Abstract
Summary: The arenaviruses are a family of negative-sense RNA viruses that cause severe human disease ranging from aseptic meningitis to hemorrhagic fever syndromes. There are currently no FDA-approved vaccines for the prevention of arenavirus disease, and therapeutic treatment is limited to the use of ribavirin and/or immune plasma for a subset of the pathogenic arenaviruses. The considerable genetic variability observed among the seven arenaviruses that are pathogenic for humans illustrates one of the major challenges for vaccine development today, namely, to overcome pathogen heterogeneity. Over the past 5 years, our group has tested several strategies to overcome pathogen heterogeneity, utilizing the pathogenic arenaviruses as a model system. Because T cells play a prominent role in protective immunity following arenavirus infection, we specifically focused on the development of human vaccines that would induce multivalent and cross-protective cell-mediated immune responses. To facilitate our vaccine development and testing, we conducted large-scale major histocompatibility complex (MHC) class I and class II epitope discovery on murine, nonhuman primate, and human backgrounds for each of the pathogenic arenaviruses, including the identification of protective HLA-restricted epitopes. Finally, using the murine model of lymphocytic choriomeningitis virus infection, we studied the phenotypic characteristics associated with immunodominant and protective T cell epitopes. This review summarizes the findings from our studies and discusses their application to future vaccine design.
INTRODUCTION
According to the National Institute of Allergy and Infectious Diseases (NIAID) “Planning for the 21st Century” report (72), one of the priorities for research in the fields of biodefense and emerging and reemerging infectious diseases is to “design new or improved vaccines that are safe and effective, with particular emphasis on multivalent and cross-protective vaccine strategies.” To date, one of the most commonly encountered challenges for vaccine development is pathogen heterogeneity. In fact, a considerable fraction of unmet vaccine needs for infectious diseases are associated with pathogens naturally displaying significant levels of genetic diversity. In particular, RNA viruses pose a substantial challenge for vaccine development because of their propensity for rapid mutation and recombination. Over the past 5 years, our group has tested several strategies to overcome pathogen heterogeneity, utilizing the pathogenic arenaviruses as a model. In the present review, we summarize our findings from the arenavirus model and discuss their application to future vaccine design. Specifically, we undertook a detailed investigation of the cell-mediated immune response to several pathogenic arenaviruses, primarily by conducting large-scale murine, nonhuman primate (NHP), and human T cell epitope discovery.
In addition to arenaviruses, which are the subject of the present review, several other RNA virus families demonstrate a significant level of genetic diversity. For instance, human immunodeficiency virus (HIV) continues to provide a daunting challenge for vaccine development due to its high mutation rate, which allows for evasion of the adaptive immune response, as well as the inherent genetic variability found among HIV clades circulating worldwide (100). Likewise, hepatitis C virus (HCV) vaccine design faces the obstacle of different genotypes that are found with differential prevalence in distinct locations (86). The four dengue virus (DENV) serotypes provide a unique challenge, as heterologous reinfection is associated with dengue hemorrhagic fever and dengue shock syndrome (42, 81). The genetic variability associated with influenza virus due to antigenic shift and/or antigenic drift is reflected in different strains and subtypes, forcing the development of yearly updated vaccines, and is also the cause of concern in the context of new influenza pandemics (25, 79). It should also be pointed out that pathogen heterogeneity is not limited to RNA viruses, as different families of DNA viruses are of concern, such as adenovirus (41) and human papillomavirus (HPV) (71), as well as bacteria such as Streptococcus pneumoniae (43).
In general, two concepts, which are not mutually exclusive, are applicable to vaccine design for genetically diverse pathogens. First, it might be possible to combine antigens or epitopes derived from each of the different pathogen serotypes in the same vaccine and thus provide effective multivalent protection. A balanced immune response against protein-based vaccines can be achieved, as individual proteins can be added at different concentrations. This approach has proven itself in the case of the currently licensed vaccines against Streptococcus pneumoniae (i.e., Pneumovax 23), which contains capsular polysaccharide antigens from 23 of the most prevalent pneumococcal serotypes (10), and HPV (i.e., Gardasil), which consists of capsid proteins derived from 4 different serotypes prominently associated with disease (19). Although successful, multivalent vaccines, specifically live attenuated virus vaccines, are often associated with significant practical challenges in both their development and manufacturing, as it is often difficult to ensure that equivalent amounts of each antigen are present within the vaccine and that a sufficient immune response is elicited against each different component (47). This has been the case for live attenuated tetravalent vaccine candidates against the four DENV serotypes, as these vaccines have not achieved a balanced attenuation and immune response to each of the four components used in the vaccine inoculum (reviewed in reference 104). Additionally, in human studies, DENV serotype-specific reactogenicity has been problematic (90). Some multivalent vaccines also lack comprehensiveness, as coverage against less frequent or newly emerged virus strains is not achieved. This is not the case for the measles, mumps, and rubella (MMR) vaccine, as it provides complete immunity to three different live attenuated viruses. However, several highly pathogenic HPV serotypes, including 33, 43, and 56, are not covered by the current vaccines, and an alarming rise in the frequency of infections due to nonvaccine pneumococcal serotypes has been reported (18).
A second vaccine approach is based on selecting antigens or epitopes that are relatively conserved between distinct pathogen serotypes. This method has the advantage of simplicity and has been considered for RNA viruses such as influenza virus (32, 89) and HIV (17). However, this approach is associated with the fundamental problem that highly conserved antigens or epitopes might not exist, might be difficult to identify, or might be incapable of inducing significant immune reactivity. Furthermore, it is currently unclear whether conserved regions within pathogens might become variable if placed under selective immune pressure. The latter point may not necessarily be applicable to pathogens that cause incidental human infection as opposed to those for which the human host is the primary reservoir.
ARENAVIRUSES AS A MODEL FOR PATHOGEN HETEROGENEITY AND VACCINE DESIGN
As reviewed elsewhere (26, 29), arenaviruses are members of the Arenaviridae family, consisting of a single genus (Arenavirus), which currently comprises at least 22 different viral species. Phylogenetically, arenaviruses are divided into two main groups: the Old World complex (i.e., Lassa virus [LASV] and lymphocytic choriomeningitis virus [LCMV]) and the New World complex (i.e., Guanarito virus [GTOV], Junin virus [JUNV], Machupo virus [MACV], Sabia virus [SABV], and Whitewater Arroyo virus [WWAV]) (26). Their genomic organization is composed of two single-stranded RNAs that are encoded in ambisense fashion; the L segment (∼7.2 kb) encodes the RNA-dependent RNA polymerase (L) and the zinc finger binding protein (Z), while the S segment encodes the nucleoprotein (NP) and the glycoprotein precursor (GPC) (GPC is posttranslationally processed into GP1, GP2, and a stable signal peptide [SSP]). Significant heterogeneity exists at the amino acid sequence level between different arenavirus species. For instance, the amino acid sequence identities range from 44 to 63% when homologous proteins of the seven Old and New World arenavirus species listed in Table 1 are compared (23). Even within a given arenavirus species, amino acid sequence identities range from 90 to 95% (23).
TABLE 1.
Primary rodent reservoirs, geographical distributions, and disease manifestations of seven pathogenic arenaviruses
| Virus | Acronyma | Primary rodent reservoir(s)b | Geographical distributionc | Human disease |
|---|---|---|---|---|
| Guanarito virus | GTOV | Zygodontomys brevicaudata | Venezuela | Venezuelan hemorrhagic fever |
| Junin virus | JUNV | Calomys musculinus | Argentina | Argentine hemorrhagic fever |
| Lassa virus | LASV | Mastomys huberti | Western coast of Africa (Republic of Guinea, Sierra Leone, Liberia, Nigeria) | Lassa fever |
| Lymphocytic choriomeningitis virus | LCMV | Mus musculus, Mus domesticus | Worldwide | Aseptic meningitis, congenital deformities, and high lethality in immunosuppressed patients |
| Machupo virus | MACV | Calomys callosus | Bolivia | Bolivian hemorrhagic fever |
| Sabia virus | SABV | Unknown | Brazil | Brazilian hemorrhagic fever |
| Whitewater arroyo virus | WWAV | Neotoma albigula | Southwestern US | Hemorrhagic fever |
Each arenavirus typically establishes a persistent, asymptomatic infection within its primary rodent reservoir. Humans can become infected by exposure to infected rodents or their infectious excreta. Person-to-person transmission can occur vertically (11), nosocomially (20), or through solid organ transplantation (35). At least seven arenaviruses (LASV and LCMV from the Old World and JUNV, MACV, GTOV, SABV, and WWAV from the New World) are pathogenic in humans and cause a variety of disease manifestations, including severe hemorrhagic fever syndromes (reviewed in reference 40), aseptic meningitis (7), and/or congenital deformities (Table 1) (11). Mortality among patients with arenaviral hemorrhagic fever ranges from 15 to 30% (62, 66). Of these, Lassa fever is regarded as the most severe threat to human health, as LASV is responsible for 100,000 to 300,000 infections and ∼5,000 deaths annually (66). Because of the morbidity and mortality associated with arenavirus infection in humans, arenaviruses that cause hemorrhagic fever require biosafety level 4 (BSL-4) containment.
Despite the pathogenic nature of arenaviruses, there are no U.S. Food and Drug Administration (FDA)-approved vaccines available. A randomized, double-blind trial involving agricultural workers in Argentina demonstrated that the live attenuated JUNV vaccine Candid 1 is an effective and safe vaccine with no serious adverse side effects. However, Candid 1 has only investigational new drug status in the United States (63). Moreover, antiviral therapies are limited to the use of hyperimmune plasma (20) or the guanosine analogue ribavirin, which can lead to adverse side effects such as thrombocytosis, severe anemia, and birth defects (34, 67). Considering the current lack of vaccines and therapeutic treatment options for preventing and/or ameliorating human arenavirus disease, the development of effective vaccines is highly desirable. Given the practical challenges associated with the development and deployment of seven different arenaviral vaccines, the development of a multivalent vaccine capable of providing protection against the major arenavirus species associated with pathogenicity in humans is considered a priority.
Here we report the results of our studies to address these issues. Because antigen-specific CD8+ T cells provide protective immunity against arenavirus infection (14), our general interest was to determine whether we could identify an HLA-restricted CD8+ T cell epitope cocktail, derived from the seven arenaviruses pathogenic in humans (Table 1), that would provide multivalent protection against these pathogens with universal population coverage. Additionally, we reasoned that the epitopes identified in our studies would also be of use as diagnostic reagents for evaluating the antigenicity of new vaccines or studying the natural history of T cell responses to arenavirus infection to determine their role in disease pathogenesis. We approached our studies along several different lines of investigation. One of our objectives was to define epitopes recognized in animal models of arenavirus infection, including epitopes restricted by common murine (H-2b and H-2d) and rhesus macaque (Mamu) major histocompatibility complex (MHC) molecules. The rationale for these studies was, in part, that the definition of epitopes restricted by murine MHC would enable us to formulate and address specific questions answerable in the murine model of LCMV infection but also potentially applicable to the human system. Furthermore, we reasoned that the definition of potential epitopes recognized by an NHP species, such as rhesus macaques, would be of interest in facilitating the preclinical and clinical evaluation of vaccine constructs destined for human testing.
In a parallel series of studies, we sought to define epitopes restricted by human (HLA) MHC molecules. As very little is known regarding the natural history of the human T cell response to arenavirus infection, identification of HLA-restricted epitopes would enable studies to define the phenotypic characteristics associated with protective T cell responses. One of the most straightforward strategies to accomplish this goal is the study of peripheral blood mononuclear cells (PBMC) derived from humans previously infected by an arenavirus. However, the paucity of available samples from humans exposed to pathogenic arenaviruses, in general, rendered this approach problematic. Therefore, we relied on an alternative strategy, namely, the use of HLA transgenic mice. Strains of mice expressing some of the most common HLA class I and class II molecules have been developed (1-3, 97). The availability of these mice allowed us to study, in a controlled experimental setting, HLA-restricted CD8+ and CD4+ T cell responses.
Following the identification of HLA-restricted epitopes, our plan was to evaluate, in HLA transgenic mice, the immunogenicity of an epitope-based vaccine incorporating arenavirus-specific determinants and to test the hypothesis that simultaneous immunity could be induced against multiple arenavirus species. We were interested in examining whether this objective could be achieved by use of conserved and/or cross-reactive epitopes or would require the inclusion of different sets of epitopes, each derived from a different arenavirus species. We designated a conserved epitope as a peptide derived from multiple arenavirus species with an identical amino acid sequence, while a peptide with an orthologous amino acid sequence between different arenaviruses was considered a cross-reactive epitope. Furthermore, we wanted to evaluate whether immunization with these epitopes could protect HLA transgenic mice from challenge with different arenavirus species. Finally, we wanted to demonstrate how the use of HLA supertype epitopes would provide balanced coverage across different HLA molecules prevalent in distinct ethnicities found worldwide. In all cases, we planned to submit the identified epitopes and associated data to the Immune Epitope Database (IEDB) (http://www.immuneepitope.org) (76).
CELL-MEDIATED IMMUNE RESPONSE AGAINST ARENAVIRUS INFECTION
CD8+ and CD4+ T cell responses have clearly been associated with reduced pathology and protection against Old World arenaviruses in animal models of infection (13, 14, 50, 56, 99, 101). Previous studies demonstrated that mice lacking CD8+ T cells or the cytolytic protein perforin are incapable of resolving a primary LCMV infection (49, 101). Studies have also shown that CD4+ T cell-deficient mice were unable to mount an effective memory CD8+ T cell response and control viremia following infection with highly virulent LCMV strains (50, 99). Furthermore, vaccine strategies aimed at generating a CD8+ T cell-mediated response confer protection against virus challenge in murine (13, 14, 56), guinea pig (5, 69), and NHP (36) models of LCMV and LASV infection. In humans, cell-mediated immunity also seems to be critical for protection against LASV infection, as neutralizing antibodies appear several weeks or months after viral clearance (28, 37), and treatment of infected patients with immune plasma does not protect against disease (65).
In contrast, antibody-mediated immunity seems to play an important role in protecting against New World arenavirus infection, as administration of convalescent-phase serum at an early infection stage significantly reduces morbidity and mortality of JUNV infection from greater than 15% to less than 1% (33, 44). T cell responses might also be involved in countering New World arenavirus infection, as, similar to the case for LASV infection, neutralizing antibodies often appear several weeks after resolution of JUNV infection (68). JUNV-specific T cell responses have also been detected in patients vaccinated with Candid 1 (63). Thus, vaccination strategies aimed at generating cell-mediated immune responses against both Old and New World arenaviruses should be considered, especially because T cell recognition of virus-infected cells is less vulnerable to viral mutation than antibody-mediated immunity (75, 96).
Identification of CD8+ and CD4+ T Cell-Mediated Immune Responses in Animal Models of Arenavirus Infection
LCMV, the prototypic member of the Arenaviridae family, is arguably one of the most well studied viruses in terms of murine CD8+ and CD4+ T cell responses (reviewed in references 52 and 87). This is in part because experimental infection of mice with LCMV can lead to several different immune response and disease outcomes that are dependent on the route of infection, the virus strain utilized, and the age of the mouse inoculated. Acute infections can be established by intraperitoneal (i.p.) inoculation of immunocompetent mice with the Armstrong clone 53b strain of LCMV, as this results in a largely asymptomatic infection that is resolved 8 to 10 days postinfection. Resolution of infection is dependent primarily on a cell-mediated immune response, specifically, perforin-mediated lysis of infected cells by CD8+ T cells (49, 101). In contrast, intracranial (i.c.) inoculation with the same virus strain results in lethal choriomeningitis. At 6 to 9 days postinfection, these animals die as a result of an immunopathological CD8+ T cell response directed against infected cells in the choroid plexus and the meninges (21). The murine i.c. infection model is highly relevant to human disease, as this is the same mechanism that causes LCMV-induced aseptic meningitis in humans (20).
Chronic infections, which are ultimately cleared, can be established in immunocompetent mice by intravenous inoculation with immunosuppressive strains of LCMV, including clone 13, Traub, WE, and WE docile (12). During a chronic LCMV infection, virus-specific CD8+ and CD4+ T cells become functionally exhausted, with impaired cytokine production and proliferation (6). Finally, life-long persistent infections can be established by inoculation of immunologically immature mice, either in utero or within 24 h of birth, with several different strains of LCMV, such as Armstrong and others.
Unlike other pathogenic arenaviruses (i.e., LASV, JUNV, MACV, GTOV, and SABV), which require BSL-4 containment, most studies involving different laboratory strains of LCMV in mice necessitate only BSL-2 facilities, as accidental inoculation (i.e., needle stick injury or aerosol exposure) with these strains typically does not result in severe hemorrhagic fever in humans. That said, it should be noted that the National Institutes of Health (NIH), which defines human pathogens by risk group (RG), lists most laboratory strains of LCMV as RG-2 (infection rarely serious), while neurotropic strains (i.e., strain Armstrong clone 53b) are classified as RG-3 (infections may be serious or lethal, but there is a low risk of spread to the community) (http://oba.od.nih.gov/oba/rac/guidelines_02/APPENDIX_B.htm#_Toc7238339). Additionally, as most human outbreaks have been associated with transmission from hamsters (22), experimental models of LCMV infection in hamsters are recommended to be conducted at BSL-3 by the Centers for Disease Control and Prevention (CDC) (http://www.cdc.gov/od/ohs/biosfty/bmbl5/BMBL_5th_Edition.pdf). Finally, BSL-3 conditions are also recommended by the CDC for activities with high potential for aerosol generation or when working with strains of LCMV shown to be lethal in NHPs (98).
We utilized the murine model of acute LCMV Armstrong infection to validate the approaches intended for use in the human studies. Indeed, we found that a significant fraction of the CD8+ CD44hi effector T cell response in C57BL/6J (H-2b) mice acutely infected with the Armstrong strain of LCMV was not accounted for by previously identified viral epitopes (56). Utilizing a combination of bioinformatic epitope prediction and overlapping peptides, we identified 19 novel epitopes (Table 2), which together with the 9 previously known epitopes accounted for the total CD8+ CD44hi response. Strikingly, 15 of the 19 new epitopes were derived from the viral L polymerase, which was not previously recognized as a target of the cellular response during arenavirus infections. Furthermore, L epitope immunization was shown to be protective against LCMV challenge. In summary, these results validated our epitope identification approach and illustrated how CD8+ T cell responses to arenaviruses are highly multispecific, thus suggesting that many different epitope candidates might also be recognized following human infection.
TABLE 2.
Murine LCMV-specific MHC class I-restricted CD8+ T cell epitopes
| Epitopea | Peptide sequence | MHC restriction | Protective capacityb | Reference(s)c |
|---|---|---|---|---|
| GPC10-18 | ALPHIIDEV | H-2d | − | 14 |
| GPC33-41 | KAVYNFATC | H-2Db | + | 14, 53, 56 |
| GPC34-41 | AVYNFATC | H-2Kb | + | 53, 56 |
| GPC44-52 | FALISFLLL | H-2Db | ND | 56 |
| GPC70-77 | GVYQFKSV | H-2Kb | ND | 64 |
| GPC92-101 | CSANNSHHYI | H-2Db | + | 56, 94 |
| GPC99-108 | HYISMGTSGL | H-2Kd | − | 95 |
| GPC118-125 | ISHNFCNL | H-2Kb | ND | 56, 94 |
| GPC166-173 | ITIQYNLT | H-2Kb | ND | 56 |
| GPC221-228 | SQTSYQYL | H-2Kb | ND | 56 |
| GPC276-286 | SGVENPGGYCL | H-2Db | − | 53, 56 |
| GPC283-291 | GYCLTKWMI | H-2Kd | +/− | 95 |
| GPC365-372 | MGVPYCNY | H-2Kb | ND | 56 |
| L156-163 | ANFKFRDL | H-2Kb | +/− | 56 |
| L313-320 | TSTEYERL | H-2Kb | ND | 56 |
| L338-346 | RQLLNLDVL | H-2Db | ND | 56 |
| L349-357 | SSLIKQSKF | H-2Kb | ND | 56 |
| L455-463 | FMKIGAHPI | H-2Db | − | 56 |
| L663-671 | VVYKLLRFL | H-2Kb | ND | 56 |
| L689-697 | KFMLNVSYL | H-2Db | ND | 56 |
| L743-751 | VFYEQMKRF | H-2Kb | ND | 56 |
| L775-782 | SSFNNGTL | H-2Kb | ND | 56 |
| L1189-1196 | MMCPFLFL | H-2Kb | ND | 56 |
| L1189-1197 | MMCPFLFLM | H-2d | ND | 14 |
| L1302-1310 | INYCIGVIF | H-2Kb | ND | 56 |
| L1369-1377 | FAAEFKSRF | H-2Kb | ND | 56 |
| L1428-1435 | NSIQRRTL | H-2Kb | ND | 56 |
| L1878-1885 | GPFQSFVS | H-2Kb | ND | 56 |
| L2062-2069 | RSIDFERV | H-2Kb | + | 56 |
| NP91-98 | VGRLSAEE | H-2k | ND | 60 |
| NP118-126 | RPQASGVYM | H-2Ld | + | 8, 82, 95 |
| NP165-175 | SSLLNNQFGTM | H-2Db | ND | 56, 93 |
| NP205-212 | YTVKYPNL | H-2Kb | + | 56, 94 |
| NP238-248 | SGYNFSLGAAV | H-2Kb | ND | 56, 93 |
| NP313-322 | WPYIACRTSI | H-2Ld | + | 78 |
| NP314-322 | PYIACRTSI | H-2Kd | +/− | 78, 95 |
| NP396-404 | FQPQNGQFI | H-2Db | + | 56, 94 |
Peptide position within the GPC, NP, or L protein of the Armstrong clone 53b strain of LCMV. Similar epitopes with nested peptide sequences have also been reported for NP165-175 and NP238-248 (64, 93).
Protective capacity was evaluated by immunizing C57BL/6J (H-2b) or BALB/c (H-2d) mice with the indicated peptide and subsequently challenging with either an i.p. inoculation of LCMV to measure a reduction in viral titers or an i.c. inoculation of a lethal LCMV dose to measure survival. ND, the protective capacity was not determined; +/−, partial protection.
Reference(s) for epitope identification and/or protection studies.
In a subsequent study, we further investigated the basis of immunodominance of LCMV-specific CD8+ T cell responses (57). Several factors have been suggested to contribute to CD8+ T cell immunodominance (reviewed in references 105 and 106). Of importance are the efficiency of antigen processing and presentation (27, 30, 74), binding affinity of peptides to MHC class I (85, 95), stability of the peptide-MHC class I complex (59), and epitope abundance on the surface of antigen-presenting cells (APCs) (39, 58). Immunodominance hierarchies have also found to be influenced by viral load and the kinetics of viral protein expression during infection (77, 93, 102, 103). Although less well defined, the kinetics of the CD8+ T cell response to antigen (61), T cell receptor (TCR) avidity (92), variations in the naïve CD8+ T cell repertoire (15, 38, 45, 58), and immunoregulatory mechanisms are also thought to play critical roles (4). How these factors are coordinated to determine CD8+ T cell immunodominance has been only partially defined.
To evaluate the role of epitope competition in immunodominance, we manipulated the number of CD8+ T cell epitopes that could be recognized during acute LCMV infection. Either decreasing total epitope numbers by infecting H-2b mice with an LCMV variant lacking dominant epitopes or wild-type LCMV infection of C57BL/6J mice lacking H-2Kb resulted in minor gamma interferon (IFN-γ) response increases for the remaining epitopes and no novel epitopes being recognized. Increasing epitope numbers by infecting H-2b F1 hybrid mice (H-2bxa, H-2bxd, H-2bxk, and H-2bxs) with LCMV, delivery of LCMV proteins by a recombinant vaccinia virus (rVACV), or epitope delivery as a pool in incomplete Freund's adjuvant (IFA) maintained the overall CD8+ T cell response pattern. Thus, we found that the immunodominance hierarchy is largely unaffected by epitope deletion or addition. We observed a positive correlation between MHC binding affinity and the magnitude of the epitope-specific CD8+ T cell response following LCMV infection. Furthermore, the naïve CD8+ T cell precursor frequency, directly measured by LCMV-specific tetramer staining, also correlated with the CD8+ T cell response hierarchy seen after LCMV infection. Thus, our results indicated that an intrinsic property of the epitope (MHC binding affinity) and an intrinsic property of the host (naïve precursor frequency) jointly dictated the immunodominance hierarchy of CD8+ T cell responses. It is difficult to determine if MHC binding affinity is more important than the T cell repertoire in shaping immunodominance hierarchies, as T cell immunogenicity is dependent on peptide-MHC binding. In agreement with previous studies (105, 106), we recently demonstrated in the VACV system that the efficiency of antigen processing, binding affinity for MHC class I, the nature of the T cell repertoire, and immunoregulatory mechanisms all play important roles in establishing immunodominance (4). In a practical sense, our results in the LCMV system suggested that a vaccine strategy based on pooling different epitopes should be feasible to elicit simultaneous responses against multiple arenavirus species.
Activation of CD4+ T cells sustains CD8+ T cell responses and is required for effective clearance of acute infection in several different viral systems, including LCMV (70). Accordingly, we proposed to define and study epitopes recognized by murine CD4+ T cells. In a primary study, we identified LCMV-specific CD4+ T cell epitopes in BALB/c mice (H-2d) acutely infected with LCMV Armstrong by utilizing overlapping peptides that spanned the entire LCMV proteome (70). These studies identified nine novel H-2d-restricted epitopes and provided the first evidence of a CD4+ T cell response against the Z protein (Table 3). Furthermore, these studies identified overlapping CD4+ and CD8+ T cell epitopes (NP116-130 and NP118-126). In a subsequent study, six H-2b-restricted epitopes (two known and four novel) were identified in LCMV-infected C57BL/6J (H-2b) mice (31). This study demonstrated a second instance of a CD4+ T cell epitope containing a nested CD8+ T cell epitope (GPC31-45 and GPC33-41, respectively). The overlap of the CD4+ and CD8+ T cell immune responses might be related to the immunodominance of particular viral regions and might be influenced by proteasomal processing, MHC binding affinity, and/or TCR repertoires (31). Taken together, these studies also demonstrated that the LCMV-specific CD4+ T cell response is more complex than previously appreciated.
TABLE 3.
Murine LCMV-specific MHC class II-restricted CD4+ T cell epitopes
| Epitopea | Peptide sequence | MHC restriction | Reference(s)c |
|---|---|---|---|
| GPC6-20 | TMFEALPHIIDEVIN | I-Ab | 31 |
| GPC31-45 | GIKAVYNFATCGIFA | I-Ab | 31 |
| GPC66-80 | DIYKGVYQFKSVEFDb | I-Ab | 31, 46, 73 |
| GPC126-140 | TSAFNKKTFDHTLMS | I-Ab | 31 |
| GPC176-190 | DAQSAQSQCRTFRGR | I-Ad | 70 |
| GPC186-200 | TFRGRVLDMFRTAFG | I-Ab | 31 |
| GPC316-330 | CDMLRLIDYNKAALS | I-Ad | 70 |
| GPC409-423 | IEQEADNMITEMLRK | I-Ak | 51 |
| NP6-20 | EVKSFQWTQALRREL | I-Ed | 70 |
| NP86-100 | KNVLKVGRLSAEELM | I-Ad | 70 |
| NP116-130 | SERPQASGVYMGNLT | I-Ad | 70 |
| NP176-190 | PSLTMACMAKQSQTP | I-Ad | 70 |
| NP311-325 | EGWPYIACRTSIVGR | I-Ab | 31, 73 |
| NP466-480 | SQNRKDIKLIDVEMT | I-Ad | 70 |
| NP496-510 | GWLCKMHTGIVRDKK | I-Ad and I-Ed | 70 |
| Z31-45 | SCKSCWQKFDSLVRC | I-Ad and I-Ed | 70 |
Peptide position within the GPC, NP, or Z protein of the Armstrong clone 53b strain of LCMV. Similar epitopes with nested peptide sequences have also been reported for GPC66-80 and NP311-325 (73).
The underlined peptide sequence indicates the optimal minimal epitope for the LCMV GPC66-80 epitope (46).
Reference(s) for epitope identification.
Finally, to further support the development of animal models of arenavirus infection, we undertook studies toward the identification of arenavirus-derived peptides with the capacity to bind rhesus macaque (Mamu) class I and II MHC molecules with high affinity. These data will enable the establishment of rigorous assays for quantification of arenavirus-specific CD8+ and CD4+ T cells and will be crucial for the development of macaque models of arenavirus infection. Protein sequences from the seven pathogenic arenaviruses listed in Table 1 were scanned, using panels of algorithms available through the IEDB analysis resource, for binding motifs of two of the most common Mamu class I (A*01 and B*17) alleles expressed by rhesus macaques. In total, 104 and 39 peptides that bound Mamu A*01 and Mamu B*17, respectively, with high affinity (50% inhibitory concentration [IC50] of ≤100 nM) were identified. For class II binding studies, we tested 298 degenerate HLA-DR binding peptides also derived from the seven pathogenic arenaviruses and found that 220 of these peptides bound to Mamu DR with high affinity. Overall, these studies identified several high-affinity Mamu binding peptides for future studies, and these data have been made available to the scientific community by deposition in the IEDB.
In summary, the studies performed in common animal models of arenavirus infection further validated our epitope identification approaches, identified several novel CD8+ and CD4+ T cell epitopes, and established that the viral L polymerase and Z proteins are targets of T cell-mediated immunity. We also demonstrated that CD8+ T cell epitopes derived from the L protein were protective against LCMV challenge, thus illustrating that epitopes from nearly every arenavirus open reading frame (ORF) should be considered for vaccine design. Furthermore, these studies also demonstrated that strict immunodominance is not necessarily applicable to arenavirus-specific T cell responses, as we defined an abundance of viral epitopes. Therefore, a multitude of epitope candidates might be available for the human studies.
Identification of Arenavirus-Specific CD8+ T Cell-Mediated Immune Responses in HLA Transgenic Mice
Two independent studies laid the foundations of our strategy for identifying HLA-restricted epitopes utilizing HLA transgenic mice. As mentioned above, these mice represent an attractive model system because they enable the identification of HLA-restricted epitopes in cases where PBMC from exposed individuals are limiting or not available. HLA transgenic mice permit the evaluation of epitope immunogenicity not only after virus infection but also following direct immunization with isolated epitopes or epitope pools. Furthermore, this model system allowed the evaluation of whether the identified epitopes could confer protection against virus challenge. The data generated from HLA transgenic mice should, however, be interpreted with caution, as it was recently demonstrated that the overlap between the repertoires of VACV-derived epitopes identified in VACV-infected HLA-A*0201 transgenic mice and HLA-A*0201-positive human Dryvax vaccinees is often incomplete (54).
A primary study, examining LASV-specific CD8+ T cell responses, identified three novel LASV GPC-derived epitopes following peptide immunization of HLA-A*0201 transgenic mice (Table 4) (13). These three CD8+ T cell epitopes displayed high binding affinity to HLA-A*0201 and were naturally processed from native LASV GPC in human HLA-A*0201-positive target cells. Furthermore, it was demonstrated that immunization with LASV GPC42-50 or GPC60-68 protected HLA-A*0201 transgenic mice against challenge with an rVACV expressing the LASV GPC. This study was a key element in the validation of our epitope identification strategy, as it showed that challenge with an rVACV expressing an arenavirus protein is a viable method to evaluate, at least in a preliminary fashion, the protective capacity of arenavirus-derived epitopes. This strategy also circumvented the need to perform initial protection evaluations utilizing natural arenavirus infection, which would generally require BSL-4 containment.
TABLE 4.
Human arenavirus-specific HLA class I-restricted CD8+ T cell epitopes
| Virus and epitopea | Peptide sequence | MHC restriction | Protective capacityb | Reference(s)c |
|---|---|---|---|---|
| GTOV L1977-1985 | ATVKNVVLR | HLA-A*1101 | ND | 55 |
| JUNV GPC18-26 | ALNIALVAV | HLA-A*0201 | ND | 55 |
| LASV | ||||
| GPC42-50 | GLVGLVTFL | HLA-A*0201 | + | 13 |
| GPC60-68 | SLYKGVYEL | HLA-A*0201 | + | 9, 13 |
| GPC111-120 | SIINHKFCNL | HLA-A*0201 | ND | 55 |
| GPC441-449 | YLISIFLHL | HLA-A*0201 | + | 9, 13 |
| LCMV | ||||
| GPC10-18 | ALPHIIDEV | HLA-A*0201 | − | 14 |
| GPC46-55 | LVSFLLLAGR | HLA-A*1101 | ND | 55 |
| GPC112-120 | FTNDSIISH | HLA-A*1101 | ND | 55 |
| GPC447-455 | YLVSIFLHL | HLA-A*0201 | + | 14 |
| NP69-77 | SLNQTVHSL | HLA-A*0201 | + | 14 |
| Z24-33 | TTYLGPLSCK | HLA-A*1101 | ND | 55 |
| Z49-58 | YLCRHCLNLL | HLA-A*0201 | − | 14, 55 |
| MACV | ||||
| GPC18-26 | ALNIALVAV | HLA-A*0201 | ND | 55 |
| NP19-27 | GLSQFTHTV | HLA-A*0201 | ND | 55 |
| NP82-90 | SIQKNTIFK | HLA-A*1101 | ND | 55 |
| NP432-440 | AMPGVLSYV | HLA-A*0201 | ND | 55 |
| Z27-36 | RTAPPSLYGR | HLA-A*1101 | ND | 55 |
| SABV | ||||
| GPC90-98 | STYYVHENK | HLA-A*1101 | + | 55 |
| GPC142-150 | GLLEWIFRA | HLA-A*0201 | + | 55 |
| NP82-90 | SSQRDTILK | HLA-A*1101 | ND | 55 |
| NP547-556 | LLPDALLFTL | HLA-A*0201 | ND | 55 |
| Z64-72 | KCLNIMLGK | HLA-A*1101 | ND | 55 |
| WWAV | ||||
| GPC42-50 | GLLQFIVFL | HLA-A*0201 | ND | 55 |
| NP274-282 | TVIKTLLEV | HLA-A*0201 | ND | 55 |
| NP439-447 | SSIIRSLPK | HLA-A*1101 | ND | 55 |
Peptide position within the GPC, NP, or L protein of prototypic strains of GTOV, JUNV, LASV, LCMV, MACV, SABV, and WWAV.
Protective capacity was evaluated by immunizing HLA-A*0201 transgenic mice with the indicated peptide individually or as a pool and subsequently challenging with either an i.p. inoculation of LCMV or the appropriate rVACV to measure a reduction in viral titers or an i.c. inoculation of a lethal LCMV dose to measure survival. ND, the protective capacity was not determined.
Refers to reference for epitope identification and/or protection studies.
In a subsequent study, utilizing strategies similar to those described above, Botten and colleagues identified four novel LCMV-derived epitopes following infection of HLA-A*0201 transgenic mice with LCMV Armstrong (14). All four LCMV-derived epitopes were naturally processed from native LCMV proteins in human HLA-A*0201-positive target cells. Furthermore, peptide immunization of the LCMV GPC447-455 epitope induced in vivo killing of peptide-pulsed target cells and protected mice against lethal i.c. challenge with LCMV. Taken together, these studies demonstrated the importance of CD8+ T cell-mediated immunity in providing protection against arenavirus infection.
The question remains, however, whether challenge with an rVACV expressing an arenavirus protein would be a sufficient mimic of a natural arenavirus infection. The data from Kotturi et al. suggested that infection of H-2b mice with either LCMV or different rVACVs, each expressing distinct LCMV proteins, yielded a similar set of CD8+ T cell epitopes and immunodominance hierarchy (56). Likewise, the studies conducted by Botten et al. demonstrated that infection of human HLA-A*0201-positive target cells with an rVACV could be used to detect naturally processed LASV- and LCMV-derived CD8+ T cell epitopes (13, 14).
Accordingly, as a tool to induce and evaluate HLA- restricted CD8+ T cell responses, we generated different rVACVs expressing a panel of proteins from the seven arenaviruses targeted by our studies (55). More specifically, we constructed rVACVs for ORFs of each arenavirus, including all four proteins (GPC, NP, L, and Z) derived from all seven arenaviruses (Table 5). The gene sequences encoding the different arenavirus proteins were derived from prototypic virus strains, and the constructs were engineered using the Western Reserve (WR) strain of VACV. In four cases, (JUNV Z, SABV L, WWAV L, and WWAV Z), rVACVs were not generated because of a lack of sequence information or technical difficulties. The remaining 24 rVACV constructs were generated, arenavirus protein expression was confirmed experimentally for each rVACV through Western blot analysis with infected cell lysates, and these constructs were utilized in the HLA epitope identification studies described below. Furthermore, each rVACV has been deposited in the Biodefense and Emerging Infections Research Resources Repository (BEI Resources; http://www.beiresources.org) (BEIR NR-15486 to NR-15509) as a resource available for the scientific community.
TABLE 5.
Panel of available rVACVs expressing proteins from seven different pathogenic arenaviruses
| Protein | Expression by rVACVa |
||||||
|---|---|---|---|---|---|---|---|
| GTOV | JUNV | LASV | LCMVb | MACV | SABV | WWAV | |
| GPC | √ | √ | √ | √ | √ | √ | √ |
| L | √ | √ | √ | √ | √ | × | × |
| NP | √ | √ | √ | √ | √ | √ | √ |
| Z | √ | × | √ | √ | √ | √ | × |
√, rVACV expressing the particular arenavirus protein was generated and used to identify HLA class I-restricted arenavirus-specific epitopes. ×, rVACV was not generated because of a lack of protein sequence information or technical difficulties. Each rVACV has been deposited at the BEI Resources (http://www.beiresources.org) (BEIR NR-15486 to NR-15509).
LCMV GPC and NP gene expression was placed under the control of the VACV P7.5K early/late promoter, while the remaining arenavirus genes were under the control of a synthetic early/late promoter, PSYN.
In the next series of experiments, we screened the GPC, L, NP, and Z protein sequences of our seven target arenavirus species for peptides predicted to bind with high affinity to HLA class I molecules. First, to avoid biases and redundancies, we sought to define a representative set of arenavirus protein sequences. Second, we sought to perform the bioinformatic epitope prediction analysis in a manner that would provide balanced coverage of the four different proteins from each virus species. Finally, we focused on epitopes conserved within known protein sequences of a given virus species and, if possible, also among protein sequences from different virus species.
To address the first issue, we developed a database containing protein sequences from the seven arenaviruses, which included a nonredundant set of 333 manually annotated protein sequences (23). Manual annotation ensured that redundant protein sequences were eliminated, and representative sequences were chosen to emphasize in vivo isolates from exposed humans. All sequence entries were linked to the NCBI and cited PubMed references. Sequence variability analyses were also performed, and the results are hosted in the arenavirus protein sequence database, available at http://epitope.liai.org:8080/projects/arena (23).
Next, we screened arenavirus sequences for six major HLA class I supertype binding motifs (A01, A02, A03/A11, A24, B07, and B44) (88), with emphasis on conserved epitopes and balanced virus coverage. To address this issue, for each HLA supertype, we selected up to 30 different candidate epitopes from each virus species and from each of the four GPC, L, NP, and Z proteins. This strategy was utilized to account for the fact that the L protein is much larger than the other arenavirus proteins, and thus, without introducing a specific “filter,” the epitope selection would have resulted in a heavy bias in terms of candidate epitopes derived from the L protein. To select the 30 candidate epitopes for each HLA supertype/virus/protein combination, we first selected all peptides with a predicted IC50 of ≤100 nM from each ORF and then ranked each candidate epitope based on intervirus conservancy (percent identity matches). In conclusion, these analyses allowed the selection of a set of candidate epitopes suitable for further evaluation in HLA transgenic mice.
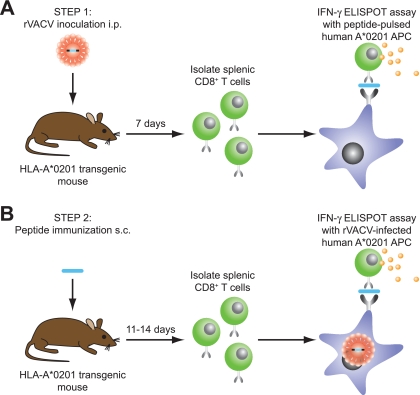
Next, we determined the antigenicities of the candidate epitopes in HLA transgenic mice. Although epitope predictions were performed for six different HLA class I supertypes, our studies in HLA transgenic mice focused on primarily defining arenavirus-specific epitopes restricted by the HLA-A02 and A03/A11 supertypes (55). To identify HLA-A*0201-restricted CD8+ T cell epitopes, HLA-A*0201 transgenic mice were infected with an rVACV expressing one of the arenavirus proteins. Seven days later, purified splenic CD8+ T cells were assayed in ex vivo IFN-γ enzyme-linked immunospot (ELISPOT) assays with the predicted epitopes pulsed in human HLA-A*0201-restricted target cells (Fig. 1). Splenic CD8+ T cells from HLA-A*0201 transgenic mice infected with wild-type VACV were utilized as negative controls. To further address which of the epitopes identified in this first set of experiments was endogenously processed by human APCs, further screenings were conducted. Specifically, HLA-A*0201 transgenic mice were immunized with peptides encoding a minimal epitope sequence, and 11 to 14 days later, CD8+ T cells were assayed for their capacity to recognize rVACV-infected human HLA-A*0201 target cells (expressing the corresponding native arenavirus antigen) in IFN-γ ELISPOT assays. Utilizing this approach, we evaluated the antigenicities of 467 unique peptides and identified 10 antigenic and endogenously processed epitopes. These epitopes were derived from six out of the seven arenavirus species studied.
FIG. 1.
Strategy for identifying HLA-A*0201-restricted arenavirus-specific CD8+ T cell epitopes. (A) In step 1, to identify antigenic peptides, HLA-A*0201 transgenic mice were inoculated i.p. with an rVACV expressing one arenavirus protein. Seven days later, purified splenic CD8+ T cells were assayed in ex vivo IFN-γ ELISPOT assays with arenavirus peptide-pulsed human Jurkat cells that express HLA-A*0201 serving as APCs. HLA-A*0201 transgenic mice inoculated i.p. with wild-type VACV-WR served as a negative control. (B) In step 2, to assess natural processing in human APCs, HLA-A*0201 transgenic mice were immunized s.c. with a single antigenic peptide identified in step 1. At 11 to 14 days postimmunization, purified splenic CD8+ T cells were assayed for IFN-γ secretion in response to human Jurkat cells infected with the rVACV expressing the appropriate arenavirus antigen. Cells infected with wild-type VACV-WR served as a negative control.
To identify HLA-A*1101-restricted CD8+ T cell epitopes, we opted for an alternative epitope identification strategy, as peptides containing positively charged amino acids at the carboxyl termini, such as those bound by the HLA-A*1101 molecule, are not effectively processed in mice (16). Accordingly, pools of 8 to 11 candidate epitopes were used for direct peptide immunization of HLA-A*1101 transgenic mice. After 11 to 14 days, splenocytes from these mice were incubated with peptide-pulsed HLA-A*1101-restricted target cells in ex vivo IFN-γ ELISPOT assays (Fig. 2). These preliminary experiments tested 527 unique HLA-A*1101 peptides in peptide-immunized transgenic mice and identified 178 potential candidate epitopes. In a second set of experiments, the 178 candidate epitopes were further tested for endogenous processing by determining whether CD8+ T cells elicited by peptide immunization recognized human HLA-A*1101-positive APCs infected with an rVACV expressing the appropriate native arenavirus protein. Using this strategy, 10 epitopes endogenously processed in human APCs from their respective native arenavirus protein were identified. These epitopes were derived from all four viral proteins and from five different arenaviruses species. In summary, the experiments described here identified 10 HLA-A*0201- and 10 HLA-A*1101-restricted arenavirus epitopes that are endogenously processed by human APCs (Table 4).
FIG. 2.
Strategy for identifying HLA-A*1101-restricted arenavirus-specific CD8+ T cell epitopes. (A) In step 1, to identify immunogenic peptides, HLA-A*1101 transgenic mice were immunized s.c. with pools of 8 to 11 of the predicted arenavirus peptides. Eleven to 14 days later, splenocytes were assayed in ex vivo IFN-γ ELISPOT assays with arenavirus peptide-pulsed lipopolysaccharide (LPS)-stimulated B lymphoblasts from the HLA-A*1101 transgenic mice serving as APCs. (B) In step 2, to identify epitopes naturally processed in human APCs, HLA-A*1101 transgenic mice were immunized s.c. with individual immunogenic arenavirus peptides defined in step 1. On day 11 to 14 postimmunization, effector splenocytes were cultured in vitro for 6 days with the immunizing peptide. CD8+ T cells were then isolated and screened for IFN-γ secretion in response to HLA-A*1101-expressing human BVR cells that had been infected with an rVACV or infected with wild-type VACV-WR.
ARENAVIRUS-DERIVED EPITOPES INDUCE CROSS-REACTIVE CD8+ T CELL RESPONSES SPANNING MULTIPLE VIRUS SPECIES
Previous studies have demonstrated T cell cross-reactivity for both Old and New World arenaviruses. It was shown in a guinea pig model that adoptive transfer of LCMV or Moepia virus (MOPV)-immune CD8+ T cells could confer protection against challenge with the highly virulent LASV (48). Likewise, in a murine model, cell transfer of LCMV-immune CD4+ and CD8+ T cells protected mice from Pichinde virus infection (83). Furthermore, in humans, T cell clones from patients who have recovered from acute LASV infection showed cross-reactive recognition of MOPV peptides (91).
Cross-reactive arenavirus epitopes might further increase the coverage afforded by the epitope set against each different arenavirus species. Having defined a set of epitopes covering a diverse set of arenavirus proteins and HLA restrictions, we next sought to address whether these epitopes could induce cross-reactive CD8+ T cell responses that recognize multiple or all seven arenavirus species targeted by our studies. To this end, we compared the amino acid sequence of each epitope to the corresponding sequences within other arenaviruses. We also included in this analysis the LASV- and LCMV-derived HLA-A*0201-restricted epitopes identified by Botten et al. (13, 14). Peptides containing two or more identical amino acids as the epitope sequence and found in ≥20% of the isolates within an arenavirus species were chosen for further analysis. HLA transgenic mice were immunized with the epitope subcutaneously (s.c.), and 11 to 14 days later splenic CD8+ T cells were tested in IFN-γ ELISPOT assays with human target cells individually pulsed with the immunizing epitope or the potentially cross-reactive peptides (Fig. 3).
FIG. 3.
Strategy for identifying arenavirus epitopes that induce cross-reactive CD8+ T cell responses upon immunization. HLA transgenic mice were immunized s.c. with individual arenavirus epitopes. Purified splenic CD8+ T cells were screened for IFN-γ secretion in response to human APCs individually pulsed with either the immunizing epitope or potentially cross-reactive arenavirus peptides.
CD8+ T cell responses induced by several epitopes were found to be cross-reactive with orthologous peptides found within other arenavirus species (55). This cross-reactivity was not widespread and was mostly limited to peptides within the Old World or New World arenavirus subgroup. Cross-reactive epitopes were associated mainly with conservative and semiconservative amino acid substitutions at either the primary position 2 anchor residue or the secondary anchor residues at positions 1, 3, and 7. In terms of response magnitude, the CD8+ T cell response to the cross-reactive peptides ranged from 20 to 100% of the epitope-specific response. To determine the percentage of the epitope-specific response, the net IFN-γ spot-forming cells (SFC) to the epitope and the cross-reactive peptide were measured through ELISPOT assays (as shown in Fig. 3). The percent response was then calculated by dividing the net SFC against the cross-reactive peptide from the net SFC against the epitope.
To provide evidence that this level of cross-reactivity was biologically meaningful, the capacity of selected epitopes to protect from heterologous challenge with a virus encoding the cross-reactive antigens was examined. Immunization with the LASV GPC441-449 epitope protected HLA-A*0201 transgenic mice from heterologous challenge with LCMV, as defined by decreased viral titers in peptide-immunized mice compared to controls (J. Botten and M. J. Buchmeier, unpublished work). Analysis of the CD8+ T cell response at the time of sacrifice demonstrated a vigorous response to the immunizing peptide (LASV GPC441-449) that was virtually identical in magnitude to the response of the cross-reactive peptide (LCMV GPC447-455).
ARENAVIRUS-DERIVED EPITOPES PROVIDE COVERAGE ACROSS DIFFERENT VIRAL SPECIES AND MAJOR ETHNIC POPULATIONS
The experiments described above defined a set of epitopes, restricted by HLA-A*0201 and HLA-A*1101 molecules, that are derived from multiple arenavirus species and viral proteins. In the analyses summarized in the present section, we explore the potential of this epitope set in terms of its capacity to provide broad coverage across different arenavirus species in distinct ethnic populations.
The number of peptides providing coverage of different arenavirus species and viral antigens is summarized in Table 6. A total of 36 epitopes and cross-reactive peptides have been defined, with an average of about five peptides per arenavirus species (range of two to eight). To estimate, in more detail, the breadth of coverage afforded by this epitope set, each arenavirus-derived peptide was tested for its capacity to bind common HLA alleles from the HLA-A02 and A03/A11 supertypes. These data, together with the frequency of the HLA-A02 and A03/A11 supertype alleles in different ethnic populations, can be used to estimate the projected population coverage afforded by the epitope set (24). Using the Population Coverage Calculation Tool available through the IEDB, we found that the epitope set provides coverage of 58.5% of the entire population (55). This coverage was fairly balanced, with an average of about three arenavirus-derived epitopes being recognized from each distinct ethnic population and with little variation in the different ethnicities analyzed. While ∼60% coverage is significant, it was not sufficient to provide coverage to the vast majority of individuals, irrespective of ethnicity. However, it should be possible to achieve essentially 100% coverage by identifying CD8+ T cell epitopes restricted by additional HLA supertypes, such as A01, A24, B07, and B44, and CD4+ T cell epitopes restricted by the HLA-DR supertype (84, 88). In conclusion, these data provide a proof-of-concept validation that an epitope set affording broad coverage across different arenavirus species and multiple HLA class I supertypes can be defined.
TABLE 6.
HLA-restricted CD8+ T cell peptides providing coverage of different arenavirus species and viral antigens
| HLA | No. of peptides providing coverage |
|||||||
|---|---|---|---|---|---|---|---|---|
| GTOV | JUNV | LASV | LCMV | MACV | SABV | WWAV | Total | |
| A*0201 | 1 | 4 | 4 | 5 | 3 | 4 | 4 | 25 |
| A*1101 | 1 | 0 | 1 | 3 | 2 | 3 | 1 | 11 |
| Total | 2 | 4 | 5 | 8 | 5 | 7 | 5 | 36a |
MULTIVALENT PROTECTIVE CAPACITY OF ARENAVIRUS-DERIVED CD8+ T CELL EPITOPES
Next, we addressed whether simultaneous CD8+ T cell responses could be induced against the various epitopes restricted by HLA-A*0201 and HLA-A*1101 and whether these responses could afford protection against multiple arenavirus species. To assay for immunogenicity, various arenavirus epitopes were pooled, based on supertype restriction, in IFA and injected into HLA-A*0201 or HLA-A*1101 transgenic mice. While the CD8+ T cell responses varied somewhat in magnitude, significant simultaneous responses were detected against all HLA-A*0201-restricted epitopes when immunized as a pool into HLA-A*0201 transgenic mice (55). Likewise, a pool of the HLA-A*1101-restricted CD8+ T cell epitopes was immunogenic when injected in HLA-A*1101 transgenic mice.
Furthermore, vaccination with a cocktail of the HLA-A*0201-restricted epitopes significantly reduced viral titers in HLA-A*0201 transgenic mice following challenge with rVACVs expressing both Old and New World arenavirus antigens (55). We also found that both epitope-specific and cross-reactive CD8+ T cell responses were significantly boosted following rVACV challenge of peptide pool-immunized mice. Thus, these findings suggest that a cell-mediated vaccine strategy might be able to protect against infection mediated by multiple arenavirus species.
CONCLUSIONS
Here we have reviewed the progress made during the last few years in a project aimed at exploring strategies to elicit multivalent CD8+ and CD4+ T cell responses directed against a panel of seven different arenavirus species that are pathogenic in humans. We have approached this task through the definition of MHC class I- and class II-restricted epitopes in mice, NHPs, and humans. Specifically, we have defined a total of 26 different HLA class I-restricted CD8+ T cell epitopes.
Our results from the HLA studies illustrate how the induction of simultaneous cell-mediated immune responses against multiple arenavirus species is possible and that such responses can protect against multiple different arenavirus species. For certain epitopes, cross-reactive CD8+ T cell recognition and protection against heterologous arenavirus challenge were observed. However, this was rather infrequent and was limited mainly to CD8+ T cell cross-reactivity among Old World or New World arenavirus species. Our results further suggest that T cell-mediated protection against numerous arenavirus species will require selection of different epitopes from multiple virus species.
By targeting epitopes restricted by common HLA supertypes, broad population coverage was achieved. Following this proof-of-concept with the HLA-A02 and A03/A11 supertypes, future studies could expand coverage of other HLA class I supertypes, including A01, A24, B07, and B44, as well as class II supertypes, such as DR. Finally, future studies examining CD8+ and CD4+ T cell responses in arenavirus-immune human donors would directly assess the degree of overlap between arenavirus-specific responses recognized in HLA transgenic mice and humans.
Our study examining the immunodominance of CD8+ T cell responses following LCMV infection demonstrated that the response hierarchy is determined, in part, by the naïve T cell repertoire. Based on these data, it seems reasonable to propose that naïve precursor frequencies could be used as a diagnostic measure of the effectiveness of a particular epitope or antigen in a vaccination setting. This would alleviate the need for obtaining lymphocytes from exposed or immunized individuals, which for arenaviruses and many other pathogens can be logistically difficult. We have also previously established that LCMV epitopes differ in their protective capacities (56, 94), and interestingly, all epitopes with a naïve precursor frequency of ∼100 cells or greater per mouse are capable of protecting mice against LCMV infection.
In closing, it should also be noted that the HLA-restricted CD8+ T cell epitopes identified through these studies represent valuable diagnostic reagents for future studies. At present, very little is known regarding the natural history of the human T cell response to arenavirus infection. While it is generally presumed that virus-specific CD8+ T cells play a protective role during human arenavirus infection, a formal demonstration of this is currently lacking. Furthermore, T cells could potentially contribute to disease pathogenesis. The epitopes identified in our studies should make it possible to carefully examine the phenotype and pattern of lymphokine secretion of arenavirus-specific CD8+ T cells during or following arenavirus infection. By conducting such analyses in patients with mild or severe disease, it should be possible to test whether the magnitude and/or polyfunctional phenotype of arenavirus-specific CD8+ T cells is predictive of disease severity. Ultimately, such studies would define the phenotypic characteristics associated with protective T cell responses and provide benchmarks for the evaluation of future arenavirus vaccines.
Acknowledgments
We thank Huynh-Hoa Bui for her assistance with the bioinformatics. We are grateful to Raphael Zellweger for helping with the graphical illustrations.
This work was supported by NIH grant NIH-AI-50840 (J.B.) and contract NO1-AI-40023 (A.S.). J.B. and B.P. were also supported, in part, by funds from the Pacific Southwest RCE AI065359.
This is La Jolla Institute for Allergy and Immunology and Kyowa Hakko Kirin California publication number 1213.
Biography
Jason Botten received his Ph.D. in Biomedical Sciences in 2000 at the University of New Mexico in Albuquerque, NM. In 2002, he joined Michael Buchmeier's group at The Scripps Research Institute in La Jolla, CA, and subsequently became an Assistant Professor in 2005. In 2008, he joined the University of Vermont College of Medicine in Burlington, VT, as an Assistant Professor. His research interests include the biology and pathogenesis of arenaviruses and hantaviruses and developing disease intervention strategies against these infectious pathogens.
John Sidney completed a B.A. in biology in 1988 at the Humboldt State University in Arcata, CA. He spent 9 years in the Immunochemistry Division at Cytel Corporation and subsequently joined Pharmexa-Epimmune, Inc., in 1997. In 2002, he joined Alessandro Sette's group in the Division of Vaccine Discovery at the La Jolla Institute for Allergy and Immunology in San Diego, CA. His main research interests are MHC class I/II peptide binding and motif predictions.
Bianca R. Mothé received her Ph.D. in Molecular and Cellular Pathology in 2002 at the University of Wisconsin in Madison, WI. She was a Staff Scientist at Pharmexa-Epimmune, Inc., between 2002 and 2003, and then in 2003 she moved to the California State University, San Marcos, CA, where she is currently an Associate Professor. Her research interests include developing macaque and ferret models of virus infection and determining the role of cell-mediated immunity in persistent virus infections.
Bjoern Peters received his Ph.D. in Biophysics and Bioinformatics in 2003 at the Humboldt University in Berlin, Germany. In 2004, he joined the La Jolla Institute for Allergy and Immunology, and he became an Assistant Member in 2008. His main research interests include the Immune Epitope Database and Analysis Resources, as well as MHC class I/II motif predictions and antibody epitope predictions for various infectious pathogens and allergens.
Alessandro Sette received his Ph.D. in Immunology in 1984 at the University of Rome in Rome, Italy. After working at the Laboratory of Pathology, Casaccia, in Rome, Italy, and the National Jewish Center for Immunology and Respiratory Medicine, he joined Cytel Corporation in 1988 and subsequently became Director of Immunochemistry in 1992. In 1996, he became Chief Scientific Officer of Pharmexa-Epimmune, Inc. He joined the La Jolla Institute for Allergy and Immunology as a Tenured Member in 2002. Currently, his research interests are to understand the immune response, measure immune activity, and develop disease intervention strategies against a number of allergies and new and emerging infectious diseases.
Maya F. Kotturi received her Ph.D. in Microbiology and Immunology in 2004 at the University of British Columbia in Vancouver, British Columbia, Canada. In 2005, she joined Alessandro Sette's group at the La Jolla Institute for Allergy and Immunology. Her current research interests include the development of immune therapies and vaccines targeting a variety of viral pathogens, including arenaviruses, dengue virus, influenza virus, and vaccinia virus, as well as IgE-mediated allergic diseases.
REFERENCES
- 1.Alexander, J., C. Oseroff, J. Sidney, and A. Sette. 2003. Derivation of HLA-B*0702 transgenic mice: functional CTL repertoire and recognition of human B*0702-restricted CTL epitopes. Hum. Immunol. 64:211-223. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, J., C. Oseroff, J. Sidney, P. Wentworth, E. Keogh, G. Hermanson, F. V. Chisari, R. T. Kubo, H. M. Grey, and A. Sette. 1997. Derivation of HLA-A11/Kb transgenic mice: functional CTL repertoire and recognition of human A11-restricted CTL epitopes. J. Immunol. 159:4753-4761. [PubMed] [Google Scholar]
- 3.Altmann, D. M., D. C. Douek, A. J. Frater, C. M. Hetherington, H. Inoko, and J. I. Elliott. 1995. The T cell response of HLA-DR transgenic mice to human myelin basic protein and other antigens in the presence and absence of human CD4. J. Exp. Med. 181:867-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Assarsson, E., J. Sidney, C. Oseroff, V. Pasquetto, H. H. Bui, N. Frahm, C. Brander, B. Peters, H. Grey, and A. Sette. 2007. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J. Immunol. 178:7890-7901. [DOI] [PubMed] [Google Scholar]
- 5.Auperin, D. D., J. J. Esposito, J. V. Lange, S. P. Bauer, J. Knight, D. R. Sasso, and J. B. McCormick. 1988. Construction of a recombinant vaccinia virus expressing the Lassa virus glycoprotein gene and protection of guinea pigs from a lethal Lassa virus infection. Virus Res. 9:233-248. [DOI] [PubMed] [Google Scholar]
- 6.Barber, D. L., E. J. Wherry, D. Masopust, B. Zhu, J. P. Allison, A. H. Sharpe, G. J. Freeman, and R. Ahmed. 2006. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439:682-687. [DOI] [PubMed] [Google Scholar]
- 7.Barton, L. L., and N. J. Hyndman. 2000. Lymphocytic choriomeningitis virus: reemerging central nervous system pathogen. Pediatrics 105:E35. [DOI] [PubMed] [Google Scholar]
- 8.Battegay, M., S. Oehen, M. Schulz, H. Hengartner, and R. M. Zinkernagel. 1992. Vaccination with a synthetic peptide modulates lymphocytic choriomeningitis virus-mediated immunopathology. J. Virol. 66:1199-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boesen, A., K. Sundar, and R. Coico. 2005. Lassa fever virus peptides predicted by computational analysis induce epitope-specific cytotoxic-T-lymphocyte responses in HLA-A2.1 transgenic mice. Clin. Diagn. Lab. Immunol. 12:1223-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogaert, D., P. W. Hermans, P. V. Adrian, H. C. Rumke, and R. de Groot. 2004. Pneumococcal vaccines: an update on current strategies. Vaccine 22:2209-2220. [DOI] [PubMed] [Google Scholar]
- 11.Bonthius, D. J., R. Wright, B. Tseng, L. Barton, E. Marco, B. Karacay, and P. D. Larsen. 2007. Congenital lymphocytic choriomeningitis virus infection: spectrum of disease. Ann. Neurol. 62:347-355. [DOI] [PubMed] [Google Scholar]
- 12.Borrow, P., and M. B. Oldstone. 1996. Lymphocytic choriomeningitis virus, p. 593-627. In N. Nathanson (ed.), Viral pathogenesis. Lippincott-Raven Publishers, Philadelphia, PA.
- 13.Botten, J., J. Alexander, V. Pasquetto, J. Sidney, P. Barrowman, J. Ting, B. Peters, S. Southwood, B. Stewart, M. P. Rodriguez-Carreno, B. Mothe, J. L. Whitton, A. Sette, and M. J. Buchmeier. 2006. Identification of protective Lassa virus epitopes that are restricted by HLA-A2. J. Virol. 80:8351-8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Botten, J., J. L. Whitton, P. Barrowman, J. Sidney, J. K. Whitmire, J. Alexander, J. P. Ting, H. H. Bui, A. Sette, and M. J. Buchmeier. 2007. HLA-A2-restricted protection against lethal lymphocytic choriomeningitis. J. Virol. 81:2307-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bousso, P., A. Casrouge, J. D. Altman, M. Haury, J. Kanellopoulos, J. P. Abastado, and P. Kourilsky. 1998. Individual variations in the murine T cell response to a specific peptide reflect variability in naive repertoires. Immunity 9:169-178. [DOI] [PubMed] [Google Scholar]
- 16.Braud, V. M., A. J. McMichael, and V. Cerundolo. 1998. Differential processing of influenza nucleoprotein in human and mouse cells. Eur. J. Immunol. 28:625-635. [DOI] [PubMed] [Google Scholar]
- 17.Brown, B. K., L. Wieczorek, E. Sanders-Buell, A. Rosa Borges, M. L. Robb, D. L. Birx, N. L. Michael, F. E. McCutchan, and V. R. Polonis. 2008. Cross-clade neutralization patterns among HIV-1 strains from the six major clades of the pandemic evaluated and compared in two different models. Virology 375:529-538. [DOI] [PubMed] [Google Scholar]
- 18.Brueggemann, A. B., R. Pai, D. W. Crook, and B. Beall. 2007. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 3:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bryan, J. T. 2007. Developing an HPV vaccine to prevent cervical cancer and genital warts. Vaccine 25:3001-3006. [DOI] [PubMed] [Google Scholar]
- 20.Buchmeier, M. J., J. C. de la Torre, and C. J. Peters. 2007. Arenaviridae: the viruses and their replication, p. 1791-1828. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Hagerstown, MD. [Google Scholar]
- 21.Buchmeier, M. J., and A. J. Zajac. 2001. Lymphocytic choriomeningitis virus, p. 575-605. In R. Ahmed and I. Chen (ed.), Persistent virus infections. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 22.Buchmeier, M. J., and A. J. Zajac. 1999. Lymphocytic choriomeningitis virus. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 23.Bui, H. H., J. Botten, N. Fusseder, V. Pasquetto, B. Mothe, M. J. Buchmeier, and A. Sette. 2007. Protein sequence database for pathogenic arenaviruses. Immunome Res. 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bui, H. H., J. Sidney, K. Dinh, S. Southwood, M. J. Newman, and A. Sette. 2006. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics 7:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charatan, F. 2009. UN warns that swine flu outbreak could turn into pandemic. BMJ 338:b1751. [DOI] [PubMed] [Google Scholar]
- 26.Charrel, R. N., X. de Lamballerie, and S. Emonet. 2008. Phylogeny of the genus Arenavirus. Curr. Opin. Microbiol. 11:362-368. [DOI] [PubMed] [Google Scholar]
- 27.Chen, W., C. C. Norbury, Y. Cho, J. W. Yewdell, and J. R. Bennink. 2001. Immunoproteasomes shape immunodominance hierarchies of antiviral CD8(+) T cells at the levels of T cell repertoire and presentation of viral antigens. J. Exp. Med. 193:1319-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clegg, J. C. 1992. Current progress towards vaccines for arenavirus-caused diseases. Vaccine 10:89-95. [DOI] [PubMed] [Google Scholar]
- 29.Clegg, J. C. 2002. Molecular phylogeny of the arenaviruses. Curr. Top. Microbiol. Immunol. 262:1-24. [DOI] [PubMed] [Google Scholar]
- 30.Crowe, S. R., S. J. Turner, S. C. Miller, A. D. Roberts, R. A. Rappolo, P. C. Doherty, K. H. Ely, and D. L. Woodland. 2003. Differential antigen presentation regulates the changing patterns of CD8+ T cell immunodominance in primary and secondary influenza virus infections. J. Exp. Med. 198:399-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dow, C., C. Oseroff, B. Peters, C. Nance-Sotelo, J. Sidney, M. Buchmeier, A. Sette, and B. R. Mothe. 2008. Lymphocytic choriomeningitis virus infection yields overlapping CD4+ and CD8+ T-cell responses. J. Virol. 82:11734-11741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ekiert, D. C., G. Bhabha, M. A. Elsliger, R. H. Friesen, M. Jongeneelen, M. Throsby, J. Goudsmit, and I. A. Wilson. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324:246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enria, D. A., and J. G. Barrera Oro. 2002. Junin virus vaccines. Curr. Top. Microbiol. Immunol. 263:239-261. [DOI] [PubMed] [Google Scholar]
- 34.Enria, D. A., A. M. Briggiler, S. Levis, D. Vallejos, J. I. Maiztegui, and P. G. Canonico. 1987. Tolerance and antiviral effect of ribavirin in patients with Argentine hemorrhagic fever. Antiviral Res. 7:353-359. [DOI] [PubMed] [Google Scholar]
- 35.Fischer, S. A., M. B. Graham, M. J. Kuehnert, C. N. Kotton, A. Srinivasan, F. M. Marty, J. A. Comer, J. Guarner, C. D. Paddock, D. L. DeMeo, W. J. Shieh, B. R. Erickson, U. Bandy, A. DeMaria, Jr., J. P. Davis, F. L. Delmonico, B. Pavlin, A. Likos, M. J. Vincent, T. K. Sealy, C. S. Goldsmith, D. B. Jernigan, P. E. Rollin, M. M. Packard, M. Patel, C. Rowland, R. F. Helfand, S. T. Nichol, J. A. Fishman, T. Ksiazek, and S. R. Zaki. 2006. Transmission of lymphocytic choriomeningitis virus by organ transplantation. N. Engl. J. Med. 354:2235-2249. [DOI] [PubMed] [Google Scholar]
- 36.Fisher-Hoch, S. P., L. Hutwagner, B. Brown, and J. B. McCormick. 2000. Effective vaccine for Lassa fever. J. Virol. 74:6777-6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher-Hoch, S. P., and J. B. McCormick. 2001. Towards a human Lassa fever vaccine. Rev. Med. Virol. 11:331-341. [DOI] [PubMed] [Google Scholar]
- 38.Flesch, I. E., W. P. Woo, Y. Wang, V. Panchanathan, Y. C. Wong, N. L. La Gruta, T. Cukalac, and D. C. Tscharke. 2010. Altered CD8(+) T cell immunodominance after vaccinia virus infection and the naive repertoire in inbred and F(1) mice. J. Immunol. 184:45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallimore, A., T. Dumrese, H. Hengartner, R. M. Zinkernagel, and H. G. Rammensee. 1998. Protective immunity does not correlate with the hierarchy of virus-specific cytotoxic T cell responses to naturally processed peptides. J. Exp. Med. 187:1647-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzalez, J. P., S. Emonet, X. de Lamballerie, and R. Charrel. 2007. Arenaviruses. Curr. Top. Microbiol. Immunol. 315:253-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gray, G. C., T. McCarthy, M. G. Lebeck, D. P. Schnurr, K. L. Russell, A. E. Kajon, M. L. Landry, D. S. Leland, G. A. Storch, C. C. Ginocchio, C. C. Robinson, G. J. Demmler, M. A. Saubolle, S. C. Kehl, R. Selvarangan, M. B. Miller, J. D. Chappell, D. M. Zerr, D. L. Kiska, D. C. Halstead, A. W. Capuano, S. F. Setterquist, M. L. Chorazy, J. D. Dawson, and D. D. Erdman. 2007. Genotype prevalence and risk factors for severe clinical adenovirus infection, United States 2004-2006. Clin. Infect. Dis. 45:1120-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guzman, M. G., G. Kouri, L. Valdes, J. Bravo, M. Alvarez, S. Vazques, I. Delgado, and S. B. Halstead. 2000. Epidemiologic studies on Dengue in Santiago de Cuba, 1997. Am. J. Epidemiol. 152:793-799. (Discussion, 804.) [DOI] [PubMed] [Google Scholar]
- 43.Hanage, W. P., C. Fraser, J. Tang, T. R. Connor, and J. Corander. 2009. Hyper-recombination, diversity, and antibiotic resistance in pneumococcus. Science 324:1454-1457. [DOI] [PubMed] [Google Scholar]
- 44.Harrison, L. H., N. A. Halsey, K. T. McKee, Jr., C. J. Peters, J. G. Barrera Oro, A. M. Briggiler, M. R. Feuillade, and J. I. Maiztegui. 1999. Clinical case definitions for Argentine hemorrhagic fever. Clin. Infect. Dis. 28:1091-1094. [DOI] [PubMed] [Google Scholar]
- 45.Hasegawa, A., C. Moriya, H. Liu, W. A. Charini, H. C. Vinet, R. A. Subbramanian, P. Sen, N. L. Letvin, and M. J. Kuroda. 2007. Analysis of TCRalphabeta combinations used by simian immunodeficiency virus-specific CD8+ T cells in rhesus monkeys: implications for CTL immunodominance. J. Immunol. 178:3409-3417. [DOI] [PubMed] [Google Scholar]
- 46.Homann, D., H. Lewicki, D. Brooks, J. Eberlein, V. Mallet-Designe, L. Teyton, and M. B. Oldstone. 2007. Mapping and restriction of a dominant viral CD4+ T cell core epitope by both MHC class I and MHC class II. Virology 363:113-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Insel, R. A. 1995. Potential alterations in immunogenicity by combining or simultaneously administering vaccine components. Ann. N. Y. Acad. Sci. 754:35-47. [DOI] [PubMed] [Google Scholar]
- 48.Jahrling, P. B., and C. J. Peters. 1986. Serology and virulence diversity among Old-World arenaviruses, and the relevance to vaccine development. Med. Microbiol. Immunol. 175:165-167. [DOI] [PubMed] [Google Scholar]
- 49.Kagi, D., B. Ledermann, K. Burki, P. Seiler, B. Odermatt, K. J. Olsen, E. R. Podack, R. M. Zinkernagel, and H. Hengartner. 1994. Cytotoxicity mediated by T cells and natural killer cells is greatly impaired in perforin-deficient mice. Nature 369:31-37. [DOI] [PubMed] [Google Scholar]
- 50.Kalams, S. A., and B. D. Walker. 1998. The critical need for CD4 help in maintaining effective cytotoxic T lymphocyte responses. J. Exp. Med. 188:2199-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kamperschroer, C., and D. G. Quinn. 1999. Quantification of epitope-specific MHC class-II-restricted T cells following lymphocytic choriomeningitis virus infection. Cell. Immunol. 193:134-146. [DOI] [PubMed] [Google Scholar]
- 52.Khanolkar, A., M. J. Fuller, and A. J. Zajac. 2002. T cell responses to viral infections: lessons from lymphocytic choriomeningitis virus. Immunol. Res. 26:309-321. [DOI] [PubMed] [Google Scholar]
- 53.Klavinskis, L. S., J. L. Whitton, E. Joly, and M. B. Oldstone. 1990. Vaccination and protection from a lethal viral infection: identification, incorporation, and use of a cytotoxic T lymphocyte glycoprotein epitope. Virology 178:393-400. [DOI] [PubMed] [Google Scholar]
- 54.Kotturi, M. F., E. Assarsson, B. Peters, H. Grey, C. Oseroff, V. Pasquetto, and A. Sette. 2009. Of mice and humans: how good are HLA transgenic mice as a model of human immune responses? Immunome Res. 5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kotturi, M. F., J. Botten, J. Sidney, H. H. Bui, L. Giancola, M. Maybeno, J. Babin, C. Oseroff, V. Pasquetto, J. A. Greenbaum, B. Peters, J. Ting, D. Do, L. Vang, J. Alexander, H. Grey, M. J. Buchmeier, and A. Sette. 2009. A multivalent and cross-protective vaccine strategy against arenaviruses associated with human disease. PLoS Pathog. 5:e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kotturi, M. F., B. Peters, F. Buendia-Laysa, Jr., J. Sidney, C. Oseroff, J. Botten, H. Grey, M. J. Buchmeier, and A. Sette. 2007. The CD8+ T-cell response to lymphocytic choriomeningitis virus involves the L antigen: uncovering new tricks for an old virus. J. Virol. 81:4928-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kotturi, M. F., I. Scott, T. Wolfe, B. Peters, J. Sidney, H. Cheroutre, M. G. von Herrath, M. J. Buchmeier, H. Grey, and A. Sette. 2008. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J. Immunol. 181:2124-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.La Gruta, N. L., K. Kedzierska, K. Pang, R. Webby, M. Davenport, W. Chen, S. J. Turner, and P. C. Doherty. 2006. A virus-specific CD8+ T cell immunodominance hierarchy determined by antigen dose and precursor frequencies. Proc. Natl. Acad. Sci. U. S. A. 103:994-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lazarski, C. A., F. A. Chaves, S. A. Jenks, S. Wu, K. A. Richards, J. M. Weaver, and A. J. Sant. 2005. The kinetic stability of MHC class II:peptide complexes is a key parameter that dictates immunodominance. Immunity 23:29-40. [DOI] [PubMed] [Google Scholar]
- 60.Lewicki, H., T. A. McKee, A. Tishon, M. Salvato, J. L. Whitton, and M. B. Oldstone. 1992. Novel LCMV-specific H-2k restricted CTL clones recognize internal viral gene products and cause CNS disease. J. Neuroimmunol. 41:15-20. [DOI] [PubMed] [Google Scholar]
- 61.Liu, F., J. L. Whitton, and M. K. Slifka. 2004. The rapidity with which virus-specific CD8+ T cells initiate IFN-gamma synthesis increases markedly over the course of infection and correlates with immunodominance. J. Immunol. 173:456-462. [DOI] [PubMed] [Google Scholar]
- 62.Maiztegui, J. I. 1975. Clinical and epidemiological patterns of Argentine haemorrhagic fever. Bull. World Health Organ. 52:567-575. [PMC free article] [PubMed] [Google Scholar]
- 63.Maiztegui, J. I., K. T. McKee, Jr., J. G. Barrera Oro, L. H. Harrison, P. H. Gibbs, M. R. Feuillade, D. A. Enria, A. M. Briggiler, S. C. Levis, A. M. Ambrosio, N. A. Halsey, and C. J. Peters. 1998. Protective efficacy of a live attenuated vaccine against Argentine hemorrhagic fever. AHF Study Group. J. Infect. Dis. 177:277-283. [DOI] [PubMed] [Google Scholar]
- 64.Masopust, D., K. Murali-Krishna, and R. Ahmed. 2007. Quantitating the magnitude of the lymphocytic choriomeningitis virus-specific CD8 T-cell response: it is even bigger than we thought. J. Virol. 81:2002-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCormick, J. B. 1986. Clinical, epidemiologic, and therapeutic aspects of Lassa fever. Med. Microbiol. Immunol. 175:153-155. [DOI] [PubMed] [Google Scholar]
- 66.McCormick, J. B., P. A. Webb, J. W. Krebs, K. M. Johnson, and E. S. Smith. 1987. A prospective study of the epidemiology and ecology of Lassa fever. J. Infect. Dis. 155:437-444. [DOI] [PubMed] [Google Scholar]
- 67.McKee, K. T., Jr., J. W. Huggins, C. J. Trahan, and B. G. Mahlandt. 1988. Ribavirin prophylaxis and therapy for experimental Argentine hemorrhagic fever. Antimicrob. Agents Chemother. 32:1304-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McKee, K. T., Jr., J. G. Oro, A. I. Kuehne, J. A. Spisso, and B. G. Mahlandt. 1993. Safety and immunogenicity of a live-attenuated Junin (Argentine hemorrhagic fever) vaccine in rhesus macaques. Am. J. Trop. Med. Hyg. 48:403-411. [DOI] [PubMed] [Google Scholar]
- 69.Morrison, H. G., S. P. Bauer, J. V. Lange, J. J. Esposito, J. B. McCormick, and D. D. Auperin. 1989. Protection of guinea pigs from Lassa fever by vaccinia virus recombinants expressing the nucleoprotein or the envelope glycoproteins of Lassa virus. Virology 171:179-188. [DOI] [PubMed] [Google Scholar]
- 70.Mothe, B. R., B. S. Stewart, C. Oseroff, H. H. Bui, S. Stogiera, Z. Garcia, C. Dow, M. P. Rodriguez-Carreno, M. Kotturi, V. Pasquetto, J. Botten, S. Crotty, E. Janssen, M. J. Buchmeier, and A. Sette. 2007. Chronic lymphocytic choriomeningitis virus infection actively down-regulates CD4+ T cell responses directed against a broad range of epitopes. J. Immunol. 179:1058-1067. [DOI] [PubMed] [Google Scholar]
- 71.Munoz, N., F. X. Bosch, S. de Sanjose, R. Herrero, X. Castellsague, K. V. Shah, P. J. Snijders, and C. J. Meijer. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518-527. [DOI] [PubMed] [Google Scholar]
- 72.NIAID. 2008. NIAID: planning for the 21st century 2008 update. U.S. Department of Health and Human Services, National Institutes of Health, National Institute of Allergy and Infectious Diseases, Bethesda, MD.
- 73.Oxenius, A., M. F. Bachmann, D. Mathis, C. Benoist, R. M. Zinkernagel, and H. Hengartner. 1997. Functional in vivo MHC class II loading by endogenously synthesized glycoprotein during viral infection. J. Immunol. 158:5717-5726. [PubMed] [Google Scholar]
- 74.Pang, K. C., M. T. Sanders, J. J. Monaco, P. C. Doherty, S. J. Turner, and W. Chen. 2006. Immunoproteasome subunit deficiencies impact differentially on two immunodominant influenza virus-specific CD8+ T cell responses. J. Immunol. 177:7680-7688. [DOI] [PubMed] [Google Scholar]
- 75.Pantaleo, G., and R. A. Koup. 2004. Correlates of immune protection in HIV-1 infection: what we know, what we don't know, what we should know. Nat. Med. 10:806-810. [DOI] [PubMed] [Google Scholar]
- 76.Peters, B., J. Sidney, P. Bourne, H. H. Bui, S. Buus, G. Doh, W. Fleri, M. Kronenberg, R. Kubo, O. Lund, D. Nemazee, J. V. Ponomarenko, M. Sathiamurthy, S. Schoenberger, S. Stewart, P. Surko, S. Way, S. Wilson, and A. Sette. 2005. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 3:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Probst, H. C., K. Tschannen, A. Gallimore, M. Martinic, M. Basler, T. Dumrese, E. Jones, and M. F. van den Broek. 2003. Immunodominance of an antiviral cytotoxic T cell response is shaped by the kinetics of viral protein expression. J. Immunol. 171:5415-5422. [DOI] [PubMed] [Google Scholar]
- 78.Rodriguez, F., M. K. Slifka, S. Harkins, and J. L. Whitton. 2001. Two overlapping subdominant epitopes identified by DNA immunization induce protective CD8(+) T-cell populations with differing cytolytic activities. J. Virol. 75:7399-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roose, K., W. Fiers, and X. Saelens. 2009. Pandemic preparedness: toward a universal influenza vaccine. Drug News Perspect. 22:80-92. [DOI] [PubMed] [Google Scholar]
- 80.Salvato, M., J. C. S. Clegg, M. D. Bowen, M. J. Buchmeier, J. P. Gonzalez, I. S. Lukashevich, C. J. Peters, R. Rico-Hesse, and V. Romanowski. 2005. Family Arenaviridae, p. 725-733. In C. M. Fauquet, A. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- 81.Sangkawibha, N., S. Rojanasuphot, S. Ahandrik, S. Viriyapongse, S. Jatanasen, V. Salitul, B. Phanthumachinda, and S. B. Halstead. 1984. Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am. J. Epidemiol. 120:653-669. [DOI] [PubMed] [Google Scholar]
- 82.Schulz, M., R. M. Zinkernagel, and H. Hengartner. 1991. Peptide-induced antiviral protection by cytotoxic T cells. Proc. Natl. Acad. Sci. U. S. A. 88:991-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Selin, L. K., S. M. Varga, I. C. Wong, and R. M. Welsh. 1998. Protective heterologous antiviral immunity and enhanced immunopathogenesis mediated by memory T cell populations. J. Exp. Med. 188:1705-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sette, A., and J. Sidney. 1999. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics 50:201-212. [DOI] [PubMed] [Google Scholar]
- 85.Sette, A., A. Vitiello, B. Reherman, P. Fowler, R. Nayersina, W. M. Kast, C. J. Melief, C. Oseroff, L. Yuan, J. Ruppert, et al. 1994. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol. 153:5586-5592. [PubMed] [Google Scholar]
- 86.Shepard, C. W., L. Finelli, and M. J. Alter. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558-567. [DOI] [PubMed] [Google Scholar]
- 87.Shin, H., and E. J. Wherry. 2007. CD8 T cell dysfunction during chronic viral infection. Curr. Opin. Immunol. 19:408-415. [DOI] [PubMed] [Google Scholar]
- 88.Sidney, J., B. Peters, N. Frahm, C. Brander, and A. Sette. 2008. HLA class I supertypes: a revised and updated classification. BMC Immunol. 9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sui, J., W. C. Hwang, S. Perez, G. Wei, D. Aird, L. M. Chen, E. Santelli, B. Stec, G. Cadwell, M. Ali, H. Wan, A. Murakami, A. Yammanuru, T. Han, N. J. Cox, L. A. Bankston, R. O. Donis, R. C. Liddington, and W. A. Marasco. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun, W., R. Edelman, N. Kanesa-Thasan, K. H. Eckels, J. R. Putnak, A. D. King, H. S. Houng, D. Tang, J. M. Scherer, C. H. Hoke, Jr., and B. L. Innis. 2003. Vaccination of human volunteers with monovalent and tetravalent live-attenuated dengue vaccine candidates. Am. J. Trop. Med. Hyg. 69:24-31. [DOI] [PubMed] [Google Scholar]
- 91.ter Meulen, J., M. Badusche, K. Kuhnt, A. Doetze, J. Satoguina, T. Marti, C. Loeliger, K. Koulemou, L. Koivogui, H. Schmitz, B. Fleischer, and A. Hoerauf. 2000. Characterization of human CD4(+) T-cell clones recognizing conserved and variable epitopes of the Lassa virus nucleoprotein. J. Virol. 74:2186-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Trautmann, L., M. Rimbert, K. Echasserieau, X. Saulquin, B. Neveu, J. Dechanet, V. Cerundolo, and M. Bonneville. 2005. Selection of T cell clones expressing high-affinity public TCRs within human cytomegalovirus-specific CD8 T cell responses. J. Immunol. 175:6123-6132. [DOI] [PubMed] [Google Scholar]
- 93.van der Most, R. G., K. Murali-Krishna, J. G. Lanier, E. J. Wherry, M. T. Puglielli, J. N. Blattman, A. Sette, and R. Ahmed. 2003. Changing immunodominance patterns in antiviral CD8 T-cell responses after loss of epitope presentation or chronic antigenic stimulation. Virology 315:93-102. [DOI] [PubMed] [Google Scholar]
- 94.van der Most, R. G., K. Murali-Krishna, J. L. Whitton, C. Oseroff, J. Alexander, S. Southwood, J. Sidney, R. W. Chesnut, A. Sette, and R. Ahmed. 1998. Identification of Db- and Kb-restricted subdominant cytotoxic T-cell responses in lymphocytic choriomeningitis virus-infected mice. Virology 240:158-167. [DOI] [PubMed] [Google Scholar]
- 95.van der Most, R. G., A. Sette, C. Oseroff, J. Alexander, K. Murali-Krishna, L. L. Lau, S. Southwood, J. Sidney, R. W. Chesnut, M. Matloubian, and R. Ahmed. 1996. Analysis of cytotoxic T cell responses to dominant and subdominant epitopes during acute and chronic lymphocytic choriomeningitis virus infection. J. Immunol. 157:5543-5554. [PubMed] [Google Scholar]
- 96.Vezys, V., A. Yates, K. A. Casey, G. Lanier, R. Ahmed, R. Antia, and D. Masopust. 2009. Memory CD8 T-cell compartment grows in size with immunological experience. Nature 457:196-199. [DOI] [PubMed] [Google Scholar]
- 97.Vitiello, A., D. Marchesini, J. Furze, L. A. Sherman, and R. W. Chesnut. 1991. Analysis of the HLA-restricted influenza-specific cytotoxic T lymphocyte response in transgenic mice carrying a chimeric human-mouse class I major histocompatibility complex. J. Exp. Med. 173:1007-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.von Herrath, M. G., and J. L. Whitton. 2001. Animal models using lymphocytic choriomeningitis virus. Curr. Protoc. Immunol. Chapter 19:Unit 19.10. [DOI] [PubMed] [Google Scholar]
- 99.von Herrath, M. G., M. Yokoyama, J. Dockter, M. B. Oldstone, and J. L. Whitton. 1996. CD4-deficient mice have reduced levels of memory cytotoxic T lymphocytes after immunization and show diminished resistance to subsequent virus challenge. J. Virol. 70:1072-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Walker, B. D., and D. R. Burton. 2008. Toward an AIDS vaccine. Science 320:760-764. [DOI] [PubMed] [Google Scholar]
- 101.Walsh, C. M., M. Matloubian, C. C. Liu, R. Ueda, C. G. Kurahara, J. L. Christensen, M. T. Huang, J. D. Young, R. Ahmed, and W. R. Clark. 1994. Immune function in mice lacking the perforin gene. Proc. Natl. Acad. Sci. U. S. A. 91:10854-10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Weidt, G., O. Utermohlen, J. Heukeshoven, F. Lehmann-Grube, and W. Deppert. 1998. Relationship among immunodominance of single CD8+ T cell epitopes, virus load, and kinetics of primary antiviral CTL response. J. Immunol. 160:2923-2931. [PubMed] [Google Scholar]
- 103.Wherry, E. J., J. N. Blattman, K. Murali-Krishna, R. van der Most, and R. Ahmed. 2003. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol. 77:4911-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Whitehead, S. S., J. E. Blaney, A. P. Durbin, and B. R. Murphy. 2007. Prospects for a dengue virus vaccine. Nat. Rev. Microbiol. 5:518-528. [DOI] [PubMed] [Google Scholar]
- 105.Yewdell, J. W. 2006. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity 25:533-543. [DOI] [PubMed] [Google Scholar]
- 106.Yewdell, J. W., and J. R. Bennink. 1999. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 17:51-88. [DOI] [PubMed] [Google Scholar]