Abstract
A cluster of low copy repeats on the proximal long arm of chromosome 15 mediate various forms of stereotyped deletions and duplication events that cause a group of neurodevelopmental disorders that are associated with autism or autism spectrum disorders (ASD). The region is subject to genomic imprinting and the behavioral phenotypes associated with the chromosome 15q11.2-q13 disorders show a parent-of-origin specific effect that suggests that increased copy number of maternally derived alleles contributes to autism susceptibility. Notably, nonimprinted, biallellically expressed genes within the interval also have been shown to be misexpressed in brains of patients with chromosome 15q11.2-q13 genomic disorders, indicating that they also likely play a role in the phenotypic outcome. This review provides an overview of the phenotypes of these disorders and their relationships with ASD and outlines the regional genes that may contribute to the autism susceptibility imparted by copy number variation of the region.
Keywords: Autism, Autism spectrum disorders, Prader Willi syndrome, Angelman Syndrome, Interstitial Duplication Chromosome 15, Isodicentric Chromosome 15, Low Copy Repeats, Imprinting
The Complexities of the Genomic Landscape of Chromosome 15q11.2-q13
Chromosome 15 has been identified as one of seven chromosomes enriched in segmental low copy repeats (LCRs) or duplicons (Bailey et al., 2002). These duplicons provide a mechanism in which LCR mediated misalignment during meiosis I leads to unequal nonallelic homologous recombination generating a series of common breakpoints (BPs) along the 15q11.2-q13(Christian et al., 1999; Robinson et al., 1993a; Robinson et al., 1998b; Robinson et al., 1993c).
The proximal three BP correspond to complex LCRs ranging in size from 50 – 400 kb and contain sequences derived from HERC2 and GOLGA8E loci (Amos-Landgraf et al., 1999; Ji et al., 2000; Makoff and Flomen, 2007)(Figure 1). While the actively transcribed HERC2 and GOLGA8E genes lie just centromeric to BP3 and BP1, respectively, numerous transcribed pseudogenes derived from these loci can be found in the vicinity of BP1, BP2 and BP3. Two more distal BP clusters (BP4 and BP5) involve a distinct set of LCRs that have limited sequence homology to the repeats at BP1-BP3. These paired LCRs are roughly 500kb in length and oriented head to head, which may facilitate the U-type crossover events that generate isodicentric chromosomes (Makoff and Flomen, 2007).
Fig 1.
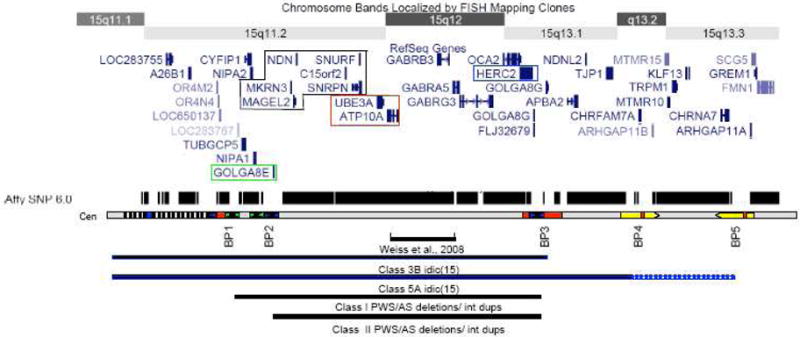
Schematic of chromosome 15q11.1-13.3 showing the position of known genes based on the UCSC genome browser. Maternally expressed transcripts highlighted in red, and paternally expressed transcripts in black. The HERC2 gene is highlighted in blue and the GOLGA8E gene is highlighted in green. (Below) The relative positions of the 5 BP clusters are shown below with sequence homology indicated by color, blue indicating regions of homology to HERC2 and green indicating regions of homology to GOLGA8E. The black and white hatching indicates a heteromorphic region near the centromere that includes a number of pseudogenes and can expand in the normal population. At least one HERC2 based repeat lies in this region. The track above the breakpoint schematic shows the density of SNP coverage for this region on the Affymetrix 6.0 whole genome array with notable gaps at the positions of the common BPs, although not all probes for detecting copy number variations are shown in the UCSC browser. The region included in Class I and Class II deletions/duplications is indicated by the black bars at the bottom with the position of the small duplication identified by Weiss et al (2008) also noted. Similarly, the region encompassed by the two most common forms of idic(15) chromosomes is indicated with solid blue line indicating a region of tetrasomy and dashed line indicating trisomy.
The complex structure and orientation of the LCRs on proximal 15q, which include both tandem and inverted repeats, contribute to a variety of rearrangements that are often stereotyped, with common blocks of genomic material that is either deleted or duplicated. Deletions of the region lead to two phenotypically distinct neurodevelopmental disorders, Prader Willi syndrome (PWS) and Angelman syndrome (AS), which have different phenotypes due to the effect of an imprinted domain between BP2 and BP3 (Buiting et al., 1995; Knoll et al., 1989). This gene rich region is under the control of a bipartite imprinting center that directs expression of a number of genes that show parent-of-origin specific expression (reviewed in (Horsthemke and Buiting, 2006). Notably, imprinted expression of some of these genes is limited to the nervous system, while some genes encode neuron-specific transcripts (Albrecht et al., 1997; Chibuk et al., 2001; Lee et al., 2003). Individuals with duplications of 15q11.2-q13 also demonstrate parent of origin differences in phenotypes, as maternally derived duplications pose the greater risk for ASD, suggesting that the autism susceptibility allele(s) at chromosome 15q11.2-q13 may be subject to imprinting.
The Deletion Syndromes and Autism Spectrum Disorders
Prader Willi Syndrome
PWS is classically characterized by hypotonia and failure to thrive in infancy, which evolves into a complex neurobehavioral phenotype accompanied by cognitive impairment, hyperphagia leading to obesity, obsessive compulsive behaviors that include hoarding and skin picking, with an increased risk of autism spectrum disorders (ASD). In addition, patients with PWS typically have hypogonadism, dysmorphic facial features, small hands and feet and may be hypopigmented (reviewed in (Cassidy et al., 2000). In the majority of cases, PWS arises by deletions on the paternal chromosome 15, either between BP1-2 (Class I) or BP2-BP3 (Class 2). Approximately 25% of patients with PWS have uniparental disomy for the maternal chromosome, which can be either isodisomic or heterodisomic (Fridman and Koiffmann, 2000). The remaining patients have imprinting errors on the paternal homolog of chromosome 15, which lead to aberrant methylation of the PWS imprinting center and downregulation of paternally expressed transcripts (Nicholls and Knepper, 2001).
Although several paternally expressed genes are known—MKRN3, MAGEL2, NDN, SNURF-SNRPN and more than seventy C/D box small nucleolar RNA genes—it is still uncertain whether a single gene or several genes are responsible for the PWS phenotype. Notably, while each of the different molecular classes of PWS lead to loss of paternally expressed genes, they have differential effects on the biallelic and maternally expressed transcripts in the region, which are likely to contribute to differences in phenotypes. Genotype-phenotype studies in PWS patients in various molecular groups have indicated that the deletion patients tend to be the most severe in presentation, both in terms of degree of learning impairment and frequency of aberrant behaviors, (Dykens and Cassidy, 1995). Notably, within the deletion group, those with class I deletions (BP1:BP3) have been found to have more significant impairments in adaptive behaviors than those with class 2 (BP2:BP3) deletions, suggesting a role of one or more of the four genes that lie with the BP1-BP2 interval (Butler et al., 2004; Milner et al., 2005).
Studies of patients with maternal UPD reveal that, while they may appear milder on some outcome measures, they are more likely than patients with deletions to manifest ASD, with studies estimating the comorbidity of ASD in PWS to be between 19–36.5% (Descheemaeker et al., 2006; Milner et al., 2005; Veltman et al., 2004). Notably, in the study conducted by Veltman et al., (2004), no significant difference in the frequency of subjects meeting the diagnostic threshold for ASD was noted in the UPD vs. the deletion groups. However, among the UPD patients, scores were significantly higher on the autism screening questionnaire applied, with deficits in social interaction driving the differences between groups. Similarly, Descheemeaecker et al. (2006) examined a cohort of PWS patients for evidence of pervasive developmental disorders (PDD) using the PDD-mental retardation scale screening questionnaire. While they found a somewhat lower incidence of PDD among their cohort than reported by Veltman (19% vs 36.5%), similar increases in scores were identified for the UPD subgroup and no evidence for an IQ effect on the incidence of PDD was detected (i.e. the frequency of PDD was the same for subjects with IQ < 70 and >70). Taken together, these studies suggest that there is an increased risk of ASD imparted by the UPD-form of PWS and implicate maternally expressed transcripts in the pathogenesis of the ASD phenotype. Additionally, patients with PWS are at increased risk of other neuropsychiatric disorders including affective and psychotic disorders (Descheemaeker et al., 2006; Milner et al., 2005; Veltman et al., 2005a; Veltman et al., 2004). It is intriguing that it is also the UPD-form of PWS that poses the greater risk for psychiatric disease in adolescence and young adulthood (Boer et al., 2002). A recent study of 119 patients with PWS revealed a prevalence of psychiatric symptoms of 64% in patients with PWS arising from maternal UPD as compared to 28.2% in the deletion cases. The disorders appear to be primarily affective disorders with a high incidence of psychotic symptoms (Soni et al., 2008) or primary psychoses (Vogels et al., 2004). In some cases, there appeared to be an evolution in the behavioral phenotype, with psychotic disorders developing in individuals who had been diagnosed with PDD in childhood (Descheemaeker et al., 2002).
Angelman Syndrome
Patients with AS have severe to profound mental retardation, microcephaly, seizures, ataxia and may also show hypopigmentation (Cassidy et al., 2000). They almost always lack speech and language is typically severely impaired with function at a level less than 2 years age equivalent (Andersen et al., 2001; Penner et al., 1993; Peters et al., 2004b). Socially, patients with AS are notable for their happy demeanors, a propensity for easy smiling and laughter and are engaging with both adults and children, making excellent eye contact and frequent social bids for communication despite the absence of expressive language (Oliver et al., 2007). AS most often arises from deletions that occur through the LCR at BP1, 2 and 3, with the critical region for AS lying just 35 kb telomeric to the PWS critical region (reviewed in (Lalande and Calciano, 2007). A subset of patients with AS have paternal uniparental disomy and a approximately 10% of patients have imprinting errors on the maternal chromosome, with demethylation of the PWS imprinting center and expression of paternal transcripts from both chromosomes. In AS, a single gene of major effect has been identified, UBE3A, which is shows imprinted expression in selected neuronal populations (Hoffman, 1997; Kishino et al., 1997; Matsuura et al., 1997). Point mutations in UBE3A occur in roughly 10% of patients with this disorder (Fang et al., 1999). Notably, genotype phenotype relationships have been less well-established in AS, although it appears that the deletion and UBE3A mutation groups are most severely affected in terms of cognition, seizure frequency and hypopigmentation, while patients with UPD and imprinting mutations are more likely to attain limited speech and have fewer seizures (Burger et al., 1996; Lossie et al., 2001).
AS is often described as falling within the autism spectrum and studies have suggested a high prevalence of comorbidity of ASD in AS (Bonati et al., 2007; Peters et al., 2004a; Steffenburg, 1996). However, these studies must be interpreted cautiously as the severity of the cognitive and language impairment in AS poses a major confound, as mental ages are typically less than 18–24 months (Andersen et al., 2001; Peters et al., 2004b). Trillingsgaard and Østergaard (2004) investigated the comorbidity of autism in 16 patients with AS using the Autism Diagnostic Observation Scale-generic (ADOS-G)(Lord et al., 1989), which revealed that 13/16 subjects met algorithm criteria for ASD (Trillingsgaard and Ostergaard, 2004). However, comparison of the social and communication interactions of the AS patients with those of a cohort of patients with idiopathic autism led the authors to conclude that the disabilities in the social and communication domains that underlie the ASD diagnosis in the AS group are better defined by developmental delay, rather than a specific deficits in social and communication skills as is typical in autism. The children simply had not reached a mental age at which reciprocal interactions and skills such as joint attention and pointing would be expected to emerge. This argument is supported by studies showing that children with AS are more likely to initiate and maintain social contact than similarly cognitively impaired children (Oliver et al., 2007), and that they are flexible in their behaviors (Didden et al., 2007). Hence, the degree of cognitive impairment in AS may lead to an overestimate of ASD using standard diagnostic measures in this group of patients (Trillingsgaard and Ostergaard, 2004).
The Duplication Syndromes
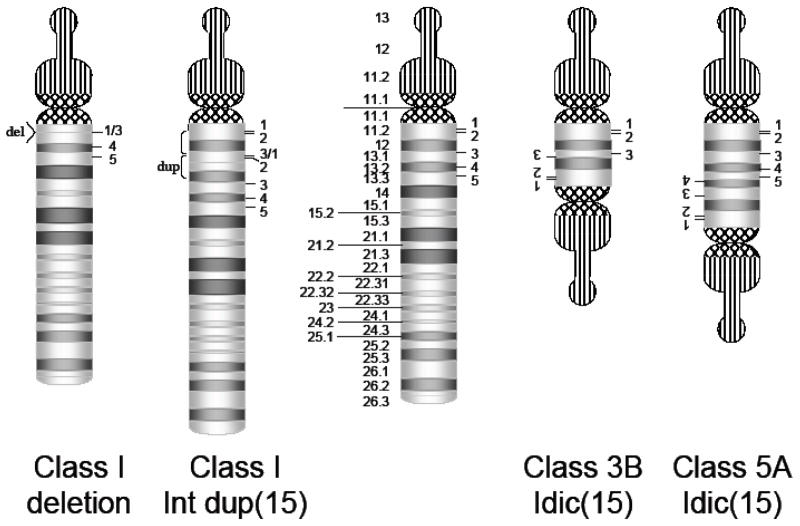
Interstitial duplications of proximal 15q11.2-13 often appear to be the reciprocal events of the PWS/AS deletions with LCR-mediated recombinations occurring through BP1-3 or BP2-3 (Repetto et al., 1998). However, more distal LCR also contribute with some duplication chromosomes and interstitial duplications and triplications extending to BP4 and BP5 (Wandstrat et al., 1998). In addition, duplications of this region can occur via U-type crossover events in meiosis (Robinson et al., 1998a), leading to the formation of a supernumerary derivative chromosome 15 that has two centromeres called an isodicentric chromosome 15 or idic(15) [also previously called inverted duplication 15] (Figure 2). Like PWS and AS, there appears to be a parent of origin effect on phenotype, with maternally inherited duplications posing the most clear-cut risk for ASD (Cook et al., 1997).
Figure 2.
Schematic of the Genomic Rearrangements of Chromosome 15. The ideogram of chromosome 15 is shown in the middle. On the left hand side, interstitial rearrangements are shown. On the far left, PWS and AS can arise by deletion of 15q11.2-q13.1. Alternatively, this region can be duplicated, shown here as a tandem duplication of the BP1:BP3 region. These lead to trisomy of the involved segments of DNA. On the right, the two most common forms of idic(15) associated with ASD are shown, Class 3B and Class 5A. The Class 3B duplications lead to tetrasomy of the involved segments of DNA, while in the class 5A idic(15) chromosomes, there is tetrasomy for the region through BP4 and trisomy for the interval between BP4 and BP5.
Interstitial duplication and triplications
Patients with maternally derived interstitial duplications of chromosome 15 present with a neurodevelopmental disorder that frequently includes autism or ASD (Bolton et al., 2001; Browne et al., 1997; Cook et al., 1997; Mao et al., 2000; Repetto et al., 1998; Roberts et al., 2002). The ASD phenotype does not appear fully penetrant, however, as not all patients with maternally derived duplications meet even the broad criteria for ASD using standardized measures of diagnosis (Boyar et al., 2001). The behavioral phenotype is complex with anxiety, emotional lability, tantrums and hyperactivity posing significant challenges for caregivers (Thomas et al., 1999). Dysmorphic features are infrequent and growth is typically normal, however hypotonia is common in infancy and gross and fine motor delays occur. Seizures, hypogonadism, gait abnormalities and episodes of unprovoked laughter have also been reported (Browne et al., 1997; Repetto et al., 1998; Thomas et al., 1999). Speech and language delays are prominent and speech apraxia, developmental language disorder and dyslexia also have been reported (Boyar et al., 2001; Cook et al., 1997). Additional features that have been identified in isolated cases have included cryptorchidism, cardiac malformations and juvenile rheumatoid arthritis, (Thomas et al., 1999).
Familial cases suggest that paternally derived duplications are associated with a more normal phenotype, as a number of seemingly unaffected mothers have transmitted paternally derived duplication chromosomes to their children (Browne et al., 1997; Cook et al., 1998; Roberts et al., 2002). There are, however, reports of paternally derived duplications associated with developmental disorders, which in at least one family, has included autism or ASD in affected siblings (Mao et al., 2000; Mohandas et al., 1999; Veltman et al., 2005b). Given that this was a single family, the possibility that this was a coincident occurrence of ASD in a family with a duplication cannot be excluded. Nonetheless, based on a limited number of cases reported, the phenotype associated with de novo paternally derived duplications appears to be widely variable - ranging from apparently normal to significantly cognitively impaired, with language and social deficits (Mohandas et al., 1999; Veltman et al., 2005b) and behaviors reminiscent of PWS (Mao et al., 2000). The basis for the discrepancies in phenotype is unclear, and given the relatively small number of cases, it is premature to make conclusions that paternally-derived duplications are benign.
By contrast, both maternally and paternally-derived interstitial triplications appear to be consistently associated with more severe neurodevelopmental phenotype, which includes hypotonia, global developmental delays with specific deficits in speech and language, severe mental retardation, and seizures (Cassidy et al., 1996; Clayton-Smith et al., 1993; Dennis et al., 2006; Long et al., 1998; Ungaro et al., 2001; Vialard et al., 2003). In addition, autism or ASD has been identified in several of these cases (Cassidy et al., 1996; Vialard et al., 2003), and behaviors suggestive of ASD are described in reports that did not include formal assessments for autism (Dennis et al., 2006). Both PWS-like (Dennis et al., 2006; Long et al., 1998; Pettigrew et al., 1987) and AS-like features (Ungaro et al., 2001) have been described, and curiously the PWS-like phenotype occurred in individual patients whose duplication chromosomes were either maternally (Dennis et al., 2006; Long et al., 1998) or paternally (Pettigrew et al., 1987) derived. The small number of these cases precludes distinction of clear phenotypic differences between those carrying maternally and paternally derived duplication chromosomes. Nonetheless, the presence of ASD among the handful of paternally derived interstitial triplication cases is of interest in terms of mechanisms that may underlie the predisposition to this aspect of the phenotype.
Isodicentric chromosome 15
As a general rule, patients with maternally derived idic(15) chromosomes that include the PWS/AS region present with similar clinical traits as those with interstitial rearrangements, albeit to a greater degree, suggesting that increasing dosage of the duplicated segment has an adverse effect on phenotype. Hypotonia in infancy is pronounced and may lead to genetic evaluation for PWS, revealing the idic(15) chromosome (Battaglia et al., 1997). In addition, patients with idic(15) frequently have minor dysmorphic features including a short upturned nose, downslanting palpebral fissures, high arched palate, incompletely folded ears and full lips (Abeliovich et al., 1995; Abuelo et al., 1995; Buoni S, 1999; Dennis et al., 2006; Grammatico et al., 1994; Mignon et al., 1996; Rineer et al., 1998; Robinson et al., 1993b; Wolpert et al., 2000b). Notably, these are often subtle, and may be missed in infancy. Various congenital anomalies have been reported in individual cases, although genitourinary malformations, joint laxity, strabismus, cortical visual impairment and hyperpigmentation have been seen in multiple cases suggesting that they may be part of the phenotype of this disorder. A few cases of hexasomy for chromosome 15q11.2-q13 have been identified. These children were severely affected with profound mental retardation and intractable epilepsy and more prominent dysmorphic features including myopathic facies, downslanting palpebral fissures and low set ears (Huang and Bartley, 2003; Mann et al., 2004; Nietzel et al., 2003; Qumsiyeh et al., 2003).
Patients with idic(15) consistently show significant developmental delays, including gross motor and fine delays, and cognitive impairment, which ranges from moderate to severe (Crolla et al., 1995; Dennis et al., 2006; Webb et al., 1998). Like the children with interstitial rearrangements, growth parameters are typically normal although both microcephaly and macrocephaly have been described (Webb et al., 1998; Wolpert et al., 2000b). Seizures, including infantile spasms, complicate the clinical course for at least half of the patients (Battaglia et al., 1997; Bingham et al., 1996; Rineer et al., 1998) and intractable epilepsy occurs in some patients with associated regression in skills (Mann et al., 2004 and Schanen, unpublished). Behavioral problems, including impulsivity, self-injurious and aggressive and/or self-injurious behaviors, hyperactivity and anxiety appearing in early childhood and often increasing in adolescence (Webb et al., 1998; Wolpert et al., 2000a; Wolpert et al., 2000b). In addition, a number of sudden, unexplained deaths among seemingly healthy individuals with idic(15) have been recently identified as well as deaths in patients with idic(15) who manifest a chronic degenerative phenotype associated with relentless seizures (Schanen and Cook, in preparation).
Although the phenotype arising from duplication of proximal 15q is complex, the disorder has gained considerable attention over recent years because these duplication chromosomes pose a substantial risk for ASD. In our ongoing study of duplications of chromosome 15 in autism, we found that 44 of 54 subjects (81%) with idic(15) chromosomes met strict criteria for autism using the combined Autism Diagnostic Interview-Revised (Lord et al., 1994) and ADOS-G criteria (Lord et al., 1989), with an additional six cases (92%) meeting the broader ASD criteria on one or both measures (Schanen, unpublished). This is consistent with data from Rineer et al. (1998), who used the Gilliam Autism Rating Scale (GARS) (Gilliam, 1995) to characterize the autistic symptoms in a cohort of children and adults with idic(15). In subjects over age 5 yrs, 20/21 had autism quotient scores above 90, suggesting a high probability of being autistic (Rineer et al., 1998). Notably, some regression in socialization appears to be part of the phenotype, as infants and toddlers with idic(15) often appear more social and make better eye contact and vocalize more than they do as older children (Borgatti et al., 2001; Mohandas et al., 1999; Rineer et al., 1998; Wolpert et al., 2000a; Wolpert et al., 2000b). Similar to the concern with AS, there is some concern that ASD may be overcalled in the lowest functioning cases of idic(15), who have mental ages under 18 mos, again younger that many of the social behaviors emerge. Speech and language delays are universal but variable in severity, with characteristic autistic speech traits such as delayed echolalia, pronoun reversal and stereotyped utterances populating the vocalizations of verbal children and adults with idic(15), while many lack functional speech (Borgatti et al., 2001; Clayton-Smith et al., 1993; Grammatico et al., 1994; Maggouta et al., 2003; Rineer et al., 1998; Wolpert et al., 2000b). Socialization is impaired in idic(15) with decreased eye contact and lack of reciprocity. They also typically display numerous repetitive and stereotyped behaviors (rocking, hand flapping, licking) that are often directed toward sensory stimulation (Borgatti et al., 2001; Wolpert et al., 2000a; Wolpert et al., 2000b). Notably, while it appears that subjects with idic(15) and ASD display core autistic traits, it is likely that there are behavioral characteristics enriched in this population that can distinguish them from other forms of autism that may provide a behavioral signature for ASD arising from the susceptibility locus on proximal 15q. Clearly, more detailed studies of the natural history of this disorder are warranted in order to fully define the behavioral profile of children with idic(15) chromosomes.
Molecular characteristics of the duplications
Cytogenetic, molecular and array comparative genomic hybridization based examination of duplication chromosomes has revealed that the interstitial duplications can arise through both intra- and interchromosomal recombination exchanges through the LCR clustered on proximal 15q (Robinson et al., 1998a). Like the PWS/AS deletions, Class I and Class 2 duplications occur, which vary in their proximal boundary and thus inclusion of the genes between BP1 and BP2 (Table 1 and Figure 2). The distal boundary for most interstitial duplications appears to reside at BP3, while in interstitial triplications, the distal BP may lie at BP4 or BP5 (Wandstrat et al., 1998). Both tandem and inverted duplication events have been recognized (Robinson et al., 1998a).
Table 1.
Classification of deletion and duplication chromosomes
| Proximal BP | Distal BP | BP1-BP2 Dosage | BP2:BP3 Dosage | BP3:BP4 Dosage | BP4:BP5 Dosage | |
|---|---|---|---|---|---|---|
| Interstitial deletions | ||||||
| Class 1 | 1 | 3 | 1 | 1 | 2 | 2 |
| Class 2 | 2 | 3 | 2 | 1 | 2 | 2 |
| Interstitial duplications | ||||||
| Class 1 | 1 | 3 | 3 | 3 | 2 | 2 |
| Class 2 | 2 | 3 | 2 | 3 | 2 | 2 |
| Idic(15) | ||||||
| Class 1 | 1 | 1 | 2 | 2 | 2 | 2 |
| Class 2A | 1 | 2 | 3 | 2 | 2 | 2 |
| Class 2B | 2 | 2 | 4 | 2 | 2 | 2 |
| Class 3A | 2 | 3 | 4 | 3 | 2 | 2 |
| Class 3B | 3 | 3 | 4 | 4 | 2 | 2 |
| Class 4A | 3 | 4 | 4 | 4 | 3 | 2 |
| Class 4B | 4 | 4 | 4 | 4 | 4 | 2 |
| Class 5A | 4 | 5 | 4 | 4 | 4 | 3 |
| Class 5B | 5 | 5 | 4 | 4 | 4 | 4 |
| Class 6 | variable | variable | variable | variable | variable | variable |
Idic(15) chromosomes that include the PWS/AS region are almost exclusively maternal in origin, with rare paternal cases occurring in mosaic form in association with the UPD form of PWS (Baumer et al., 2001; Saitoh et al., 2007). A number of different structural forms have been identified that can be classified by their distal BP position (Table 1). Class 1 and 2 idic(15) chromosomes, defined as idic(15) that do not hybridize with probes to the PWS/AS critical region, are small, heterochromatic derivative chromosomes that appear to have no phenotypic consequences. Interestingly, they have been reported in association with UPD in patients with PWS and AS, suggesting they may lead to meiotic segregation errors (Robinson et al., 1993d). Notably, Class 2 idic(15) chromosomes, which would include the genes between BP1 and BP2 have not been specifically described, however it is intriguing to consider the possibility that they might have phenotypic consequences given the proposed role of genes in the interval in development and the differences in phenotype in Class I versus Class 2 PWS deletion patients and the observation that CYFIP1 was overexpressed in lymphoblasts from patients with larger idic(15) chromosomes (Butler et al., 2004; Nishimura et al., 2007). Most of the idic(15) identified in association with neurodevelopmental disorders fall into the remaining classes, with Classes 3B and 5A being the predominant forms (Figure 2) (Wang et al., 2004). Class 3B idic(15) chromosomes are symmetric, extending to BP3 on each end and result in tetrasomy for the genomic region encompassed by the duplication chromosome. Roughly half of the idic(15) chromosomes characterized by array CGH fell into this class (Sahoo et al., 2005; Wang et al., 2004). Class 5 idic(15) arise through BP5 exchanges, and the asymmetric form (Class 5A; BP4:BP5) appear to be other major form of idic(15) chromosome, again accounting for almost half of the cases in the study by Wang et al (2004). Class 6 idic(15) chromosomes are atypical rearrangements that involve non-LCR based BP (Wang et al., 2004; Wang et al., 2008). While it is likely that this structural variability impacts phenotypic outcome, each of these molecular groups is associated with cognitive impairment, seizures and risk for ASD, suggesting that the genes that are the major contributors to those aspects of the phenotype lie in the region shared by most duplication chromosomes.
The basis for the extreme bias in identification of maternally derived idic(15) chromosomes remains enigmatic. Likely explanations include a combination of a tendency for the meiotic error to occur in the maternal germline, an ascertainment bias based severity of symptoms arising from maternal duplications and perhaps selection against paternally derived idic(15) chromosomes in early development potentially through pregnancy loss or loss of the idic(15) chromosome. Rare cases of paternally derived interstitial triplications implies that paternal trisomy for the PWS/AS region is not lethal (Browne et al., 1997; Cassidy et al., 1996; Mao et al., 2000; Mohandas et al., 1999; Ungaro et al., 2001) although further investigation is required to fully understand the consequences of excess paternal dosage.
Prevalence of duplications of chromosome 15 in autism
Systematic screening for chromosomal abnormalities in cohorts of patients with autism using cytogenetic and array-based strategies has revealed that copy number variation (CNV) at this region contributes significantly to the etiology of ASD (Christian et al., 2008; Schroer et al., 1998; Sebat et al., 2007). Most estimates of prevalence fall in the 0.5–3% range, depending on the criteria used for sample ascertainment and method used for identifying the duplications (Browne et al., 1997; Cook et al., 1997; Schroer et al., 1998; Sebat et al., 2007; Weiss et al., 2008). The likelihood of identifying a duplication clearly increases if the patient is also significantly cognitively impaired and has gross motor delays and/or seizures. Notably, interstitial duplications are being increasingly recognized with the use of array based strategies for diagnosis of CNV in children with developmental disorders. In addition, both typical and smaller, atypical duplications have been identified using high resolution tools to assess CNV among well characterized autism samples that have been widely used for linkage studies (Sebat et al., 2007; Szatmari et al., 2007; Ullmann et al., 2007; Weiss et al., 2008).
Neurologically relevant 15q11.2-13 genes with altered expression in 15q11.2-q13 deletions and duplications
While the 13 MB region encompassed by the scope of 15q11.2-q13 deletion and duplications syndromes contains at least 30 characterized genes, the overwhelming focus of gene expression studies has been on the genes showing imprinted expression, as these are presumably the most likely to show parent-of-origin specific gene expression differences. The smallest of the regional duplications detected by CNV analysis includes only the ATP10A and GABRB3 genes, implicating them as critical genes for the ASD phenotype, although the data needs to be interpreted cautiously (Weiss et al., 2008). In light of recent results showing that nonimprinted GABAA receptor genes (GABRB3, GABRA5, GABRG3) from 15q11.2-q13 can show parental expression differences in the absence of a normal biparental contribution (Hogart et al., 2007), the expectation of maternal only expression alterations may need to be reexamined. In this section, several of the imprinted and nonimprinted genes likely to have pathologically relevant dysregulation in 15q11.2-q13 duplications and ASD are discussed.
UBE3A and ATP10A
The maternally expressed UBE3A encodes an ubiquitin E3 ligase enzyme that directs ubiquitin molecules transferred from E1 and E2 pathway enzymes to specific substrates (Ciechanover et al., 1994). UBE3A loss of function mutations or deficiency due to maternal deletion or imprinting mutation cause AS (Kishino et al., 1997; Matsuura et al., 1997; Moncla et al., 1999) and engineered Ube3a mutations in mouse models recapitulate several features of AS, including motor dysfunction and hippocampal impairments (Jiang et al., 1998; Miura et al., 2002). A Drosophila model of dUBE3A overexpression identified a novel downstream target Rho-GEF Pbl/ECT2 that showed abnormal cellular localization in mutant mouse brain (Reiter et al., 2006). UBE3A was shown to be transcriptionally active from the duplication chromosome in lymphocytes (Herzing et al., 2002) and overexpression of UBE3A is also observed in 15q11.2-q13 maternal duplication lymphoblast samples (Baron et al., 2006). These results from 15q11.2-q13 duplication cell lines suggest that increased UBE3A expression is likely also to occur in the brains of these patients, but the phenotypic consequences are currently unclear. PWS cortical tissues with maternal UPD express twice the normal level of UBE3A compared to controls (Hogart et al., 2007) and PWS UPD patients have more autistic behaviors than those with paternal 15q11.2-q13 deletions (Descheemaeker et al., 2006; Milner et al., 2005; Veltman et al., 2004). These combined results suggest that gene dosage directed UBE3A overexpression in 15q11.2-q13 duplications could have a major impact on the ASD phenotype.
ATP10A encodes a type IV P-type ATPase that when maternally deleted in mice leads to obesity and type II diabetes (Dhar et al., 2004). While maternal inheritance of the obesity phenotype implies that Atp10a is imprinted, discrepancies in imprinting in different mouse strains suggests that imprinting of Atp10a may be less robust than Ube3a and potentially dependent on genetic modifiers (Kashiwagi et al., 2003; Kayashima et al., 2003a; Kayashima et al., 2003b). In humans, ATP10A is proposed to be imprinted similarly to UBE3A (Herzing et al., 2001), however imprinting was also observed in lymphoblasts from patients with AS (Meguro et al., 2001), unlike the biallelic expression in blood observed for UBE3A (Sutcliffe et al., 1997). While some genetic association data suggests that ATP10A may contribute to autism (Nurmi et al., 2003a), a causative role for ATP10A in the pathogenesis of 15q11.2-q13 chromosome disorders remains to be proven.
SNRPN and snoRNAs
The SNRPN locus on chromosome 15q11.2-q13 encodes highly complex paternally expressed imprinted transcripts that extend from upstream of the SNRPN promoter throughout UBE3A in the antisense orientation (Landers et al., 2004; Runte et al., 2001). The 5′ end of SNRPN contains the imprinting center involved in marking the maternal and paternal chromosomes through differential DNA methylation (Sutcliffe et al., 1994). Two small spliceosome proteins, SNURF and SmN, small nucleoprotein N, encoded by the 5′ exons, (Gray et al., 1999) have minimal influence on the phenotype of PWS, as mice with disruptions of the protein are phenotypically normal (Bressler et al., 2001; Yang et al., 1998). Splicing of downstream noncoding exons produces several classes of small nucleolar RNAs (snoRNAs) (Runte et al., 2001) that are proposed to be involved in mRNA processing of neurologically relevant genes including the serotonin receptor 2C (Cavaillé et al., 2000; Kishore and Stamm, 2006). Mice with deletions of the cluster of MBII-85 snoRNA sequences (orthologous to human HBII-85) within the large SNRPN transcript exhibit early post-natal growth deficiencies and late-onset hyperphagia suggesting that these noncoding RNAs contribute to the phenotype of PWS (Ding et al., 2008; Skryabin et al., 2007).
NECDIN and MAGEL2
NECDIN (NDN) and the related gene MAGEL2 are paternally expressed imprinted genes primarily expressed in post-mitotic neurons (Jay et al., 1997; Lee et al., 2000; Lee et al., 2005). The mouse knockout model of Ndn has shown conflicting results as one model yielded early postnatal lethality due to respiratory distress, while another Ndn deficient mouse failed to display any apparent phenotypes (Gérard et al., 1999; Tsai et al., 1999). Elimination of Magel2 in mice results in neonatal growth deficiencies, metabolic abnormalities, and alterations in activity, food intake, and fertility (Bischof et al., 2007). While neither of these genes seems to fully account for the phenotypes observed in PWS, it is clear that expression abnormalities of these genes could have profound effects on many neurological processes including respiration and circadian outputs.
GABAA Receptor Subunit Genes
GABA, the major inhibitory neurotransmitter in the brain, functions to hyperpolarize the membrane through binding to ionotropic pentameric GABAA receptors. Chromosome 15q11.2-q13 contains three GABAA receptor subunit genes (GABR), GABRB3, GABRA5, and GABRG3 that are normally biallelically expressed in mouse and human (Buettner et al., 2004; Hogart et al., 2007; Nicholls, 1993). Although the 15q11.2-q13 GABR genes are not imprinted, the increased severity of PWS and AS deletion patients compared to UPD patients (Varela et al., 2004; Varela et al., 2005) implicates a role for GABR genes in these disorders. Mice lacking Gabra5 and Gabrg3 appear normal (Culiat et al., 1994), while Gabrb3 knockout mice have profound neurological phenotypes including seizures, sleep abnormalities, hyperactivity, hypersensitivity to touch stimuli, learning and memory deficits, and defects in social and exploratory behaviors (DeLorey et al., 1998; DeLorey et al., 2008; Hashemi et al., 2007; Homanics et al., 1997). Genetic linkage and association studies in idiopathic autism cohorts have repeatedly found significant evidence for a susceptibility allele in GABRB3 (Buxbaum et al., 2002; Cook et al., 1998; Martin et al., 2000; McCauley et al., 2004; Nurmi et al., 2003b; Shao et al., 2003). Additionally, significantly reduced GABRB3 protein levels were commonly observed in postmortem brain samples from idiopathic autistic individuals (Samaco et al., 2005) and normal biallelic GABR expression was commonly disrupted in autistic individuals with protein expression deficits (Hogart et al., 2007). The effect of maternal 15q11.2-q13 duplications on GABR gene expression in brain is currently unknown, however genetic and molecular evidence suggests that GABRB3 expression abnormalities may contribute to the autism component of these chromosomal disorders.
CYFIP1
Although located outside the critical deletion region for PWS and AS, the nonimprinted gene CYFIP1 may contribute to the increased severity observed in patients with larger 15q11.2-q13 deletions and duplications. CYFIP1, the cytoplasmic FMRP interacting protein 1, was characterized by its physical interaction with the protein product of the Fragile X syndrome gene FMR1 (Schenck et al., 2001). Additionally CYFIP1, also called Sra-1, interacts with the Rac1 small GTPase (Kobayashi et al., 1998) and is proposed to play a role in bridging cytoskeleton remodeling with translation (Schenck et al., 2003). Similar to UBE3A, CYFIP1 has been shown to be overexpressed in lymphoblast cell lines from 15q11.2-q13 duplication patients (Nishimura et al., 2007). Interestingly, overexpression of CYFIP1 and knockdown of FMR1 in a human neuronal cell line resulted in similar effects on two downstream target genes, JAKMIP1 and GPR155 (Nishimura et al., 2007), suggesting that abnormalities in these pathways may contribute to some phenotypic overlap between 15q11.2-q13 duplication syndromes and Fragile X syndrome. Furthermore, a Prader-Willi obesity and hyperphagia phenotype was recently described in a subset of Fragile X patients with comorbid autism and reduced transcription of nonimprinted 15q11.2-q13 gene CYFIP, further suggesting overlap between Fragile X and 15q11.2-q13 duplication and deletion syndromes (Nowicki et al., 2007).
Epigenetic factors regulating 15q11.2-q13
Chromosome 15q11.2-q13 parental imprinting patterns and long-range gene expression effects are mediated by a bi-partite imprinting control region (ICR) upstream of the large paternal transcription unit from SNRPN through UBE3A. The PWS-ICR is hypermethylated on the silent maternal allele and deletion of this region on the paternal chromosome is sufficient to cause PWS (Sutcliffe et al., 1994). An AS-ICR located 35 kb upstream of the PWS-ICR is required for establishing the maternal methylation mark at the PWS-ICR, as deletion of this region in AS patients result in imprinting errors (Buiting et al., 2001; Buiting et al., 1995). In addition to methylation, other parental epigenetic marks define the PWS-ICR, including DNAse I hypersensitivity sites and histone H3 and H4 acetylation on the active paternal allele, and matrix attachment and histone H3K9 methylation on the silent maternal allele (Greally J, 1999; Rodriguez-Jato et al., 2005; Wu et al., 2006). Interestingly, methyl CpG binding protein 2 (MeCP2) binds to the PWS-ICR and to multiple additional intergenic sites throughout 15q11.2-q13 (Yasui et al., 2007), potentially acting to organize long range chromatin interactions of the region. While MeCP2 is not required for maintaining imprinted expression of 15q11.2-q13 genes, MeCP2 deficiency results in reduced expression of both UBE3A and GABRB3 in some studies (Hogart et al., 2007; Makedonski et al., 2005; Samaco et al., 2005). Individuals with MECP2 mutations have Rett syndrome, a neurodevelopmental disorder with significant phenotypic overlaps with AS and autism in terms of language impairment and stereotyped mannerisms (Amir et al., 1999; Watson et al., 2001).
Nuclear organization and homologous pairing of 15q11.2-q13 alleles in human neurons
Chromosome dosage changes to 15q11.2-q13 may also impact the normal balance of homologous chromosome 15 interactions previously observed in both lymphocytes and neurons (LaSalle and Lalande, 1996; Thatcher et al., 2005). Similar to X chromosome counting mechanisms that require transient homologous X chromosome pairing during development for proper X chromosome inactivation in females (Xu et al., 2006), some gene expression patterns within 15q11.2-q13 appear to require interactions between both parental chromosomes. Homologous pairing of maternal and paternal 15q11.2-q13 alleles in neurons is a developmentally regulated process and dependent on the binding of MeCP2 (Thatcher et al., 2005). Homologous pairing of 15q11.2-q13 is deficient in a subset of Rett syndrome and autism brains and may contribute to the dysregulation of GABRB3 expression observed in these human brain samples (Samaco et al., 2005; Thatcher and LaSalle, 2006). These observations have lead to the hypothesis that 15q11.2-q13 homologous pairing may be required for optimal biallelic expression of GABRB3. Examination of a brain sample from a mosaic complex der(15) patient by FISH has revealed that der(15) alleles can pair with the normal parental chromosome 15s (Wang et al., 2008), suggesting that additional copies of this regions can disrupt the normal balance of homologous pairing of 15q11.2-q13 alleles in the brain and potentially impact GABRB3 expression in a manner not predicted by gene dosage.
Future directions
Although maternal duplications of 15q11.2-q13 are the most common cytogenetic cause of autism, not all individuals with duplications have classic autism, so understanding the molecular basis of the clinical heterogeneity is imperative. The clinical heterogeneity among individuals with similar duplications suggests that genetic or epigenetic modifiers influence gene expression of 15q11.2-q13 genes or the downstream targets of these genes. Generation of a mouse model with excess maternal dosage for the syntenic region to 15q11.2-q13 (7qB4) is critical to begin to elucidate how the duplication affects gene expression in the developing mammalian brain. While mice provide controlled models for human disorders, differences in genetic backgrounds and biology between humans and mice may prove challenging in creating a good model for a variable human disorder (as proven by the difficulties in creating an appropriate mouse model for PWS). Complementary molecular and pathological studies of human brain samples are also necessary to critically evaluate how extra 15q11.2-q13 dosage affects neurological function. Although the maternally expressed imprinted gene UBE3A may be predicted to drive the pathogenesis of 15q11.2-q13 duplications, the complex regulation and trans effects observed in this locus suggest that any level of abnormal parental dosage may impair normal gene expression.
Acknowledgments
The authors are supported by grants from the National Institutes of Health [NIH F31 MH078377 (AH), NIH R01 HD48799 (JML), NIH P3P01 HD35470 (NCS), NIH R01 HD37874 (NCS), P20-RR020173 (NCS)], and Nemours (NCS and DW). We are indebted to the Isodicentric Exchange, Advocacy and Support Group and families with idic(15) for their participation in the ongoing research into the investigation of the phenotype of patients with idic(15) and interstitial duplication 15 chromosomes. The authors also thank Suzanne Cassidy for providing detailed information on an unpublished case interstitial triplication chromosome 15 and the relationship with PWS and ASD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Abeliovich D, Dagan J, Werner M, Lerer I, Shapira Y, Meiner V. Simultaneous formation of inv dup(15) and dup(15q) in a girl with developmental delay: origin of the abnormal chromosomes. Eur J Hum Genet. 1995;3:49–55. doi: 10.1159/000472273. [DOI] [PubMed] [Google Scholar]
- Abuelo D, Mark HF, Bier JA. Developmental delay caused by a supernumerary chromosome, inv dup (15), identified by fluorescent in situ hybridization. Clin Pediatr (Phila) 1995;34:223–226. doi: 10.1177/000992289503400410. [DOI] [PubMed] [Google Scholar]
- Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, Eichele G, Beaudet AL. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet. 1997;17:75–78. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstrat AE, Cassidy SB, Driscoll DJ, Rogan PK, Schwartz S, Nicholls RD. Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet. 1999;65:370–386. doi: 10.1086/302510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen WH, Rasmussen RK, Stromme P. Levels of cognitive and linguistic development in Angelman syndrome: a study of 20 children. Logoped Phoniatr Vocol. 2001;26:2–9. [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Baron CA, Tepper CG, Liu SY, Davis RR, Wang NJ, Schanen NC, Gregg JP. Genomic and functional profiling of duplicated chromosome 15 cell lines reveal regulatory alterations in UBE3A-associated ubiquitin-proteasome pathway processes. Hum Mol Genet. 2006;15:853–869. doi: 10.1093/hmg/ddl004. [DOI] [PubMed] [Google Scholar]
- Battaglia A, Gurrieri F, Bertini E, Bellacosa A, Pomponi MG, Paravatou-Petsotas M, Mazza S, Neri G. The inv dup(15) syndrome: a clinically recognizable syndrome with altered behavior, mental retardation, and epilepsy. Neurology. 1997;48:1081–1086. doi: 10.1212/wnl.48.4.1081. [DOI] [PubMed] [Google Scholar]
- Baumer A, Wiedemann U, Hergersberg M, Schinzel A. A novel MSP/DHPLC method for the investigation of the methylation status of imprinted genes enables the molecular detection of low cell mosaicisms. Hum Mutat. 2001;17:423–430. doi: 10.1002/humu.1118. [DOI] [PubMed] [Google Scholar]
- Bingham PM, Spinner NB, Sovinsky L, Zackai EH, Chance PF. Infantile spasms associated with proximal duplication of chromosome 15q. Pediatr Neurol. 1996;15:163–165. doi: 10.1016/0887-8994(96)00119-1. [DOI] [PubMed] [Google Scholar]
- Bischof JM, Stewart CL, Wevrick R. Inactivation of the mouse Magel2 gene results in growth abnormalities similar to Prader-Willi syndrome. Hum Mol Genet. 2007;16:2713–2719. doi: 10.1093/hmg/ddm225. [DOI] [PubMed] [Google Scholar]
- Boer H, Holland A, Whittington J, Butler J, Webb T, Clarke D. Psychotic illness in people with Prader Willi syndrome due to chromosome 15 maternal uniparental disomy. Lancet. 2002;359:135–136. doi: 10.1016/S0140-6736(02)07340-3. [DOI] [PubMed] [Google Scholar]
- Bolton P, Dennis NR, Browne CE, Thomas NS, Veltman MWM, Thompson RJ, Jacobs P. The phenotypic manifestations of interstitial duplications of proximal 15q with special reference to the autistic spectrum disorders. American Journal of Medical Genetics. 2001;105:675–685. doi: 10.1002/ajmg.1551. [DOI] [PubMed] [Google Scholar]
- Bonati MT, Russo S, Finelli P, Valsecchi MR, Cogliati F, Cavalleri F, Roberts W, Elia M, Larizza L. Evaluation of autism traits in Angelman syndrome: a resource to unfold autism genes. Neurogenetics. 2007;8:169–178. doi: 10.1007/s10048-007-0086-0. [DOI] [PubMed] [Google Scholar]
- Borgatti R, Piccinelli P, Passoni D, Dalprà L, Miozzo M, Micheli R, Gagliardi C, Balottin U. Relationship between clinical and genetic features in “inverted duplicated chromosome 15” patients. Pediatric Neurology. 2001;24:111–116. doi: 10.1016/s0887-8994(00)00244-7. [DOI] [PubMed] [Google Scholar]
- Boyar FZ, Whitney MM, Lossie AC, Gray BA, Keller KL, Stalker HJ, Zori RT, Geffken G, Mutch J, Edge PJ, et al. A family with a grand-maternally derived interstitial duplication of proximal 15q. Clin Genet. 2001;60:421–430. doi: 10.1034/j.1399-0004.2001.600604.x. [DOI] [PubMed] [Google Scholar]
- Bressler J, Tsai TF, Wu MY, Tsai SF, Ramirez MA, Armstrong D, Beaudet AL. The SNRPN promoter is not required for genomic imprinting of the Prader-Willi/Angelman domain in mice. Nat Genet. 2001;28:232–240. doi: 10.1038/90067. [DOI] [PubMed] [Google Scholar]
- Browne CE, Dennis NR, Maher E, Long FL, Nicholson JC, Sillibourne J, Barber JC. Inherited interstitial duplications of proximal 15q: genotype-phenotype correlations. American Journal of Human Genetics. 1997;61:1342–1352. doi: 10.1086/301624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner VL, Longmate JA, Barish ME, Mann JR, Singer-Sam J. Analysis of imprinting in mice with uniparental duplication of proximal chromosomes 7 and 15 by use of a custom oligonucleotide microarray. Mamm Genome. 2004;15:199–209. doi: 10.1007/s00335-003-2322-8. [DOI] [PubMed] [Google Scholar]
- Buiting K, Barnicoat A, Lich C, Pembrey M, Malcolm S, Horsthemke B. Disruption of the bipartite imprinting center in a family with Angelman syndrome. Am J Hum Genet. 2001;68:1290–1294. doi: 10.1086/320120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buiting K, Saitoh S, Gross S, Dittrich B, Schwartz S, Nicholls RD, Horsthemke B. Inherited microdeletions in the Angelman and Prader-Willi syndromes define an imprinting centre on human chromosome 15. Nat Genet. 1995;9:395–400. doi: 10.1038/ng0495-395. [DOI] [PubMed] [Google Scholar]
- Buoni SSL, Farnetani M, Pucci L, Fois A. The Syndrome of Inv Dup(15): Clinical, Electroencephalographic, and imaging findings. Journal of child neurology. 1999;15:380–385. doi: 10.1177/088307380001500605. [DOI] [PubMed] [Google Scholar]
- Burger J, Kunze J, Sperling K, Reis A. Phenotypic differences in Angelman syndrome patients: imprinting mutations show less frequently microcephaly and hypopigmentation than deletions. Am J Med Genet. 1996;66:221–226. doi: 10.1002/(SICI)1096-8628(19961211)66:2<221::AID-AJMG19>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Butler MG, Bittel DC, Kibiryeva N, Talebizadeh Z, Thompson T. Behavioral differences among subjects with Prader-Willi syndrome and type I or type II deletion and maternal disomy. Pediatrics. 2004;113:565–573. doi: 10.1542/peds.113.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum JD, Silverman JM, Smith CJ, Greenberg DA, Kilifarski M, Reichert J, Cook EH, Jr, Fang Y, Song CY, Vitale R. Association between a GABRB3 polymorphism and autism. Mol Psychiatry. 2002;7:311–316. doi: 10.1038/sj.mp.4001011. [DOI] [PubMed] [Google Scholar]
- Cassidy SB, Conroy J, Becker LA, Schwartz S. Paternal Triplication of 15q11-q13 in a Hypotonic, Developmentally Delayed Child without Prader-Willi or Angelman Syndrome. American journal of Medical Genetics. 1996;62:205–212. [Google Scholar]
- Cassidy SB, Dykens E, Williams CA. Prader-Willi and Angelman syndromes: sister imprinted disorders. Am J Med Genet. 2000;97:136–146. doi: 10.1002/1096-8628(200022)97:2<136::aid-ajmg5>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Cavaillé J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Hüttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [Comment In: Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14035–7 UI: 20570424] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibuk TK, Bischof JM, Wevrick R. A necdin/MAGE-like gene in the chromosome 15 autism susceptibility region: expression, imprinting, and mapping of the human and mouse orthologues. BMC Genet. 2001;2:22. doi: 10.1186/1471-2156-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian SL, Brune CW, Sudi J, Kumar RA, Liu S, Karamohamed S, Badner JA, Matsui S, Conroy J, McQuaid D, et al. Novel Submicroscopic Chromosomal Abnormalities Detected in Autism Spectrum Disorder. Biol Psychiatry. 2008 doi: 10.1016/j.biopsych.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH. Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13) Hum Mol Genet. 1999;8:1025–1037. doi: 10.1093/hmg/8.6.1025. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Shkedy D, Oren M, Bercovich B. Degradation of the tumor suppressor protein p53 by the ubiquitin-mediated proteolytic system requires a novel species of ubiquitin-carrier protein, E2. J Biol Chem. 1994;269:9582–9589. [PubMed] [Google Scholar]
- Clayton-Smith J, Webb T, Cheng XJ, Pembrey ME, Malcolm S. Duplication of chromosome 15 in the region 15q11-13 in a patient with developmental delay and ataxia with similarities to Angelman syndrome. J Med Genet. 1993;30:529–531. doi: 10.1136/jmg.30.6.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook EH, Jr, Courchesne RY, Cox NJ, Lord C, Gonen D, Guter SJ, Lincoln A, Nix K, Haas R, Leventhal BL, Courchesne E. Linkage-disequilibrium mapping of autistic disorder, with 15q11-13 markers. American Journal of Human Genetics. 1998;62:1077–1083. doi: 10.1086/301832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook EH, Jr, Lindgren V, Leventhal BL, Courchesne R, Lincoln A, Shulman C, Lord C, Courchesne E. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet. 1997;60:928–934. [PMC free article] [PubMed] [Google Scholar]
- Crolla JA, Harvey JF, Sitch FL, Dennis NR. Supernumerary marker 15 chromosomes: a clinical, molecular and FISH approach to diagnosis and prognosis. Hum Genet. 1995;95:161–170. doi: 10.1007/BF00209395. [DOI] [PubMed] [Google Scholar]
- Culiat CT, Stubbs LJ, Montgomery CS, Russell LB, Rinchik EM. Phenotypic consequences of deletion of the gamma 3, alpha 5, or beta 3 subunit of the type A gamma-aminobutyric acid receptor in mice. Proc Natl Acad Sci U S A. 1994;91:2815–2818. doi: 10.1073/pnas.91.7.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLorey TM, Handforth A, Anagnostaras SG, Homanics GE, Minassian BA, Asatourian A, Fanselow MS, Delgado-Escueta A, Ellison GD, Olsen RW. Mice lacking the beta3 subunit of the GABAA receptor have the epilepsy phenotype and many of the behavioral characteristics of Angelman syndrome. J Neurosci. 1998;18:8505–8514. doi: 10.1523/JNEUROSCI.18-20-08505.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLorey TM, Sahbaie P, Hashemi E, Homanics GE, Clark JD. Gabrb3 gene deficient mice exhibit impaired social and exploratory behaviors, deficits in non-selective attention and hypoplasia of cerebellar vermal lobules: a potential model of autism spectrum disorder. Behav Brain Res. 2008;187:207–220. doi: 10.1016/j.bbr.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis NR, Veltman MW, Thompson R, Craig E, Bolton PF, Thomas NS. Clinical findings in 33 subjects with large supernumerary marker(15) chromosomes and 3 subjects with triplication of 15q11-q13. Am J Med Genet A. 2006;140:434–441. doi: 10.1002/ajmg.a.31091. [DOI] [PubMed] [Google Scholar]
- Descheemaeker MJ, Govers V, Vermeulen P, Fryns JP. Pervasive developmental disorders in Prader-Willi syndrome: the Leuven experience in 59 subjects and controls. Am J Med Genet A. 2006;140:1136–1142. doi: 10.1002/ajmg.a.31235. [DOI] [PubMed] [Google Scholar]
- Descheemaeker MJ, Vogels A, Govers V, Borghgraef M, Willekens D, Swillen A, Verhoeven W, Fryns JP. Prader-Willi syndrome: new insights in the behavioural and psychiatric spectrum. J Intellect Disabil Res. 2002;46:41–50. doi: 10.1046/j.1365-2788.2002.00354.x. [DOI] [PubMed] [Google Scholar]
- Dhar MS, Sommardahl CS, Kirkland T, Nelson S, Donnell R, Johnson DK, Castellani LW. Mice heterozygous for Atp10c, a putative amphipath, represent a novel model of obesity and type 2 diabetes. J Nutr. 2004;134:799–805. doi: 10.1093/jn/134.4.799. [DOI] [PubMed] [Google Scholar]
- Didden R, Sigafoos J, Green VA, Korzilius H, Mouws C, Lancioni GE, O’Reilly MF, Curfs LM. Behavioural flexibility in individuals with Angelman syndrome, Down syndrome, non-specific intellectual disability and Autism spectrum disorder. J Intellect Disabil Res. 2007 doi: 10.1111/j.1365-2788.2008.01055.x. [DOI] [PubMed] [Google Scholar]
- Ding F, Li HH, Zhang S, Solomon NM, Camper SA, Cohen P, Francke U. SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PLoS ONE. 2008;3:e1709. doi: 10.1371/journal.pone.0001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykens EM, Cassidy SB. Correlates of maladaptive behavior in children and adults with Prader-Willi syndrome. Am J Med Genet. 1995;60:546–549. doi: 10.1002/ajmg.1320600612. [DOI] [PubMed] [Google Scholar]
- Fang P, Lev-Lehman E, Tsai TF, Matsuura T, Benton CS, Sutcliffe JS, Christian SL, Kubota T, Halley DJ, Meijers-Heijboer H, et al. The spectrum of mutations in UBE3A causing Angelman syndrome. Hum Mol Genet. 1999;8:129–135. doi: 10.1093/hmg/8.1.129. [DOI] [PubMed] [Google Scholar]
- Fridman C, Koiffmann CP. Origin of uniparental disomy 15 in patients with Prader-Willi or Angelman syndrome. Am J Med Genet. 2000;94:249–253. doi: 10.1002/1096-8628(20000918)94:3<249::aid-ajmg12>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Gérard M, Hernandez L, Wevrick R, Stewart CL. Disruption of the mouse necdin gene results in early post-natal lethality. Nature Genetics. 1999;23:199–202. doi: 10.1038/13828. [see comments] [DOI] [PubMed] [Google Scholar]
- Gilliam J. The Gilliam Autism Rating Scale. Austin, TX: Pro-Ed; 1995. pp. 1–31. [Google Scholar]
- Grammatico P, Di Rosa C, Roccella M, Falcolini M, Pelliccia A, Roccella F, Del Porto G. Inv dup(15): contribution to the clinical definition of phenotype. Clin Genet. 1994;46:233–237. doi: 10.1111/j.1399-0004.1994.tb04232.x. [DOI] [PubMed] [Google Scholar]
- Gray TA, Saitoh S, Nicholls RD. An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc Natl Acad Sci U S A. 1999;96:5616–5621. doi: 10.1073/pnas.96.10.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greally JGT, Gabriel J, Song L, Zemel S, Nicholls R. Conserved characteristics of heterochromatin-forming DNA at the 15q11-q13 imprinting center. PNAS. 1999;96:14430–14435. doi: 10.1073/pnas.96.25.14430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi E, Sahbaie P, Davies MF, Clark JD, DeLorey TM. Gabrb3 gene deficient mice exhibit increased risk assessment behavior, hypotonia and expansion of the plexus of locus coeruleus dendrites. Brain Res. 2007;1129:191–199. doi: 10.1016/j.brainres.2006.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzing LB, Cook EH, Jr, Ledbetter DH. Allele-specific expression analysis by RNA-FISH demonstrates preferential maternal expression of UBE3A and imprint maintenance within 15q11- q13 duplications. Hum Mol Genet. 2002;11:1707–1718. doi: 10.1093/hmg/11.15.1707. [DOI] [PubMed] [Google Scholar]
- Herzing LB, Kim SJ, Cook EH, Jr, Ledbetter DH. The human aminophospholipid-transporting ATPase gene ATP10C maps adjacent to UBE3A and exhibits similar imprinted expression. American Journal of Human Genetics. 2001;68:1501–1505. doi: 10.1086/320616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman AR. Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nat Genet. 1997;17:12–13. doi: 10.1038/ng0997-12. [letter] [DOI] [PubMed] [Google Scholar]
- Hogart A, Nagarajan RP, Patzel KA, Yasui DH, Lasalle JM. 15q11-13 GABAA receptor genes are normally biallelically expressed in brain yet are subject to epigenetic dysregulation in autism-spectrum disorders. Hum Mol Genet. 2007;16:691–703. doi: 10.1093/hmg/ddm014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homanics GE, DeLorey TM, Firestone LL, Quinlan JJ, Handforth A, Harrison NL, Krasowski MD, Rick CE, Korpi ER, Makela R, et al. Mice devoid of gamma-aminobutyrate type A receptor beta3 subunit have epilepsy, cleft palate, and hypersensitive behavior. Proc Natl Acad Sci U S A. 1997;94:4143–4148. doi: 10.1073/pnas.94.8.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsthemke B, Buiting K. Imprinting defects on human chromosome 15. Cytogenet Genome Res. 2006;113:292–299. doi: 10.1159/000090844. [DOI] [PubMed] [Google Scholar]
- Huang B, Bartley J. Partial hexasomy of chromosome 15. Am J Med Genet A. 2003;121:277–280. doi: 10.1002/ajmg.a.20182. [DOI] [PubMed] [Google Scholar]
- Jay P, Rougeulle C, Massacrier A, Moncla A, Mattei MG, Malzac P, Roeckel N, Taviaux S, Lefranc JL, Cau P, et al. The human necdin gene, NDN, is maternally imprinted and located in the Prader-Willi syndrome chromosomal region. Nat Genet. 1997;17:357–361. doi: 10.1038/ng1197-357. [DOI] [PubMed] [Google Scholar]
- Ji Y, Rebert NA, Joslin JM, Higgins MJ, Schultz RA, Nicholls RD. Structure of the highly conserved HERC2 gene and of multiple partially duplicated paralogs in human. Genome Res. 2000;10:319–329. doi: 10.1101/gr.10.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- Kashiwagi A, Meguro M, Hoshiya H, Haruta M, Ishino F, Shibahara T, Oshimura M. Predominant maternal expression of the mouse Atp10c in hippocampus and olfactory bulb. J Hum Genet. 2003;48:194–198. doi: 10.1007/s10038-003-0009-3. [DOI] [PubMed] [Google Scholar]
- Kayashima T, Ohta T, Niikawa N, Kishino T. On the conflicting reports of imprinting status of mouse ATP10a in the adult brain: strain-background-dependent imprinting? J Hum Genet. 2003a;48:492–493. doi: 10.1007/s10038-003-0061-z. author reply 494. [DOI] [PubMed] [Google Scholar]
- Kayashima T, Yamasaki K, Yamada T, Sakai H, Miwa N, Ohta T, Yoshiura K, Matsumoto N, Nakane Y, Kanetake H, et al. The novel imprinted carboxypeptidase A4 gene ( CPA4) in the 7q32 imprinting domain. Hum Genet. 2003b;112:220–226. doi: 10.1007/s00439-002-0891-3. [DOI] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- Knoll JH, Nicholls RD, Magenis RE, Graham JM, Jr, Lalande M, Latt SA. Angelman and Prader-Willi syndromes share a common chromosome 15 deletion but differ in parental origin of the deletion. Am J Med Genet. 1989;32:285–290. doi: 10.1002/ajmg.1320320235. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Kuroda S, Fukata M, Nakamura T, Nagase T, Nomura N, Matsuura Y, Yoshida-Kubomura N, Iwamatsu A, Kaibuchi K. p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J Biol Chem. 1998;273:291–295. doi: 10.1074/jbc.273.1.291. [DOI] [PubMed] [Google Scholar]
- Lalande M, Calciano MA. Molecular epigenetics of Angelman syndrome. Cell Mol Life Sci. 2007;64:947–960. doi: 10.1007/s00018-007-6460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landers M, Bancescu DL, Le Meur E, Rougeulle C, Glatt-Deeley H, Brannan C, Muscatelli F, Lalande M. Regulation of the large (approximately 1000 kb) imprinted murine Ube3a antisense transcript by alternative exons upstream of Snurf/Snrpn. Nucleic Acids Res. 2004;32:3480–3492. doi: 10.1093/nar/gkh670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSalle JM, Lalande M. Homologous association of oppositely imprinted chromosomal domains. Science. 1996;272:725–728. doi: 10.1126/science.272.5262.725. [DOI] [PubMed] [Google Scholar]
- Lee S, Kozlov S, Hernandez L, Chamberlain SJ, Brannan CI, Stewart CL, Wevrick R. Expression and imprinting of MAGEL2 suggest a role in Prader-willi syndrome and the homologous murine imprinting phenotype. Human Molecular Genetics. 2000;9:1813–1819. doi: 10.1093/hmg/9.12.1813. [DOI] [PubMed] [Google Scholar]
- Lee S, Walker CL, Karten B, Kuny SL, Tennese AA, O’Neill MA, Wevrick R. Essential role for the Prader-Willi syndrome protein necdin in axonal outgrowth. Hum Mol Genet. 2005;14:627–637. doi: 10.1093/hmg/ddi059. [DOI] [PubMed] [Google Scholar]
- Lee S, Walker CL, Wevrick R. Prader-Willi syndrome transcripts are expressed in phenotypically significant regions of the developing mouse brain. Gene Expr Patterns. 2003;3:599–609. doi: 10.1016/s1567-133x(03)00113-3. [DOI] [PubMed] [Google Scholar]
- Long FL, Duckett DP, Billam LJ, Williams DK, Crolla JA. Triplication of 15q11-q13 with inv dup(15) in a female with developmental delay. J Med Genet. 1998;35:425–428. doi: 10.1136/jmg.35.5.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Rutter M, Goode S, Heemsbergen J, Jordan H, Mawhood L, Schopler E. Autism diagnostic observation schedule: a standardized observation of communicative and social behavior. J Autism Dev Disord. 1989;19:185–212. doi: 10.1007/BF02211841. [DOI] [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- Lossie AC, Whitney MM, Amidon D, Dong HJ, Chen P, Theriaque D, Hutson A, Nicholls RD, Zori RT, Williams CA, Driscoll DJ. Distinct phenotypes distinguish the molecular classes of Angelman syndrome. J Med Genet. 2001;38:834–845. doi: 10.1136/jmg.38.12.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggouta F, Roberts SE, Dennis NR, Veltman MW, Crolla JA. A supernumerary marker chromosome 15 tetrasomic for the Prader-Willi/Angelman syndrome critical region in a patient with a severe phenotype. J Med Genet. 2003;40:e84. doi: 10.1136/jmg.40.7.e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makedonski K, Abuhatzira L, Kaufman Y, Razin A, Shemer R. MeCP2 deficiency in Rett syndrome causes epigenetic aberrations at the PWS/AS imprinting center that affects UBE3A expression. Hum Mol Genet. 2005;14:1049–1058. doi: 10.1093/hmg/ddi097. [DOI] [PubMed] [Google Scholar]
- Makoff AJ, Flomen RH. Detailed analysis of 15q11-q14 sequence corrects errors and gaps in the public access sequence to fully reveal large segmental duplications at breakpoints for Prader-Willi, Angelman and inv dup(15) syndromes. Genome Biol. 2007;8:R114. doi: 10.1186/gb-2007-8-6-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann SM, Wang NJ, Liu DH, Wang L, Schultz RA, Dorrani N, Sigman M, Schanen NC. Supernumerary tricentric derivative chromosome 15 in two boys with intractable epilepsy: another mechanism for partial hexasomy. Hum Genet. 2004;115:104–111. doi: 10.1007/s00439-004-1127-5. [DOI] [PubMed] [Google Scholar]
- Mao R, Jalal SM, Snow K, Michels VV, Szabo SM, Babovic-Vuksanovic D. Characteristics of two cases with dup(15)(q11.2-q12): one of maternal and one of paternal origin. Genet Med. 2000;2:131–135. doi: 10.1097/00125817-200003000-00003. [DOI] [PubMed] [Google Scholar]
- Martin ER, Menold MM, Wolpert CM, Bass MP, Donnelly SL, Ravan SA, Zimmerman A, Gilbert JR, Vance JM, Maddox LO, et al. Analysis of linkage disequilibrium in gamma-aminobutyric acid receptor subunit genes in autistic disorder. Am J Med Genet. 2000;96:43–48. doi: 10.1002/(sici)1096-8628(20000207)96:1<43::aid-ajmg9>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- McCauley JL, Olson LM, Delahanty R, Amin T, Nurmi EL, Organ EL, Jacobs MM, Folstein SE, Haines JL, Sutcliffe JS. A linkage disequilibrium map of the 1-Mb 15q12 GABA(A) receptor subunit cluster and association to autism. Am J Med Genet B Neuropsychiatr Genet. 2004;131:51–59. doi: 10.1002/ajmg.b.30038. [DOI] [PubMed] [Google Scholar]
- Meguro M, Kashiwagi A, Mitsuya K, Nakao M, Kondo I, Saitoh S, Oshimura M. A novel maternally expressed gene, ATP10C, encodes a putative aminophospholipid translocase associated with Angelman syndrome. Nat Genet. 2001;28:19–20. doi: 10.1038/ng0501-19. [DOI] [PubMed] [Google Scholar]
- Mignon C, Malzac P, Moncla A, Depetris D, Roeckel N, Croquette MF, Mattei MG. Clinical heterogeneity in 16 patients with inv dup 15 chromosome: cytogenetic and molecular studies, search for an imprinting effect. Eur J Hum Genet. 1996;4:88–100. doi: 10.1159/000472176. [DOI] [PubMed] [Google Scholar]
- Milner KM, Craig EE, Thompson RJ, Veltman MW, Thomas NS, Roberts S, Bellamy M, Curran SR, Sporikou CM, Bolton PF. Prader-Willi syndrome: intellectual abilities and behavioural features by genetic subtype. J Child Psychol Psychiatry. 2005;46:1089–1096. doi: 10.1111/j.1469-7610.2005.01520.x. [DOI] [PubMed] [Google Scholar]
- Miura K, Kishino T, Li E, Webber H, Dikkes P, Holmes GL, Wagstaff J. Neurobehavioral and electroencephalographic abnormalities in Ube3a maternal-deficient mice. Neurobiol Dis. 2002;9:149–159. doi: 10.1006/nbdi.2001.0463. [DOI] [PubMed] [Google Scholar]
- Mohandas TK, Park JP, Spellman RA, Filiano JJ, Mamourian AC, Hawk AB, Belloni DR, Noll WW, Moeschler JB. Paternally derived de novo interstitial duplication of proximal 15q in a patient with developmental delay. Am J Med Genet. 1999;82:294–300. [PubMed] [Google Scholar]
- Moncla A, Malzac P, Livet MO, Voelckel MA, Mancini J, Delaroziere JC, Philip N, Mattei JF. Angelman syndrome resulting from UBE3A mutations in 14 patients from eight families: clinical manifestations and genetic counselling. J Med Genet. 1999;36:554–560. [PMC free article] [PubMed] [Google Scholar]
- Nicholls RD. Genomic imprinting and candidate genes in the Prader-Willi and Angelman syndromes. Curr Opin Genet Dev. 1993;3:445–456. doi: 10.1016/0959-437x(93)90119-a. [published erratum appears in Curr Opin Genet Dev 1993 Oct;3(5):802] [DOI] [PubMed] [Google Scholar]
- Nicholls RD, Knepper JL. Genome organization, function and imprinting in Prader-Willi and Angelman syndromes. Annual Review of Genomics and Human Genetics. 2001;2:153–175. doi: 10.1146/annurev.genom.2.1.153. [DOI] [PubMed] [Google Scholar]
- Nietzel A, Albrecht B, Starke H, Heller A, Gillessen-Kaesbach G, Claussen U, Liehr T. Partial hexasomy 15pter-->15q13 including SNRPN and D15S10: first molecular cytogenetically proven case report. J Med Genet. 2003;40:e28. doi: 10.1136/jmg.40.3.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Martin CL, Vazquez-Lopez A, Spence SJ, Alvarez-Retuerto AI, Sigman M, Steindler C, Pellegrini S, Schanen NC, Warren ST, Geschwind DH. Genome-wide expression profiling of lymphoblastoid cell lines distinguishes different forms of autism and reveals shared pathways. Hum Mol Genet. 2007;16:1682–1698. doi: 10.1093/hmg/ddm116. [DOI] [PubMed] [Google Scholar]
- Nowicki ST, Tassone F, Ono MY, Ferranti J, Croquette MF, Goodlin-Jones B, Hagerman RJ. The Prader-Willi phenotype of fragile X syndrome. J Dev Behav Pediatr. 2007;28:133–138. doi: 10.1097/01.DBP.0000267563.18952.c9. [DOI] [PubMed] [Google Scholar]
- Nurmi EL, Amin T, Olson LM, Jacobs MM, McCauley JL, Lam AY, Organ EL, Folstein SE, Haines JL, Sutcliffe JS. Dense linkage disequilibrium mapping in the 15q11-q13 maternal expression domain yields evidence for association in autism. Mol Psychiatry. 2003a;8:624–634. 570. doi: 10.1038/sj.mp.4001283. [DOI] [PubMed] [Google Scholar]
- Nurmi EL, Dowd M, Tadevosyan-Leyfer O, Haines JL, Folstein SE, Sutcliffe JS. Exploratory subsetting of autism families based on savant skills improves evidence of genetic linkage to 15q11-q13. J Am Acad Child Adolesc Psychiatry. 2003b;42:856–863. doi: 10.1097/01.CHI.0000046868.56865.0F. [DOI] [PubMed] [Google Scholar]
- Oliver C, Horsler K, Berg K, Bellamy G, Dick K, Griffiths E. Genomic imprinting and the expression of affect in Angelman syndrome: what’s in the smile? J Child Psychol Psychiatry. 2007;48:571–579. doi: 10.1111/j.1469-7610.2007.01736.x. [DOI] [PubMed] [Google Scholar]
- Penner KA, Johnston J, Faircloth BH, Irish P, Williams CA. Communication, Cognition, and Social Interaction in the Angelman Syndrome. American Journal of Medical Genetics. 1993;46:34–39. doi: 10.1002/ajmg.1320460108. [DOI] [PubMed] [Google Scholar]
- Peters SU, Beaudet AL, Madduri N, Bacino CA. Autism in Angelman syndrome: implications for autism research. Clin Genet. 2004a;66:530–536. doi: 10.1111/j.1399-0004.2004.00362.x. [DOI] [PubMed] [Google Scholar]
- Peters SU, Goddard-Finegold J, Beaudet AL, Madduri N, Turcich M, Bacino CA. Cognitive and adaptive behavior profiles of children with Angelman syndrome. Am J Med Genet A. 2004b;128:110–113. doi: 10.1002/ajmg.a.30065. [DOI] [PubMed] [Google Scholar]
- Pettigrew AL, Gollin SM, Greenberg F, Riccardi VM, Ledbetter DH. Duplication of proximal 15q as a cause of Prader-Willi syndrome. Am J Med Genet. 1987;28:791–802. doi: 10.1002/ajmg.1320280403. [DOI] [PubMed] [Google Scholar]
- Qumsiyeh MB, Rafi SK, Sarri C, Grigoriadou M, Gyftodimou J, Pandelia E, Laskari H, Petersen MB. Double supernumerary isodicentric chromosomes derived from 15 resulting in partial hexasomy. Am J Med Genet A. 2003;116:356–359. doi: 10.1002/ajmg.a.10050. [DOI] [PubMed] [Google Scholar]
- Reiter LT, Seagroves TN, Bowers M, Bier E. Expression of the Rho-GEF Pbl/ECT2 is regulated by the UBE3A E3 ubiquitin ligase. Hum Mol Genet. 2006;15:2825–2835. doi: 10.1093/hmg/ddl225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repetto GM, White LM, Bader PJ, Johnson D, Knoll JH. Interstitial duplications of chromosome region 15q11q13: clinical and molecular characterization. Am J Med Genet. 1998;79:82–89. doi: 10.1002/(sici)1096-8628(19980901)79:2<82::aid-ajmg2>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Rineer S, Finucane B, Simon EW. Autistic symptoms among children and young adults with isodicentric chromosome 15. Am J Med Genet. 1998;81:428–433. doi: 10.1002/(sici)1096-8628(19980907)81:5<428::aid-ajmg12>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Roberts S, Dennis N, Browne C, Willatt L, Woods C, Croos I, Jacobs P, Thomas N. Characterisation of interstitial duplications and triplacations of chromosome 15q11-q13. Human Genetics. 2002;110:227–234. doi: 10.1007/s00439-002-0678-6. [DOI] [PubMed] [Google Scholar]
- Robinson WP, Bernasconi F, Mutirangura A, Ledbetter DH, Langlois S, Malcolm S, Morris MA, Schinzel AA. Nondisjunction of chromosome 15: origin and recombination. Am J Hum Genet. 1993a;53:740–751. [PMC free article] [PubMed] [Google Scholar]
- Robinson WP, Binkert F, Gine R, Vazquez C, Muller W, Rosenkranz W, Schinzel A. Clinical and molecular analysis of five inv dup(15) patients. Eur J Hum Genet. 1993b;1:37–50. doi: 10.1159/000472386. [DOI] [PubMed] [Google Scholar]
- Robinson WP, Dutly F, Nicholls RD, Bernasconi F, Penaherrera M, Michaelis RC, Abeliovich D, Schinzel AA. The mechanisms involved in formation of deletions and duplications of 15q11-q13. J Med Genet. 1998a;35:130–136. doi: 10.1136/jmg.35.2.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson WP, Kuchinka BD, Bernasconi F, Petersen MB, Schulze A, Brondum-Nielsen K, Christian SL, Ledbetter DH, Schinzel AA, Horsthemke B, et al. Maternal meiosis I non-disjunction of chromosome 15: dependence of the maternal age effect on level of recombination. Hum Mol Genet. 1998b;7:1011–1019. doi: 10.1093/hmg/7.6.1011. [DOI] [PubMed] [Google Scholar]
- Robinson WP, Spiegel R, Schinzel AA. Deletion breakpoints associated with the Prader-Willi and Angelman syndromes (15q11-q13) are not sites of high homologous recombination. Hum Genet. 1993c;91:181–184. doi: 10.1007/BF00222722. [DOI] [PubMed] [Google Scholar]
- Robinson WP, Wagstaff J, Bernasconi F, Baccichetti C, Artifoni L, Franzoni E, Suslak L, Shih LY, Aviv H, Schinzel AA. Uniparental disomy explains the occurrence of the Angelman or Prader-Willi syndrome in patients with an additional small inv dup(15) chromosome. J Med Genet. 1993d;30:756–760. doi: 10.1136/jmg.30.9.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Jato S, Nicholls RD, Driscoll DJ, Yang TP. Characterization of cis- and trans-acting elements in the imprinted human SNURF-SNRPN locus. Nucleic Acids Res. 2005;33:4740–4753. doi: 10.1093/nar/gki786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runte M, Huttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- Sahoo T, Shaw CA, Young AS, Whitehouse NL, Schroer RJ, Stevenson RE, Beaudet AL. Array-based comparative genomic hybridization analysis of recurrent chromosome 15q rearrangements. Am J Med Genet A. 2005;139:106–113. doi: 10.1002/ajmg.a.31000. [DOI] [PubMed] [Google Scholar]
- Saitoh S, Hosoki K, Takano K, Tonoki H. Mosaic paternally derived inv dup(15) may partially rescue the Prader-Willi syndrome phenotype with uniparental disomy. Clin Genet. 2007;72:378–380. doi: 10.1111/j.1399-0004.2007.00860.x. [DOI] [PubMed] [Google Scholar]
- Samaco RC, Hogart A, LaSalle JM. Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Hum Mol Genet. 2005;14:483–492. doi: 10.1093/hmg/ddi045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Langmann C, Harden N, Mandel JL, Giangrande A. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron. 2003;38:887–898. doi: 10.1016/s0896-6273(03)00354-4. [DOI] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci U S A. 2001;98:8844–8849. doi: 10.1073/pnas.151231598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer RJ, Phelan MC, Michaelis RC, Crawford EC, Skinner SA, Cuccaro M, Simensen RJ, Bishop J, Skinner C, Fender D, Stevenson RE. Autism and maternally derived aberrations of chromosome 15q. American Journal of Medical Genetics. 1998;76:327–336. doi: 10.1002/(sici)1096-8628(19980401)76:4<327::aid-ajmg8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Cuccaro ML, Hauser ER, Raiford KL, Menold MM, Wolpert CM, Ravan SA, Elston L, Decena K, Donnelly SL, et al. Fine mapping of autistic disorder to chromosome 15q11-q13 by use of phenotypic subtypes. Am J Hum Genet. 2003;72:539–548. doi: 10.1086/367846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skryabin BV, Gubar LV, Seeger B, Pfeiffer J, Handel S, Robeck T, Karpova E, Rozhdestvensky TS, Brosius J. Deletion of the MBII-85 snoRNA gene cluster in mice results in postnatal growth retardation. PLoS Genet. 2007;3:e235. doi: 10.1371/journal.pgen.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni S, Whittington J, Holland AJ, Webb T, Maina EN, Boer H, Clarke D. The phenomenology and diagnosis of psychiatric illness in people with Prader-Willi syndrome. Psychol Med. 2008:1–10. doi: 10.1017/S0033291707002504. [DOI] [PubMed] [Google Scholar]
- Steffenburg S, Gillberg CL, Steffenburg U, Kyllerman M. Autism in Angelman syndrome: A population-based study. Pediatr Neurol. 1996;14:131–136. doi: 10.1016/0887-8994(96)00011-2. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JS, Jiang YH, Galijaard RJ, Matsuura T, Fang P, Kubota T, Christian SL, Bressler J, Cattanach B, Ledbetter DH, Beaudet AL. The E6-Ap ubiquitin-protein ligase (UBE3A) gene is localized within a narrowed Angelman syndrome critical region. Genome Res. 1997;7:368–377. doi: 10.1101/gr.7.4.368. [letter] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JS, Nakao M, Christian S, Orstavik KH, Tommerup N, Ledbetter DH, Beaudet AL. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat Genet. 1994;8:52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher KN, LaSalle JM. Dynamic changes in Histone H3 lysine 9 acetylation localization patterns during neuronal maturation require MeCP2. Epigenetics. 2006;1:24–31. doi: 10.4161/epi.1.1.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher KN, Peddada S, Yasui DH, Lasalle JM. Homologous pairing of 15q11-13 imprinted domains in brain is developmentally regulated but deficient in Rett and autism samples. Hum Mol Genet. 2005;14:785–797. doi: 10.1093/hmg/ddi073. [DOI] [PubMed] [Google Scholar]
- Thomas NS, Browne CE, Oley C, Healey S, Crolla JA. Investigation of a cryptic interstitial duplication involving the Prader-Willi/Angelman syndrome critical region. Hum Genet. 1999;105:384–387. doi: 10.1007/s004390051120. [DOI] [PubMed] [Google Scholar]
- Trillingsgaard A, Ostergaard JR. Autism in Angelman syndrome: an exploration of comorbidity. Autism. 2004;8:163–174. doi: 10.1177/1362361304042720. [DOI] [PubMed] [Google Scholar]
- Tsai TF, Armstrong D, Beaudet AL. Necdin-deficient mice do not show lethality or the obesity and infertility of Prader-Willi syndrome. Nat Genet. 1999;22:15–16. doi: 10.1038/8722. [DOI] [PubMed] [Google Scholar]
- Ullmann R, Turner G, Kirchhoff M, Chen W, Tonge B, Rosenberg C, Field M, Vianna-Morgante AM, Christie L, Krepischi-Santos AC, et al. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat. 2007;28:674–682. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- Ungaro P, Christian SL, Fantes JA, Mutirangura A, Black S, Reynolds J, Malcolm S, Dobyns WB, Ledbetter DH. Molecular characterisation of four cases of intrachromosomal triplication of chromosome 15q11-q14. J Med Genet. 2001;38:26–34. doi: 10.1136/jmg.38.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela MC, Kok F, Otto PA, Koiffmann CP. Phenotypic variability in Angelman syndrome: comparison among different deletion classes and between deletion and UPD subjects. Eur J Hum Genet. 2004;12:987–992. doi: 10.1038/sj.ejhg.5201264. [DOI] [PubMed] [Google Scholar]
- Varela MC, Kok F, Setian N, Kim CA, Koiffmann CP. Impact of molecular mechanisms, including deletion size, on Prader-Willi syndrome phenotype: study of 75 patients. Clin Genet. 2005;67:47–52. doi: 10.1111/j.1399-0004.2005.00377.x. [DOI] [PubMed] [Google Scholar]
- Veltman MW, Craig EE, Bolton PF. Autism spectrum disorders in Prader-Willi and Angelman syndromes: a systematic review. Psychiatr Genet. 2005a;15:243–254. doi: 10.1097/00041444-200512000-00006. [DOI] [PubMed] [Google Scholar]
- Veltman MW, Thompson RJ, Craig EE, Dennis NR, Roberts SE, Moore V, Brown JA, Bolton PF. A paternally inherited duplication in the Prader-Willi/Angelman syndrome critical region: a case and family study. J Autism Dev Disord. 2005b;35:117–127. doi: 10.1007/s10803-004-1039-1. [DOI] [PubMed] [Google Scholar]
- Veltman MW, Thompson RJ, Roberts SE, Thomas NS, Whittington J, Bolton PF. Prader-Willi syndrome--a study comparing deletion and uniparental disomy cases with reference to autism spectrum disorders. Eur Child Adolesc Psychiatry. 2004;13:42–50. doi: 10.1007/s00787-004-0354-6. [DOI] [PubMed] [Google Scholar]
- Vialard F, Mignon-Ravix C, Parain D, Depetris D, Portnoi MF, Moirot H, Mattei MG. Mechanism of intrachromosomal triplications 15q11-q13: a new clinical report. Am J Med Genet A. 2003;118:229–234. doi: 10.1002/ajmg.a.10164. [DOI] [PubMed] [Google Scholar]
- Vogels A, De Hert M, Descheemaeker MJ, Govers V, Devriendt K, Legius E, Prinzie P, Fryns JP. Psychotic disorders in Prader-Willi syndrome. Am J Med Genet A. 2004;127:238–243. doi: 10.1002/ajmg.a.30004. [DOI] [PubMed] [Google Scholar]
- Wandstrat AE, Leana-Cox J, Jenkins L, Schwartz S. Molecular cytogenetic evidence for a common breakpoint in the largest inverted duplications of chromosome 15. American Journal of Human Genetics. 1998;62:925–936. doi: 10.1086/301777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang NJ, Liu D, Parokonny AS, Schanen NC. High-resolution molecular characterization of 15q11-q13 rearrangements by array comparative genomic hybridization (array CGH) with detection of gene dosage. Am J Hum Genet. 2004;75:267–281. doi: 10.1086/422854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang NJ, Parokonny AS, Thatcher KN, Driscoll J, Malone BM, Dorrani N, Sigman M, LaSalle JM, Schanen NC. Multiple forms of atypical rearrangements generating supernumerary derivative chromosome 15. BMC Genet. 2008;9:2. doi: 10.1186/1471-2156-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P, Black G, Ramsden S, Barrow M, Super M, Kerr B, Clayton-Smith J. Angelman syndrome phenotype associated with mutations in MECP2, a gene encoding a methyl CpG binding protein. J Med Genet. 2001;38:224–228. doi: 10.1136/jmg.38.4.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb T, Hardy CA, King M, Watkiss E, Mitchell C, Cole T. A clinical, cytogenetic and molecular study of ten probands with supernumerary inv dup (15) marker chromosomes. Clin Genet. 1998;53:34–43. doi: 10.1034/j.1399-0004.1998.531530107.x. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Wolpert C, Pericak-Vance MA, Abramson RK, Wright HH, Cuccaro ML. Autistic symptoms among children and young adults with isodicentric chromosome 15. Am J Med Genet. 2000a;96:128–129. doi: 10.1002/(sici)1096-8628(20000207)96:1<128::aid-ajmg25>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Wolpert CM, Menold MM, Bass MP, Qumsiyeh MB, Donnelly SL, Ravan SA, Vance JM, Gilbert JR, Abramson RK, Wright HH, et al. Three probands with autistic disorder and isodicentric chromosome 15. Am J Med Genet. 2000b;96:365–372. doi: 10.1002/1096-8628(20000612)96:3<365::aid-ajmg25>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Wu MY, Chen KS, Bressler J, Hou A, Tsai TF, Beaudet AL. Mouse imprinting defect mutations that model Angelman syndrome. Genesis. 2006;44:12–22. doi: 10.1002/gene.20179. [DOI] [PubMed] [Google Scholar]
- Xu N, Tsai CL, Lee JT. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- Yang T, Adamson TE, Resnick JL, Leff S, Wevrick R, Francke U, Jenkins NA, Copeland NG, Brannan CI. A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nature Genetics. 1998;19:25–31. doi: 10.1038/ng0598-25. [DOI] [PubMed] [Google Scholar]
- Yasui DH, Peddada S, Bieda MC, Vallero RO, Hogart A, Nagarajan RP, Thatcher KN, Farnham PJ, Lasalle JM. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci U S A. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]