala, and to a lesser extent the putamen (Figure 5). While IBASPM appears to have underestimated the caudate and putamen and overestimated the hippocampus on average, it is difficult to consider this systematic due to the wide variation of the volumetric error in these structures. A notable exception is the right amygdala, in which IBASPM exhibited a similar degree of variability to FreeSurfer (FS: ±203 mm3; IBASPM: ±212 mm3) while achieving a smaller mean difference in measurement relative to AAM tracing. An examination of Bland-Altman plots revealed that the amount or direction of error did not vary systematically with the volume of the structure except the putamen as measured by FreeSurfer and the amygdala as measured by IBASPM.
Figure 5.
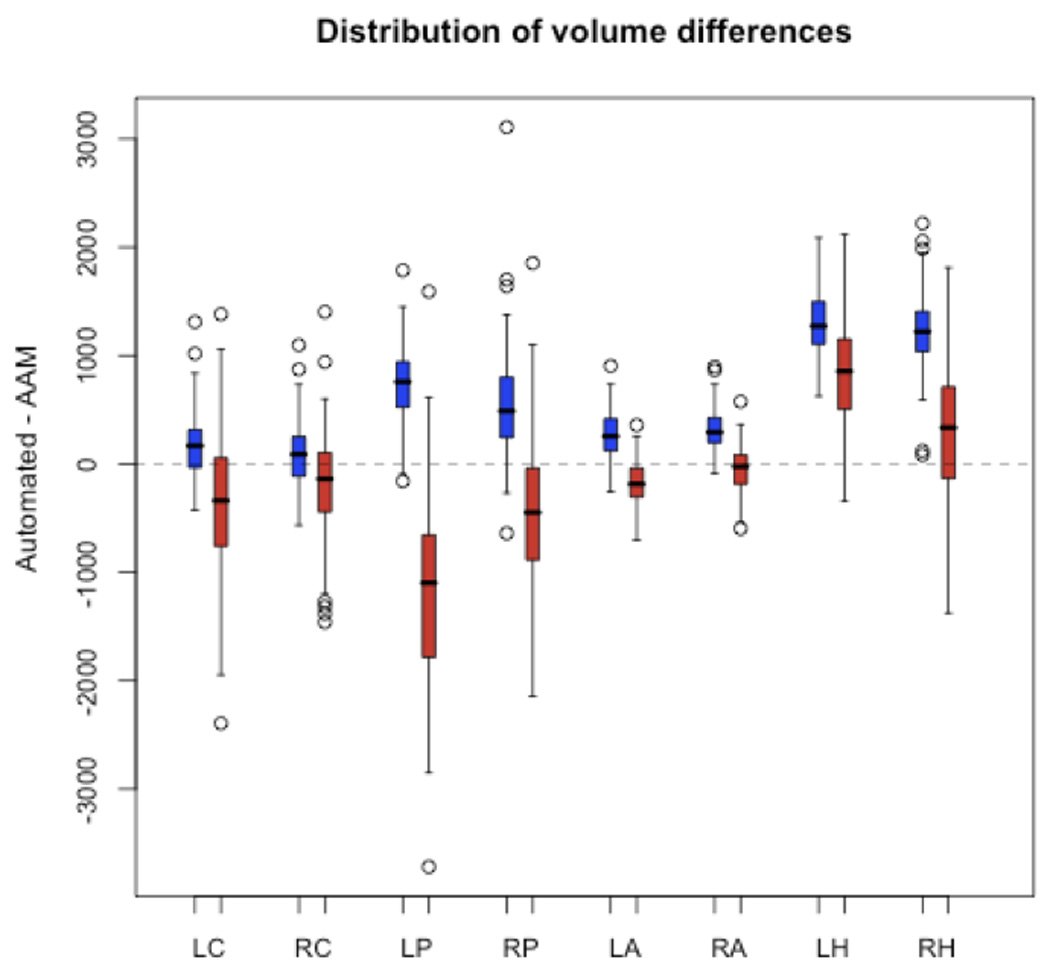
Distributions of volumetric error (v(AAM)-v(automated)) reveal that FreeSurfer systematically overestimated the volumes of the hippocampus and, to a lesser extent, the putamen and amygdala. IBASPM volumetric error was more variable in all structures except the amygdala, and only in the left putamen and left hippocampus can a trend toward systematic error be seen.
Correlation with clinical data
AAM-obtained segmentations demonstrated four statistically significant (p<0.05) correlations between the nuclei volumes and clinical measures of disease severity (Table 2). Relative to the AAM data set, two common and two unique correlations with clinical variables were identified by FreeSurfer while IBASPM identified one common correlation and four unique correlations. FreeSurfer and IBASPM did not identify any significant correlations in common (Table 3a).
Table 2.
Correlations between structure volumes and clinical measures of HIV progression. Bold offset indicates significant correlation (p<0.05). Highlights indicate a correlation significantly different from AAM in that comparison. Red highlight denotes a case in which one of the three methods yielded a significant correlation and the results this comparison would have varied depending on the method used. For example, HIV Duration vs. right putamen volume: FreeSurfer data alone yielded a significant correlation between these variables; the correlation found with AAM data was not significantly different from the FreeSurfer correlation, and thus these methods would have yielded similar results. The IBASPM correlation was significantly different from both correlations, and thus would yield different results. Grey highlight denotes a case where one method's correlation was significantly different from AAM, but no significant correlations were present, and thus the reported results would have been similar regardless of the method used.
| LC | RC | LP | RP | LA | RA | LB | RII | ||
|---|---|---|---|---|---|---|---|---|---|
| AAM | −0.058 | −0.105 | −0.079 | −0.093 | 0.084 | 0.061 | −0.010 | −0.010 | |
| Nadir CD4‡ | FreeSurfer | 0.005 | −0.082 | −0.065 | −0.123 | 0.100 | 0.041 | 0.072 | 0.091 |
| IBASPM | 0.019 | −0.032 | 0.044 | −0.024 | 0.165 | 0.086 | 0.224 | 0.210 | |
| AAM | 0.034 | 0.023 | −0.102 | −0.158 | 0.130 | −0.119 | −0.029 | 0.073 | |
| Age | FreeSurfer | 0.065 | 0.059 | −0.151 | −0.147 | −0.089 | −0.106 | −0.037 | −0.087 |
| IBASPM | 0.121 | 0.052 | 0.039 | 0.008 | 0.103 | 0.081 | 0.013 | −0.067 | |
| AAM | 0.015 | 0.027 | −0.165 | −0.173 | 0.170 | 0.066 | 0.060 | 0.083 | |
| HIV Duration | FreeSurfer | 0.014 | 0.028 | −0.183 | −0.206 | 0.065 | 0.089 | 0.117 | 0.113 |
| IBASPM | −0.063 | −0.084 | 0.059 | 0.062 | 0.184 | 0.125 | 0.157 | 0.068 | |
| AAM | 0.054 | −0.012 | −0.037 | −0.005 | 0.103 | −0.007 | 0.116 | 0.104 | |
| CD4 Count‡ | FreeSurfer | 0.094 | 0.076 | −0.04 | −0.084 | 0.000 | 0.017 | 0.161 | 0.073 |
| IBASPM | −0.021 | 0.021 | −0.002 | −0.053 | 0.101 | −0.044 | 0.035 | −0.039 | |
| AAM | −0.094 | −0.146 | −0.221 | −0.271 | −0.174 | −0.242 | −0.249 | −0.239 | |
| ADC Stage* | FreeSurfer | −0.084 | −0209 | −0.22 | −0.214 | −0.181 | −0.214 | −0.163 | −0.170 |
| IBASPM | −0.208 | −0.186 | −0.23 | −0.256 | 0.169 | −0.011 | −0.138 | −0.184 | |
| AAM | −0.325 | −0.261 | −0.069 | 0.022 | −0.277 | −0.201 | −0.06 | −0.157 | |
| Plasma VL** | FreeSurfer | −0.274 | −0.269 | −0.092 | −0.013 | −0.046 | −0.046 | 0.063 | −0.008 |
| IBASPM | −0.205 | −0.133 | −0.055 | −0.039 | 0.08 | 0201 | 0.194 | 0.286 | |
Clinical data natural log transformed;
polyserial correlation calculated;
biserial correlation calculated.
Table 3.
Survey of agreement between AAM and automated segmentation when correlated with clinical measures of HIV severity, defined by (a) both correlations achieving significance in a given comparison, e.g. AAM and IBASPM when correlating ADC stage with right putamen volumes; or (b) the magnitude of the two correlations being similar to one another (i.e. not statistically different) when at least one method achieved significance, e.g. AAM and FreeSurfer in the above comparison.
| (a) | |||
|---|---|---|---|
| AAM | FS | IBASPM | |
| AAM | 4 | 2 | 1 |
| FS | 2 | 4 | 0 |
| IBASPM | 1 | 0 | 5 |
| (b) | |||
|---|---|---|---|
| AAM | FS | IBASPM | |
| AAM | 4 | 4 | 1 |
| FS | 3 | 4 | 1 |
| IBASPM | 3 | 2 | 5 |
Tests for significant differences (p<0.05) between correlations found by the three methods in each comparison are summarized summarized in Table 3b. FreeSurfer correlations with clinical covariates were not significantly different in three of the four significant correlations found in the AAM data. Moreover, the two unique FreeSurfer correlations described in the previous analysis were not significantly higher than those found with AAM data. Similarly, IBASPM correlations were not significantly different from three of the four correlations found with AAM data. However, all four of the correlations identified by IBASPM that were not supported by significant AAM correlations were also significantly higher.
Discussion
Performance characteristics of FreeSurfer and IBASPM
Past validation studies examining automated segmentation methods have varied widely in the measures they have used. The analyses in this study were chosen in an attempt to apply the full range of metrics that have appeared in various combinations in prior publications. Moreover, each metric characterizes a slightly different aspect of segmentation performance and must be considered in relation to one another in order to adequately interpret the results of an analysis. For example, absolute volume difference does not assess spatial overlap, which requires a Dice coefficient. Calculation of sensitivity characterizes the ability of a method to identify true positives, but it does not account for false positives and thus must be interpreted in the context of the Dice coefficient. Additionally, measures of correlation are required to present the direction of error and determine the degree to which this over- or under-estimation is systematic across a group of patients. When these measures are considered as a whole, an accurate picture of segmentation performance can be assembled.
The performances of FreeSurfer and IBASPM were not consistent across the metrics described above. FreeSurfer mean volumetric results were closer to those of AAM tracings in five of eight structures, but FreeSurfer spatial overlap was higher in all structures examined. Furthermore, FreeSurfer displayed higher sensitivity in seven of eight structures. Though these results may seem incompatible, they merely suggest that while FreeSurfer may have greater mean volumetric differences when compared to IBASPM when measuring some structures, it more accurately characterizes the actual shape and location of all the structures examined.
The higher correlation between FreeSurfer and AAM volumes across all structures suggests that the error in FreeSurfer measurements is more predictable in nature, whereas the direction of IBASPM volumetric error may vary widely within a group of patients. A measurement error distribution plot (Figure 5) confirms that FreeSurfer systematically overestimated the volumes of the putamen, amygdala, and hippocampus in both hemispheres. While this trend may also appear to be present in IBASPM measures of the left putamen and left hippocampus, the large variability of measurement error in these structures precludes the error being considered systematic.
This last observation highlights a noteworthy difference in the relative reliability of FreeSurfer and IBASPM measurements. While neither method emerged as categorically superior to the other in measures of accuracy, the standard deviation of FreeSurfer data was lower than that of IBASPM in every comparison. This may be due to the more localized areas of anatomical variance observed in FreeSurfer segmentations across all structures. FreeSurfer tended to overestimate the inferior surface of the caudate head and body, often including the nucleus accumbens and stria terminalis in the caudate segmentation. The vast majority of FreeSurfer putamen overestimation was attributable to expansion of the lateral aspect to include parts of the external capsule and claustrum. The border between the amygdala and hippocampus was frequently erratic in FreeSurfer segmentations, and the tail of the hippocampus was often expanded to include the inferior horn of the lateral ventricles on the lateral aspect and adjacent cortex on the medial aspect. IBASPM error was less systematic, and much of the variation and discrepancy in volumetric measurements and spatial overlap is likely due to the holes and spiny projections observed in all structures, particularly the putamen and hippocampus (Figures 6 and 7).
Figure 6.
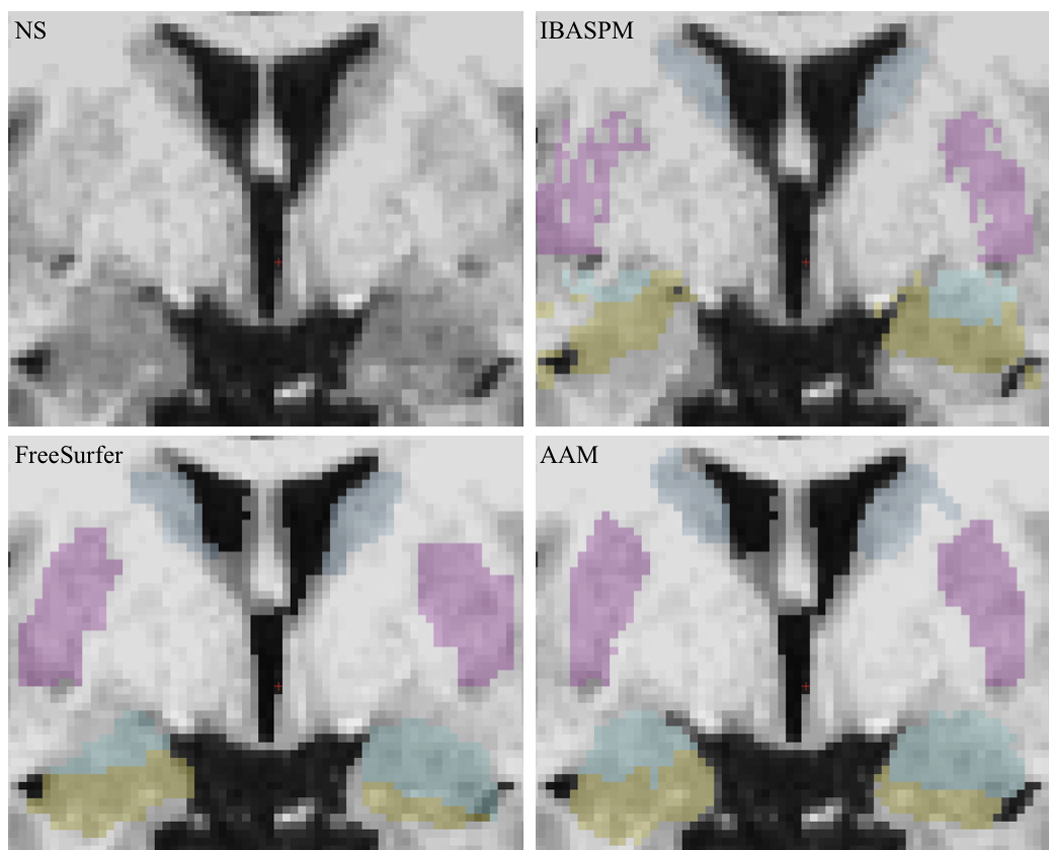
Segmentation screenshots from a randomly selected subject.
Figure 7.
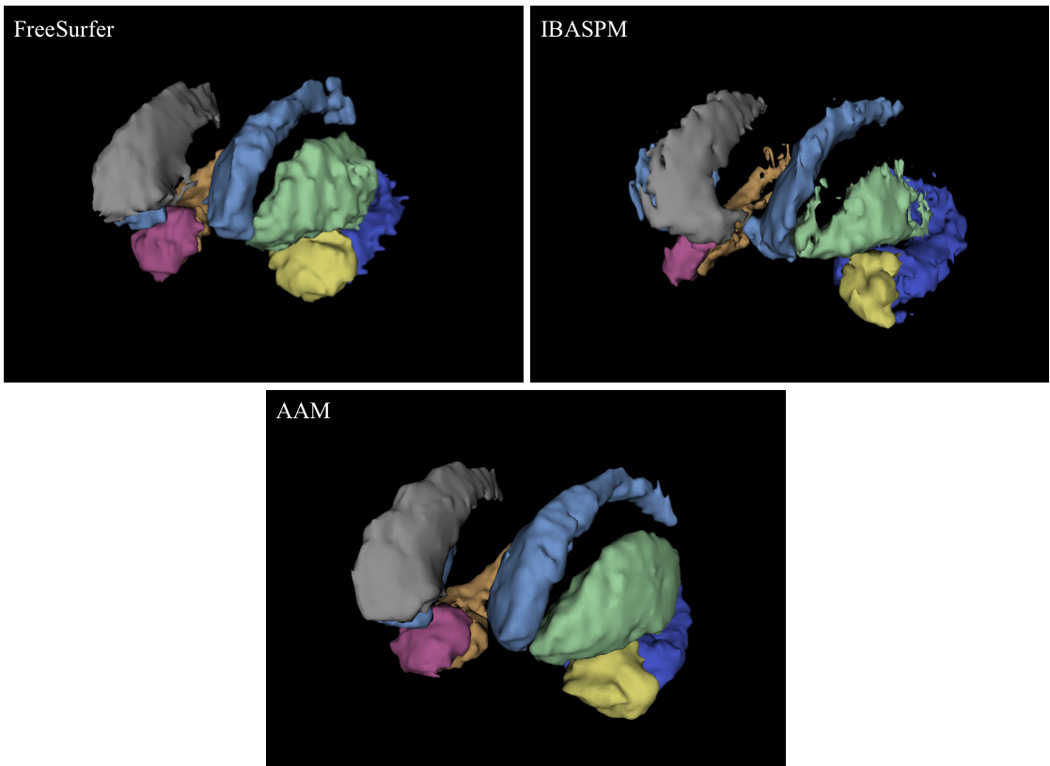
3D models of segmentations from a randomly selected subject. The segmentations produced by both automated methods exhibit rough edges, projections, and even holes in structures that were not present in the AAM segmentation. These errors were particularly prevalent in the hippocampus and putamen. Caudate - light blue/grey; putamen - green/blue; amygdala - yellow/pink; hippocampus - deep blue/orange. Models are in radiological orientation.
While not heavily emphasized in previous validation studies, the variability of error can significantly affect the utility of any automated segmentation method. Data containing systematic errors can still capture trends and may be of some use when correlated with other variables of interest. Moreover, if systematic error is sufficiently consistent, it can even be minimized or eliminated through post-hoc corrections. While this may not be the case for the data presented here, the significantly higher degree of predictability in FreeSurfer error (i.e., systematic over-estimation of volumes) is worth considering when planning investigations that utilize this tool.
Correlation with clinical measures
In our data set, neither FreeSurfer nor IBASPM volumetric data yielded significant correlations with clinical covariates similar to those found using AAM-obtained data (Table 3a). Four significant correlations were found between AAM segmentations and clinical variables, of which FreeSurfer data yielded two and IBASPM data one. Moreover, two additional significant correlations were found in FreeSurfer data, and four in IBASPM data, that were not found using the AAM segmented data set. These findings suggest that, at least in a data set with similar characteristics, the method of volumetric measurement will seriously influence the findings of any analyses attempting to relate volume measures to other relevant covariates, and that FreeSurfer and IBASPM are not equivalent substitutes for AAM segmentation in such studies.
The significance of the correlations described above is influenced by factors that will be unique to each data set, and thus our conclusions may not be generalizable to other studies. For this purpose it is more instructive to consider the degree to which correlations given by the three methods in each comparison differ from one another. For this analysis, we define a method to be similar to another in a given comparison if the correlations yielded by both methods were not significantly different at the 0.05 level in a pair-wise analysis, with the assumption that similar correlations may both achieve significance depending on the sample, presence/exclusion of outliers, etc. When examining comparisons in which AAM-obtained volumes were significantly correlated with a clinical covariate, both FreeSurfer and IBASPM performed equally, yielding similar correlations in three of four cases. However, in the comparisons when the data from either automated method yielded uniquely significant correlations, AAM-obtained data was similarly correlated in both FreeSurfer cases and none of the four IBASPM cases. Moreover, FreeSurfer correlations were significantly different from AAM in five additional cases - and IBASPM in four - where significance was not achieved by any of the three methods. These cases are equally important, as at least one method may disagree under conditions of greater statistical power.
The discrepancies between default FreeSurfer and IBASPM output applied to clinical covariates are noteworthy, as this a common application of automated volumetry in diseased patient cohorts. The presence of significant correlations in such analyses is often taken as de facto proof of their validity, leading to potentially erroneous conclusions. However, there is potential that these discrepancies may be overcome through optimization of the automated processing pipeline. The vast majority of significant differences between FreeSurfer/IBASPM and AAM correlations occurred in the amygdala and hippocampus, which were shown to be the most difficult structures for the automated tools to accurately segment. It has been suggested that the accuracy and reliability of automated segmentation in these structures could be enhanced through refinements such as more accurate sub-cortical registration (Tae et al. 2009). If such modifications could reduce the variability of measurement error alone, the ability of an automated tool to accurately characterize relationships with clinical covariates could be vastly improved. Regardless, these findings highlight the importance of visually inspecting automated segmentation output, even when volumetric data is not being directly assessed or reported.
Concordance with previous publications
Previous studies comparing FreeSurfer output with manual tracing have focused primarily on the hippocampus, with one group addressing the amygdala as well; moreover, no study has yet been performed using an HIV-infected cohort, not to mention multi-site data in this disease context. However, our findings generally agree with those of previous publications (Table 4). The two notable exceptions are the measures of absolute volume difference reported in Morey et al. (2009) in both the amygdala and hippocampus, which they found to be much smaller than those reported here. One contributing factor is the formula used to calculate this metric; while Morey et al. used the average of FreeSurfer and manual volumes as their denominator, we chose to divide by the AAM traced volume alone in order to yield descriptives that were easily interpreted as the percentage of the AAM traced structure that was over- or under-estimated; using our formula, the numbers reported by Morey et al. would have been larger. Additional factors include scan resolution (3T in Morey et al. versus 1.5T here) and disease specific factors affecting structure volumes in our patient population, as Morey et al. examined healthy patients.
Table 4.
Results of previous FreeSurfer validation studies in the hippocampus (a) and the amygdala (b), as well as IBASPM in the hippocampus (c), compared with findings presented here. LH - left hippocampus; RH - right hippocampus. Standard deviation of results included where available.
| Dice Coefificient | Abs. Volume Difference | ||||
|---|---|---|---|---|---|
| LH | RH | LH | RH | ||
| Present study | 0.744 (±0.065) | 0.749 (±0.069) | 30.05% (±6.17) | 28.64% (±7.13) | |
| (a) | Morey et al. | 0.82 (±0.015) | 0.82 (±0.028) | 7% (±3.0) | 9% (±2.7) |
| Tae et al.* | - | - | 40% | 35% | |
| Cherbuin et al. | - | - | 23% | 29% | |
| (b) | Present study | 0.744 (±0.064) | 0.744 (±0.071) | 17.95% (±10.20) | 19.14% (±10.30) |
| Morey et al. | 0.75 (±0.032) | 0.72 (±0.040) | 7% (±3.0) | 9% (±2.7) | |
| (c) | Present study | 0.527 (±0.123) | 0.374 (±0.112) | 21.74% (±10.35) | 15.43% (±10.61) |
| Tae et al* | - | - | 21% | 2% | |
Percentages from Tae et al. 2008 are calculated based on mean FreeSurfer/IBASPM and manually obtained volume voxel counts using our formula for absolute volume difference described above, and are therefore approximate.
Relatively fewer studies have examined IBASPM performance in subcortical structures. Tae et al. (2008) investigated both FreeSurfer and IBASPM hippocampal measurements in chronic Major Depressive Disorder patients. Similar to our results, they found IBASPM errors in hippocampal measurements to be smaller on average than those of FreeSurfer but more widely distributed. The FreeSurfer volumetric errors reported were similar in magnitude to those presented here (Table 4). The authors cite a high frequency of registration errors in the hippocampus as a potential source of the errors in IBASPM output, which based on our own visual inspection may likely pertain to the discrepancies observed in the present study as well.
Previous studies have also utilized shape-based analyses to precisely characterize the localization of discrepancies between automated methods and manual tracing, though these have only addressed FreeSurfer. Morey et al. (2009) found that the majority of FreeSurfer hippocampal overestimation in their results was localized to the anterior-medial surface, with additional overestimation occurring to a lesser degree along the tail region. This latter finding is supported by Shen et al. (2009), who found that FreeSurfer-segmented hippocampi had larger tails with more erratic surfaces. In the amygdala, Morey et al. found that FreeSurfer expanded the anterior and posterio-lateral surfaces, which accounted for the majority of overestimation. The results of both studies agree with the error localization we observed when inspecting FreeSurfer output as described above.
Optimization of automated volumetry
In spite of significant discrepancies in performance, FreeSurfer and IBASPM both hold great promise. For this study, both packages were implemented using the default settings recommended by their developers. However, a number of options are available to customize and optimize these techniques. Through active email forums and online wikis (FreeSurfer: http://surfer.nmr.mgh.harvard.edu/fswiki; IBASPM: https://www.jiscmail.ac.uk/cgi-bin/discuss.cgi?LMGT1=SPM), users can seek the advice of other users, as well as the program creators, and alert the community to potential pitfalls of each program. Additionally, the creators of both packages can be contacted directly for assistance in optimizing each routine for the needs of an individual study. For instance, in order to create the starting point for the semi-automated segmentations used in this study, we utilized a pipeline specifically modified by FreeSurfer’s creators to be more robust at detecting the putamen given the characteristics of our data. Other authors have suggested that improvement of the registration methods employed by both programs to be more accurate in sub-cortical regions may drastically improve automated segmentations (Tae et al. 2008, Tae et al. 2009). Other optimizations may address the reference atlas underlying automated segmentation decisions; as a follow-up to the present study, we are currently pursuing approaches to create patient- and population-specific atlases for prospective studies using FreeSurfer.
The results presented here underscore the importance of optimization when using FreeSurfer or IBASPM in volumetric studies. With careful modification, higher levels of accuracy can be achieved with automated volumetric packages (Han and Fischl 2007, Yeo et al. 2008). In spite of these improvements, users are still responsible for manually inspecting the output of either package to ensure that accurate and reliable results are being obtained and correcting mistakes in automated segmentations as necessary. Such intervention is particularly critical in studies attempting to correlate volumetric data with clinical measures, as the present results have demonstrated the potential for unique trends to appear when using unedited volumetric data from either software package. Recently, Beutner et al. described a Markov Chain Monte Carlo approach for estimating uncertainty in brain region segmentation obtained from fully automated methods. In synthetic and real data sets, their method reported higher uncertainty for images with lower quality or pathology, both of which negatively affect automated segmentation performance (Beutner et al. 2009). This method may provide an additional tool for researchers to identify inaccurate segmentations and streamline the process of manual data inspection in large cohorts.
The procedure used for AAM segmentation in the present study is one example of how the power of automated volumetry can be harnessed while still ensuring accuracy via post-processing manual intervention. While we demonstrated high reliability relative to manual traces from raw, unsegmented data, we were able to arrive at these segmentations in a fraction of the time. For instance, while manual segmentation from scratch can require up to two hours per structure per hemisphere, tracing times in our study averaged twenty to thirty minutes, due in large part to processing optimizations such as customized atlases that we will be reporting in an upcoming publication.
Limitations
A potential shortcoming of the present study is the dependence on accurate AAM tracings. Variance is naturally introduced when a patient group of this magnitude is manually corrected, especially in structures with poorly defined borders, such as the hippocampus and amygdala. To control this variance to the greatest possible extent, the tracing of each structure was overseen by a trained expert (caudate - JD, putamen - JP, amygdala/hippocampus - TR) who had demonstrated reliability when re-tracing a subset of patient scans (Cronbach’s α > 0.92). The anatomical boundaries used for tracing were based on previously described protocols (Westmoreland and Cretsinger (a, b), Pantel et al. 2000, Pulsipher et al. 2007), and the level of intra-rater reliability achieved was equal to or greater than that described in prior publications (Mori et al. 2008, Tae et al. 2008, Wu et al. 2006). Moreover, our AAM-obtained volumes agree well with those described in previous publications. For instance, Cherbuin et al. (2009) reported average manual measurements of 2992 mm3 (± 335) and 3068 mm3 (± 340) in the left and right hippocampus, respectively, as compared to 3006 mm3 (± 420) and 3079 mm3 (± 459) in our data. We attribute the increased variability in our measurements to a number of potential factors due to HIV neuropathology and/or multisite acquisition, including a greater range of structure volumes and blurring of grey/white matter boundaries.
Additionally, the conversion of AAM and FreeSurfer segmentation from coronal to axial space resulted in slight alteration of the segmentations due to the resampling procedure for computing spatial overlap. However, this is currently the only way to make spatial comparisons between the output of these two programs. The conversion tools used for this step replicate previously published methods (Klauschen et al. 2009, Morey et al. 2009) and were recommended by the FreeSurfer developers in public FreeSurfer forums.
Conclusion
The goal of this study was to characterize the performance of two widely used automated brain MRI-based volumetry packages in a manner that would significantly augment the existing literature by expanding the scope of the analyses to include additional structures within a unique patient cohort. To our knowledge, this is the first paper addressing reliability and validity of automated subcortical volumetry in the context of HIV. Although one previous publication has addressed automated volumetry in HIV patients, these investigators focused exclusively on the ventricles and did not utilize FreeSurfer of SPM (Carmichael et al 2007). Our data set is among the largest to date when comparing these automated methods, second only to that of Cherbuin et al. (2009), though we broaden their findings by including a wider range of structures. Moreover, the fact that our data were acquired at multiple sites across the United States suggests factors affecting inter-site variance (i.e., small differences in scan parameters, scanner upgrades, etc) inherent in large cohort studies (i.e., Alzheimer’s Disease Neuroimaging Initiative, drug studies) may impact the performance of automated volumetric methods. Finally, we expanded our analyses from those of other studies to include four important subcortical structures and used a wide range of metrics to fully capture the accuracy and reliability of the automated packages examined.
Our results demonstrate that default (i.e. unoptimized) subcortical volumetry with FreeSurfer or SPM may be prone to significant errors when presented with data collected from multiple sites or a diseased patient population. In order to ensure accurate analyses, it is imperative that researchers employing either of these tools manually inspect the output and pursue opportunities to optimize the routines as necessary for their particular research context. Furthermore, as suggested by the developers of these tools, the full range of parameters implemented must be specifically reported to ensure credibility and reproducibility of results and to guide further development of the segmentation package being used.
Future Directions
Though FreeSurfer and IBASPM are intended to yield similar segmentations and volumetric results, the underlying algorithms are different in many respects. A detailed analysis of the differences in processing pipelines, such as tracing protocols used for atlas creation, atlas patient composition, registration algorithms, and statistical application of the prior probabilities represented in the reference atlases, would help to elucidate the source of the discrepancies reported here and in prior publications. These investigations may also open new avenues to the development of optimizations for these packages. As both tools are continually improved and techniques in the field of atlas-based segmentation progress, additional studies examining the reliability and validity of these methods will no doubt be needed until a truly robust, accurate, and efficient method emerges.
Acknowledgments
We greatly acknowledge the following HIV Neuroimaging Consortium sites for the data used in this study: Stanford University, University of California Los Angeles, UCLA Harbor, University of California San Diego, University of Colorado, University of Pittsburgh, University of Rochester. We also acknowledge the support of the following funding sources: R01 NS036524 and K23 MH073416.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alemán-Gómez Y, Melie-García L, Valdés-Hernandez P. IBASPM: Toolbox for automatic parcellation of brain structures. Presented at the 12th Annual Meeting of the Organization for Human Brain Mapping; June 11–15, 2006; Florence, Italy: Available on CD-Rom in NeuroImage; 2006. [Google Scholar]
- Andersen AH, Zhang Z, Avison MJ, Gash DM. Automated segmentation of multispectral brain MR images. J Neurosci Methods. 2002;122:13–23. doi: 10.1016/s0165-0270(02)00273-x. [DOI] [PubMed] [Google Scholar]
- Archibald SL, Masliah E, Fennema-Notestine C, Marcotte TD, Ellis RJ, McCutchan JA, Heaton RK, Grant I, Mallory M, Miller A, Jernigan TL. Correlation of in vivo neuroimaging abnormalities with postmortem human immunodeficiency virus encephalitis and dendritic loss. Arch Neurol. 2004;61:369–376. doi: 10.1001/archneur.61.3.369. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston K. Multimodal image coregistration and partitioning--a unified framework. Neuroimage. 1997;6:209–217. doi: 10.1006/nimg.1997.0290. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Andersson JL, Friston KJ. High-dimensional image registration using symmetric priors. Neuroimage. 1999;9:619–628. doi: 10.1006/nimg.1999.0437. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Beutner KR, Prasad G, Fletcher E, DeCarli C, Carmichael OT. Estimating uncertainty in brain region delineations. Information Processing in Medical Imaging. In Lecture Notes in Computer Science. 2009;5636:479–490. doi: 10.1007/978-3-642-02498-6_40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman AM, Schupf N, Manly JJ, Luchsinger JA, Andrews H, Tang MX, Reitz C, Small SA, Mayeux R, DeCarli C, Brown TR. Brain morphology in older african americans, caribbean hispanics, and whites from northern manhattan. Archives of Neurology. 2008;65:1053–1061. doi: 10.1001/archneur.65.8.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael OT, Aizenstein HA, Davis SW, Becker JT, Thompson PM, Meltzer CC, Liu Y. Atlas-Based hippocampus segmentation in alzheimer's disease and mild cognitive impairment. Neuroimage. 2005;27:979–990. doi: 10.1016/j.neuroimage.2005.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael OT, Kuller LH, Lopez OL, Thompson PM, Dutton RA, Lu A, Lee SE, Lee JY, Aizenstein HJ, Meltzer CC, Liu Y, Toga AW, Becker JT. Cerebral ventricular changes associated with transitions between normal cognitive function, mild cognitive impairment, and dementia. Alzheimer Disease & Associated Disorders. 2007;21:14–24. doi: 10.1097/WAD.0b013e318032d2b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbuin N, Anstey KJ, Réglade-Meslin C, Sachdev PS, Greenlee MW. In vivo hippocampal measurement and memory: A comparison of manual tracing and automated segmentation in a large community-based sample. PLoS ONE. 2009;4:e5265. doi: 10.1371/journal.pone.0005265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csapo I, Price J, Russell T, Dewey J, Sem E, McCaffrey D, Guttman CR, Navia B, Tate DF. Effect of patient population specific atlases on automatic segmentation of subcortical structures in freesurfer [Abstract]. Proceedings of the International Society for Magnetic Resonance in Medicine, 17Th Meeting and Scientific Exhibition.2009. [Google Scholar]
- Csernansky JG, Joshi S, Wang L, Haller JW, Gado M, Miller JP, Grenander U, Miller MI. Hippocampal morphometry in schizophrenia by high dimensional brain mapping. Proc Natl Acad Sci USA. 1998;95:11406–11411. doi: 10.1073/pnas.95.19.11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elderkin-Thompson V, Ballmaier M, Hellemann G, Pham D, Kumar A. Executive function and MRI prefrontal volumes among healthy older adults. Neuropsychology. 2008;22:626–637. doi: 10.1037/0894-4105.22.5.626. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- FreeSurfer. [Accessed 9/15/2008]; Availabile at http://surfer.nmr.mgh.harvard.edu/.
- FreeSurfer Wiki. [Accessed 10/05/2008]; Available at http://surfer.nmr.mgh.harvard.edu/fswiki.
- González-Scarano F, Martín-García J. The neuropathogenesis of AIDS. Nat Rev Immunol. 2005;5:69–81. doi: 10.1038/nri1527. [DOI] [PubMed] [Google Scholar]
- Hall M, Whaley R, Robertson K, Hamby S, Wilkins J, Hall C. The correlation between neuropsychological and neuroanatomic changes over time in asymptomatic and symptomatic HIV-1-infected individuals. Neurology. 1996;46:1697–1702. doi: 10.1212/wnl.46.6.1697. [DOI] [PubMed] [Google Scholar]
- Han X, Fischl B. Atlas renormalization for improved brain MR image segmentation across scanner platforms. IEEE Trans Med Imaging. 2007;26:479–486. doi: 10.1109/TMI.2007.893282. [DOI] [PubMed] [Google Scholar]
- Heckemann RA, Hajnal JV, Aljabar P, Rueckert D, Hammers A. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuroimage. 2006;33:115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- Hickie I, Naismith S, Ward PB, Turner K, Scott E, Mitchell P, Wilhelm K, Gordon P. Reduced hippocampal volumes and memory loss in patients with early- and late-onset depression. Br J Psychiatry. 2005;186:197–202. doi: 10.1192/bjp.186.3.197. [DOI] [PubMed] [Google Scholar]
- IBASPM. [Accessed 11/06/2008];Individual Brain Atlases using Statistical Parametric Mapping. Available at http://www.thomaskoenig.ch/Lester/ibaspm.htm.
- Iosifescu DV, Shenton ME, Warfield SK, Kikinis R, Dengler J, Jolesz FA, McCarley RW. An automated registration algorithm for measuring MRI subcortical brain structures. Neuroimage. 1997;6:13–25. doi: 10.1006/nimg.1997.0274. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, Bentley MD, Twomey CK, Zinsmeister AR. MR imaging-based volume measurements of the hippocampal formation and anterior temporal lobe: Validation studies. Radiology. 1990;176:205–209. doi: 10.1148/radiology.176.1.2353093. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, Theodore WH, Cook M, McCarthy G. MRI-based hippocampal volumetrics: Data acquisition, normal ranges, and optimal protocol. Magnetic Resonance Imaging. 1995;13:1057. doi: 10.1016/0730-725x(95)02013-j. [DOI] [PubMed] [Google Scholar]
- Jongen C, Biessels GJ. Structural brain imaging in diabetes: A methodological perspective. European Journal of Pharmacology. 2008;585:208–218. doi: 10.1016/j.ejphar.2007.11.085. [DOI] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Han X, Salat D, van der Kouwe A, Quinn B, Pacheco J, Albert M, Killiany R, Blacker D, Maguire P, Rosas D, Makris N, Gollub R, Dale A, Dickerson BC, Fischl B. MRI-derived measurements of human subcortical, ventricular and intracranial brain volumes: Reliability effects of scan sessions, acquisition sequences, data analyses, scanner upgrade, scanner vendors and field strengths. Neuroimage. 2009;46:177–192. doi: 10.1016/j.neuroimage.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauschen F, Goldman A, Barra V, Meyer-Lindenberg A, Lundervold A. Evaluation of automated brain MR image segmentation and volumetry methods. Hum Brain Mapp. 2009;30:1310–1327. doi: 10.1002/hbm.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarski JZ, McIntyre RS, Kennedy SH, Rafi-Tari S, Soczynska JK, Ketter TA. Volumetric neuroimaging investigations in mood disorders: Bipolar disorder versus major depressive disorder. Bipolar Disorders. 2008;10:1–37. doi: 10.1111/j.1399-5618.2008.00435.x. [DOI] [PubMed] [Google Scholar]
- Liu T, Chu WC, Young G, Li K, Yeung BH, Guo L, Man GC, Lam WM, Wong ST, Cheng JC. MR analysis of regional brain volume in adolescent idiopathic scoliosis: Neurological manifestation of a systemic disease. J Magnetic Resonance Im. 2008;27:732–736. doi: 10.1002/jmri.21321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey RA, Petty CM, Xu Y, Hayes JP, Wagner HR, Lewis DV, LaBar KS, Styner M, McCarthy G. A comparison of automated segmentation and manual tracing for quantifying hippocampal and amygdala volumes. Neuroimage. 2009;45:855–866. doi: 10.1016/j.neuroimage.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Oishi K, Jiang H, Jiang L, Li X, Akhter K, Hua K, Faria AV, Mahmood A, Woods R, Toga AW, Pike GB, Neto PR, Evans A, Zhang J, Huang H, Miller MI, van Zijl P, Mazziotta J. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. Neuroimage. 2008;40:570–582. doi: 10.1016/j.neuroimage.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel J, O’Leary DS, Cretsinger K, Bockholt HJ, Keefe H, Magnotta VA, Andreasen NC. A new method for the in vivo volumetric measurement of the human hippocampus with high neuroanatomical accuracy. Hippocampus. 2000;10:752–758. doi: 10.1002/1098-1063(2000)10:6<752::AID-HIPO1012>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Paul R, Cohen R, Navia B, Tashima K. Relationships between cognition and structural neuroimaging findings in adults with human immunodeficiency virus type-1. Neurosci Biobehav Rev. 2002;26:353–359. doi: 10.1016/s0149-7634(02)00006-4. [DOI] [PubMed] [Google Scholar]
- Perantie DC, Wu J, Koller JM, Lim A, Warren SL, Black KJ, Sadler M, White NH, Hershey T. Regional brain volume differences associated with hyperglycemia and severe hypoglycemia in youth with type 1 diabetes. Diabetes Care. 2007;30:2331–2337. doi: 10.2337/dc07-0351. [DOI] [PubMed] [Google Scholar]
- Powell S, Magnotta VA, Johnson H, Jammalamadaka VK, Pierson R, Andreasen NC. Registration and machine learning-based automated segmentation of subcortical and cerebellar brain structures. Neuroimage. 2008;39:238–247. doi: 10.1016/j.neuroimage.2007.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulsipher DT, Seidenberg M, Morton JJ, Geary E, Parrish J, Hermann B. MRI volume loss of subcortical structures in unilateral temporal lobe epilepsy. Epilepsy Behav. 2007;11:442–449. doi: 10.1016/j.yebeh.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragin AB, Wu Y, Storey P, Cohen BA, Edelman RR, Epstein LG. Diffusion tensor imaging of subcortical brain injury in patients infected with human immunodeficiency virus. J Neurovirol. 2005;11:292–298. doi: 10.1080/13550280590953799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KR, Smurzynski M, Parsons TD, Wu K, Bosch RJ, Wu J, McArthur JC, Collier AC, Evans SR, Ellis RJ. The prevalence and incidence of neurocognitive impairment in the HAART era. AIDS. 2007;21:1915–1921. doi: 10.1097/QAD.0b013e32828e4e27. [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Mirza M, Adisetiyo V, Hojatkashani C, Salamon G, Narr KL, Poldrack RA, Bilder RM, Toga AW. Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage. 2008;39:1064–1080. doi: 10.1016/j.neuroimage.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Firpi HA, Saykin AJ, West JD. Parametric surface modeling and registration for comparison of manual and automated segmentation of the hippocampus. Hippocampus. 2009;19:588–595. doi: 10.1002/hipo.20613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPM Forums. [Accessed 11/06/2008]; Available at https://www.jiscmail.ac.uk/cgi-bin/discuss.cgi?LMGT1=SPM.
- Sporer B, Linke R, Seelos K, Paul R, Klopstock T, Pfister HW. HIV-induced chorea: evidence for basal ganglia dysregulation by SPECT. J Neurol. 2005;252:356–358. doi: 10.1007/s00415-005-0626-1. [DOI] [PubMed] [Google Scholar]
- Stout JC, Ellis RJ, Jernigan TL, Archibald SL, Abramson I, Wolfson T, McCutchan JA, Wallace MR, Atkinson JH, Grant I. Progressive cerebral volume loss in human immunodeficiency virus infection: a longitudinal volumetric magnetic resonance imaging study. HIV Neurobehavioral Research Center Group. Arch Neurol. 1998;55:161–168. doi: 10.1001/archneur.55.2.161. [DOI] [PubMed] [Google Scholar]
- Styner M, Lieberman JA, Pantazis D, Gerig G. Boundary and medial shape analysis of the hippocampus in schizophrenia. Medical Image Analysis. 2004;8:197–203. doi: 10.1016/j.media.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Tae WS, Kim SS, Lee KU, Nam E-C, Kim KW. Validation of hippocampal volumes measured using a manual method and two automated methods (FreeSurfer and IBASPM) in chronic major depressive disorder. Neuroradiology. 2008;50:569–581. doi: 10.1007/s00234-008-0383-9. [DOI] [PubMed] [Google Scholar]
- Tae WS, Kim SS, Lee KU, Nam EC, Kim KW. Validation of hippocampal volumes measured with one manual and two automated methods using FreeSurfer and IBASPM in chronic major depressive disorder. Neuroradiology. 2009;51:203–204. doi: 10.1007/s00234-008-0383-9. [DOI] [PubMed] [Google Scholar]
- Teipel SJ, Meindl T, Grinberg L, Heinsen H, Hampel H. Novel MRI techniques in the assessment of dementia. Eur J Nuc Med Mol Im. 2008;35 Suppl 1:S58–S69. doi: 10.1007/s00259-007-0703-z. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Mega MS, Woods RP, Zoumalan CI, Lindshield CJ, Blanton RE, Moussai J, Holmes CJ, Cummings JL, Toga AW. Cortical change in Alzheimer's disease detected with a disease-specific population-based brain atlas. Cereb Cortex. 2001;11:1–16. doi: 10.1093/cercor/11.1.1. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Dutton RA, Hayashi KM, Toga AW, Lopez OL, Aizenstein HJ, Becker JT. Thinning of the cerebral cortex visualized in HIV/AIDS reflects CD4+ T lymphocyte decline. Proc Natl Acad Sci USA. 2005;102:15647–15652. doi: 10.1073/pnas.0502548102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Dutton RA, Hayashi KM, Lu A, Lee SE, Lee JY, Lopez OL, Aizenstein HJ, Toga AW, Becker JT. 3D mapping of ventricular and corpus callosum abnormalities in HIV/AIDS. Neuroimage. 2006;31:12–23. doi: 10.1016/j.neuroimage.2005.11.043. [DOI] [PubMed] [Google Scholar]
- Tiehuis AM, van der Graaf Y, Visseren FL, Vincken KL, Biessels GJ, Appelman AP, Kappelle LJ, Mali WP. Diabetes increases atrophy and vascular lesions on brain MRI in patients with symptomatic arterial disease. Stroke. 2008;39:1600–1613. doi: 10.1161/STROKEAHA.107.506089. [DOI] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in IBASPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Udupa JK, Leblanc VR, Zhuge Y, Imielinska C, Schmidt H, Currie LM, Hirsch BE, Woodburn J. A framework for evaluating image segmentation algorithms. Comput Med Imaging Graph. 2006;30:75–87. doi: 10.1016/j.compmedimag.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Van Leemput KV, Maes F, Vandermeulen D, Suetens P. Automated model-based tissue classification of MR images of the brain. IEEE Trans Med Imaging. 1999;18:897–908. doi: 10.1109/42.811270. [DOI] [PubMed] [Google Scholar]
- Walhovd KB, Fjell AM, Reinvang I, Lundervold A, Dale AM, Eilertsen DE, Quinn BT, Salat D, Makris N, Fischl B. Effects of age on volumes of cortex, white matter and subcortical structures. Neurobiol Aging. 2005;26:1261–1270. doi: 10.1016/j.neurobiolaging.2005.05.020. discussion 1275–1268. [DOI] [PubMed] [Google Scholar]
- Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): An algorithm for the validation of image segmentation. IEEE Transactions on Medical Imaging. 2004;23:903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Centre for Neuroimaging. [Accessed 10/15/2008]; Available at http://www.fil.ion.acl.uk/spm/.
- Wessels AM, Rombouts SA, Remijnse PL, Boom Y, Scheltens P, Barkhof F, Heine RJ, Snoek FJ. Cognitive performance in type 1 diabetes patients is associated with cerebral white matter volume. Diabetologia. 2007;50:1763–1769. doi: 10.1007/s00125-007-0714-0. [DOI] [PubMed] [Google Scholar]
- Westmoreland P, Cretsinger K. [Accessed 9/15/2008];(a) Caudate Tracing Guidelines. Available at http://www.psychiatry.uiowa.edu/mhcrc/pdf/papers/caudate.pdf.
- Westmoreland P, Cretsinger K. [Accessed 9/15/2008];(b) Putamen Tracing Guidelines. Available at http://www.psychiatry.uiowa.edu/mhcrc/pdf/papers/putamen.pdf.
- Wu M, Carmichael O, Lopez-Garcia P, Carter CS, Aizenstein HJ. Quantitative comparison of AIR, IBASPM, and the fully deformable model for atlas-based segmentation of functional and structural MR images. Hum Brain Mapp. 2006;27:747–754. doi: 10.1002/hbm.20216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BTT, Sabuncu MR, Desikan R, Fischl B, Golland P. Effects of registration regularization and atlas sharpness on segmentation accuracy. Med Image Anal. 2008;12:603–615. doi: 10.1016/j.media.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi H, Ruest T, Schoenahl F, Montandon ML. Comparative assessment of statistical brain MR image segmentation algorithms and their impact on partial volume correction in PET. Neuroimage. 2006;32:1591–1607. doi: 10.1016/j.neuroimage.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Zhang YJ. A survey on evaluation methods for image segmentation. Pattern Recognition. 1996;29:1335–1346. [Google Scholar]


