Abstract
Summary: Within the last 15 years, members of the bacterial genus Acinetobacter have risen from relative obscurity to be among the most important sources of hospital-acquired infections. The driving force for this has been the remarkable ability of these organisms to acquire antibiotic resistance determinants, with some strains now showing resistance to every antibiotic in clinical use. There is an urgent need for new antibacterial compounds to combat the threat imposed by Acinetobacter spp. and other intractable bacterial pathogens. The essential processes of chromosomal DNA replication, transcription, and cell division are attractive targets for the rational design of antimicrobial drugs. The goal of this review is to examine the wealth of genome sequence and gene knockout data now available for Acinetobacter spp., highlighting those aspects of essential systems that are most suitable as drug targets. Acinetobacter spp. show several key differences from other pathogenic gammaproteobacteria, particularly in global stress response pathways. The involvement of these pathways in short- and long-term antibiotic survival suggests that Acinetobacter spp. cope with antibiotic-induced stress differently from other microorganisms.
INTRODUCTION
Species of the bacterial genus Acinetobacter are becoming increasingly important as a source of hospital-acquired infections (31, 185, 204). Acinetobacter spp. are ubiquitous nonmotile gammaproteobacteria, typified by metabolic versatility and a capacity for natural transformation (172, 204). The species of most clinical relevance is A. baumannii; however, pathogenic strains of A. lwoffi and A. baylyi have also been described (38, 185, 215).
A. baumannii is most commonly associated with pneumonia or bacteremia, although the incidence of soft tissue infections and meningitis appears to be increasing (204). Mortality rates associated with A. baumannii infection are a matter of some contention, as there is a tendency for A. baumannii to infect patients who are critically ill and already have a poor prognosis (204). It is clear, however, that patients with A. baumannii infections stay significantly longer in hospitals than uninfected patients and represent a significant burden on health care systems worldwide (204). Exacerbating this, A. baumannii can be highly persistent within the hospital setting, due in part to its resistance to disinfectants (262) and remarkable insensitivity to desiccation (122). Of greatest concern, however, is the rapid emergence of multidrug resistance (65). A. baumannii strains that are resistant to every antibiotic compound in clinical use, including recently approved drugs such as tigecyclin, have now been reported (65, 187, 203). In the face of the growing threat posed by these and other intractable Gram-negative pathogens, there is an urgent need for new classes of antibiotic compounds with novel mechanisms of action (245).
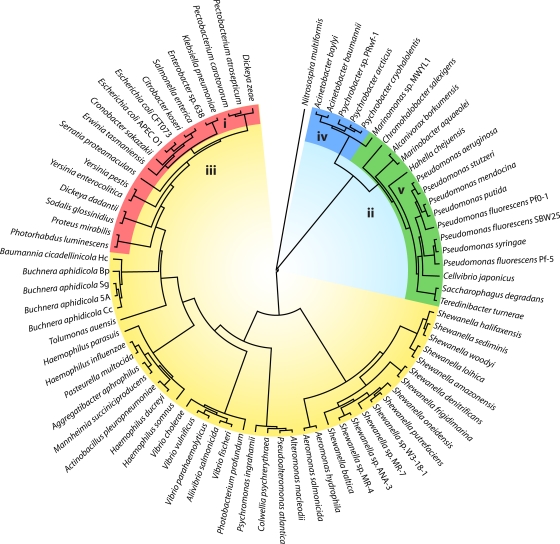
Antibiotics act by inhibiting the essential working parts of bacterial cells, with the most common targets being ribosomal protein synthesis, cell wall biosynthesis, and primary metabolic pathways. The processes of chromosomal DNA replication, transcription and cell division are relatively unexplored as drug targets. They could, however, be considered attractive targets for rational design of novel antimicrobial compounds, as they are (i) essential for the propagation of bacterial cells and therefore infection, (ii) conserved across all bacterial species, and (iii) in many ways distinct from the equivalent processes in eukaryotic cells (150). Acinetobacter represents an appealing system for studying these processes. In addition to their direct clinical relevance, Acinetobacter spp. show some similarities to other problematic pathogens, such as Pseudomonas aeruginosa, that are not shared by the more thoroughly studied model bacterium Escherichia coli (15). At the same time, the Acinetobacter genus has diverged significantly from other gammaproteobacteria, as highlighted by a phylogenetic tree based on the highly conserved sequences of 16S rRNA genes (Fig. 1) (116, 188, 213, 222, 265, 266). As such, Acinetobacter sp. proteins generally show considerable sequence divergence from equivalent proteins in other organisms and can therefore be used to highlight the most conserved aspects of essential biological systems. Of course, these are the most attractive targets for the discovery or rational design of broad-spectrum antibiotics.
FIG. 1.
Subbranch of a phylogenetic tree based on 16S rRNA sequences. 16S rRNA sequences encoded by the rrsA gene were from gammaproteobacteria with completely sequenced genomes and were filtered to remove identical sequences prior to tree construction. The tree was constructed using the neighborhood-joining tree method in Geneious (Biomatters, Auckland, New Zealand), using the Jukes-Cantor genetic distance model and employing the bootstrap method with 10,000 replicates. The sequence of rrsA from the betaproteobacterium Nitrosospira multiformis was included as an outgroup. Colored segments indicate the phylogenetic distribution of particular genetic characteristics: (i) dnaC, holE, tus, amiC, and envC genes present on chromosome; (ii) long Pseudomonas-type ψ subunit of DNA polymerase III; (iii) short E. coli-type ψ subunit of DNA polymerase III, rnhA and dnaQ genes adjacent and transcribed in opposite directions, yacL gene present; (iv) rnhA-dnaQ fusion gene present and no homologs of ftsE, ftsX, sulA, or lexA on chromosome (includes Acinetobacter spp.); (v) rnhA and dnaQ genes adjacent and transcribed in same direction and amiC gene present.
Currently, complete genome sequences are available for seven Acinetobacter strains: the highly transformable laboratory strain A. baylyi ADP1 (15); a human body louse symbiont, A. baumannii SDF (248); and the human pathogens A. baumannii strains ATCC 17978 (233), AYE (248), ACICU (115), AB0057 (4), and AB307-0294 (4). At least eight additional genome sequence projects are currently in progress or at draft assembly stage at the time of writing of this review. In addition, the Genoscope group has produced a complete single-gene deletion library for A. baylyi ADP1, allowing the essentiality of individual genes to be assessed (61). These resources provide a firm starting point for the investigation of essential biological processes in these organisms. The goal of this review is to examine this recently available information, primarily by extrapolating knowledge from well-studied model organisms, in the context of DNA replication, transcription, and cell division, with a view toward highlighting unusual aspects and identification of candidate enzymes and complexes most suitable for future drug discovery and design.
DNA REPLICATION
Our knowledge of bacterial DNA replication draws extensively from studies of the model organism E. coli (16, 209, 226, 242). Over the last 5 decades, each of the individual DNA replication proteins have been identified, for most their three-dimensional structures (or those of their domains) have been determined, and many protein-protein and protein-DNA interactions have been mapped (Table 1).
TABLE 1.
Conservation of chromosomal DNA replication proteins in Acinetobacter spp.
| Protein | Function | Protein length (amino acids)a | % Sequence identity (residue range) |
Essentialitye | Homologs with known structuresb (residue range of sequence match; % sequence identity) | Known protein-protein interactions in E. coli | References | ||
|---|---|---|---|---|---|---|---|---|---|
| A. baylyi vs E. colib | A. baumannii vs E. colid | A. baylyi vs A. baumanniib | |||||||
| DnaA | Initiator | 465 | 46 (2-465) | 47 (2-465) | 91 (1-465) | A, P, E | E. coli (2-108; 32), Thermatoga maritima (109-432; 36) | DnaB, Hda, DiaA, Dps, HU | 1, 14, 40, 41, 142, 145, 196 |
| DnaB | DNA helicase | 481 | 51 (24-480) | 51 (24-480) | 92 (1-481) | A, P, E | Geobacillus stearothermophilus (29-473; 42) | DnaA, DnaC, DnaG, τ, Rep | 1, 12, 82, 83, 97, 191 |
| DnaC | Helicase loader | —f | — | — | — | E | Aquifex aoelicus (no homolog) | DnaB | 82, 181 |
| DnaE | α subunit, polymerase III core | 1,187 | 48 (2-1145) | 48 (2-1145) | 87 (1-1187) | A, P, E | E. coli (2-901; 53), Thermus aquaticus (1-1010; 38) | ɛ, β, τ | 13, 55, 125, 149, 157, 192, 197, 206, 241 |
| DnaG | DNA primase | 629 | 39 (3-472) | 40 (3-464) | 81 (1-629) | A, E | G. stearothermophilus (3-95; 51), E. coli (165-472; 44) | DnaB, SSB | 12, 51, 191, 275 |
| DnaN | β sliding clamp | 382 | 44 (1-382) | 45 (1-382) | 91 (1-382) | A, P, E | E. coli (1-382; 44) | α, δ, Hda, UmuC, UmuD, DinB1, MutS, MutL, DNA ligase, PolA, PolB | 55, 70, 87, 127, 142, 158, 159 |
| DnaQ (fusion with RnhA) | ɛ subunit, polymerase III core | 450 | 46 (223-447) | 46 (232-456) | 78 (1-450) | A, P | E. coli (1-136 RNase HI; 49), E. coli (223-393; 52) | α, θ | 99, 137, 197, 206 |
| DnaX | τ and γ subunits, clamp loader complex | 711 | 56 (2-360) | 55 (2-363) | 66 (1-711) | A, P, E | E. coli (2-360; 56) | δ, δ′, α, γ, ψ | 83, 88, 125, 126, 231, 240 |
| HolA | δ subunit, clamp loader complex | 331 | 26 (34-303) | 29 (46-308) | 83 (1-327) | A, P, E | E. coli (34-303; 26) | τ, γ, δ′, β | 88, 126, 127, 231 |
| HolB | δ′ subunit, clamp loader complex | 331 | 28 (15-189) | 31 (26-170) | 72 (11-328) | A, P, E | E. coli (15-189; 30) | τ, γ, δ | 88, 126, 231 |
| HolC | χ subunit, clamp loader complex | 135 | 23 (1-127) | 21 (1-127) | 68 (1-135) | D | E. coli (1-127; 23) | ψ, SSB | 96, 263, 275 |
| HolD | ψ subunit, clamp loader complex | 204 | 7 (1-204)c | 7 (1-204)c | 39 (1-204)c | A, P | E. coli (not detected) | χ, γ/τ | 96, 231 |
| HolE | θ subunit, polymerase III core | — | — | — | — | D | E. coli (no homolog) | ɛ | 144, 208 |
| SSB | Single-stranded-DNA binding | 192 | 49 (2-192) | 64 (2-114) | 76 (1-192) | A, P, E | E. coli (2-192; 46) | DnaG, χ, RecQ, TopB, UmuC, RecJ, PriA, RecO, exonuclease I, GroEL | 9, 30, 86, 100, 135, 148, 162, 229, 243, 263, 275 |
| Tus | Terminator | — | — | — | — | D | E. coli (no homolog) | DnaB | 182, 183, 189 |
| Hda | Initiation repressor | 235 | 30 (29-229) | 29 (29-226) | 88 (1-235) | P | Shewanella amazonensis (3-157; 28) | β, DnaA | 137, 142 |
| PolA | DNA polymerase I | 920 | 49 (3-920) | 48 (3-923) | 83 (1-920) | A | T. aquaticus (2-920; 39), E. coli (324-920; 55) | β | 17, 154, 159 |
| LigA | DNA ligase | 676 | 50 (10-673) | 51 (16-684) | 86 (1-674) | A, P, E | E. coli (10-673; 50) | β | 159, 186 |
In E. coli and many other bacteria, DNA replication is initiated at a single site on the circular chromosome, the origin of replication or oriC, which contains a series of 9-bp sequence repeats known as DnaA boxes (180). Binding of multiple DnaA molecules to oriC leads to separation of the two template DNA strands at a nearby AT-rich region. One molecule of the replicative helicase DnaB is loaded onto each of the resulting single DNA strands, and each proceeds to unwind the parental DNA duplex to create replication forks that move away from the origin in opposite directions. DNA primase associates with DnaB and constructs short RNA primers, onto which the multisubunit DNA polymerase III holoenzyme (Pol III HE) builds each new DNA strand. DNA synthesis by Pol III is carried out only in the 5′-to-3′ direction; thus, synthesis of one strand (the leading strand) is continuous while synthesis of the other strand (the lagging strand) is discontinuous. The DnaB helicase translocates on the lagging strand at each fork. DNA replication continues bidirectionally around the circular chromosome until the two replication forks meet in the terminus region, located approximately opposite the origin (182, 189). This eventually yields two copies of the bacterial chromosome, each containing one strand from the parental chromosome and one nascent strand.
With the availability of complete genome sequences for more than 900 bacterial strains, we can now extrapolate our solid understanding of E. coli DNA replication into genera outside the Enterobacteriaceae, such as Acinetobacter. While both are members of the gammaproteobacteria, the Acinetobacter genus is relatively remote from E. coli (see the phylogenetic tree in Fig. 1). The sequences of Acinetobacter DNA replication proteins have diverged significantly from those of other organisms, in some cases beyond the detection level of standard homology searches. While this in some ways restricts the level of information that can be extrapolated from the E. coli system, it also provides strong indications of which aspects of DNA replication are conserved and which are not. Acinetobacter spp. display several peculiarities among their replication components, some of which are discussed in detail below. Some of these, such as the fusion of rnhA and dnaQ genes in a single cistron as previously noted by Barbe and colleagues (15), are unique to members of Acinetobacter and the related genus Psychrobacter. Others highlight the previously identified relationship between the Acinetobacter and Pseudomonas genera (15), with the two groups often displaying common protein features despite significant divergence at the DNA sequence level.
Initiation of Replication in Acinetobacter
All Acinetobacter spp. for which genome sequences have been completed contain a single circular chromosome, with the origin of replication (oriC) located between the rpmH and dnaA loci (15). This arrangement is also seen in Pseudomonas spp. (270) and contrasts with the case for E. coli, whose origin maps between the gidA (mnmG) and mioC genes, some 40 kb from rpmH and dnaA. For each Acinetobacter sp., the replication initiator protein DnaA is relatively well conserved (46% identity with E. coli DnaA [Table 1]), particularly within the N-terminal DnaB-interacting domain (domain I), the C-terminal AAA+ ATPase domain (domain III), and the DNA-binding domain (domain IV) (134). Domain II, identified as a flexible linker in the E. coli protein, has diverged beyond recognition in Acinetobacter DnaA proteins.
Initiation of DNA replication in E. coli is regulated by a paralog of DnaA known as Hda (138, 180). When bound to a DNA-associated β sliding clamp, Hda causes the active ATP-bound form of DnaA to hydrolyze its substrate and thus become inactive, preventing unscheduled reinitiation of replication (142). Genes encoding proteins with moderate homology to E. coli Hda (30% sequence identity) are found within the Acinetobacter sp. genomes. Although they are frequently annotated as “putative chromosomal replication initiator, DnaA type,” these proteins likely perform a role analogous to that of Hda, i.e., suppression of untimely initiation of DNA replication.
Historically, the study of DNA replication has focused on two model organisms: the Gram-negative E. coli (a gammaproteobacterium) and the Gram-positive Bacillus subtilis (a firmicute). Both these organisms utilize AAA+ ATPase family helicase loader proteins to load the replicative DNA helicase (DnaB in E. coli and DnaC in B. subtilis) onto the template DNA at origins of replication or during replication restart after fork blockage (59). In E. coli, helicase loading is carried out by DnaC, which complexes and thereby inactivates DnaB until its activity is required and dissociates upon helicase loading (226). In B. subtilis, three proteins, DnaB, DnaD, and DnaI, are used; the last is a homolog of E. coli DnaC (26, 117). No homologs of E. coli DnaC or B. subtilis DnaB, -D, or -I are identifiable within the Acinetobacter sp. genomes, with the exception of prophage-associated DnaC homologs in A. baumannii strains AYE and ACICU. Homology searches across those bacteria with completed genome sequences show the phylogenetic distribution of DnaC to be restricted to the Enterobacteriaceae (Fig. 1). The DnaB, -D, and -I helicase-loading proteins are similarly restricted to the Firmicutes, which include the genera Bacillus, Staphylococcus, and Mycoplasma. This implies that Acinetobacter spp., and for that matter all other bacteria outside the Enterobacteriaceae and the Firmicutes, either are capable of loading the replicative helicase without additional loader proteins or have helicase loaders whose sequences are not related to the E. coli and B. subtilis proteins. In support of the former scenario, the dnaB (helicase) gene from the epsilonproteobacterium Helicobacter pylori has been found to successfully complement temperature-sensitive dnaC mutations in E. coli (236). This suggests that H. pylori DnaB not only can be loaded without the activity of DnaC in vivo but also can be assembled successfully into an E. coli replisome. Further evidence comes from the observation that DnaB proteins from Pseudomonas spp. can be loaded onto the origin of a broad-host-range plasmid in vitro, using only DnaA, i.e., without assistance from a loader protein (35). Since the E. coli DnaA and DnaC proteins are structurally related (58, 77, 181, 226), it is possible that DnaA (or a DnaA homolog) loads DnaB in other organisms, not just at oriC but also during replication restart. Further work is clearly required to better understand the process of helicase loading in organisms outside the Enterobacteriaceae and Firmicutes.
A Highly Diverged Pseudomonas-Type ψ Subunit
Most of the 10 components of the Pol III HE complex are readily identifiable in Acinetobacter spp. based on homology searches with the E. coli proteins (Table 1). Nevertheless, such searches could not locate a homolog of the holD gene, which encodes the ψ subunit of the clamp loader complex in E. coli (7). A heterodimer formed between the χ and ψ subunits binds to the clamp loader complex [(τ/γ)3δδ′] of DNA polymerase III through a region at the N terminus of ψ (96, 231). The ψχ complex mediates the handoff of the primed template molecule from the DnaG primase via single-stranded DNA-binding protein (SSB) to the clamp loader complex (τ3δδ′), thus representing a functional link between the primosome complex (DnaB-DnaG) and Pol III HE within the replisome (275). Originally thought to represent a simple “bridge” between the clamp loader complex and the χ subunit, the ψ subunit has now been shown to play a significant role in loading of the β clamp onto its DNA template in E. coli (7).
During their work on the Pseudomonas replisome, McHenry's group noted that Pseudomonas spp. also lacked an identifiable homolog of the E. coli holD gene (120). This group found, however, that addition of purified ψχ complex from E. coli dramatically stimulated the activity of a minimal Pseudomonas replication complex (αɛ-τδδ′-β) in vitro. As this stimulation was not observed following the addition of Pseudomonas χ subunit alone, this strongly suggested the existence of a cryptic ψ subunit in Pseudomonas, which the group later identified and characterized (121). The Pseudomonas-type ψ subunit was found to be considerably longer than its E. coli counterpart (282 versus 137 residues). Despite this, multiple-sequence alignments revealed three distinct conserved regions corresponding to helices α1 and α4 of the E. coli ψ and a region at the N terminus known to interact with domain III of τ/γ (7, 96).
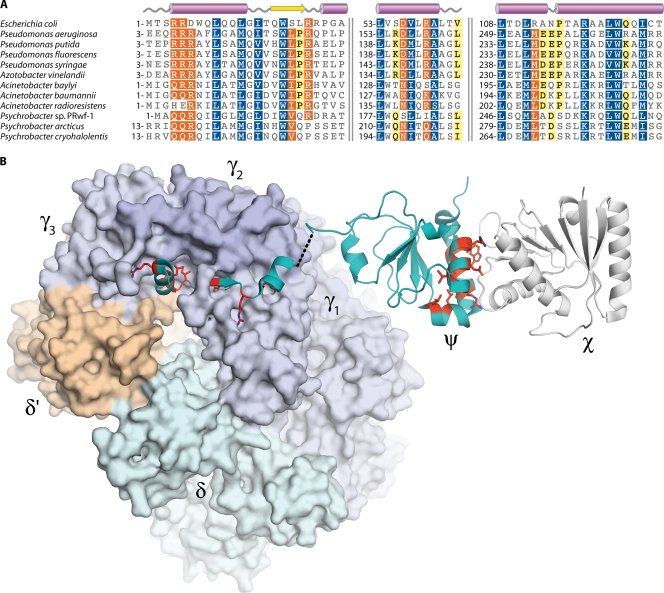
We have used hidden Markov model (HMM)-based homology searches of the A. baylyi ADP1 genome with the program HHsearch (234, 235) to reveal a poorly conserved homolog of P. aeruginosa ψ (hypothetical protein ACIAD1326, 17% sequence identity). Homologs of this putative ψ protein are found within each of the available A. baumannii genome sequences (62% sequence identity with A. baylyi ψ [Table 1]). From multiple-sequence alignments, it appears that the open reading frame within the A. baylyi gene should begin 11 residues earlier than in the published genome annotation. Using these newly identified homologs as input for HHsearch, it was further possible to detect extremely weak, but significant, homology between these proteins and the E. coli ψ subunit (for the A. baylyi protein, E value = 1.8 × 10−4; 13% overall sequence identity). These newly identified Acinetobacter ψ proteins maintain the three conserved regions (Fig. 2A) that were previously identified within the E. coli and Pseudomonas ψ subunits (121). When the conserved residues are mapped onto the E. coli clamp loader (γ3δδ′ψ) and ψχ structures (96, 231), they fall within the N-terminal clamp loader-interacting region and the χ-contacting helices α1 and α4 (Fig. 2B). When these ψ types are mapped to a phylogenetic tree for the gammaproteobacteria (Fig. 1), a decisive split is seen, in which Pseudomonas, Acinetobacter, and their relatives use the longer Pseudomonas-type ψ subunit, while members of the Enterobacteriaceae, Vibrio, Haemophilus, and Shewanella have the shorter E. coli-type protein. Interestingly, gene knockout studies show that the holD gene is dispensable in E. coli (11) but is essential in both A. baylyi ADP1 (61) and P. aeruginosa PA01 (118) (Table 1). This might suggest that the longer, Pseudomonas-type ψ subunits have an additional, as-yet-unidentified function in vivo.
FIG. 2.
Sequence conservation within the ψ subunit of DNA polymerase III. (A) Multiple alignment of ψ amino acid sequences. Numbers indicate the sequence position of the first residue in each alignment block; pairs of vertical lines denote discontinuities in the sequences. White characters with blue shading indicate positions with homology across all sequences. White characters with orange shading indicate residues conserved in 80% of sequences, while black characters with yellow shading indicate 60% conservation. The sequence alignment was produced using the MUSCLE algorithm (72) within the Geneious software package (Biomatters). (B) Crystal structures of the E. coli γ3δδ′ψ and χψ complexes. Residues conserved in the A. baylyi ψ subunit are colored red and have side chains shown. (Produced with PyMOL [64] using data from Protein Data Bank entries 3GLI [231] and 1EM8 [96].)
An Unusual Primosome Complex: Unique Features of Acinetobacter DNA Primase
DNA polymerase III cannot synthesize new DNA strands de novo and first requires the construction of a short RNA primer by the DnaG primase (226); this occurs every second or two during Okazaki fragment synthesis on the lagging strands (67). During this step, DnaG binds to the replicative helicase DnaB, forming a complex known as the primosome. This interaction formed by DnaB and DnaG has been shown to modulate the activities of both enzymes (39).
DnaB consists of two domains: a primase-binding domain at the N terminus and an ATP-utilizing DNA helicase domain at the C terminus (12). Bacterial primases contain three distinct domains: a zinc-binding domain at the N terminus, a central RNA polymerase (RNAP) domain, and a C-terminal helicase-binding domain that is structurally related to the N-terminal domain of DnaB (51, 78, 191, 237, 241, 258). The structural basis for the primosome interaction was revealed recently in crystal structures of the complex between DnaB and the C-terminal domain of DnaG from Geobacillus stearothermophilus (12). DnaB was seen to form a 3-fold symmetric hexamer, mediated by the formation of a ring of N-terminal primase-binding domains. The C-terminal helicase-binding domains of three DnaG molecules were found to associate with this ring, with DnaG residues forming a network of hydrogen bonds, which were later shown to play a role in modulating DnaB activity (12, 39).
In Acinetobacter spp., the enzymatic sites of DnaB and DnaG are well conserved. The domains responsible for forming the primosome complex, however, have diverged significantly. In fact, the DnaG helicase-binding domain has diverged so greatly that it bears no resemblance to any other protein in the current sequence databases, including DnaG proteins from members of the closely related genus Psychrobacter. Therefore, it is not currently possible to model the Acinetobacter primosome complex based on the G. stearothermophilus structure. In contrast, the DnaG helicase-binding domains of other Gram-negative organisms, namely, E. coli and P. aeruginosa, do show detectable sequence homology with the G. stearothermophilus domain. As such, primosome complexes could conceivably be modeled for these organisms using the G. stearothermophilus structure as a template.
The DnaG proteins of Acinetobacter spp. and Psychrobacter spp. are further complicated by a large insertion (50 to 100 residues) between the N-terminal zinc-binding domain and the central RNA polymerase domain. Based on secondary structure and disorder predictions (155, 221), this insertion appears to be disordered and thus probably acts as a linker region. Increased flexibility between the zinc-binding and RNA polymerase domains may have significant implications for primase activity, as (i) the zinc-binding domain is believed to act as an anchor that recognizes priming sites in DNA and regulates RNA primer length and (ii) primase zinc-binding domains have been demonstrated to function in trans with other primase molecules in E. coli (50). A thorough biochemical and structural investigation of these unusual DnaG proteins is required to fully elucidate the similarities to and differences from the better-studied E. coli enzyme.
Highly Conserved Protein Interaction Modules within the β Sliding Clamp and SSB
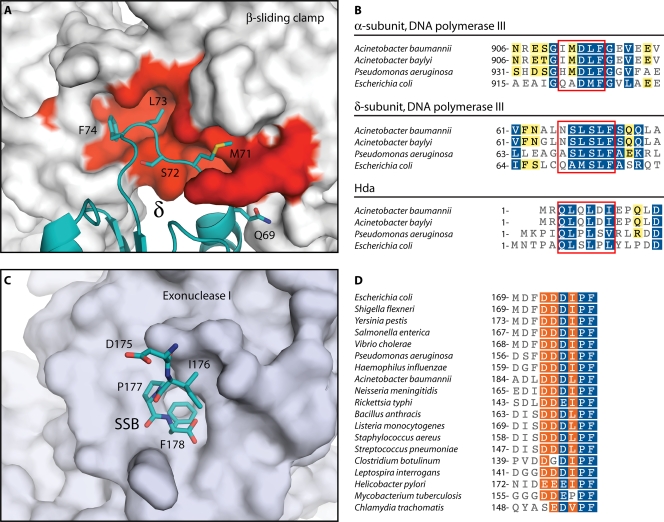
In addition to binding the Pol III δ subunit during clamp loading/unloading (127) and the α polymerase subunit (261), greatly increasing its processivity, the E. coli β sliding clamp also binds to DNA polymerases I, II, IV, and V; DNA ligase; Hda; and the mismatch repair proteins MutS and MutL (55, 159, 260). Each of these proteins binds to a hydrophobic groove (127) on the β sliding clamp by way of a pentameric or hexameric peptide motif, which is often located close to the N or C terminus (55). In Acinetobacter spp., the residues comprising the protein-binding groove of the β sliding clamp are highly conserved (Fig. 3A). Variants of the β-binding motifs (QxD/SLF or QLxLxL) are also conserved within several Acinetobacter proteins, including the α subunits of Pol III (IMDLF) and Hda (QLQLDI) (Fig. 3B). In E. coli, a β-binding motif within the Pol III δ subunit (QAMSLF) is utilized in the loading of the β sliding clamp by the clamp loader complex (Fig. 3B) (127). Despite sharing only weak overall sequence homology with the E. coli δ (26% identity for the A. baylyi protein), this motif is also maintained in Acinetobacter sp. δ subunits (NSLSLF). This suggests that despite significant sequence divergence in δ and other components of the clamp loader complex in Acinetobacter spp. (Table 1), loading of the β sliding clamp onto DNA is likely to utilize a mechanism similar to that of the E. coli system.
FIG. 3.
Highly conserved protein interaction modules in the Acinetobacter replisome. (A) Structure of the E. coli βδ complex (127). The solvent-accessible surface of β is shown in gray and red. Red indicates residues that are conserved in A. baylyi. The δ subunit is colored teal, and side chains are shown for the β-binding consensus residues. (B) Portions of sequence alignments for DNA replication proteins containing β-binding motifs. Boxes indicate β-binding motifs. Residues are highlighted as in Fig. 2. (C) Structure of E. coli exonuclease I bound to the C-terminal peptide of SSB (162). The solvent-accessible surface of exonuclease I is shown in gray. The C-terminal peptide of SSB is shown in teal. (D) Portion of a sequence alignment for SSB proteins. In panels B and D, white characters with blue shading indicate positions with homology across all sequences, white characters with orange shading indicate residues conserved in 80% of sequences, and black characters with yellow shading indicate 60% conservation. Sequence alignments were produced using the MUSCLE algorithm (72) within the Geneious software package (Biomatters). (Panels A and C were produced with PyMOL [64] using data from Protein Data Bank entries IJQL [127] and 3C94 [162], respectively.)
The hydrophobic groove on the β sliding clamp and its binding motifs in other proteins are strongly conserved across bacterial species (27, 260). An analogous mechanism is also shared by the PCNA sliding clamp of archaea and eukaryotes (209), although the consensus sequence recognized by PCNA (QxxLxxFF) is different from that of the bacterial β-binding motifs (55). The strong conservation of this protein-binding mechanism across bacteria, together with the essentiality of the interactions formed, makes the protein-binding groove of the β sliding clamp (Fig. 3B) an attractive target for the rational design of novel antibiotic compounds. Promisingly, the O'Donnell group has recently described a small molecule that binds within the β clamp binding groove and is capable of disrupting key protein-protein interactions in diverse bacteria without inhibiting analogous interactions in the eukaryotic PCNA system (87).
A second replication protein, the single-stranded DNA-binding protein (SSB), also interacts with many other proteins by way of a conserved peptide motif (229). In addition to interacting with the DnaG primase and the χ subunit of Pol III HE during DNA replication, SSB also binds to a range of DNA repair enzymes (9, 30, 86, 100, 135, 148, 162, 225, 229, 243, 263, 275). These interactions are mediated by the final six to nine residues at the SSB C terminus, which in E. coli has the sequence MDFDDDIPF (229, 275). The crystal structure of the terminal SSB peptide in complex with exonuclease I (Fig. 3C) was recently determined by Keck's group (162). The structure and additional biochemical studies (163) revealed a key role for the final two residues of SSB in binding to a hydrophobic pocket in exonuclease I. While no high-resolution structures have been reported for any other proteins in complex with the SSB peptide (230), it is anticipated that its binding to other proteins occurs in a similar fashion; i.e., the terminal residues of SSB will be inserted into specific pockets on the protein surfaces.
The SSB C-terminal peptide represents a universal system for tying together processes transacted on single-stranded DNA (229). Even in genetically remote organisms, such as Acinetobacter spp., the final six residues of SSB are strongly conserved (Fig. 3D). This conservation extends, in fact, to SSB proteins from a wide range of bacterial species, indicating that this system is probably used universally across all of the bacteria (Fig. 3D) (229).
The β clamp and SSB constitute major protein interaction hubs in bacteria. Each is known to interact with at least 10 other proteins, forming interactions with different partners at different times. In doing so, the β clamp and SSB tie together functional processes required for genome maintenance by recruiting other proteins to their sites of action on double- and single-stranded DNA, respectively. Many of these interacting partners are essential across a wide range of bacteria (277). At the heart of these functional linkages are short, highly conserved peptide motifs that could be exploited as the basis for the design of novel antibiotics. At the individual level, inhibitors of β clamp and SSB interactions should be capable of simultaneously disrupting several functions critical to the survival of a bacterial cell. Furthermore, as these modules show strong sequence conservation across diverse bacterial genera, inhibitors should also have broad-spectrum activity. Finally, and perhaps most crucially, the fact that numerous proteins interact at a single site on each of these hubs might help to preclude the opportunity for resistance to such inhibitors. The accumulation of point mutations that decrease the affinity of inhibitory compounds for their target is a common theme in antibiotic resistance (168). Presumably, in the case of protein-protein interaction sites, any mutation occurring on one interacting partner would need to be compensated for with a complementary mutation on the other partner to maintain the integrity of the interaction site. For this to occur in the β-clamp and SSB protein-binding sites, it would necessitate the unlikely acquisition of simultaneous compensatory mutations in the binding sites of every one of the many proteins that bind at these sites. These protein interaction hubs thus represent an unutilized and relatively unexplored resource for the future discovery of novel antibiotic drugs.
CELL DIVISION
Cell division is an appealing target for the design of antibacterial drugs, as it is also essential for the viability of bacterial populations and, as many of the proteins involved are external to the cytoplasm, it is a target relatively accessible to attack with small-molecule inhibitors. In addition, the majority of proteins have little or no homology to human proteins. In contrast to the case for the processes of DNA replication and transcription, the majority of our knowledge on cell division has emerged only over the past 10 to 15 years. Most studies so far have focused on two rod-shaped organisms, B. subtilis and E. coli, both of which divide precisely at the cell center. Currently, at least 12 proteins are known to be involved in the cell division process, and these are distributed between the cytoplasm, cell membranes, and periplasmic space (76, 89, 103, 156, 259). For the majority of these proteins, their exact biochemical functions remain unknown. Three-dimensional structures of the entire proteins or parts of them have been solved for many, however, and protein-protein interactions are now being determined and characterized (Table 2).
TABLE 2.
Conservation of cell division proteins in Acinetobacter spp.
| Protein | Function | Protein locationa | Protein length (amino acids)b | % Sequence identity (residue range) |
Essentialitye | Homologs with known structuresb (residue range of sequence match; % sequence identity) | Known protein-protein interactions in E. coli | Reference(s) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| A. baylyi vs E. colic | A. baumannii vs E. colid | A. baylyi vs A. baumanniic | ||||||||
| FtsA | Tethers FtsZ to membrane, stabilizes Z rings at membrane | c | 420 | 29 (8-409) | 30 (8-409) | 88 (1-420) | A, E, P | Thermotoga maritima (10-351; 23) | FtsZ | 101, 207, 249, 257 |
| FtsB (DivIC) | Unknown; role in septal peptidoglycan synthesis? | m | 110 | 41 (6-77) | 38 (25-96) | 91 (6-110) | A, E, P | —f | FtsQ, FtsL | 28, 92 |
| FtsE | Unknown; similar to ABC transporter | c | — | — | — | — | E | — | FtsZ, FtsX | 46, 218, 227 |
| FtsI/PBP3 (PBPB) | Cross-links septal peptidoglycan | m | 621 | 40 (15-607) | 39 (7-598) | 89 (10-617) | A, E, P | — | FtsQ, FtsN | 66, 69 |
| FtsK | Role in chromosome segregation and septum formation | m | 1018 | 60 (528-1017) | 59 (516-1009) | 77 (1-1018) | A, E, P | Pseudomonas aeruginosa (527-942; 59), E. coli (528-1017; 59) | FtsZ, FtsL, FtsQ | 66, 169 |
| FtsL | Unknown; role in septal peptidoglycan synthesis? | m | 110 | 24 (26-100) | 26 (28-100) | 88 (29-109) | A, E, P | — | FtsQ, FtsB, FtsW, FtsK | 28, 66, 69 |
| FtsN | Unknown | m | 200 | 23 (44-197) | 16 (3-206) | 87 (1-200) | A, E, P | E. coli (120-200; 19) | FtsQ, FtsI, FtsW | 52, 53, 66, 69, 268 |
| FtsQ (DivIB) | Unknown; role in septal peptidoglycan synthesis? | m | 284 | 25 (61-261) | 26 (90-259) | 81 (1-284) | A, E, P | Yersinia enterocolitica (59-266; 24), E. coli (61-267; 23) | FtsB, FtsL, FtsI, FtsN, FtsW, FtsK | 28, 34, 69, 220, 250 |
| FtsW | Septal peptidoglycan synthesis | m | 399 | 42 (32-388) | 42 (31-387) | 85 (2-399) | A, E, P | — | FtsI, FtsL, FtsN, FtsQ | 66, 69 |
| FtsX | Unknown; similar to ABC transporter | m | — | — | — | — | E | — | FtsE | 218, 227 |
| FtsZ | First protein to form midcell ring in cell division | c | 389 | 47 (31-386) | 48 (31-388) | 90 (1-389) | A, E, P | P. aeruginosa (31-322; 55), B. subtilis (35-389; 46), Mycobacterium tuberculosis (31-322; 53), Methanococcus jannaschii (5-322; 42), A. aeolicus (31-320; 43) | FtsA, ZapA, ZapB, ZipA, FtsE, FtsK, MinC, SulA, ClpX, SlmA | 22, 46, 66, 71, 79, 95, 98, 101, 108, 110, 151, 160, 161, 165, 179, 193, 194, 207, 228, 257 |
| ZapA | Promotes FtsZ polymerization and protofilament bundling | c | 94 | 21 (7-94) | 26 (1-88) | 80 (1-92) | D | Pseudomonas aeruginosa (1-93; 16) | FtsZ | 95, 160 |
| ZapB | Promotes FtsZ polymerization | c | — | — | — | — | D | — | FtsZ | 71 |
| ZipA | Stabilizes Z rings at membrane | m | 325 | 28 (185-310) | 27 (205-330) | 62 (1-324) | E | E. coli (183-323; 26) | FtsZ | 98, 101, 179 |
| MinC | cell division inhibitor | c | 240 | 40 (111-230) | 41 (122-229) | 78 (1-240) | D | T. maritima (111-213; 28) | MinD, FtsZ | 47, 108-110, 193, 228 |
| MinD | Membrane ATPase of the MinC-MinD-MinE system | m | 278 | 64 (9-278) | 65 (9-278) | 95 (1-278) | E | Archaeglobus fulgidis (9-277; 30), Pyrococcus furiosus (9-256; 30) | MinC, MinE | 48, 104, 108-110, 224 |
| MinE | Cell division topological specificity factor | c | 90 | 33 (9-88) | 32 (9-88) | 92 (1-90) | E | E. coli (36-90; 28) | MinD | 62, 110, 147, 214, 278 |
| SlmA | Spatial regulation of Z-ring formation (nucleoid occlusion) | c | — | — | — | — | D | — | FtsZ | 22 |
| SulA | FtsZ assembly inhibitor | c | — | — | — | — | D | P. aeruginosa (no homolog) | FtsZ | 49, 110 |
| ClpX | FtsZ assembly inhibitor | m | 436 | 70 (10-408) | 71 (10-408) | 91 (1-436) | D | Helicobacter pylori (54-417; 58) | FtsZ | 32, 79, 146 |
| AmiC | Amidase; required for septal wall degradation and cell separation | p | — | — | — | — | D | — | — | 20 |
| EnvC | Murein hydrolase; required for septal wall degradation and cell separation | p | — | — | — | — | D | — | — | 21 |
c, cytoplasmic; p, periplasmic; m, membrane associated.
Acinetobacter baylyi protein (15).
A, essential in Acinetobacter baylyi (61); P, essential in Pseudomonas aeruginosa (118); E, essential in Escherichia coli (11); D, dispensable gene.
—, not applicable, no structure determined, or not present in Acinetobacter spp.
The process of cell division involves the ingrowth of the cell envelope layers to form a septum at the cell center between two newly replicated chromosomes. This septum then splits to form two new daughter cells. The first step in cell division is the assembly of the tubulin-like protein FtsZ into a structure known as the Z ring, precisely at midcell (103). This assembly involves ZipA in E. coli (98) and the membrane-attaching protein FtsA in both E. coli and B. subtilis (54, 103, 255). The Z ring marks the future site of cell division and acts as a “landing pad” for the other proteins involved in the division process. Once in place, the remaining cell division proteins are recruited to the Z ring, forming a complex collectively known as the divisome (103). In E. coli these later-assembling proteins include FtsE, -X, -K, -Q, -L, -B, -W, -I, and -N; AmiC; and EnvC (255, 259). Homologs of all of these proteins except FtsN are found in B. subtilis.
Homologs of most of these proteins can also be found within the genomes of Acinetobacter spp., although in most cases their sequences have diverged substantially from those of their E. coli counterparts (Table 2). Interestingly however, no homologs could be found for FtsE, FtsX, or SulA, which are conserved in almost all other gammaproteobacteria. FtsE and -X together form a membrane-associated ABC transporter of unknown cellular function that is essential for the survival of E. coli only under low-ionic-strength conditions (218). In B. subtilis this protein complex has recently been linked to the initiation of sporulation (85). SulA is an inhibitor of FtsZ polymerization that is induced as part of the SOS response in E. coli, where it serves to temporarily halt cell division in the event of DNA damage (49, 112). As discussed in a later section, the lack of SulA in Acinetobacter spp. has strong implications for the mechanisms underlying global stress responses in these organisms.
Assembly of FtsZ at Midcell
FtsZ is the most conserved bacterial cell division protein and is an essential player in the cell division process (103). Homologs of FtsZ have been identified in almost all prokaryotes as well as in plant chloroplasts and the mitochondria of lower eukaryotes (165, 253). In E. coli, the ftsZ gene resides in the dcw cluster, from which ftsQ, -A, and -Z are cotranscribed (63, 84, 271). Analysis of the available Acinetobacter sp. genomes suggests the same to be true for this genus, with ftsQ, -A, and -Z being adjacent to each other in the chromosome. The sequences of FtsZ from Acinetobacter spp. are relatively well conserved (Table 2), particularly within the core domain, which contains the GTP-binding motif and is important for the polymerization of FtsZ into the Z ring. Amino acid residues involved in GTP hydrolysis and formation of the active site (190, 257) are also conserved.
Assembly of FtsZ into the Z ring at precisely midcell requires a balance among several proteins that interact directly with FtsZ to regulate its assembly (3, 54, 156). These proteins either promote or inhibit FtsZ polymerization and are thought to act together to support Z-ring assembly at the right place and time. In E. coli, FtsZ assembly is promoted by FtsA, ZipA, and ZapA and -B and is inhibited by MinC, ClpX, SulA, and SlmA (3, 54, 156).
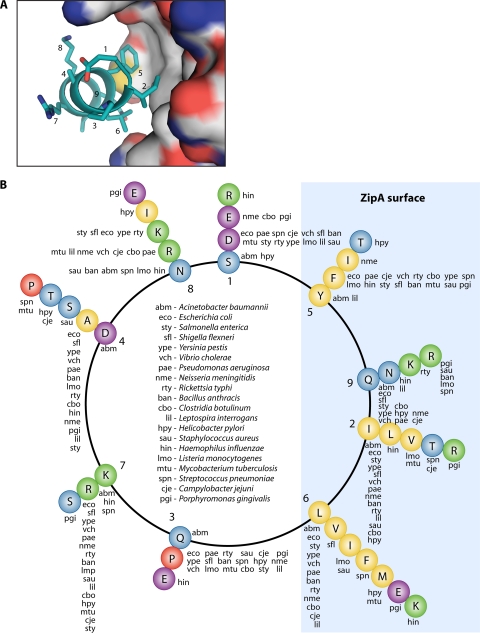
At the C terminus of FtsZ is a nine-amino-acid consensus sequence, often referred to as the C-terminal core domain, which has been shown to be involved in interactions with both ZipA and FtsA (101, 165, 179, 267). In E. coli and most other gammaproteobacteria, the sequence of this region is DIPAFLRKQ. The Somers group has determined the crystal structure of the C-terminal core domain in complex with ZipA (Fig. 4A). The nonapeptide protein-binding motif of FtsZ was shown to form an amphipathic helical structure in which residues I374, F377, and L378 form the primary contacts with ZipA. Earlier studies using alanine-scanning mutagenesis showed that these same three residues provide nearly all of the binding strength in the interaction with ZipA in E. coli (179). Residues D373 and P375 of E. coli FtsZ are also important in the interaction with ZipA, however, presumably setting the C-terminal region in the correct conformation for binding (101).
FIG. 4.
The FtsZ-ZipA interaction. (A) Diagram of the FtsZ C-terminal peptide in contact with the ZipA surface (179). The solvent-accessible surface of ZipA is colored according to atom type: C, white; N, blue; O, red; S, yellow. Numbering indicates positions within the conserved nine-residue motif. (Produced with PyMOL [64] using data from Protein Data Bank entry 1F47 [179].) (B) Helical projection of the FtsZ C-terminal peptide. Letters at each position indicate the residue occupying that position in different bacterial species.
By projecting the nonapeptide motifs from the FtsZ proteins of diverse bacteria onto a helical wheel, it can be seen that most maintain an amphipathic helix structure with hydrophobic residues at positions 2, 5, and 6; Asp at position 1; and Pro at position 3 (Fig. 4B). The fact that this motif is conserved in Gram-positive organisms (which lack ZipA) supports the idea that the interaction of FtsZ with other proteins, most notably FtsA, also involves the C-terminal amphipathic helix. Interestingly, while hydrophobic residues are maintained at positions 2, 5, and 6 of the FtsZ C-terminal peptides in Acinetobacter spp. (SIQDYLKNQ), the conserved Asp and Pro residues found in other organisms at positions 1 and 3 have been replaced with Ser and Gln. It has previously been shown that site-specific mutation of these positions (D373 and P375) in E. coli FtsZ results in disrupted binding to ZipA (101). Importantly, P375 mutants also show reduced binding to FtsA (165). The unusual replacement of this residue with Gln in Acinetobacter spp. might suggest a somewhat different binding mode for FtsZ to both ZipA and FtsA in these organisms.
Complexation of FtsZ with FtsA, ZipA, ZapA, and ZapB represents the starting point of the cell division pathway in E. coli. As such, these interactions mediated by the FtsZ C-terminal helix have been viewed as attractive targets for the design of novel antibiotics. Wyeth, in particular, has invested considerable effort in the development of antagonists of the FtsZ-ZipA interaction and has discovered several compounds that bind to ZipA at the FtsZ interaction site (123, 124, 179, 223, 244, 247). Importantly however, none of these have led to a useful antagonist of the FtsZ-ZipA interaction, despite the opportunities for structure-guided design afforded by crystal structures of ZipA in complex with these ligands (123, 124, 223, 244, 247). The difficulty in expanding these lead compounds lies with the fact that FtsZ binds into a shallow, and relatively featureless, hydrophobic groove on the ZipA surface (179). The broad interacting surface provides few opportunities to incorporate polar contacts between ZipA and the designed ligands. The few attempts to do so have yielded little or no increase in binding strength (247). Wyeth has since abandoned its efforts to develop an FtsZ-ZipA antagonist.
Wyeth's inability to successfully extend apparently promising lead compounds into useful inhibitors of this interaction highlights the difficulty associated with targeting protein-protein interactions using structure-aided design. The broad and relatively shallow interaction surface of ZipA ultimately decreases the utility of this interaction as a target. Furthermore, analysis of the C-terminal helices of FtsZ proteins from diverse bacteria suggests only a vague requirement for hydrophobic residues on the interacting face of the helix (Fig. 4B). This implies that there is significant variation in the shape of FtsZ-binding sites in proteins from different bacteria, further complicating drug design efforts. Importantly, as the binding site for FtsA within FtsZ overlaps significantly with the binding site for ZipA, this might imply that the FtsZ-FtsA interaction, also viewed as a potential drug target, could be equally difficult to target.
Recruitment of Membrane-Associated Proteins
Following formation of the Z ring, the later-assembling membrane-associated cell division proteins are recruited to the midcell site and drive the process of septum formation (89, 254). These proteins represent appealing targets for the development of novel antibacterials, as regions essential for their function are largely located outside the (inner) cell membrane, making them more accessible to inhibitors. Their potential as antibacterial targets is also increased because most of these proteins are essential and the majority lack homologs in eukaryotes (156). The core set of essential membrane-associated cell division proteins have now been identified (103). However, few biochemical data are yet available for these proteins, and many are still to be structurally defined owing to the inherent difficulties associated with in vitro analyses of membrane proteins. All of the core membrane cell division proteins are identifiable in Acinetobacter spp. on the basis of HMM-based homology searches with E. coli proteins (Table 2).
Of the membrane-associated cell division proteins in E. coli, FtsQ interacts with the greatest number of other proteins, with two-hybrid studies and coimmunoprecipitation experiments revealing interactions with FtsA, -K, -X, -I, -W, -B, -L, and -N (28, 66, 69, 136). Separate regions within the periplasmic portion of FtsQ are required for localization of the protein to the division site and recruitment of downstream proteins (37, 250). The periplasmic region forms a modular two-domain structure that crystallizes as a dimer yet is monomeric in solution (250). Localization to the Z ring requires interaction with upstream proteins such as FtsK and is mediated largely by the α domain (residues 58 to 125) (250). Acinetobacter FtsQ proteins are only moderately conserved, sharing 26% overall sequence identity with E. coli FtsQ (Table 2). Residues previously shown to be required for localization of E. coli FtsQ (V92, Q108, V111, and K113) (37, 90, 250) map to the outermost tip of the α domain and are conserved within Acinetobacter FtsQ proteins (equivalent positions are L95, R110, V113, and R115).
Immunoprecipitation experiments have demonstrated that in E. coli the three proteins FtsL, -B, and -Q assemble away from the midcell site and arrive as a complex (28). Recruitment of these proteins by FtsQ involves the β domain (residues 128 to 260), also within the periplasmic portion (250). Residues required for these interactions map to the final two strands of a central β sheet, at the interface of the crystallographic dimer. The corresponding positions in Acinetobacter FtsQ proteins are relatively well conserved, suggesting the recruitment of FtsB and -L to share a mechanism with E. coli. Further studies of the structural basis of FtsQ interactions are required to identify potential targets for the rational design of novel antibacterials. In the future, it might then be possible to design compounds that disrupt the localization of FtsQ to the Z ring or its recruitment of downstream proteins such as FtsB and -L.
Peptidoglycan Recycling and Response to β-Lactam Antibiotics
Recent studies have revealed that A. baylyi strains carrying mutations in the peptidoglycan-recycling enzyme UDP-N-acetylmuramate:l-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase (murein peptide ligase; Mpl) are hypersensitive to β-lactam antibiotics (91). In contrast, mpl-deficient strains of E. coli are no more sensitive to β-lactam compounds than wild-type strains (91). Deletion of the entire mpl gene results in nonviability in A. baylyi ADP1 under the growth conditions used in the high-throughput gene knockout study (61). The equivalent gene is dispensable for growth under similar conditions in E. coli (11, 170). Interestingly, an A. baylyi strain carrying an mpl mutation is also sensitive to UV-induced DNA damage (36). Aside from transport proteins, no proteins involved in cell envelope synthesis have been linked with the DNA damage response in E. coli (36).
Both UV damage and β-lactam compounds trigger the LexA-mediated SOS response in E. coli (80, 173). This leads to upregulation of the cell division inhibitor SulA, leading to temporary arrest of cell division, allowing time for DNA repair processes to occur (143). As discussed below, Acinetobacter spp. lack not only SulA but also the SOS regulator protein LexA. The sensitivity of mpl-deficient A. baylyi to both β-lactams and UV irradiation might therefore reflect an inability to arrest cell division in response to DNA damage in Acinetobacter spp. It has previously been shown that Acinetobacter spp. undergo reductive division following nutrient starvation; i.e., a large bacilliform cell divides to yield smaller coccoid cells (119). It is tempting to speculate that reductive division represents a general stress response in Acinetobacter and that the observed β-lactam/UV sensitivity of mpl-deficient strains might reflect a defect in remodeling peptidoglycan in the transition to a coccoid morphology. This is supported by the identification of mutations in other peptidoglycan-recycling enzymes that lead to β-lactam sensitivity in Acinetobacter spp. (91), although one cannot rule out more direct inhibition of each of these enzymes by β-lactam compounds.
It has been suggested that if suitable inhibitors of Mpl can be found, this might lead to novel antibiotics that are selective for Acinetobacter spp. and act to increase the efficacy of β-lactam compounds (91). The Joint Center for Structural Genomics has recently determined the crystal structure of Psychrobacter arcticum Mpl (Protein Data Bank accession no. 3HN7), highlighting the previously noted relationship with MurC enzymes (177). P. arcticum Mpl shows relatively high sequence conservation with Acinetobacter Mpl proteins (56% sequence identity). It might now be possible to rationally design inhibitors of Acinetobacter Mpl proteins based on the P. arcticum structure.
RNA TRANSCRIPTION
Bacterial transcription and transcriptional regulation have been the subjects of intense analysis since the concurrent isolation of DNA-dependent RNA polymerase (RNAP) from E. coli by four separate laboratories during the early 1960s (114). The process can be divided into three distinct phases: initiation, elongation, and termination (25). First, RNAP in complex with a promoter specificity component, or σ factor, recognizes and unwinds the template DNA at a specific promoter sequence upstream of the gene to be transcribed (25). RNAP then begins synthesizing mRNA, at first producing a series of short abortive transcripts before clearing the promoter and entering a highly processive elongation stage. Transcription is completed either by intrinsic termination at a specific sequence in the template DNA or by Rho factor-dependent termination, which occurs at poorly conserved Rho utilization sites (43). Each stage of transcription is highly regulated, either by RNAP-associated transcription factor proteins or by specific RNA or DNA sequences that interact with the RNAP transcription complex (74). By specifically regulating the transcription of individual genes (or groups of genes), the cellular levels of each gene product can be regulated and adjusted in response to a variety of cellular and environmental stimuli.
The Acinetobacter sp. genomes sequenced to date originate from strains that occupy three distinct environmental niches: a free-living soil organism, a human body louse symbiont, and a series of multidrug-resistant and drug-susceptible clinical isolates (4, 15, 115, 233, 248). Thus, as well as being able to assess the overall conservation of transcription components in Acinetobacter spp. (Table 3), we are now in a position to identify accessory factors likely to be involved with gene regulation during pathogenesis and antibiotic-induced stress.
TABLE 3.
Conservation of RNA transcription proteins in Acinetobacter spp.
| Protein | Function | Protein length (amino acids)a | % Sequence identity (residue range) |
Essentialityd | Homologs with known structuresb (residue range of sequence match; % sequence identity) | Known protein-protein interactions in E. coli | Reference(s) | ||
|---|---|---|---|---|---|---|---|---|---|
| A. baylyi vs E. colib | A. baumannii vs E. colic | A. baylyi vs A. baumanniib | |||||||
| RpoA | RNAP, α subunit | 335 | 66 (1-324) | 66 (1-324) | 99 (1-335) | A, P, E | E. coli (1-235; 60), Thermus aquaticus (24-312; 39) | RpoB, RpoC, RpoZ | 33, 276 |
| RpoB | RNAP, β subunit | 1,362 | 70 (1-1360) | 70 (1-1360) | 92 (1-1362) | A, P, E | Thermus thermophilus (13-219; 44/357-938; 52/1055-1357; 53) | RpoA, RpoC, RpoD, RpoH, RpoN, RpoZ, NusA, Mfd | 60, 198, 269, 276 |
| RpoC | RNAP, β′ subunit | 1,413 | 71 (17-1386) | 70 (1-1370) | 97 (1-1380) | A, P, E | T. aquaticus (37-1044; 50/1145-1374; 47) | RpoA, RpoB, RpoD, RpoH, RpoN, RpoZ, NusG | 174, 178, 276 |
| RpoD | RNAP sigma 70 factor | 629 | 60 (5-628) | 59 (5-289) | 95 (1-628) | A, P, E | T. thermophilus (97-182; 34/359-627; 61) | RpoB, RpoC | 272, 276 |
| RpoE | RNA polymerase sigma 70 factor, ECF subfamily | 206 | 26 (31-197) | 25 (55-195) | 100 (1-206) | E | —e | RpoB, RpoC | 94 |
| RpoH | RNAP sigma factor 32 | 289 | 58 (5-288) | 59 (5-289) | 95 (1-288) | A, P, E | — | RpoB, RpoC | 94 |
| RpoN | RNAP sigma factor 54 | 482 | 46 (1-482) | 47 (1-482) | 81 (1-482) | D | A. aeolicus (429-480; 61) | RpoB, RpoC | 68, 94 |
| RpoS | RNAP sigma factor 38 | — | — | — | — | D | — | RpoB, RpoC | 94 |
| RpoZ | RNAP, ω subunit | 93 | 54 (1-64) | 54 (1-64) | 93 (1-93) | D | T. thermophilus (1-67; 30) | RpoA, RpoB, RpoC | 276 |
| GreA | Transcription elongation factor, cleaves 3′ nucleotide of paused mRNA | 158 | 68 (1-155) | 70 (1-146) | 93 (1-146) | D | E. coli (1-158; 68) | RpoC | 152, 238 |
| GreB | Transcription elongation factor and transcript cleavage | 157 | 61 (1-155) | 67 (1-159) | 58 (1-158) | D | E. coli (1-155; 61) | RpoC | 152, 252 |
| DksA | DnaK suppressor protein | 179 | 62 (41-179) | 63 (42-186) | 87 (7-186) | D | E. coli (41-179; 62) | RpoC | 201, 205 |
| NusA | Transcription elongation factor NusA (N utilization substance protein A) | 494 | 52 (1-493) | 53 (1-493) | 94 (1-494) | A, P, E | T. maritima (1-344; 40) | RpoB, phage λ N protein | 212, 264, 269 |
| NusB | Transcription termination, L factor (N utilization substance protein B) | 149 | 44 (19-145) | 44 (19-145) | 91 (1-148) | P | E. coli (19-145; 44) | NusE | 5, 152 |
| NusE | 30S ribosomal protein S10 | 108 | 83 (6-108) | 83 (1-108) | 99 (1-108) | A, P, E | E. coli (6-108; 83) | NusB | 164 |
| NusG | Transcription antitermination protein | 177 | 61 (2-176) | 59 (2-176) | 96 (1-177) | A, P, E | E. coli (2-176; 61) | RpoC, Rho | 153, 178, 232, 239 |
| Mfd | Transcription-repair coupling protein | 1,171 | 50 (30-1165) | 50 (14-1147) | 86 (1-1153) | D | E. coli (30-1163; 50) | RpoB | 60, 198 |
| Rho | Transcription termination factor Rho | 422 | 77 (1-418) | 77 (1-418) | 98 (1-421) | E, P | E. coli (1-418; 77) | NusG | 153, 232, 246 |
Architecture of Transcription Complexes and Suitability for Drug Design
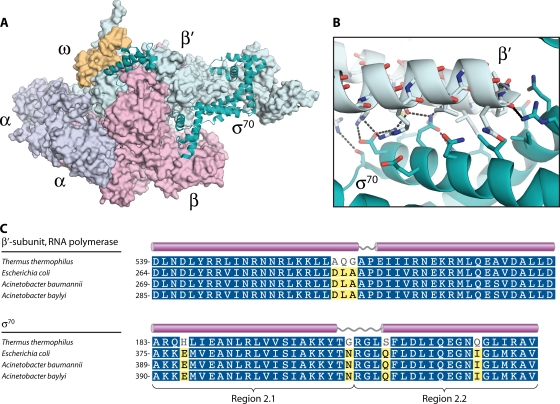
The RNAP core contains five protein subunits, assembled in the order α → α2 → α2β → α2ββ′ → α2ββ′ω (24). To initiate transcription (152), the ∼400-kDa core must first associate with a σ factor to form the RNAP holoenzyme (α2ββ′ωσ). The architecture of the RNAP complex has been elucidated through extensive biochemical and biophysical analyses in E. coli (reviewed in reference 24) and crystal structures of the Thermus aquaticus RNAP core (276) and Thermus thermophilus holoenzyme (251). The overall topology of RNAP resembles a “crab claw,” in which the large β and β′ subunits comprise the “pincers” of the claw (276) (Fig. 5A). In addition to providing a structural brace for RNAP, the β and β′ subunits interact to create a cleft that opens into the main channel of the enzyme. Deep inside the base of this cleft resides the catalytically active center, which is marked by a stably bound Mg2+ ion.
FIG. 5.
Architecture of RNA polymerase and its interaction with σ70. (A) Structure of the Thermus thermophilus RNAP holoenzyme (251). The solvent-accessible surface is shown for RNAP core subunits (α2ββ′ω), and σ70 is shown in ribbon representation. (B) Primary interaction surface between the coiled-coil region within the β′ subunit and conserved regions 2.1 and 2.2 of σ70. Dashed lines indicate hydrogen bonds. (C) Sequence alignments for the coiled-coil region of the β′ subunit and conserved regions 2.1 and 2.2 of σ70. White characters with blue shading indicate positions with homology across all sequences, while black characters with yellow shading indicate 60% conservation. Sequence alignments were produced using the MUSCLE algorithm (72) within the Geneious software package (Biomatters). (Panels A and B were produced with PyMOL [64] using data from Protein Data Bank entry 1IW7 [251].)
Genes encoding RNAP subunits are readily identifiable in Acinetobacter spp. and maintain a relatively high level of sequence conservation (54 to 71% identity at the amino acid level) with equivalent E. coli subunits (Table 3). Within the identified α, β, β′, ω, and σ subunits, key functional regions (e.g., sequence motifs A to H of the β′ subunit [132]) are highly conserved. The relatively high conservation of RNAP subunits despite the relative genetic remoteness of the Acinetobacter genus within the gammaproteobacteria (Fig. 1) highlights the potential utility of RNAP as a target for broad-spectrum antibacterial compounds.
RNAP has been identified as the target for several classes of transcription inhibitors, including the clinically important rifamycin family (256). High-resolution crystal structures of RNAP in complex with members of the rifamycin, streptolydigin, sorangicin, and lipiarmycin inhibitor families have shed light on their modes of action (256). Each of these compounds was found to bind at distinct locations within the DNA-binding and secondary channels of RNAP, away from the active site. This suggests that they bind and interfere with regulatory sites within the enzyme, acting either to sterically block nucleoside triphosphates (NTPs) from entering the active site or by allosteric effects. Most compounds that inhibit RNAP activity exhibit a broad-spectrum bactericidal effect. It has been suggested that RNAP is an underutilized resource and remains a promising target for the discovery of new antibiotics (for reviews, see references 42 and 256).
While several antimicrobial compounds have been found to target RNAP directly, very few target the highly conserved protein-protein interactions that govern the formation of higher transcription complexes. In vivo, the RNAP core forms a series of transient complexes with other protein cofactors: σ70 and other σ factors, antitermination factors (NusA, -B, -E, and -G), transcript cleavage factors (GreA and -B), and regulatory factors (e.g., DksA) (152). Ultimately, these interactions underpin the regulation of gene expression and the transcription of rRNA and thus play important roles in cell proliferation.
The primary interaction surface between σ70 and the RNAP core has been mapped to the coiled-coil motif of the β′ subunit and the conserved region 2.2 of σ70 (272). The structural basis for the RNAP-σ70 interaction was demonstrated by the crystal structure of the T. thermophilus holoenzyme (251). The interaction surface (130) is relatively polar and is formed primarily through a network of side chain-side chain and side chain-main chain hydrogen bonds (Fig. 5B). If novel antagonists of this site could be found, they should interfere with assembly of the functional RNAP holoenzyme and thus inhibit transcription in vivo. The β′ and σ70 residues that mediate this interaction in E. coli are identical in the Acinetobacter sp. proteins (Fig. 5C) and are strongly conserved among all other bacteria (130).
Using an enzyme-linked immunosorbent assay (ELISA)-based approach, the Leonetti group has screened a library of small molecules for inhibitors of the assembly of σ70 onto the RNAP core (8). Several inhibitors of holoenzyme assembly were identified, with some demonstrating bactericidal activity, particularly against Gram-positive organisms. Interestingly, while E. coli proteins were used for the initial screen and were inhibited in vitro, no antimicrobial activity was observed for E. coli or its fellow Gram-negative bacterium P. aeruginosa. Activity was seen, however, against an E. coli mutant strain with increased outer membrane permeability, suggesting that the lack of activity against the wild-type bacterium stems from limited cellular uptake of the compounds. Nonetheless, this work demonstrates the potential for protein-protein interactions within transcription complexes as targets for antimicrobial compounds.
High-throughput screening approaches could be similarly used to identify antagonists of other RNAP-interacting proteins such as NusA and -G, both of which are essential in E. coli, P. aeruginosa, and A. baylyi (Table 3). Traditionally these interactions would not be considered good targets, as the interaction surfaces lack clear grooves, within which an inhibitor might form more extensive protein contacts. However, recent developments in anticancer therapies with stapled peptide antagonists of NOTCH complex formation indicate that (i) interaction surfaces lacking deep clefts or grooves can be targeted and (ii) prevention of the formation of active transcription complexes represents a potentially valuable therapeutic approach (176).
Transcription complexes involved in rRNA synthesis, often called antitermination complexes, require recruitment of many protein transcription factors, such as NusA, -B, -E, and -G, and also require nucleic acid elements of the RNA leader of rRNA genes called boxB, -A, and -C (202). An antitermination complex cannot correctly form without the full complement of protein and RNA elements. The boxA sequences are highly conserved across the eubacteria (consensus, 5′-GCUCUUU[A/G][A/G]A; Acinetobacter consensus, 5′-GAUCAUUAAG) and are integral to the formation of the NusB-NusE heterodimer that binds RNA polymerase (93). More recently it has been shown that NusA binds boxC (consensus, 5′-UGUGU[U/G][U/G]; Acinetobacter consensus, 5′-UGUGUGG), which is involved in antitermination complex formation (10, 23). Targeting these protein-nucleic acid interactions may also be a valid approach for the development of new antibiotics, as preventing antitermination complex formation would severely disrupt cell proliferation. Indeed rRNA gene dosage is closely related to the rate of cell proliferation, with fast-growing species such as B. subtilis, E. coli, and Vibrio natriegens containing 7 to 13 rRNA operons per chromosome, while the slow-growing organism Mycobacterium tuberculosis contains a single rRNA gene (152). Acinetobacter species contain five to seven rRNA operons and are known to grow rapidly, suggesting that rRNA synthesis may be a valid target for bacteriostatic or bactericidal agent development. It may, for example, be possible to design nucleic acid mimetic drugs, such as peptide nucleic acid (PNA) or locked nucleic acid (LNA) (217) to competitively inhibit antitermination complex formation through interaction with boxA or C sequences. Although cell permeability remains a significant hurdle to the effective use of PNAs and LNAs as drugs, they can nevertheless be used to discover pharmacophores for the design of more cell-permeant compounds (217).
Alternative Sigma Factors and Their Regulation
Bacteria respond to changing environmental conditions by (often dramatically) altering the expression of stress response genes. This regulation is effected primarily at the level of transcription, in particular through the differential use of σ factors within the RNAP holoenzyme (167). During exponential growth in rich media, σ70, the housekeeping σ factor, directs transcription of most essential genes. Following application of a particular cellular stress, an alternative σ factor with specific affinity for a certain promoter sequence can replace σ70 and thus direct transcription toward the specific set of genes required under the stressed conditions. Under normal conditions, these alternative σ factors are repressed, either by rapid degradation or by their sequestration by anti-σ factors.
A wide disparity is observed in the number of σ factors encoded by bacteria; e.g., E. coli has 7 different σ factors, B. subtilis has 17, and Streptomyces coelicolor has 63 (152). Alternative σ factors identified in E. coli include σ28, σ38, σ32, σ54, and the extracytoplasmic function σ factors (ECFs) σE and σFecI (80, 175). Of these, only homologs of σ32 and σ54 are identifiable in Acinetobacter spp., along with one to five ECF σ factors (Table 4).
TABLE 4.
Escherichia coli alternative σ factors and regulators and their homologs in Pseudomonas aeruginosa and Acinetobacter spp.
| Function(s) |
E. coli proteins |
Homologs (% sequence identitya) |
||||||
|---|---|---|---|---|---|---|---|---|
|
P. aeruginosa |
A. baylyi |
A. baumannii |
||||||
| σ factor | Anti-σ factor(s)/regulator(s) | σ factor | Anti-σ factor(s)/regulator(s) | σ factor | Anti-σ factor(s)/regulator(s) | σ factor | Anti-σ factor(s)/regulator(s) | |
| Cell envelope stress response, mucoid phenotype | RpoE (σE) | RseA | PA0762 (AlgU) (65) | PA0763 (MucA) (34) | ACIAD2627 (24) | —b | ABAYE0897 (24) | — |
| development | RseB | PA0764 (MucB) (30) | ACIAD0935 (26) | ACIAD0936 (17) | ||||
| Regulation of flagellum assembly | RpoF (σ28) | FlgM | PA1455 (53) | PA3351 (23) | — | — | — | — |
| Heat shock response | RpoH (σ32) | — | PA0376 (59) | — | ACIAD1311 (58) | — | ABAYE1217 (59) | — |
| Regulation of nitrogen metabolism, metabolism of aromatic compounds, pathogenesis | RpoN (σ54) | RpoX (Hpf) | PA4462 (54) | PA4463 (54) | ACIAD0657 (46) | ACIAD0658 (34) | ABAYE3136 (47) | ABAYE3135 (32) |
| Regulation of stationary phase and stress response proteins | RpoS (σ38) | — | PA3622 (76) | — | — | — | — | — |
| Iron uptake, unidentified functions | FecI | FecR | PA3899 (52) | PA3900 (36) | ACIAD1161 (34) | ACIAD1162 (19) | AB57_0985/ACICU_00873 (35) | AB57_0986/ACICU00874 (30) |
| PA1912 (49) | PA1911 (37) | ACIAD1778 (43) | ACIAD1779 (27) | |||||
| PA0472 (46) | PA0471 (30) | ACIAD1469 (33) | ACIAD1471 (25) | |||||
| PA2468 (50) | PA2467 (37) | |||||||
| PA2050 (45) | PA2051 (28) | |||||||
| PA4896 (42) | PA4895 (36) | |||||||
| PA2093 (38) | PA2094 (26) | |||||||
| PA1300 (36) | PA1301 (28) | |||||||
| PA2387 (34) | PA2388 (24) | |||||||
| PA3410 (28) | PA3409 (41) | |||||||
| PA2426 (31) | — | |||||||
| PA0149 (26) | PA0150 (35) | |||||||
| PA2896 (31) | — | |||||||
| PA0675 (25) | PA0676 (26) | |||||||
| PA1776 (23) | — | |||||||
Sequence identity with equivalent E. coli proteins.
—, not applicable or protein not present in organism.
σ32 (RpoH) is the primary regulator of the heat shock response in bacteria. In E. coli, a temperature shift to >42°C results in the induction of a subset of more than 20 σ32-regulated heat shock genes, including the molecular chaperones DnaJ and -K and GroEL and -ES. These proteins serve to stabilize expressed proteins in the cell cytoplasm, preventing misfolding and aggregation, which are especially common during periods of heat stress. It has been shown that the strain A. calcoaceticus 69-V responds to heat shock by upregulating a similar set of proteins, including DnaK, GroEL, and HtpG (18). This suggests that the σ32 regulon in Acinetobacter spp. is probably similar to that in E. coli.
The σ54 family is unusual in that, unlike all other σ factors, its members display no sequence homology with the housekeeping factor σ70. In E. coli, σ54 was originally implicated in the regulation of nitrogen metabolism. However, many additional regulatory roles have since been attributed to this protein (211, 219). While no systematic attempt has yet been made to characterize the σ54 regulon in Acinetobacter spp., σ54 has been shown to be required for the utilization of phenol and benzyl alkanoates as metabolic substrates in A. baylyi (73, 133). The expression of ComP, a pilin-like protein essential for transformational competence in A. baylyi ADP1, is present at the highest levels during late stationary phase and also is likely to be regulated by σ54 (210). It was recently found that genes under the control of the σ54-regulated Pu promoter from Pseudomonas putida could be expressed exogenously in A. baylyi ADP1 (111). Expression patterns mirrored those seen in its natural host, suggesting that there is at least some compatibility between the σ54 regulatory systems of Acinetobacter spp. and Pseudomonas spp.
ECF sigma factors are extremely divergent, and in general sequence analysis alone is insufficient to enable prediction of their function (106). E. coli contains two σ factors which group within the ECF subclass: σE and σFecI. Analysis of the genome sequence of A. baylyi ADP1 reveals that this organism contains three genes with some similarity to E. coli σFecI, which is involved in a signaling cascade leading to the cellular uptake of iron, in the form of ferric citrate (Table 4). In the A. baylyi ADP1 chromosome, all three of these sequences reside upstream of a gene that encodes a FecR-like anti-σ factor and a membrane receptor protein. In contrast, A. baumannii strains contain either none or only one of these proteins, suggesting that alternative iron uptake pathways are important in its pathogenesis (279).
In E. coli, σE is induced following heat shock and during the early stationary phase and regulates the expression of around 200 genes, overseeing the cell envelope stress response (57). It is essential in E. coli, even under apparently nonstressed conditions, and loss of σE activity has been shown to result in loss of cell envelope integrity (105). Remarkably, Salmonella enterica serovar Typhimurium σE, which shares 99% sequence identity with E. coli σE, is nonessential but plays a critical role in virulence (113). The equivalent protein in P. aeruginosa, AlgU, regulates genes involved with the mucoid phenotype associated with cystic fibrosis (211). Clearly, there is much variation in the cellular functions regulated by σE regulons in different bacterial species, although all are linked by their site of action at the cell envelope.
The scope of the σE regulon in Acinetobacter spp. has yet to be examined. Interestingly, however, it has been noted that the AdeABC multidrug efflux pump, which is associated with resistance to several classes of antibiotics in A. baumannii, is likely to be transcribed from an ECF-regulated promoter (166). Given that some strains of A. baumannii do not contain any discernible σFecI homologs, it is highly likely that the adeABC operon is transcribed by RNAP-σE in Acinetobacter spp. No homologs of the anti-σ factor RseA/MucA or the periplasmic protein RseB/MucB, which negatively regulate the activity of σE (273), were found in the published Acinetobacter sp. genomes. The genome of A. baylyi ADP1 contains a second homolog of σE (ACIAD0935); the function of this protein is not yet clear (Table 4).
The absence of σ28 (or RpoF) from the genomes of Acinetobacter spp. comes as no surprise. The σ28 factor is a regulator of flagellar biosynthesis (200), and as members of a nonmotile genus, Acinetobacter spp. do not carry genes for flagellum assembly. Intriguingly, the σ38 (or RpoS) sigma factor, which is thought to be the master stress response regulator in gammaproteobacteria (107), is also missing from Acinetobacter spp. This σ factor controls the induction of a subset of more than 100 genes involved in stress resistance in E. coli, including those whose products resist oxidative stress, acidic pH, ethanol, and hyperosmolarity (107). Although σ38 is undetectable in exponential-phase cultures, its levels reach 30% of that of σ70 in stationary phase, allowing it to more effectively compete with σ70 for binding to core RNAP (129). This is further enhanced by the concurrent induction of Rsd, an anti-σ factor that binds to σ70, preventing it from interacting with the RNAP core (274). The gene encoding Rsd is similarly absent from the genomes of Acinetobacter spp. The apparent lack of a σ38 system in Acinetobacter (and the related genus Psychrobacter) is unique within gammaproteobacteria (excluding symbiotic organisms with reduced genomes). The potential implications of a lack of σ38 in the global stress response and adaptation to antibiotics in these organisms are discussed below.
Pathogenic Acinetobacter spp. Are Enriched in Transcription Factors
A recent comparative genomics study of Acinetobacter strains has revealed a set of 475 genes that are found in the genomes of all completely sequenced pathogen strains but not in the nonpathogenic A. baylyi ADP1 genome (4). This set, termed the pan-A. baumannii accessory genes, includes 59 that encode proteins predicted to be transcription factors based on sequence homology. The LuxR, TetR, MerR, AraC, MarR, GntR, AcrR, ArsR, BadM/Rrf2, XRE, and HxlR families of transcription factors are represented, suggesting that Acinetobacter pathogenesis involves a complex transcriptional network that regulates the expression of proteins with diverse biochemical functions. Further investigation is required to examine the precise roles of these transcription factors in pathogenesis.
AN UNUSUAL DNA DAMAGE RESPONSE
Several genes associated with DNA replication, transcription, and cell division are not found within the genomes of Acinetobacter spp., despite being conserved in virtually all other gammaproteobacteria. In particular, the error-prone polymerase DinB (DNA polymerase II), the stress response factor σ38, and the cell division proteins SulA, FtsE, and FtsX appear to be uniquely missing in Acinetobacter and Psychrobacter spp. (Table 5).
TABLE 5.
DNA damage response proteins in Acinetobacter spp.
| Protein | Function and comment |
|---|---|
| E. coli DNA damage proteins not found in Acinetobacter spp. | |
| DinB | DNA polymerase II; error-prone polymerase involved in SOS mutagenesis |
| σ38 | Stationary phase/stress response σ factor |
| SulA | Cell division inhibitor; induced during SOS response |
| FtsEX | Transporter of unknown function; induced during SOS response |
| LexA | Transcriptional regulator; primary regulator of SOS response |
| Recognized DNA damage response proteins in Acinetobacter spp. | |
| UmuC | DNA polymerase V subunit; truncated to UmuC* (first 64 residues of UmuC) in A. baylyi ADP1 |
| UmuDAb | DNA polymerase V subunit; 50% longer than E. coli UmuD protein, contains LexA-like protease recognition site, regulates expression of DdrR in response to DNA damage |
| DdrR | Unknown function; upregulated following induction of DNA damage in A. baylyi ADP1, found only in Acinetobacter spp. |
| RecA | Recombination protein A; upregulated following induction of DNA damage in A. baylyi ADP1, required for DNA damage-inducible regulation of DdrR |
While σ38 is best known for its roles in stationary phase and global stress response regulation, its role in the response to DNA damage in E. coli was recently described (171). In the same organism, dinB and sulA are both induced as part of the SOS DNA damage response mediated by the transcriptional regulator LexA. Together with the fact that acinetobacters also lack LexA, these missing genes indicate an unusual DNA damage response in Acinetobacter spp.
The SOS response has been well characterized in E. coli (143). The response is triggered by DNA damage, some classes of antibiotics, or the disruption or overexpression of certain genes (14, 143, 195). Detection of one of these triggers leads to RecA-dependent autoproteolysis of LexA, a repressor that governs the transcription of around 30 genes in E. coli (29). While some variation is seen among the members of the LexA regulon in different species, secondary DNA polymerases (e.g., DinB and UmuD) and cell division inhibitors, such as SulA, are almost always upregulated (2, 44, 45, 56, 128, 143). Promoters governing the expression of these proteins are derepressed, and therefore upregulated, upon cleavage of LexA (143). As a result, cell division is temporarily arrested and DNA damage is repaired with the aid of repair and recombination systems.
Thus far, only two DNA damage-inducible loci have been identified in Acinetobacter spp. (Table 5). Not surprisingly, DNA damage in A. baylyi ADP1 (then known as A. calcoaceticus) was found to result in upregulation of the recA gene, which is associated with DNA repair and recombination in bacteria (216). Unlike in most other organisms, however, this regulation was independent of the RecA protein, suggesting a novel mechanism for the DNA damage response in this organism. The second gene found to be upregulated following DNA damage in A. baylyi ADP1 is ddrR, which has no identifiable sequence homologs in the current sequence databases and is of unknown function (102). Regulation of ddrR was found to be RecA dependent and appears to involve an unusual UmuD (DNA polymerase V) protein, UmuDAb, that contains a LexA-like domain at the N terminus. Disruption of this novel regulator led to a decrease in the expression of ddrR, suggesting that it acts as an activator rather than a repressor, in contrast to LexA. In A. baylyi ADP1, the umuDAb gene forms part of an unusual umuDC operon in which the umuC gene has been truncated, apparently as a result of transposon activity (102). umuDAb and the truncated umuC gene are constitutively expressed from this operon. A. baumannii strains each appear to contain an umuDAb-like gene in an operon with a full-length umuC gene.
The σ38-mediated stress and LexA-mediated SOS responses are activated not only by DNA damage but also by common drug classes: β-lactams, quinolones, and folate biosynthesis inhibitors (143, 184). Activation of these responses leads to upregulation of mutation-inducing error-prone DNA polymerases and increased rates of lateral gene transfer (80). The σ38 and SOS responses are now thought to play a significant role in the evolution of antibiotic resistance in bacteria by increasing the scope for adaptive evolution through increased rates of mutation and gene transfer (75, 80, 81, 143). This sets up an apparent paradox for some Acinetobacter spp.: they lack the σ38 and LexA stress responses yet have acquired resistance to all antibiotics in clinical use. In fact, it has been shown that unlike most other bacteria, A. baylyi ADP1 does not show an induction of mutagenesis following irradiation with UV light (19). This suggests either that alternate uncharacterized stress response mechanisms support the acquisition of drug resistance determinants in Acinetobacter spp. or that the pathways leading to mutation and/or lateral gene transfer are constitutively enabled. The high level of transformability associated with the laboratory strain A. baylyi ADP1 could be taken as evidence for the latter scenario; however, high-level natural competence has not yet been demonstrated for any other Acinetobacter strain. Given the rapid emergence of drug resistance in Acinetobacter spp. and their increasing significance as pathogens, it is important to develop an understanding of antibiotic-induced stress responses in these organisms, as they are clearly different from those that have been studied to date.
PERSPECTIVES
This review examines three essential biological systems within Acinetobacter spp., i.e., DNA replication, cell division, and RNA transcription, with a view to future exploitation of these systems as targets for rational design of novel antimicrobial compounds. Of course, these are not the only potential targets for drug design in Acinetobacter spp. Ribosomal protein synthesis, for example, has not been covered here, yet it is one of the most common targets for antibacterial compounds. Compared with replication, cell division, and transcription proteins, ribosomal proteins and rRNA molecules are very highly conserved across the bacteria and in fact even within the eukaryotes and archaea. Acinetobacter spp. show little variation within these components relative to other, better-studied bacteria. Primary metabolism and cell wall structures, which are also common drug targets, are not covered here, as information about these systems is difficult to extract from genome sequence information. Our analysis has, however, demonstrated the merits of using genome sequence data to extrapolate information about fundamental biological processes from model organisms to clinically relevant, but more poorly understood, bacteria. This analysis has allowed us to identify highly conserved aspects of DNA replication, cell division, and transcription in these organisms, which could have potential as targets for the rational design of novel antimicrobials. We recognize, of course, that a great deal more work is required to assess the actual drug susceptibilities of these systems, even before the challenges of developing inhibitors that are nontoxic to humans, are available at the site of infection, are capable of being taken up by bacterial pathogens, and then exhibit a bacteriostatic effect can be addressed.
While research into the fundamental aspects of Acinetobacter biology is still in its infancy, it is already clear that these bacteria have many unusual attributes. Here we have brought to the surface some peculiar characteristics of their DNA replication machinery. These observations imply that chromosomal replication in these organisms might involve subtle mechanistic differences from E. coli and other bacteria. Within the cell division proteins, interruption of the gene encoding the peptidoglycan-recycling enzyme Mpl has been shown to produce a β-lactam-sensitive phenotype in A. baylyi ADP1. Such readily observable phenotypes are rare for knockouts of peptidoglycan-recycling genes (199). Although the reason for this is currently unknown, it does suggest that A. baylyi ADP1 might prove to be a useful model organism for studying peptidoglycan recycling in Gram-negative bacteria.
RNAP and other components of the basic transcription machinery in Acinetobacter spp. are similar to those found in other bacteria. Some variation is seen, however, in the use of transcriptional regulators. Acinetobacter spp. appear to contain a somewhat minimized set of alternative σ factors, with some strains containing only σE, σ32, σ54, and one ECF-type σ factor. Interestingly, Acinetobacter spp. also lack many other canonical stress response proteins, including the DNA damage response regulator LexA. They do, however, contain a large number of putative DNA-binding repressors and activators, many of which are of unknown function. The numbers and types of these transcriptional regulators vary among Acinetobacter species, suggesting that they play specific roles in adaptation of each species to its environment. This is evident in the fact that the genomes of pathogenic Acinetobacter spp. are highly enriched for genes encoding transcriptional regulators. Further research into transcriptional stress responses in individual Acinetobacter species is vital, as such responses are likely to play a central role in the development of antibiotic resistance in these organisms, which is perhaps the foremost problem associated with treating Acinetobacter sp. infections in the clinical setting.
Already it is clear that Acinetobacter spp. have diverged significantly from other gammaproteobacteria over time and have developed unique solutions for coping with evolutionary pressures. While this presents challenges for conducting Acinetobacter research, the tremendous utility of A. baylyi ADP1 as a laboratory organism provides an exciting opportunity to understand many processes and pathways in these organisms, thereby increasing our understanding of bacterial evolution in the context of a group of bacteria that are still emerging as human pathogens.
Acknowledgments
Our research in these areas was supported by a project grant (to P.J.L., E.J.H., and N.E.D.) from the Australian National Health and Medical Research Council and by an Australian Research Council Australian Professorial Fellowship to N.E.D.
Biography
 Andrew Robinson obtained a B.Sc. in Biological Chemistry with First Class Honors from Macquarie University (Sydney, Australia) in 2003. He then commenced postgraduate studies in the laboratory of Associate Professor Bridget Mabbutt at Macquarie, receiving his Ph.D. in 2008. His graduate studies focused on the structures of integron-associated proteins and their roles in adaptation in bacteria. He joined the laboratory of Professor Nick Dixon at the University of Wollongong in 2007, where he is characterizing structures and functions of DNA replication proteins from Acinetobacter spp. He has a keen interest in the evolution of bacterial proteins and the development of antibiotic resistance.
Andrew Robinson obtained a B.Sc. in Biological Chemistry with First Class Honors from Macquarie University (Sydney, Australia) in 2003. He then commenced postgraduate studies in the laboratory of Associate Professor Bridget Mabbutt at Macquarie, receiving his Ph.D. in 2008. His graduate studies focused on the structures of integron-associated proteins and their roles in adaptation in bacteria. He joined the laboratory of Professor Nick Dixon at the University of Wollongong in 2007, where he is characterizing structures and functions of DNA replication proteins from Acinetobacter spp. He has a keen interest in the evolution of bacterial proteins and the development of antibiotic resistance.
 Anthony J. Brzoska graduated with First Class Honors from the University of Technology, Sydney, in 2001, before commencing a Ph.D. in the laboratory of Dr. Neville Firth and Professor Ron Skurray at the University of Sydney in 2003. His Ph.D. studies focused on the molecular and functional analysis of par, an active partitioning system from the staphylococcal multiresistance plasmid pSK41. In 2007, he joined the laboratory of Associate Professor Peter Lewis at the University of Newcastle, where his current work focuses on the determination of novel protein-protein interactions in transcription and DNA replication in Acinetobacter spp. and Staphylococcus aureus.
Anthony J. Brzoska graduated with First Class Honors from the University of Technology, Sydney, in 2001, before commencing a Ph.D. in the laboratory of Dr. Neville Firth and Professor Ron Skurray at the University of Sydney in 2003. His Ph.D. studies focused on the molecular and functional analysis of par, an active partitioning system from the staphylococcal multiresistance plasmid pSK41. In 2007, he joined the laboratory of Associate Professor Peter Lewis at the University of Newcastle, where his current work focuses on the determination of novel protein-protein interactions in transcription and DNA replication in Acinetobacter spp. and Staphylococcus aureus.
 Kylie M. Turner undertook her postgraduate studies at the University of Sydney, Australia, in the laboratory of Dr. Lesley Wright and Dr. Julianne Djordjevic and received her Ph.D. in 2007. The focus of her graduate studies was the biochemical and structural characterization of the cryptococcal virulence factor phospholipase B. She commenced her postdoctoral work in Associate Professor Elizabeth Harry's laboratory at the Institute for the Biotechnology of Infectious Diseases at the University of Technology, Sydney, in 2007, where her research focused on the identification of novel cell division proteins in Acinetobacter spp., Bacillus subtilis, and Staphylococcus aureus. She has a strong interest in the discovery of novel antimicrobials.
Kylie M. Turner undertook her postgraduate studies at the University of Sydney, Australia, in the laboratory of Dr. Lesley Wright and Dr. Julianne Djordjevic and received her Ph.D. in 2007. The focus of her graduate studies was the biochemical and structural characterization of the cryptococcal virulence factor phospholipase B. She commenced her postdoctoral work in Associate Professor Elizabeth Harry's laboratory at the Institute for the Biotechnology of Infectious Diseases at the University of Technology, Sydney, in 2007, where her research focused on the identification of novel cell division proteins in Acinetobacter spp., Bacillus subtilis, and Staphylococcus aureus. She has a strong interest in the discovery of novel antimicrobials.
 Ryan Withers obtained a bachelor's degree in biotechnology at the University of Newcastle in 2007. He is currently undertaking Ph.D. studies in molecular microbiology in Peter Lewis' laboratory, focusing on characterization of transcription factors in Acinetobacter baylyi.
Ryan Withers obtained a bachelor's degree in biotechnology at the University of Newcastle in 2007. He is currently undertaking Ph.D. studies in molecular microbiology in Peter Lewis' laboratory, focusing on characterization of transcription factors in Acinetobacter baylyi.
 Elizabeth J. Harry received her Ph.D. in the laboratory of Professor Gerry Wake from the University of Sydney in Australia. Her postdoctoral work was in Professor Richard Losick's laboratory at Harvard University. She was an Australian Research Council Research Fellow at the University of Sydney until 2004, when she was appointed an Associate Professor and principal investigator at the Institute for the Biotechnology of Infectious Diseases (IBID) at the University of Technology, Sydney. An enduring focus of her research is on the mechanism of bacterial cell division and its regulation. A more recent interest is the discovery and mechanism of action of antibacterials.
Elizabeth J. Harry received her Ph.D. in the laboratory of Professor Gerry Wake from the University of Sydney in Australia. Her postdoctoral work was in Professor Richard Losick's laboratory at Harvard University. She was an Australian Research Council Research Fellow at the University of Sydney until 2004, when she was appointed an Associate Professor and principal investigator at the Institute for the Biotechnology of Infectious Diseases (IBID) at the University of Technology, Sydney. An enduring focus of her research is on the mechanism of bacterial cell division and its regulation. A more recent interest is the discovery and mechanism of action of antibacterials.
 Peter J. Lewis obtained a B.Sc.(Hons) in Biochemistry at the University of Sheffield, United Kingdom, in 1986. Following a move to Australia, he gained a Ph.D. in Biochemistry at the University of Sydney in 1991. He returned to the United Kingdom in 1992 as an SERC/NATO postdoctoral fellow with Professor Jeff Errington, working on the regulation of sporulation, chromosome segregation, and subcellular protein localization in Bacillus subtilis. In 2000 he returned to Australia to establish an independent research group at the University of Newcastle. Currently, his research program focuses on bacterial transcription regulation, transcription complex composition, structure, and subcellular dynamics.
Peter J. Lewis obtained a B.Sc.(Hons) in Biochemistry at the University of Sheffield, United Kingdom, in 1986. Following a move to Australia, he gained a Ph.D. in Biochemistry at the University of Sydney in 1991. He returned to the United Kingdom in 1992 as an SERC/NATO postdoctoral fellow with Professor Jeff Errington, working on the regulation of sporulation, chromosome segregation, and subcellular protein localization in Bacillus subtilis. In 2000 he returned to Australia to establish an independent research group at the University of Newcastle. Currently, his research program focuses on bacterial transcription regulation, transcription complex composition, structure, and subcellular dynamics.
 Nicholas E. Dixon was awarded a Ph.D. in biochemistry by the University of Queensland, Australia, in 1978, and carried out postdoctoral studies in chemistry at the Australian National University (ANU), Canberra, and held Fulbright and C.J. Martin Fellowships at Stanford University, where he worked with Professor Arthur Kornberg. He returned to Australia as a Queen Elizabeth II Fellow and was appointed to the faculty of the Research School of Chemistry, ANU, in 1986. In 2006, he moved to the University of Wollongong, Australia, where he is currently an Australian Research Council Professorial Fellow in biological chemistry. His interests are in mechanistic and structural aspects of bacterial DNA replication.
Nicholas E. Dixon was awarded a Ph.D. in biochemistry by the University of Queensland, Australia, in 1978, and carried out postdoctoral studies in chemistry at the Australian National University (ANU), Canberra, and held Fulbright and C.J. Martin Fellowships at Stanford University, where he worked with Professor Arthur Kornberg. He returned to Australia as a Queen Elizabeth II Fellow and was appointed to the faculty of the Research School of Chemistry, ANU, in 1986. In 2006, he moved to the University of Wollongong, Australia, where he is currently an Australian Research Council Professorial Fellow in biological chemistry. His interests are in mechanistic and structural aspects of bacterial DNA replication.
REFERENCES
- 1.Abe, Y., T. Jo, Y. Matsuda, C. Matsunaga, T. Katayama, and T. Ueda. 2007. Structure and function of DnaA N-terminal domains: specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J. Biol. Chem. 282:17816-17827. [DOI] [PubMed] [Google Scholar]
- 2.Abella, M., S. Campoy, I. Erill, F. Rojo, and J. Barbé. 2007. Cohabitation of two different lexA regulons in Pseudomonas putida. J. Bacteriol. 189:8855-8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams, D. W., and J. Errington. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7:642-653. [DOI] [PubMed] [Google Scholar]
- 4.Adams, M. D., K. Goglin, N. Molyneaux, K. M. Hujer, H. Lavender, J. J. Jamison, I. J. MacDonald, K. M. Martin, T. Russo, A. A. Campagnari, A. M. Hujer, R. A. Bonomo, and S. R. Gill. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190:8053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altieri, A. S., M. J. Mazzulla, D. A. Horita, R. H. Coats, P. T. Wingfield, A. Das, D. L. Court, and R. A. Byrd. 2000. The structure of the transcriptional antiterminator NusB from Escherichia coli. Nat. Struct. Biol. 7:470-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson, S. G., C. R. Williams, M. O'Donnell, and L. B. Bloom. 2007. A function for the ψ subunit in loading the Escherichia coli DNA polymerase sliding clamp. J. Biol. Chem. 282:7035-7045. [DOI] [PubMed] [Google Scholar]
- 8.André, E., L. Bastide, S. Michaux-Charachon, A. Gouby, P. Villain-Guillot, J. Latouche, A. Bouchet, M. Gualtiéri, and J.-P. Leonetti. 2006. Novel synthetic molecules targeting the bacterial RNA polymerase assembly. J. Antimicrob. Chemother. 57:245-251. [DOI] [PubMed] [Google Scholar]
- 9.Arad, G., A. Hendel, C. Urbanke, U. Curth, and Z. Livneh. 2008. Single-stranded DNA-binding protein recruits DNA polymerase V to primer termini on RecA-coated DNA. J. Biol. Chem. 283:8274-8282. [DOI] [PubMed] [Google Scholar]
- 10.Arnvig, K. B., S. Pennell, B. Gopal, and M. J. Colston. 2004. A high-affinity interaction between NusA and the rrn nut site in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 101:8325-8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey, S., W. K. Eliason, and T. A. Steitz. 2007. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science 318:459-463. [DOI] [PubMed] [Google Scholar]
- 13.Bailey, S., R. A. Wing, and T. A. Steitz. 2006. The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell 126:893-904. [DOI] [PubMed] [Google Scholar]
- 14.Banack, T., N. Clauson, N. Ogbaa, J. Villar, D. Oliver, and W. Firshein. 2005. Overexpression of the Hda DnaA-related protein in Escherichia coli inhibits multiplication, affects membrane permeability, and induces the SOS response. J. Bacteriol. 187:8507-8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbe, V., D. Vallenet, N. Fonknechten, A. Kreimeyer, S. Oztas, L. Labarre, S. Cruveiller, C. Robert, S. Duprat, P. Wincker, L. N. Ornston, J. Weissenbach, P. Marlière, G. N. Cohen, and C. Médigue. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32:5766-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beck, J. L., T. Urathamakul, S. J. Watt, M. M. Sheil, P. M. Schaeffer, and N. E. Dixon. 2006. Proteomic dissection of DNA polymerization. Expert Rev. Proteomics 3:197-211. [DOI] [PubMed] [Google Scholar]
- 17.Beese, L. S., and T. A. Steitz. 1991. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 10:25-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benndorf, D., N. Loffhagen, and W. Babel. 1999. Induction of heat shock proteins in response to primary alcohols in Acinetobacter calcoaceticus. Electrophoresis 20:781-789. [DOI] [PubMed] [Google Scholar]
- 19.Berenstein, D. 1987. UV-inducible DNA repair in Acinetobacter calcoaceticus. Mutat. Res. 183:219-224. [DOI] [PubMed] [Google Scholar]
- 20.Bernhardt, T. G., and P. A. J. de Boer. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48:1171-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernhardt, T. G., and P. A. J. de Boer. 2004. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol. Microbiol. 52:1255-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernhardt, T. G., and P. A. J. de Boer. 2005. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beuth, B., S. Pennell, K. B. Arnvig, S. R. Martin, and I. A. Taylor. 2005. Structure of a Mycobacterium tuberculosis NusA-RNA complex. EMBO J. 24:3576-3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borukhov, S., and E. Nudler. 2008. RNA polymerase: the vehicle of transcription. Trends Microbiol. 16:126-134. [DOI] [PubMed] [Google Scholar]
- 25.Browning, D. F., and S. J. Busby. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2:57-65. [DOI] [PubMed] [Google Scholar]
- 26.Bruand, C., M. Farache, S. McGovern, S. D. Ehrlich, and P. Polard. 2001. DnaB, DnaD and DnaI proteins are components of the Bacillus subtilis replication restart primosome. Mol. Microbiol. 42:245-256. [DOI] [PubMed] [Google Scholar]
- 27.Bruck, I., R. E. Georgescu, and M. O'Donnell. 2005. Conserved interactions in the Staphylococcus aureus DNA PolC chromosome replication machine. J. Biol. Chem. 280:18152-18162. [DOI] [PubMed] [Google Scholar]
- 28.Buddelmeijer, N., and J. Beckwith. 2004. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol. Microbiol. 52:1315-1327. [DOI] [PubMed] [Google Scholar]
- 29.Butala, M., D. Žgur-Bertok, and S. J. W. Busby. 2009. The bacterial LexA transcriptional repressor. Cell Mol. Life Sci. 66:82-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cadman, C. J., and P. McGlynn. 2004. PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 32:6378-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calhoun, J. H., C. K. Murray, and M. M. Manring. 2008. Multidrug-resistant organisms in military wounds from Iraq and Afghanistan. Clin. Orthop. Relat. Res. 466:1356-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camberg, J. L., J. R. Hoskins, and S. Wickner. 2009. ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc. Natl. Acad. Sci. U. S. A. 106:10614-10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campbell, E. A., N. Korzheva, A. Mustaev, K. Murakami, S. Nair, A. Goldfarb, and S. A. Darst. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901-912. [DOI] [PubMed] [Google Scholar]
- 34.Carson, M. J., J. Barondess, and J. Beckwith. 1991. The FtsQ protein of Escherichia coli: membrane topology, abundance, and cell division phenotypes due to overproduction and insertion mutations. J. Bacteriol. 173:2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caspi, R., M. Pacek, G. Consiglieri, D. R. Helinski, A. Toukdarian, and I. Konieczny. 2001. A broad host range replicon with different requirements for replication initiation in three bacterial species. EMBO J. 20:3262-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chakravorty, A., M. Klovstad, G. Peterson, R. E. Lindeman, and L. A. Gregg-Jolly. 2008. Sensitivity of an Acinetobacter baylyi mpl mutant to DNA damage. Appl. Environ. Microbiol. 74:1273-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen, J. C., M. Minev, and J. Beckwith. 2002. Analysis of ftsQ mutant alleles in Escherichia coli: complementation, septal localization, and recruitment of downstream cell division proteins. J. Bacteriol. 184:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen, T.-L., L.-K. Siu, Y.-T. Lee, C.-P. Chen, L.-Y. Huang, R. C.-C. Wu, W.-L. Cho, and C.-P. Fung. 2008. Acinetobacter baylyi as a pathogen for opportunistic infection. J. Clin. Microbiol. 46:2938-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chintakayala, K., M. A. Larson, M. A. Griep, S. H. Hinrichs, and P. Soultanas. 2008. Conserved residues of the C-terminal p16 domain of primase are involved in modulating the activity of the bacterial primosome. Mol. Microbiol. 68:360-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chodavarapu, S., M. M. Felczak, J. Rouvière Yaniv, and J. M. Kaguni. 2008. Escherichia coli DnaA interacts with HU in initiation at the E. coli replication origin. Mol. Microbiol. 67:781-792. [DOI] [PubMed] [Google Scholar]
- 41.Chodavarapu, S., R. Gomez, M. Vicente, and J. M. Kaguni. 2008. Escherichia coli Dps interacts with DnaA protein to impede initiation: a model of adaptive mutation. Mol. Microbiol. 67:1331-1346. [DOI] [PubMed] [Google Scholar]
- 42.Chopra, I. 2007. Bacterial RNA polymerase: a promising target for the discovery of new antimicrobial agents. Curr. Opin. Invest. Drugs 8:600-607. [PubMed] [Google Scholar]
- 43.Ciampi, M. S. 2006. Rho-dependent terminators and transcription termination. Microbiology 152:2515-2528. [DOI] [PubMed] [Google Scholar]
- 44.Cirz, R. T., M. B. Jones, N. A. Gingles, T. D. Minogue, B. Jarrahi, S. N. Peterson, and F. E. Romesberg. 2007. Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J. Bacteriol. 189:531-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cirz, R. T., B. M. O'Neill, J. A. Hammond, S. R. Head, and F. E. Romesberg. 2006. Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic ciprofloxacin. J. Bacteriol. 188:7101-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corbin, B. D., Y. Wang, T. K. Beuria, and W. Margolin. 2007. Interaction between cell division proteins FtsE and FtsZ. J. Bacteriol. 189:3026-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cordell, S. C., R. E. Anderson, and J. Löwe. 2001. Crystal structure of the bacterial cell division inhibitor MinC. EMBO J. 20:2454-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cordell, S. C., and J. Löwe. 2001. Crystal structure of the bacterial cell division regulator MinD. FEBS Lett. 492:160-165. [DOI] [PubMed] [Google Scholar]
- 49.Cordell, S. C., E. J. H. Robinson, and J. Löwe. 2003. Crystal structure of the SOS cell division inhibitor SulA and in complex with FtsZ. Proc. Natl. Acad. Sci. U. S. A. 100:7889-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corn, J. E., P. J. Pease, G. L. Hura, and J. M. Berger. 2005. Crosstalk between primase subunits can act to regulate primer synthesis in trans. Mol. Cell 20:391-401. [DOI] [PubMed] [Google Scholar]
- 51.Corn, J. E., J. G. Pelton, and J. M. Berger. 2008. Identification of a DNA primase template tracking site redefines the geometry of primer synthesis. Nat. Struct. Mol. Biol. 15:163-169. [DOI] [PubMed] [Google Scholar]
- 52.Dai, K., Y. Xu, and J. Lutkenhaus. 1993. Cloning and characterization of ftsN, an essential cell division gene in Escherichia coli isolated as a multicopy suppressor of ftsA12(Ts). J. Bacteriol. 175:3790-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dai, K., Y. Xu, and J. Lutkenhaus. 1996. Topological characterization of the essential Escherichia coli cell division protein FtsN. J. Bacteriol. 178:1328-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dajkovic, A., and J. Lutkenhaus. 2006. Z ring as executor of bacterial cell division. J. Mol. Microbiol. Biotechnol. 11:140-151. [DOI] [PubMed] [Google Scholar]
- 55.Dalrymple, B. P., K. Kongsuwan, G. Wijffels, N. E. Dixon, and P. A. Jennings. 2001. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl. Acad. Sci. U. S. A. 98:11627-11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.da Rocha, R. P., A. C. de Miranda Paquola, M. do Valle Marques, C. F. M. Menck, and R. S. Galhardo. 2008. Characterization of the SOS regulon of Caulobacter crescentus. J. Bacteriol. 190:1209-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dartigalongue, C., D. Missiakas, and S. Raina. 2001. Characterization of the Escherichia coli σE regulon. J. Biol. Chem. 276:20866-20875. [DOI] [PubMed] [Google Scholar]
- 58.Davey, M. J., D. Jeruzalmi, J. Kuriyan, and M. O'Donnell. 2002. Motors and switches: AAA+ machines within the replisome. Nat. Rev. Mol. Cell Biol. 3:826-835. [DOI] [PubMed] [Google Scholar]
- 59.Davey, M. J., and M. O'Donnell. 2003. Replicative helicase loaders: ring breakers and ring makers. Curr. Biol. 13:R594-R596. [DOI] [PubMed] [Google Scholar]
- 60.Deaconescu, A. M., A. L. Chambers, A. J. Smith, B. E. Nickels, A. Hochschild, N. J. Savery, and S. A. Darst. 2006. Structural basis for bacterial transcription-coupled DNA repair. Cell 124:507-520. [DOI] [PubMed] [Google Scholar]
- 61.de Berardinis, V., D. Vallenet, V. Castelli, M. Besnard, A. Pinet, C. Cruaud, S. Samair, C. Lechaplais, G. Gyapay, C. Richez, M. Durot, A. Kreimeyer, F. Le Fèvre, V. Schächter, V. Pezo, V. Döring, C. Scarpelli, C. Médigue, G. N. Cohen, P. Marlière, M. Salanoubat, and J. Weissenbach. 2008. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol. Syst. Biol. 4:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641-649. [DOI] [PubMed] [Google Scholar]
- 63.de la Fuente, A., P. Palacios, and M. Vicente. 2001. Transcription of the Escherichia coli dcw cluster: evidence for distal upstream transcripts being involved in the expression of the downstream ftsZ gene. Biochimie 83:109-115. [DOI] [PubMed] [Google Scholar]
- 64.DeLano, W. L. 2002. The PyMOL molecular graphics system. Delano Scientific, San Carlos, CA.
- 65.Dijkshoorn, L., A. Nemec, and H. Seifert. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939-951. [DOI] [PubMed] [Google Scholar]
- 66.Di Lallo, G., M. Fagioli, D. Barionovi, P. Ghelardini, and L. Paolozzi. 2003. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology 149:3353-3359. [DOI] [PubMed] [Google Scholar]
- 67.Dixon, N. E. 2009. DNA replication: prime-time looping. Nature 462:854-855. [DOI] [PubMed] [Google Scholar]
- 68.Doucleff, M., L. T. Malak, J. G. Pelton, and D. E. Wemmer. 2005. The C-terminal RpoN domain of σ54 forms an unpredicted helix-turn-helix motif similar to domains of σ70. J. Biol. Chem. 280:41530-41536. [DOI] [PubMed] [Google Scholar]
- 69.D'Ulisse, V., M. Fagioli, P. Ghelardini, and L. Paolozzi. 2007. Three functional subdomains of the Escherichia coli FtsQ protein are involved in its interaction with the other division proteins. Microbiology 153:124-138. [DOI] [PubMed] [Google Scholar]
- 70.Duzen, J. M., G. C. Walker, and M. D. Sutton. 2004. Identification of specific amino acid residues in the E. coli β processivity clamp involved in interactions with DNA polymerase III, UmuD and UmuD′. DNA Repair (Amsterdam) 3:301-312. [DOI] [PubMed] [Google Scholar]
- 71.Ebersbach, G., E. Galli, J. Møller-Jensen, J. Löwe, and K. Gerdes. 2008. Novel coiled-coil cell division factor ZapB stimulates Z ring assembly and cell division. Mol. Microbiol. 68:720-735. [DOI] [PubMed] [Google Scholar]
- 72.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ehrt, S., L. N. Ornston, and W. Hillen. 1994. RpoN (σ54) is required for conversion of phenol to catechol in Acinetobacter calcoaceticus. J. Bacteriol. 176:3493-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Erie, D. A. 2002. The many conformational states of RNA polymerase elongation complexes and their roles in the regulation of transcription. Biochim. Biophys. Acta 1577:224-239. [DOI] [PubMed] [Google Scholar]
- 75.Erill, I., S. Campoy, and J. Barbé. 2007. Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol. Rev. 31:637-656. [DOI] [PubMed] [Google Scholar]
- 76.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Erzberger, J. P., M. M. Pirruccello, and J. M. Berger. 2002. The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J. 21:4763-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fass, D., C. E. Bogden, and J. M. Berger. 1999. Crystal structure of the N-terminal domain of the DnaB hexameric helicase. Structure 7:691-698. [DOI] [PubMed] [Google Scholar]
- 79.Flynn, J. M., S. B. Neher, Y.-I. Kim, R. T. Sauer, and T. A. Baker. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11:671-683. [DOI] [PubMed] [Google Scholar]
- 80.Foster, P. L. 2007. Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 42:373-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Galhardo, R. S., P. J. Hastings, and S. M. Rosenberg. 2007. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 42:399-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Galletto, R., M. J. Jezewska, and W. Bujalowski. 2003. Interactions of the Escherichia coli DnaB helicase hexamer with the replication factor the DnaC protein. Effect of nucleotide cofactors and the ssDNA on protein-protein interactions and the topology of the complex. J. Mol. Biol. 329:441-465. [DOI] [PubMed] [Google Scholar]
- 83.Gao, D., and C. S. McHenry. 2001. τ binds and organizes Escherichia coli replication proteins through distinct domains. Domain IV, located within the unique C terminus of τ, binds the replication fork helicase, DnaB. J. Biol. Chem. 276:4441-4446. [DOI] [PubMed] [Google Scholar]
- 84.Garrido, T., M. Sánchez, P. Palacios, M. Aldea, and M. Vicente. 1993. Transcription of ftsZ oscillates during the cell cycle of Escherichia coli. EMBO J. 12:3957-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garti-Levi, S., R. Hazan, J. Kain, M. Fujita, and S. Ben-Yehuda. 2008. The FtsEX ABC transporter directs cellular differentiation in Bacillus subtilis. Mol. Microbiol. 69:1018-1028. [DOI] [PubMed] [Google Scholar]
- 86.Genschel, J., U. Curth, and C. Urbanke. 2000. Interaction of E. coli single-stranded DNA binding protein (SSB) with exonuclease I. The carboxy-terminus of SSB is the recognition site for the nuclease. Biol. Chem. 381:183-192. [DOI] [PubMed] [Google Scholar]
- 87.Georgescu, R. E., O. Yurieva, S.-S. Kim, J. Kuriyan, X.-P. Kong, and M. O'Donnell. 2008. Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp. Proc. Natl. Acad. Sci. U. S. A. 105:11116-11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Glover, B. P., and C. S. McHenry. 2000. The DnaX-binding subunits δ′ and ψ are bound to γ and not τ in the DNA polymerase III holoenzyme. J. Biol. Chem. 275:3017-3020. [DOI] [PubMed] [Google Scholar]
- 89.Goehring, N. W., and J. Beckwith. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15:R514-R526. [DOI] [PubMed] [Google Scholar]
- 90.Goehring, N. W., I. Petrovska, D. Boyd, and J. Beckwith. 2007. Mutants, suppressors, and wrinkled colonies: mutant alleles of the cell division gene ftsQ point to functional domains in FtsQ and a role for domain 1C of FtsA in divisome assembly. J. Bacteriol. 189:633-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gomez, M. J., and A. A. Neyfakh. 2006. Genes involved in intrinsic antibiotic resistance of Acinetobacter baylyi. Antimicrob. Agents Chemother. 50:3562-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gonzalez, M. D., and J. Beckwith. 2009. Divisome under construction: distinct domains of the small membrane protein FtsB are necessary for interaction with multiple cell division proteins. J. Bacteriol. 191:2815-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Greive, S. J., A. F. Lins, and P. H. von Hippel. 2005. Assembly of an RNA-protein complex. Binding of NusB and NusE (S10) proteins to boxA RNA nucleates the formation of the antitermination complex involved in controlling rRNA transcription in Escherichia coli. J. Biol. Chem. 280:36397-36408. [DOI] [PubMed] [Google Scholar]
- 94.Gruber, T. M., and C. A. Gross. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57:441-466. [DOI] [PubMed] [Google Scholar]
- 95.Gueiros-Filho, F. J., and R. Losick. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gulbis, J. M., S. L. Kazmirski, J. Finkelstein, Z. Kelman, M. O'Donnell, and J. Kuriyan. 2004. Crystal structure of the chi:psi sub-assembly of the Escherichia coli DNA polymerase clamp-loader complex. Eur. J. Biochem. 271:439-449. [DOI] [PubMed] [Google Scholar]
- 97.Guy, C. P., J. Atkinson, M. K. Gupta, A. A. Mahdi, E. J. Gwynn, C. J. Rudolph, P. B. Moon, I. C. van Knippenberg, C. J. Cadman, M. S. Dillingham, R. G. Lloyd, and P. McGlynn. 2009. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol. Cell 36:654-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hale, C. A., and P. A. J. de Boer. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88:175-185. [DOI] [PubMed] [Google Scholar]
- 99.Hamdan, S., P. D. Carr, S. E. Brown, D. L. Ollis, and N. E. Dixon. 2002. Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure 10:535-546. [DOI] [PubMed] [Google Scholar]
- 100.Han, E. S., D. L. Cooper, N. S. Persky, V. A. Sutera, Jr., R. D. Whitaker, M. L. Montello, and S. T. Lovett. 2006. RecJ exonuclease: substrates, products and interaction with SSB. Nucleic Acids Res. 34:1084-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Haney, S. A., E. Glasfeld, C. Hale, D. Keeney, Z. He, and P. de Boer. 2001. Genetic analysis of the Escherichia coli FtsZ·ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J. Biol. Chem. 276:11980-11987. [DOI] [PubMed] [Google Scholar]
- 102.Hare, J. M., S. N. Perkins, and L. A. Gregg-Jolly. 2006. A constitutively expressed, truncated umuDC operon regulates the recA-dependent DNA damage induction of a gene in Acinetobacter baylyi strain ADP1. Appl. Environ. Microbiol. 72:4036-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Harry, E., L. Monahan, and L. Thompson. 2006. Bacterial cell division: the mechanism and its precison. Int. Rev. Cytol. 253:27-94. [DOI] [PubMed] [Google Scholar]
- 104.Hayashi, I., T. Oyama, and K. Morikawa. 2001. Structural and functional studies of MinD ATPase: implications for the molecular recognition of the bacterial cell division apparatus. EMBO J. 20:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hayden, J. D., and S. E. Ades. 2008. The extracytoplasmic stress factor, σE, is required to maintain cell envelope integrity in Escherichia coli. PLoS One 3:e1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47-110. [DOI] [PubMed] [Google Scholar]
- 107.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hu, Z., and J. Lutkenhaus. 2000. Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ. J. Bacteriol. 182:3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hu, Z., C. Saez, and J. Lutkenhaus. 2003. Recruitment of MinC, an inhibitor of Z-ring formation, to the membrane in Escherichia coli: role of MinD and MinE. J. Bacteriol. 185:196-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang, J., C. Cao, and J. Lutkenhaus. 1996. Interaction between FtsZ and inhibitors of cell division. J. Bacteriol. 178:5080-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang, W. E., A. C. Singer, A. J. Spiers, G. M. Preston, and A. S. Whiteley. 2008. Characterizing the regulation of the Pu promoter in Acinetobacter baylyi ADP1. Environ. Microbiol. 10:1668-1680. [DOI] [PubMed] [Google Scholar]
- 112.Huisman, O., and R. D'Ari. 1981. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature 290:797-799. [DOI] [PubMed] [Google Scholar]
- 113.Humphreys, S., A. Stevenson, A. Bacon, A. B. Weinhardt, and M. Roberts. 1999. The alternative sigma factor, σE, is critically important for the virulence of Salmonella typhimurium. Infect. Immun. 67:1560-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hurwitz, J. 2005. The discovery of RNA polymerase. J. Biol. Chem. 280:42477-42485. [DOI] [PubMed] [Google Scholar]
- 115.Iacono, M., L. Villa, D. Fortini, R. Bordoni, F. Imperi, R. J. P. Bonnal, T. Sicheritz-Ponten, G. De Bellis, P. Visca, A. Cassone, and A. Carattoli. 2008. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob. Agents Chemother. 52:2616-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ibrahim, A., P. Gerner-Smidt, and W. Liesack. 1997. Phylogenetic relationship of the twenty-one DNA groups of the genus Acinetobacter as revealed by 16S ribosomal DNA sequence analysis. Int. J. Syst. Bacteriol. 47:837-841. [DOI] [PubMed] [Google Scholar]
- 117.Ioannou, C., P. M. Schaeffer, N. E. Dixon, and P. Soultanas. 2006. Helicase binding to DnaI exposes a cryptic DNA-binding site during helicase loading in Bacillus subtilis. Nucleic Acids Res. 34:5247-5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.James, G. A., D. R. Korber, D. E. Caldwell, and J. W. Costerton. 1995. Digital image analysis of growth and starvation responses of a surface-colonizing Acinetobacter sp. J. Bacteriol. 177:907-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jarvis, T. C., A. A. Beaudry, J. M. Bullard, N. Janjic, and C. S. McHenry. 2005. Reconstitution of a minimal DNA replicase from Pseudomonas aeruginosa and stimulation by non-cognate auxiliary factors. J. Biol. Chem. 280:7890-7900. [DOI] [PubMed] [Google Scholar]
- 121.Jarvis, T. C., A. A. Beaudry, J. M. Bullard, U. Ochsner, H. G. Dallmann, and C. S. McHenry. 2005. Discovery and characterization of the cryptic ψ subunit of the pseudomonad DNA replicase. J. Biol. Chem. 280:40465-40473. [DOI] [PubMed] [Google Scholar]
- 122.Jawad, A., H. Seifert, A. M. Snelling, J. Heritage, and P. M. Hawkey. 1998. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J. Clin. Microbiol. 36:1938-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jennings, L. D., K. W. Foreman, T. S. Rush, III, D. H. H. Tsao, L. Mosyak, S. L. Kincaid, M. N. Sukhdeo, A. G. Sutherland, W. Ding, C. H. Kenny, C. L. Sabus, H. Liu, E. G. Dushin, S. L. Moghazeh, P. Labthavikul, P. J. Petersen, Ma. Tuckman, and A. V. Ruzin. 2004. Combinatorial synthesis of substituted 3-(2-indolyl)piperidines and 2-phenyl indoles as inhibitors of ZipA-FtsZ interaction. Bioorg. Med. Chem. 12:5115-5131. [DOI] [PubMed] [Google Scholar]
- 124.Jennings, L. D., K. W. Foreman, T. S. Rush, III, D. H. H. Tsao, L. Mosyak, Y. Li, M. N. Sukhdeo, W. Ding, E. G. Dushin, C. H. Kenny, S. L. Moghazeh, P. J. Petersen, A. V. Ruzin, M. Tuckman, and A. G. Sutherland. 2004. Design and synthesis of indolo[2,3-a]quinolizin-7-one inhibitors of the ZipA-FtsZ interaction. Bioorg. Med. Chem. Lett. 14:1427-1431. [DOI] [PubMed] [Google Scholar]
- 125.Jergic, S., K. Ozawa, N. K. Williams, X.-C. Su, D. D. Scott, S. M. Hamdan, J. A. Crowther, G. Otting, and N. E. Dixon. 2007. The unstructured C-terminus of the τ subunit of Escherichia coli DNA polymerase III holoenzyme is the site of interaction with the α subunit. Nucleic Acids Res. 35:2813-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jeruzalmi, D., M. O'Donnell, and J. Kuriyan. 2001. Crystal structure of the processivity clamp loader gamma (γ) complex of E. coli DNA polymerase III. Cell 106:429-441. [DOI] [PubMed] [Google Scholar]
- 127.Jeruzalmi, D., O. Yurieva, Y. Zhao, M. Young, J. Stewart, M. Hingorani, M. O'Donnell, and J. Kuriyan. 2001. Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell 106:417-428. [PubMed] [Google Scholar]
- 128.Jin, H., D. M. Retallack, S. J. Stelman, C. D. Hershberger, and T. Ramseier. 2007. Characterization of the SOS response of Pseudomonas fluorescens strain DC206 using whole-genome transcript analysis. FEMS Microbiol. Lett. 269:256-264. [DOI] [PubMed] [Google Scholar]
- 129.Jishage, M., A. Iwata, S. Ueda, and A. Ishihama. 1996. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol. 178:5447-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Johnston, E. B., P. J. Lewis, and R. Griffith. 2009. The interaction of Bacillus subtilis σA with RNA polymerase. Protein Sci. 18:2287-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Reference deleted.
- 132.Jokerst, R. S., J. R. Weeks, W. A. Zehring, and A. L. Greenleaf. 1989. Analysis of the gene encoding the largest subunit of RNA polymerase II in Drosophila. Mol. Gen. Genet. 215:266-275. [DOI] [PubMed] [Google Scholar]
- 133.Jones, R. M., and P. A. Williams. 2001. areCBA is an operon in Acinetobacter sp. strain ADP1 and is controlled by AreR, a σ54-dependent regulator. J. Bacteriol. 183:405-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kaguni, J. M. 2006. DnaA: controlling the initiation of bacterial DNA replication and more. Annu. Rev. Microbiol. 60:351-375. [DOI] [PubMed] [Google Scholar]
- 135.Kantake, N., M. V. V. M. Madiraju, T. Sugiyama, and S. C. Kowalczykowski. 2002. Escherichia coli RecO protein anneals ssDNA complexed with its cognate ssDNA-binding protein: a common step in genetic recombination. Proc. Natl. Acad. Sci. U. S. A. 99:15327-15332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Karimova, G., N. Dautin, and D. Ladant. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187:2233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Katayanagi, K., M. Miyagawa, M. Matsushima, M. Ishikawa, S. Kanaya, M. Ikehara, T. Matsuzaki, and K. Morikawa. 1990. Three-dimensional structure of ribonuclease H from E. coli. Nature 347:306-309. [DOI] [PubMed] [Google Scholar]
- 138.Kato, J., and T. Katayama. 2001. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20:4253-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Katoh, K., K. Kuma, H. Toh, and T. Miyata. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Katoh, K., K. Misawa, K. Kuma, and T. Miyata. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Katoh, K., and H. Toh. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9:286-298. [DOI] [PubMed] [Google Scholar]
- 142.Kawakami, H., M. Su'etsugu, and T. Katayama. 2006. An isolated Hda-clamp complex is functional in the regulatory inactivation of DnaA and DNA replication. J. Struct. Biol. 156:220-229. [DOI] [PubMed] [Google Scholar]
- 143.Kelley, W. L. 2006. Lex marks the spot: the virulent side of SOS and a closer look at the LexA regulon. Mol. Microbiol. 62:1228-1238. [DOI] [PubMed] [Google Scholar]
- 144.Keniry, M. A., A. Y. Park, E. A. Owen, S. M. Hamdan, G. Pintacuda, G. Otting, and N. E. Dixon. 2006. Structure of the θ subunit of Escherichia coli DNA polymerase III in complex with the ɛ subunit. J. Bacteriol. 188:4464-4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Keyamura, K., N. Fujikawa, T. Ishida, S. Ozaki, M. Su'etsugu, K. Fujimitsu, W. Kagawa, S. Yokoyama, H. Kurumizaka, and T. Katayama. 2007. The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes. Genes Dev. 21:2083-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kim, D. Y., and K. K. Kim. 2003. Crystal structure of ClpX molecular chaperone from Helicobacter pylori. J. Biol. Chem. 278:50664-50670. [DOI] [PubMed] [Google Scholar]
- 147.King, G. F., Y.-L. Shih, M. W. Maciejewski, N. P. S. Bains, B. Pan, S. L. Rowland, G. P. Mullen, and L. I. Rothfield. 2000. Structural basis for the topological specificity function of MinE. Nat. Struct. Biol. 7:1013-1017. [DOI] [PubMed] [Google Scholar]
- 148.Laine, P. S., and R. R. Meyer. 1992. Interaction of the heat shock protein GroEL of Escherichia coli with single-stranded DNA-binding protein: suppression of ssb-113 by groEL46. J. Bacteriol. 174:3204-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lamers, M. H., R. E. Georgescu, S.-G. Lee, M. O'Donnell, and J. Kuriyan. 2006. Crystal structure of the catalytic α subunit of E. coli replicative DNA polymerase III. Cell 126:881-892. [DOI] [PubMed] [Google Scholar]
- 150.Lange, R. P., H. H. Locher, P. C. Wyss, and R. L. Then. 2007. The targets of currently used antibacterial agents: lessons for drug discovery. Curr. Pharm. Des. 13:3140-3154. [DOI] [PubMed] [Google Scholar]
- 151.Leung, A. K. W., E. L. White, L. J. Ross, R. C. Reynolds, J. A. DeVito, and D. W. Borhani. 2004. Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches. J. Mol. Biol. 342:953-970. [DOI] [PubMed] [Google Scholar]
- 152.Lewis, P. J., G. P. Doherty, and J. Clarke. 2008. Transcription factor dynamics. Microbiology 154:1837-1844. [DOI] [PubMed] [Google Scholar]
- 153.Li, J., S. W. Mason, and J. Greenblatt. 1993. Elongation factor NusG interacts with termination factor ρ to regulate termination and antitermination of transcription. Genes Dev. 7:161-172. [DOI] [PubMed] [Google Scholar]
- 154.Li, Y., S. Korolev, and G. Waksman. 1998. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 17:7514-7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Linding, R., R. B. Russell, V. Neduva, and T. J. Gibson. 2003. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 31:3701-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lock, R. L., and E. J. Harry. 2008. Cell-division inhibitors: new insights for future antibiotics. Nat. Rev. Drug Discov. 7:324-338. [DOI] [PubMed] [Google Scholar]
- 157.López de Saro, F. J., R. E. Georgescu, and M. O'Donnell. 2003. A peptide switch regulates DNA polymerase processivity. Proc. Natl. Acad. Sci. U. S. A. 100:14689-14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.López de Saro, F. J., M. G. Marinus, P. Modrich, and M. O'Donnell. 2006. The β sliding clamp binds to multiple sites within MutL and MutS. J. Biol. Chem. 281:14340-14349. [DOI] [PubMed] [Google Scholar]
- 159.López de Saro, F. J., and M. O'Donnell. 2001. Interaction of the β sliding clamp with MutS, ligase, and DNA polymerase I. Proc. Natl. Acad. Sci. U. S. A. 98:8376-8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Low, H. H., M. C. Moncrieffe, and J. Löwe. 2004. The crystal structure of ZapA and its modulation of FtsZ polymerisation. J. Mol. Biol. 341:839-852. [DOI] [PubMed] [Google Scholar]
- 161.Löwe, J., and L. A. Amos. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203-206. [DOI] [PubMed] [Google Scholar]
- 162.Lu, D., and J. L. Keck. 2008. Structural basis of Escherichia coli single-stranded DNA-binding protein stimulation of exonuclease I. Proc. Natl. Acad. Sci. U. S. A. 105:9169-9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lu, D., M. A. Windsor, S. H. Gellman, and J. L. Keck. 2009. Peptide inhibitors identify roles for SSB C-terminal residues in SSB/exonuclease I complex formation. Biochemistry 48:6764-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Luo, X., H.-H. Hsiao, M. Bubunenko, G. Weber, D. L. Court, M. E. Gottesman, H. Urlaub, and M. C. Wahl. 2008. Structural and functional analysis of the E. coli NusB-S10 transcription antitermination complex. Mol. Cell 32:791-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ma, X., and W. Margolin. 1999. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 181:7531-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Marchand, I., L. Damier-Piolle, P. Courvalin, and T. Lambert. 2004. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 48:3298-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Marles-Wright, J., and R. J. Lewis. 2007. Stress responses of bacteria. Curr. Opin. Struct. Biol. 17:755-760. [DOI] [PubMed] [Google Scholar]
- 168.Martinez, J. L., A. Fajardo, L. Garmendia, A. Hernandez, J. F. Linares, L. Martínez-Solano, and M. B. Sánchez. 2009. A global view of antibiotic resistance. FEMS Microbiol. Rev. 33:44-65. [DOI] [PubMed] [Google Scholar]
- 169.Massey, T. H., C. P. Mercogliano, J. Yates, D. J. Sherratt, and J. Löwe. 2006. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol. Cell 23:457-469. [DOI] [PubMed] [Google Scholar]
- 170.Mengin-Lecreulx, D., J. van Heijenoort, and J. T. Park. 1996. Identification of the mpl gene encoding UDP-N-acetylmuramate:l-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J. Bacteriol. 178:5347-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Merrikh, H., A. E. Ferrazzoli, A. Bougdour, A. Olivier-Mason, and S. T. Lovett. 2009. A DNA damage response in Escherichia coli involving the alternative sigma factor, RpoS. Proc. Natl. Acad. Sci. U. S. A. 106:611-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Metzgar, D., J. M. Bacher, V. Pezo, J. Reader, V. Döring, P. Schimmel, P. Marlière, and V. de Crécy-Lagard. 2004. Acinetobacter sp. ADP1: an ideal model organism for genetic analysis and genome engineering. Nucleic Acids Res. 32:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Miller, C., L. E. Thomsen, C. Gaggero, R. Mosseri, H. Ingmer, and S. N. Cohen. 2004. SOS response induction by β-lactams and bacterial defense against antibiotic lethality. Science 305:1629-1631. [DOI] [PubMed] [Google Scholar]
- 174.Minakhin, L., S. Bhagat, A. Brunning, E. A. Campbell, S. A. Darst, R. H. Ebright, and K. Severinov. 2001. Bacterial RNA polymerase subunit ω and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc. Natl. Acad. Sci. U. S. A. 98:892-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Missiakas, D., and S. Raina. 1998. The extracytoplasmic function sigma factors: role and regulation. Mol. Microbiol. 28:1059-1066. [DOI] [PubMed] [Google Scholar]
- 176.Moellering, R. E., M. Cornejo, T. N. Davis, C. Del Bianco, J. C. Aster, S. C. Blacklow, A. L. Kung, D. G. Gilliland, G. L. Verdine, and J. E. Bradner. 2009. Direct inhibition of the NOTCH transcription factor complex. Nature 462:182-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mol, C. D., A. Brooun, D. R. Dougan, M. T. Hilgers, L. W. Tari, R. A. Wijnands, M. W. Knuth, D. E. McRee, and R. V. Swanson. 2003. Crystal structures of active fully assembled substrate- and product-bound complexes of UDP-N-acetylmuramic acid:l-alanine ligase (MurC) from Haemophilus influenzae. J. Bacteriol. 185:4152-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Mooney, R. A., K. Schweimer, P. Rösch, M. Gottesman, and R. Landick. 2009. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J. Mol. Biol. 391:341-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mosyak, L., Y. Zhang, E. Glasfeld, S. Haney, M. Stahl, J. Seehra, and W. S. Somers. 2000. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 19:3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mott, M. L., and J. M. Berger. 2007. DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 5:343-354. [DOI] [PubMed] [Google Scholar]
- 181.Mott, M. L., J. P. Erzberger, M. M. Coons, and J. M. Berger. 2008. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell 135:623-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mulcair, M. D., P. M. Schaeffer, A. J. Oakley, H. F. Cross, C. Neylon, T. M. Hill, and N. E. Dixon. 2006. A molecular mousetrap determines polarity of termination of DNA replication in E. coli. Cell 125:1309-1319. [DOI] [PubMed] [Google Scholar]
- 183.Mulugu, S., A. Potnis, Shamsuzzaman, J. Taylor, K. Alexander, and D. Bastia. 2001. Mechanism of termination of DNA replication of Escherichia coli involves helicase-contrahelicase interaction. Proc. Natl. Acad. Sci. U. S. A. 98:9569-9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Murakami, K., T. Ono, D. Viducic, S. Kayama, M. Mori, K. Hirota, K. Nemoto, and Y. Miyake. 2005. Role for rpoS gene of Pseudomonas aeruginosa in antibiotic tolerance. FEMS Microbiol. Lett. 242:161-167. [DOI] [PubMed] [Google Scholar]
- 185.Murray, C. K., and D. R. Hospenthal. 2008. Acinetobacter infection in the ICU. Crit. Care Clin. 24:237-248. [DOI] [PubMed] [Google Scholar]
- 186.Nandakumar, J., P. A. Nair, and S. Shuman. 2007. Last stop on the road to repair: structure of E. coli DNA ligase bound to nicked DNA-adenylate. Mol. Cell 26:257-271. [DOI] [PubMed] [Google Scholar]
- 187.Navon-Venezia, S., A. Leavitt, and Y. Carmeli. 2007. High tigecycline resistance in multidrug-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 59:772-774. [DOI] [PubMed] [Google Scholar]
- 188.Nemec, A. 1996. Taxonomy of the genus Acinetobacter. Epidemiol. Mikrobiol. Imunol. 45:23-29. (In Czech.) [PubMed] [Google Scholar]
- 189.Neylon, C., A. V. Kralicek, T. M. Hill, and N. E. Dixon. 2005. Replication termination in Escherichia coli: structure and antihelicase activity of the Tus-Ter complex. Microbiol. Mol. Biol. Rev. 69:501-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nogales, E., K. H. Downing, L. A. Amos, and J. Löwe. 1998. Tubulin and FtsZ form a distinct family of GTPases. Nat. Struct. Biol. 5:451-458. [DOI] [PubMed] [Google Scholar]
- 191.Oakley, A. J., K. V. Loscha, P. M. Schaeffer, E. Liepinsh, G. Pintacuda, M. C. J. Wilce, G. Otting, and N. E. Dixon. 2005. Crystal and solution structures of the helicase-binding domain of Escherichia coli primase. J. Biol. Chem. 280:11495-11504. [DOI] [PubMed] [Google Scholar]
- 192.Oakley, A. J., P. Prosselkov, G. Wijffels, J. L. Beck, M. C. J. Wilce, and N. E. Dixon. 2003. Flexibility revealed by the 1.85 Å crystal structure of the β sliding-clamp subunit of Escherichia coli DNA polymerase III. Acta Crystallogr. D 59:1192-1199. [DOI] [PubMed] [Google Scholar]
- 193.Okuno, T., M. Ogoh, H. Tanina, N. Funasaki, and K. Kogure. 2009. Direct monitoring of interaction between Escherichia coli proteins, MinC and monomeric FtsZ, in solution. Biol. Pharm. Bull. 32:1473-1475. [DOI] [PubMed] [Google Scholar]
- 194.Oliva, M. A., D. Trambaiolo, and J. Löwe. 2007. Structural insights into the conformational variability of FtsZ. J. Mol. Biol. 373:1229-1242. [DOI] [PubMed] [Google Scholar]
- 195.O'Reilly, E. K., and K. N. Kreuzer. 2004. Isolation of SOS constitutive mutants of Escherichia coli. J. Bacteriol. 186:7149-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ozaki, S., H. Kawakami, K. Nakamura, N. Fujikawa, W. Kagawa, S.-Y. Park, S. Yokoyama, H. Kurumizaka, and T. Katayama. 2008. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J. Biol. Chem. 283:8351-8362. [DOI] [PubMed] [Google Scholar]
- 197.Ozawa, K., S. Jergic, A. Y. Park, N. E. Dixon, and G. Otting. 2008. The proofreading exonuclease subunit ɛ of Escherichia coli DNA polymerase III is tethered to the polymerase subunit α via a flexible linker. Nucleic Acids Res. 36:5074-5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Park, J.-S., M. T. Marr, and J. W. Roberts. 2002. E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell 109:757-767. [DOI] [PubMed] [Google Scholar]
- 199.Park, J. T., and T. Uehara. 2008. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 72:211-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Park, K., S. Choi, M. Ko, and C. Park. 2001. Novel σF-dependent genes of Escherichia coli found using a specified promoter consensus. FEMS Microbiol. Lett. 202:243-250. [DOI] [PubMed] [Google Scholar]
- 201.Paul, B. J., M. M. Barker, W. Ross, D. A. Schneider, C. Webb, J. W. Foster, and R. L. Gourse. 2004. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118:311-322. [DOI] [PubMed] [Google Scholar]
- 202.Paul, B. J., W. Ross, T. Gaal, and R. L. Gourse. 2004. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 38:749-770. [DOI] [PubMed] [Google Scholar]
- 203.Peleg, A. Y., B. A. Potoski, R. Rea, J. Adams, J. Sethi, B. Capitano, S. Husain, E. J. Kwak, S. V. Bhat, and D. L. Paterson. 2007. Acinetobacter baumannii bloodstream infection while receiving tigecycline: a cautionary report. J. Antimicrob. Chemother. 59:128-131. [DOI] [PubMed] [Google Scholar]
- 204.Peleg, A. Y., H. Seifert, and D. L. Paterson. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Perederina, A., V. Svetlov, M. N. Vassylyeva, T. H. Tahirov, S. Yokoyama, I. Artsimovitch, and D. G. Vassylyev. 2004. Regulation through the secondary channel—structural framework for ppGpp-DksA synergism during transcription. Cell 118:297-309. [DOI] [PubMed] [Google Scholar]
- 206.Perrino, F. W., S. Harvey, and S. M. McNeill. 1999. Two functional domains of the ɛ subunit of DNA polymerase III. Biochemistry 38:16001-16009. [DOI] [PubMed] [Google Scholar]
- 207.Pichoff, S., and J. Lutkenhaus. 2007. Identification of a region of FtsA required for interaction with FtsZ. Mol. Microbiol. 64:1129-1138. [DOI] [PubMed] [Google Scholar]
- 208.Pintacuda, G., A. Y. Park, M. A. Keniry, N. E. Dixon, and G. Otting. 2006. Lanthanide labeling offers fast NMR approach to 3D structure determinations of protein-protein complexes. J. Am. Chem. Soc. 128:3696-3702. [DOI] [PubMed] [Google Scholar]
- 209.Pomerantz, R. T., and M. O'Donnell. 2007. Replisome mechanics: insights into a twin DNA polymerase machine. Trends Microbiol. 15:156-164. [DOI] [PubMed] [Google Scholar]
- 210.Porstendörfer, D., O. Gohl, F. Mayer, and B. Averhoff. 2000. ComP, a pilin-like protein essential for natural competence in Acinetobacter sp. strain BD413: regulation, modification, and cellular localization. J. Bacteriol. 182:3673-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Potvin, E., F. Sanschagrin, and R. C. Levesque. 2008. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 32:38-55. [DOI] [PubMed] [Google Scholar]
- 212.Prasch, S., S. Schwarz, A. Eisenmann, B. M. Wöhrl, K. Schweimer, and P. Rösch. 2006. Interaction of the intrinsically unstructured phage λ N protein with Escherichia coli NusA. Biochemistry 45:4542-4549. [DOI] [PubMed] [Google Scholar]
- 213.Rainey, F. A., E. Lang, and E. Stackebrandt. 1994. The phylogenetic structure of the genus Acinetobacter. FEMS Microbiol. Lett. 124:349-353. [DOI] [PubMed] [Google Scholar]
- 214.Ramos, D., T. Ducat, J. Cheng, N. F. Eng., J. R. Dillon, and N. K. Goto. 2006. Conformation of the cell division regulator MinE: evidence for interactions between the topological specificity and anti-MinCD domains. Biochemistry 45:4593-4601. [DOI] [PubMed] [Google Scholar]
- 215.Rathinavelu, S., Y. Zavros, and J. L. Merchant. 2003. Acinetobacter lwoffii infection and gastritis. Microbes Infect. 5:651-657. [DOI] [PubMed] [Google Scholar]
- 216.Rauch, P. J., R. Palmen, A. A. Burds, L. A. Gregg-Jolly, J. R. van der Zee, and K. J. Hellingwerf. 1996. The expression of the Acinetobacter calcoaceticus recA gene increases in response to DNA damage independently of RecA and of development of competence for natural transformation. Microbiology 142:1025-1032. [DOI] [PubMed] [Google Scholar]
- 217.Ray, A., and B. Nordén. 2000. Peptide nucleic acid (PNA): its medical and biotechnical applications and promise for the future. FASEB J. 14:1041-1060. [DOI] [PubMed] [Google Scholar]
- 218.Reddy, M. 2007. Role of FtsEX in cell division of Escherichia coli: viability of ftsEX mutants is dependent on functional SufI or high osmotic strength. J. Bacteriol. 189:98-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Reitzer, L., and B. L. Schneider. 2001. Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 65:422-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Robson, S. A., and G. F. King. 2006. Domain architecture and structure of the bacterial cell division protein DivIB. Proc. Natl. Acad. Sci. U. S. A. 103:6700-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Rost, B., G. Yachdav, and J. Liu. 2004. The PredictProtein server. Nucleic Acids Res. 32:W321-W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Rudant, E., P. Bouvet, P. Courvalin, and T. Lambert. 1999. Phylogenetic analysis of proteolytic Acinetobacter strains based on the sequence of genes encoding aminoglycoside 6′-N-acetyltransferases. Syst. Appl. Microbiol. 22:59-67. [DOI] [PubMed] [Google Scholar]
- 223.Rush, T. S., III, J. A. Grant, L. Mosyak, and A. Nicholls. 2005. A shape-based 3-D scaffold hopping method and its application to a bacterial protein-protein interaction. J. Med. Chem. 48:1489-1495. [DOI] [PubMed] [Google Scholar]
- 224.Sakai, N., M. Yao, H. Itou, N. Watanabe, F. Yumoto, M. Tanokura, and I. Tanaka. 2001. The three-dimensional structure of septum site-determining protein MinD from Pyrococcus horikoshii OT3 in complex with Mg-ADP. Structure 9:817-826. [DOI] [PubMed] [Google Scholar]
- 225.Savvides, S. N., S. Raghunathan, K. Fütterer, A. G. Kozlov, T. M. Lohman, and G. Waksman. 2004. The C-terminal domain of full-length E. coli SSB is disordered even when bound to DNA. Protein Sci. 13:1942-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Schaeffer, P. M., M. J. Headlam, and N. E. Dixon. 2005. Protein-protein interactions in the eubacterial replisome. IUBMB Life 57:5-12. [DOI] [PubMed] [Google Scholar]
- 227.Schmidt, K. L., N. D. Peterson, R. J. Kustusch, M. C. Wissel, B. Graham, G. J. Phillips, and D. S. Weiss. 2004. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J. Bacteriol. 186:785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Shen, B., and J. Lutkenhaus. 2009. The conserved C-terminal tail of FtsZ is required for the septal localization and division inhibitory activity of MinCC/MinD. Mol. Microbiol. 72:410-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Shereda, R. D., A. G. Kozlov, T. M. Lohman, M. M. Cox, and J. L. Keck. 2008. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 43:289-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Shereda, R. D., N. J. Reiter, S. E. Butcher, and J. L. Keck. 2009. Identification of the SSB binding site on E. coli RecQ reveals a conserved surface for binding SSB's C terminus. J. Mol. Biol. 386:612-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Simonetta, K. R., S. L. Kazmirski, E. R. Goedken, A. J. Cantor, B. A. Kelch, R. McNally, S. N. Seyedin, D. L. Makino, M. O'Donnell, and J. Kuriyan. 2009. The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell 137:659-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Skordalakes, E., and J. M. Berger. 2003. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell 114:135-146. [DOI] [PubMed] [Google Scholar]
- 233.Smith, M. G., T. A. Gianoulis, S. Pukatzki, J. J. Mekalanos, L. N. Ornston, M. Gerstein, and M. Snyder. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 21:601-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Söding, J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951-960. [DOI] [PubMed] [Google Scholar]
- 235.Söding, J., A. Biegert, and A. N. Lupas. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244-W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Soni, R. K., P. Mehra, G. Mukhopadhyay, and S. K. Dhar. 2005. Helicobacter pylori DnaB helicase can bypass Escherichia coli DnaC function in vivo. Biochem. J. 389:541-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Soultanas, P. 2005. The bacterial helicase-primase interaction: a common structural/functional module. Structure 13:839-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Stebbins, C. E., S. Borukhov, M. Orlova, A. Polyakov, A. Goldfarb, and S. A. Darst. 1995. Crystal structure of the GreA transcript cleavage factor from Escherichia coli. Nature 373:636-640. [DOI] [PubMed] [Google Scholar]
- 239.Steiner, T., J. T. Kaiser, S. Marinkoviç, R. Huber, and M. C. Wahl. 2002. Crystal structures of transcription factor NusG in light of its nucleic acid- and protein-binding activities. EMBO J. 21:4641-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Su, X.-C., S. Jergic, M. A. Keniry, N. E. Dixon, and G. Otting. 2007. Solution structure of domains IVa and V of the τ subunit of Escherichia coli DNA polymerase III and interaction with the α subunit. Nucleic Acids Res. 35:2825-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Su, X.-C., P. M. Schaeffer, K. V. Loscha, P. H. P. Gan, N. E. Dixon, and G. Otting. 2006. Monomeric solution structure of the helicase-binding domain of Escherichia coli DnaG primase. FEBS J. 273:4997-5009. [DOI] [PubMed] [Google Scholar]
- 242.Sugino, A., and H. Araki. 2006. Historical view of DNA replication studies, with special reference to Japan. IUBMB Life 58:323-327. [DOI] [PubMed] [Google Scholar]
- 243.Suski, C., and K. J. Marians. 2008. Resolution of converging replication forks by RecQ and topoisomerase III. Mol. Cell 30:779-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Sutherland, A. G., J. Alvarez, W. Ding, K. W. Foreman, C. H. Kenny, P. Labthavikul, L. Mosyak, P. J. Petersen, T. S. Rush III, A. Ruzin, D. H. H. Tsao, and K. L. Wheless. 2003. Structure-based design of carboxybiphenylindole inhibitors of the ZipA-FtsZ interaction. Org. Biomol. Chem. 1:4138-4140. [DOI] [PubMed] [Google Scholar]
- 245.Talbot, G. H. 2008. What is in the pipeline for Gram-negative pathogens? Expert Rev. Anti-Infect. Ther. 6:39-49. [DOI] [PubMed] [Google Scholar]
- 246.Thomsen, N. D., and J. M. Berger. 2009. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell 139:523-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Tsao, D. H. H., A. G. Sutherland, L. D. Jennings, Y. Li, T. S. Rush III, J. C. Alvarez, W. Ding, E. G. Dushin, R. G. Dushin, S. A. Haney, C. H. Kenny, A. K. Malakian, R. Nilakantan, and L. Mosyak. 2006. Discovery of novel inhibitors of the ZipA/FtsZ complex by NMR fragment screening coupled with structure-based design. Bioorg. Med. Chem. 14:7953-7961. [DOI] [PubMed] [Google Scholar]
- 248.Vallenet, D., P. Nordmann, V. Barbe, L. Poirel, S. Mangenot, E. Bataille, C. Dossat, S. Gas, A. Kreimeyer, P. Lenoble, S. Oztas, J. Poulain, B. Segurens, C. Robert, C. Abergel, J.-M. Claverie, D. Raoult, C. Médigue, J. Weissenbach, and S. Cruveiller. 2008. Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS One 3:e1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.van den Ent, F., and J. Löwe. 2000. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 19:5300-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.van den Ent, F., T. M. F. Vinkenvleugel, A. Ind, P. West, D. Veprintsev, N. Nanninga, T. den Blaauwen, and J. Löwe. 2008. Structural and mutational analysis of the cell division protein FtsQ. Mol. Microbiol. 68:110-123. [DOI] [PubMed] [Google Scholar]
- 251.Vassylyev, D. G., S. Sekine, O. Laptenko, J. Lee, M. N. Vassylyeva, S. Borukhov, and S. Yokoyama. 2002. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417:712-719. [DOI] [PubMed] [Google Scholar]
- 252.Vassylyeva, M. N., V. Svetlov, A. D. Dearborn, S. Klyuyev, I. Artsimovitch, and D. G. Vassylyev. 2007. The carboxy-terminal coiled-coil of the RNA polymerase β′-subunit is the main binding site for Gre factors. EMBO Rep. 8:1038-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Vaughan, S., B. Wickstead, K. Gull, and S. G. Addinall. 2004. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J. Mol. Evol. 58:19-29. [DOI] [PubMed] [Google Scholar]
- 254.Vicente, M., and A. I. Rico. 2006. The order of the ring: assembly of Escherichia coli cell division components. Mol. Microbiol. 61:5-8. [DOI] [PubMed] [Google Scholar]
- 255.Vicente, M., A. I. Rico, R. Martínez-Arteaga, and J. Mingorance. 2006. Septum enlightenment: assembly of bacterial division proteins. J. Bacteriol. 188:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Villain-Guillot, P., L. Bastide, M. Gualtieri, and J.-P. Leonetti. 2007. Progress in targeting bacterial transcription. Drug Discov. Today 12:200-208. [DOI] [PubMed] [Google Scholar]
- 257.Wang, X., J. Huang, A. Mukherjee, C. Cao, and J. Lutkenhaus. 1997. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol. 179:5551-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Weigelt, J., S. E. Brown, C. S. Miles, N. E. Dixon, and G. Otting. 1999. NMR structure of the N-terminal domain of E. coli DnaB helicase: implications for structure rearrangements in the helicase hexamer. Structure 7:681-690. [DOI] [PubMed] [Google Scholar]
- 259.Weiss, D. S. 2004. Bacterial cell division and the septal ring. Mol. Microbiol. 54:588-597. [DOI] [PubMed] [Google Scholar]
- 260.Wijffels, G., B. Dalrymple, K. Kongsuwan, and N. E. Dixon. 2005. Conservation of eubacterial replicases. IUBMB Life 57:413-419. [DOI] [PubMed] [Google Scholar]
- 261.Wing, R. A., S. Bailey, and T. A. Steitz. 2008. Insights into the replisome from the structure of a ternary complex of the DNA polymerase III α-subunit. J. Mol. Biol. 382:859-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wisplinghoff, H., R. Schmitt, A. Wöhrmann, D. Stefanik, and H. Seifert. 2007. Resistance to disinfectants in epidemiologically defined clinical isolates of Acinetobacter baumannii. J. Hosp. Infect. 66:174-181. [DOI] [PubMed] [Google Scholar]
- 263.Witte, G., C. Urbanke, and U. Curth. 2003. DNA polymerase III χ subunit ties single-stranded DNA binding protein to the bacterial replication machinery. Nucleic Acids Res. 31:4434-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Worbs, M., G. P. Bourenkov, H. D. Bartunik, R. Huber, and M. C. Wahl. 2001. An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol. Cell 7:1177-1189. [DOI] [PubMed] [Google Scholar]
- 265.Yamamoto, S., P. J. M. Bouvet, and S. Harayama. 1999. Phylogenetic structures of the genus Acinetobacter based on gyrB sequences: comparison with the grouping by DNA-DNA hybridization. Int. J. Syst. Bacteriol. 49:87-95. [DOI] [PubMed] [Google Scholar]
- 266.Yamamoto, S., and S. Harayama. 1996. Phylogenetic analysis of Acinetobacter strains based on the nucleotide sequences of gyrB genes and on the amino acid sequences of their products. Int. J. Syst. Bacteriol. 46:506-511. [DOI] [PubMed] [Google Scholar]
- 267.Yan, K., K. H. Pearce, and D. J. Payne. 2000. A conserved residue at the extreme C-terminus of FtsZ is critical for the FtsA-FtsZ interaction in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 270:387-392. [DOI] [PubMed] [Google Scholar]
- 268.Yang J.-C., F. van den Ent, D. Neuhaus, J. Brevier, and J. Löwe. 2004. Solution structure and domain architecture of the divisome protein FtsN. Mol. Microbiol. 52:651-660. [DOI] [PubMed] [Google Scholar]
- 269.Yang, X., S. Molimau, G. P. Doherty, E. B. Johnston, J. Marles-Wright, R. Rothnagel, B. Hankamer, R. J. Lewis, and P. J. Lewis. 2009. The structure of bacterial RNA polymerase in complex with the essential transcription elongation factor NusA. EMBO Rep. 10:997-10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Yee, T. W., and D. W. Smith. 1990. Pseudomonas chromosomal replication origins: a bacterial class distinct from Escherichia coli-type origins. Proc. Natl. Acad. Sci. U. S. A. 87:1278-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Yi, Q.-M., S. Rockenbach, J. E. Ward, Jr., and J. Lutkenhaus. 1985. Structure and expression of the cell division genes ftsQ, ftsA and ftsZ. J. Mol. Biol. 184:399-412. [DOI] [PubMed] [Google Scholar]
- 272.Young, B. A., L. C. Anthony, T. M. Gruber, T. M. Arthur, E. Heyduk, C. Z. Lu, M. M. Sharp, T. Heyduk, R. R. Burgess, and C. A. Gross. 2001. A coiled-coil from the RNA polymerase β′ subunit allosterically induces selective nontemplate strand binding by σ70. Cell 105:935-944. [DOI] [PubMed] [Google Scholar]
- 273.Yu, H., M. J. Schurr, and V. Deretic. 1995. Functional equivalence of Escherichia coli σE and Pseudomonas aeruginosa AlgU: E. coli rpoE restores mucoidy and reduces sensitivity to reactive oxygen intermediates in algU mutants of P. aeruginosa. J. Bacteriol. 177:3259-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Yuan, A. H., B. D. Gregory, J. S. Sharp, K. D. McCleary, S. L. Dove, and A. Hochschild. 2008. Rsd family proteins make simultaneous interactions with regions 2 and 4 of the primary sigma factor. Mol. Microbiol. 70:1136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Yuzhakov, A., Z. Kelman, and M. O'Donnell. 1999. Trading places on DNA—a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell 96:153-163. [DOI] [PubMed] [Google Scholar]
- 276.Zhang, G., E. A. Campbell, L. Minakhin, C. Richter, K. Severinov, and S. A. Darst. 1999. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell 98:811-824. [DOI] [PubMed] [Google Scholar]
- 277.Zhang, R., and Y. Lin. 2009. DEG 5.0, a database of essential genes in both prokaryotes and eukaryotes. Nucleic Acids Res. 37:D455-D458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Zhao, C.-R., P. A. J. de Boer, and L. I. Rothfield. 1995. Proper placement of the Escherichia coli division site requires two functions that are associated with different domains of the MinE protein. Proc. Natl. Acad. Sci. U. S. A. 92:4313-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Zimbler, D. L., W. F. Penwell, J. A. Gaddy, S. M. Menke, A. P. Tomaras, P. L. Connerly, and L. A. Actis. 2009. Iron acquisition functions expressed by the human pathogen Acinetobacter baumannii. Biometals 22:23-32. [DOI] [PubMed] [Google Scholar]