Abstract
Summary: Sex is shrouded in mystery. Not only does it preferentially occur in the dark for both fungi and many animals, but evolutionary biologists continue to debate its benefits given costs in light of its pervasive nature. Experimental studies of the benefits and costs of sexual reproduction with fungi as model systems have begun to provide evidence that the balance between sexual and asexual reproduction shifts in response to selective pressures. Given their unique evolutionary history as opisthokonts, along with metazoans, fungi serve as exceptional models for the evolution of sex and sex-determining regions of the genome (the mating type locus) and for transitions that commonly occur between outcrossing/self-sterile and inbreeding/self-fertile modes of reproduction. We review here the state of the understanding of sex and its evolution in the fungal kingdom and also areas where the field has contributed and will continue to contribute to illuminating general principles and paradigms of sexual reproduction.
INTRODUCTION
Sexual development is common in eukaryotic organisms from yeasts to humans. However, the question as to why sexual reproduction is so pervasive is a conundrum in evolutionary biology. The cost of sexual development might lead to detrimental effects during evolution. For example, sexual populations suffer a 2-fold cost compared to asexual populations (271). A sexual population consists of two genders, one (female) which can produce offspring and the other (male) which cannot. However, in an asexual population, any individual can produce their own offspring. Thus, while in sexual populations, two parents contribute to produce one progeny, in asexual populations, a single individual produces one progeny. Another way to view the 2-fold cost of sex is that in sexual populations, one parent transmits only one-half of their genes to any given progeny, whereas in asexual reproduction, the full genome complement is transmitted from parent to progeny. A further cost associated with sex involves locating a mating partner (271).
How is sex beneficial in evolution? For more than a century, the basic tenets for the benefits of sexual reproduction have posited that sex might serve to purge the genome of deleterious mutations, to produce recombinant progeny better suited to the environment, or both. In The Masterpiece of Nature: the Evolution and Genetics of Sexuality, Bell proposed one explanation (17). He suggested the “Red Queen hypothesis” based on a passage in Lewis Carroll's book Through the Looking-Glass: “Now here, you see, it takes all the running you can do to keep in the same place” (34a). In this view, sexual reproduction, like Alice, enables sexual organisms to keep pace with both changes in their environment and threats from their own genome instability.
In sexual reproduction, two compatible partners undergo a fusion of their genetic traits followed by recombination, meiosis, and mitosis to produce genetically recombinant progeny. In multicellular eukaryotes, for example, in humans, meiosis produces gametes, sperm and oocytes, which fuse to produce the zygote that develops to complete the sexual cycle. Therefore, one outcome of sexual development is genetically divergent progeny as a result of the crossing-over and recombination that occur during meiosis. This allows species to better adapt to given environments and could purge harmful mutations (226, 277). There are many empirical examples that sexual recombination increases genetic diversity to increase the chances for enhanced fitness (82). There is an intriguing observation regarding the benefit of sex, in which New Zealand mudsnails have evolved to utilize sexual reproduction to escape natural parasitic pathogens but are asexual in the absence of the pathogen (135). Therefore, sexual development might have contributed to maintain living organisms during long-term evolution. The genetics orchestrating sexual development can provide clues to unveil the evolutionary paradoxes of sex.
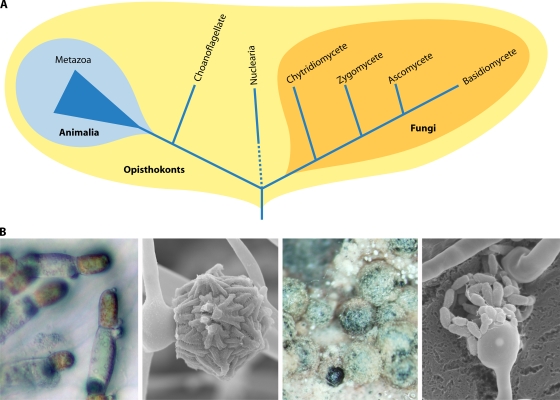
Fungi are evolutionarily closely related to metazoans, and together, the animal and fungal kingdoms form the opisthokonts (10, 301) (Fig. 1). The kingdom Fungi is further subgrouped into four phyla: Ascomycota, Basidiomycota, Zygomycota, and Chytridiomycota. The ascomycetes and basidiomycetes are known to be derived fungi that form a monophyletic group known as the dikarya, whereas the other two groups are basal fungi that diverged earlier. Current molecular phylogenetic analyses (the Fungal Tree of Life [AFTOL] project) revealed that the two basal fungal groups are polyphyletic and that as many as eight or nine phyla populate the fungal kingdom (124). For simplicity, in this review, we employ the traditional four-phylum classification.
FIG. 1.
Phylogeny of fungi and metazoans in the eukaryotic opisthokonts and sexual structures of four major fungal phyla. (A) The animal and fungal kingdoms are derived from a common ancestor, forming a clade called the opisthokonts. The opisthokonts contain several unicellular lineages (i.e., choanoflagellates and nuclearia). Within the four fungal phyla, the chytridiomycetes and zygomycetes form the basal lineages, and the ascomycetes and basidiomycetes are more recently diverged as the monophyletic dikarya. (B) The four major phyla have their own characteristics in sexual development. From left to right are sexual hyphae of the chytridiomycete Allomyces macrogynus, in which the male is orange and the female is hyaline, from which flagellated gametes arise; a zygospore of Mucor circinelloides, a pathogenic zygomycete; cleistothecia during sexual development in the ascomycete Aspergillus nidulans, which harbor sexual spores; and basidia and sexual spore chains of the human-pathogenic basidiomycete Cryptococcus neoformans.
The recent Fungal Tree of Life project supports the Chytridiomycota and Zygomycota as early-diverged fungi (124, 177), although the two lineages are not monophyletic (122, 282). Animals and fungi evolved from a single-celled and flagellate ancestor, from which the choanoflagellate outgroup to the metazoans also descends (151). Interestingly, chytrids also conserve the opisthokont-related flagella (14). Thus, flagellate zoospores of chytrids are a shared ancestral trait with metazoan and premetazoan lineages (124). Other fungi lost the flagella, possibly when aquatic fungi exited the oceans and became terrestrial. Multiple other losses have occurred in the other fungal lineages and prominently in the Ascomycota and Basidiomycota, involving, for example, RNA interference (RNAi) or light-sensory capacity (124). Analysis of the genome of the zygomycete Rhizopus oryzae revealed a series of genes thought to be animal specific, including those encoding the GTPase Rab32 and the Ras-like GTPase Ral, that are conserved in R. oryzae but not in the ascomycetes or basidiomycetes (178). Therefore, the two basal lineages retain shared common traits (symplesiomorphy) with the metazoans that are not shared with the dikarya. As such, basal fungi provide a novel evolutionary window into both the last common ancestor and the evolutionary trajectory of other fungi and metazoans.
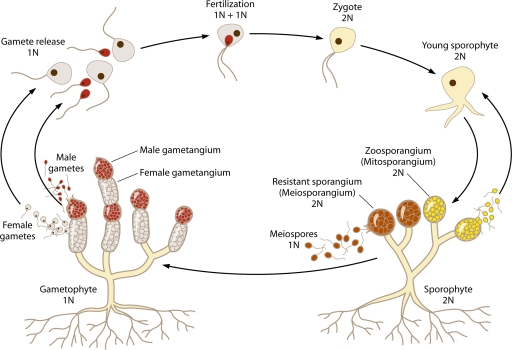
Fungal sex includes three steps similar to those for multicellular eukaryotes. Two compatible mating partners recognize each other and undergo cell fusion (plasmogamy). Thereafter, two parental nuclei undergo fusion (karyogamy). Interestingly, nuclear fusion occurs immediately after cell fusion in the two basal fungal lineages and some ascomycetes, whereas for other ascomycetes and the basidiomycetes, nuclear fusion is delayed after cell fusion. In the latter lineages, a dikaryotic stage with two paired nuclei persists after cell fusion, especially in the basidiomycetes, which have stable dikaryotic hyphae during the majority of their hyphal growth state; only later do the two nuclei fuse, and the third step in sex, meiosis, occurs to produce haploid recombinant progeny (3).
Many fungi are known to be sexual organisms, and diverse patterns of sexual recombination occur throughout the four phyla. Most early-diverged chytridiomycetes have motile gametes with flagella. However, in the other more-derived lineages, as these fungi left the oceans and became terrestrialized, they lost the flagella and instead evolved to disperse their spores aerially. Zygomycete sex occurs when two compatible hyphae fuse and the fused hyphae form a sexual spore called the zygospore. Zygospores are exposed; however, sexual spores of ascomycetes are enveloped within an ascus (plural, asci), and in some further-derived ascomycetes, multiple asci are sealed in ascocarps and protected. Basidiomycete hyphae are mostly dikaryotic during sex. At the termini of the hyphae, a bulb-like structure, the basidium, forms, where nuclear fusion followed by meiosis occurs. Unlike ascomycete sexual spores that are enveloped within asci, basidiomycete sexual spores are not enveloped and are instead exposed on the surface of the basidium (3).
Fungal sexual development is orchestrated by a genetic locus called the mating type or MAT locus. Each group of fungi evolved a different strategy for sex determination. Even subgroups within a phylum have evolved distinct patterns of genetic regulation of sex. Thus, the evolutionary trajectory of sex in fungi is an intriguing subject, which further helps to elucidate the basis of sexual development and the evolution of sex. There is a series of recent and classic reviews discussing the MAT locus in a wide variety of fungi from the fundamental model mating system Saccharomyces cerevisiae to the mating and virulence of human-pathogenic fungi and to the mushroom fungi (8, 31, 80, 99, 111, 137, 140, 148, 161, 192, 207, 253). Here we seek to cover the topic of sexual development throughout the fungal kingdom, including a focus on recent advances in sex in basal fungal lineages. We will discuss the sexual development of fungi, transitions between self-fertility (homothallism) and cross-fertility (heterothallism), and the mating type locus of fungal groups, including the Ascomycota, Basidiomycota, and the basal fungal lineages the Zygomycota, microsporidia, and the Chytridiomycota (Fig. 1). From the fungi, much has been and remains to be learned about sex-determinant/sex chromosome evolution and transitions in patterns of sexual reproduction that provide a unique vantage point from which to understand similar transitions that commonly occur throughout the eukaryotic tree of life.
SEX IN ASCOMYCETES
Some ascomycetous fungi sporulate both sexually and asexually. Asexual spores are exposed and unsealed. However, sexual spores (ascospores) are enveloped within an ascus. Asci contain four or eight ascospores depending on the species. In euascomycetes (or Pezizomycotina), the asci reside in an ascocarp, which is classified into different structures known as a cleistothecium (closed ascocarp found in Aspergillus nidulans and dermatophytes), a perithecium (an opened-bottle-like structure found in Neurospora spp.), an apothecium (completely open structure found in cup fungi such as morels), and ascostroma (a cavity-like structure that is called a locule found in sooty molds). Unlike the euascomycetes, hemiascomycetes (or Saccharomycotina, i.e., Saccharomyces cerevisiae and Candida spp.) lack an ascocarp. Ascomycete sexual development includes cell fusion, nuclear fusion, and meiosis. Ascomycete sex is orchestrated by the MAT locus, which encodes key transcription factor genes that govern all identity and developmental fate.
In this section, we discuss the Saccharomyces cerevisiae MAT locus as a paradigmatic fungal MAT locus, the evolution of the MAT locus and sexual development in the Candida clade, and the MAT locus of euascomycetes, including Aspergillus spp. and the dermatophytes.
S. cerevisiae as the Mating Paradigm
S. cerevisiae is a single-celled ascomycete and is further subgrouped into the hemiascomycetes. Unlike euascomycetes, S. cerevisiae does not produce an ascocarp during sexual development and instead forms a naked ascus containing four haploid meiotic progeny. S. cerevisiae grows mostly as a yeast and can exist as either a haploid or a diploid, but the predominant ploidy isolated from nature is diploid. No fungal species has been more intensively studied than S. cerevisiae. Its mating and genetics of sexual development are also well studied, and thus, S. cerevisiae provides a foundation from which to understand sexual reproduction in the fungal kingdom. There are two haploid mating type cells, a and α, in S. cerevisiae. Each cell type has a differentiated system for cell-cell recognition and the regulation of cell-specific gene expression.
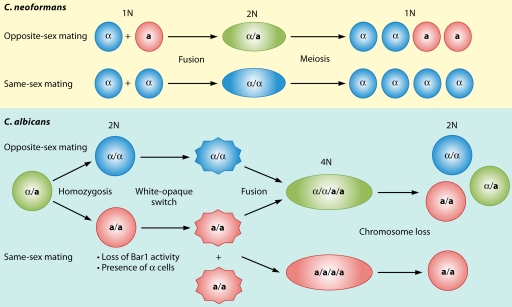
The identity of cell type and sexuality are orchestrated by a specific genomic locus called the MAT locus (Fig. 2A) (102). a cells harbor the MATa allele, containing the a1 gene, whose product is an HD2 class homeodomain (HD) transcription factor. In α cells, the MATα locus carries the α1 gene, encoding an alpha box transcription factor, and the α2 gene encodes an HD1 class homeodomain transcription factor. In the α haploid cell type, both α1 and α2 genes regulate the expression of α and a cell-specific genes, respectively (Fig. 2A). For example, the α1 and α2 genes in the MATα locus upregulate STE3, encoding the a factor pheromone receptor, and downregulate STE2, encoding the α factor pheromone receptor, respectively. In contrast, a1 plays no role in defining the a haploid cell type; the a cell type is the default, and cells lacking MAT entirely mate as a cells (102).
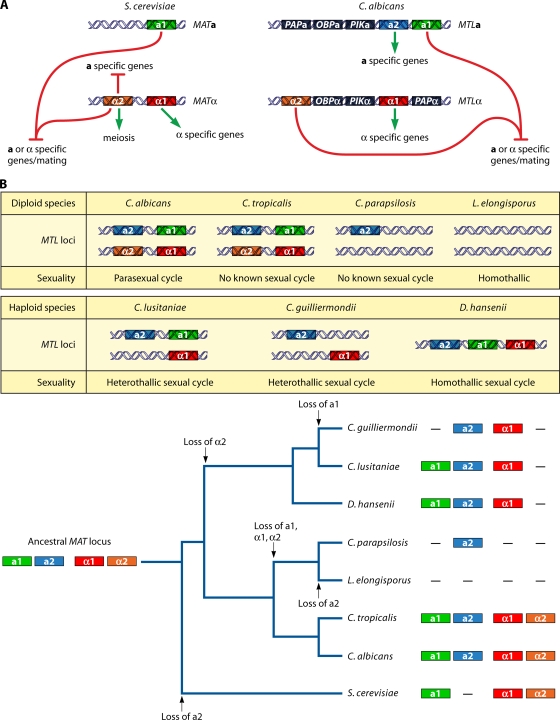
FIG. 2.
MAT and MTL loci of S. cerevisiae and C. albicans and the evolutionary trajectory of the MTL locus and sexuality within the Candida clade. (A) The MAT locus of S. cerevisiae and the MTL locus of C. albicans share similar architectures. Compared to S. cerevisiae, C. albicans has additional genes, a2, PAP, OBP, and PIK, in the MTL locus. (B) Compositions of the MTL loci of diploid and haploid Candida spp. vary. During speciation within the Candida clade, multiple independent losses of the ancestral MAT locus components a1, a2, α1, and α2 and transitions in sexuality have occurred (see also reference 274). The additional genes are not presented. The tree was redrawn based on a six-gene phylogeny (50).
Mating occurs when a cells encounter α cells. Haploid a cells produce a factor to signal α cells; similarly, haploid α cells produce α factor to signal a cells. Upon responses to a compatible mating partner, both cells develop projections called shmoos, and cell fusion occurs between the two a and α cell shmoo tips followed by nuclear fusion to form an a/α diploid. Thus, there are three cell types (a, α, and a/α diploid) found in the S. cerevisiae population. In the diploid cell, a1 and α2 form a heterodimer that serves as a transcriptional repressor of haploid-specific genes, and a/α cells do not mate. Instead, the a/α diploid cells undergo meiosis and generate haploid meiotic progeny, two α and two a cells in an ascus (Fig. 2A) (72, 73, 102, 126). These foundations of mating in S. cerevisiae provide a basic understanding of cell-to-cell recognition and the genetics of sexual development in other fungi.
The presence of two mating types and the stable inheritance of two different MAT loci to progeny are sufficient to confer heterothallism (mating between two different mating type partners) in S. cerevisiae. However, this yeast also evolved the ability to switch mating type and undergo a self-fertile sexual cycle (homothallism) (reviewed in reference 92). Two silenced loci called HML (homology to MAT left) and HMR (homology to MAT right) underlie the basis of mating type switching (280, 281). Cells of both mating types encode additional silenced copies of HML(α) and HMR(a). Recombination between MATa and HML(α) or MATα and HMR(a) results in a mating type switch. The key enzyme enabling this process is the HO (homothallic switching) endonuclease, which cleaves the MAT locus, causing a gene conversion between the MAT locus and the HML or HMR locus (210, 91, 92). Thus, a cell of a single mating type can undergo mating by this process; following cell divisions, HO is activated in the mother cells (but not in the daughter cells), evoking a mating type switch event to result in a pair of cells of opposite mating type and thereby enabling mating between the mother and daughter cells.
When an S. cerevisiae α cell switches mating type, HO cleaves at MAT, and recombination can then occur with either HMR(a) or HML(α). Switches with HMR(a) will lead to a productive mating-type-switching event, whereas switches with HML(α) lead to a futile cycle where α information at MAT is replaced with α information from HML (an α cell nonproductively switches to an α cell). Thus, one may expect the result of switching to be 50% productive and 50% futile. However, a biased ratio is observed, where mating type switching for both a and α cells is productive ∼90% of the time and only a minority of switch events represent cryptic futile switches from α to α or a to a (91). How does this system work at a molecular level? Wu and Haber found a cis-acting “recombination enhancer” (RE) sequence that lies on the left arm of chromosome III between HML and MAT (315), where α2 binds along with Mcm1 and the α2-Mcm1 complex juxtaposes nucleosomes across the RE (314), resulting in less active recombination in the left arm, leading to a preference for MATα switching with HMR(a). In a cells, α2 is lacking, and therefore, the RE is active and HML(α) is the preferential donor (315).
S. cerevisiae has a well-defined sexual cycle in which cells are either homothallic (self-fertile) or heterothallic (self-sterile). The basis for homothallism involves mating type switching evoked by the expression of the HO endonuclease and the subsequent mating of cells of the opposite mating type derived from a progenitor cell of one mating type. In nature, the vast majority of S. cerevisiae isolates are diploid and are thus capable of sporulation to produce fertile haploid progeny. Interestingly, environmental isolates of S. cerevisiae are either homothallic (HO) or heterothallic (ho), and this is attributable to naturally occurring mutations in the HO gene (reviewed in reference 193). Some isolates are even heterozygous for ho mutations and, thus, HO/ho, sporulating to give rise to two progeny (HO) that switch and self to return to the diploid state and two haploid (ho) progeny that do not switch and are fertile (55). Studies establishing the central tenets of yeast genetics by Winge and Lindegren were at one time contentious, as one group was studying a natural homothallic isolate and the other was studying a natural heterothallic isolate (208, 209). It is quite remarkable that the naturally occurring population of the well-defined species S. cerevisiae consists of homothallic/self-fertile and heterothallic/self-sterile individuals. Thus, not only are transitions between homothallism and heterothallism common throughout the fungal kingdom, but both patterns can also occur contemporaneously among members of the population. These distinct patterns of sexual reproduction may contribute to the relative levels of outcrossing and inbreeding that occur within a population and also enable recessive mutations that arise to be rapidly homozygosed and expressed in diploid isolates in the population. Transitions between the two modes of reproduction may occur in response to differing environmental selective pressures, and given that these two patterns occur frequently and even simultaneously in the case of multiple fungal species, those environmental conditions favoring one or the other may be rapidly fluctuating or present within or near similar environmental niches.
S. cerevisiae mating type switching evolved from an ancestral heterothallic state with two MAT idiomorphs by the sequential acquisition of silent mating type cassettes and the recruitment of the HO endonuclease. Detailed molecular, genetic, and phylogenetic analyses have revealed the evolutionary trajectory of MAT and the origins of mating type switching by analysis of related and diverged species of the hemiascomycetes (32). Comparisons of species related to S. cerevisiae, such as Candida glabrata and Saccharomyces castelli, reveal the presence of both an active MAT locus and two silent mating type cassettes and the HO endonuclease gene. In the most distantly related species compared, such as Candida albicans or Yarrowia lipolytica, there are no silent mating type cassettes. More closely related, yet still relatively distant, yeasts such as Kluveromyces lactis harbor the three mating type cassettes (one active and two silenced) but no recognizable HO endonuclease. Mating type switching can occur in K. lactis, and classical studies (101) and current models suggest that these mating type switch events occur via mitotic recombination in the absence of a recognizable HO endonuclease or HO cleavage site in MAT, as is known to occur at a low frequency even in S. cerevisiae ho mutant strains. Recent findings by Barsoum et al. revealed the underlying mechanisms involved in K. lactis, in which a transposable element (TE) plays a key role in mating type switching (15). In α cells, mating type switching to a occurs through recombination between the silenced HMRa locus and MATα3. The α3 gene encodes a transposase homolog uniquely found in the K. lactis MATα locus. The MATα3 locus is excised by an α3 transposase homolog that functions with the Mts1 protein that binds 5′ to α3. The excised MATα3 gene is eventually lost, and the DNA lesion in the genome undergoes a gene conversion through a recombination between left (L) and right (R) repeats found in the flanking regions of both MATα and HMRa. Thus, MATα undergoes gene conversion from HMRa to become MATa. In a cells, mating type switching to α occurs through the binding of Mts1 to the MATa locus, leading to a DNA lesion followed by a gene conversion between MATa and the HMRα locus (15). Thus, along with S. cerevisiae and Schizosaccharomyces pombe (see below), K. lactis then represents a third example in which high-efficiency mating type switching evoked by an initiating DNA lesion evolved via independent underlying molecular mechanisms.
HO itself is thought to have been conscripted to a novel biological role in mating type switching from an ancestral role as an intein-encoded endonuclease that promoted the mobility of an ancestral host genetic element. An interesting further feature is how the silencing of the HMR and HML versions of MAT first evolved. Given that HML and HMR are near but not at the telomeres, one possible scenario is that the gene duplication and triplication that first gave rise to extraneous copies of MAT placed these genes at true subtelomeric genomic positions and that telomeric silencing sufficed to quell expression sufficiently for mating to still occur with reasonable efficiency. Subsequently, a more sophisticated and efficient silencing mechanism may have evolved, involving the Sir proteins and the movement of HMR and HML near but not within the subtelomeric regions. Alternatively, an ancestral homothallic species may have harbored two active MAT alleles, similar to extant homothallic species such as Aspergillus nidulans and Neosartorya fischeri (discussed below) in which both MAT alleles are contained in the genome, and mating type switching evolved subsequently.
S. cerevisiae serves as the paradigm for the understanding of homothallism in fungi, and it is quite striking that another model yeast, the fission yeast Schizosaccharomyces pombe, is also homothallic and undergoes mating type switching. What is even more striking is that mating type switching evolved independently in the two diverged model yeasts (6). Both species harbor an active and two silent mating type cassettes, but the genes resident within MAT, their organization and the organization of the silent mating type cassettes, and the mechanisms both of silencing and of mating type switching differ considerably in molecular detail. From a phylogenetic and evolutionary perspective, mating type switching as a mechanism of self-fertile homothallic reproduction apparently evolved twice independently in the fungal kingdom, and other examples may remain to be discovered. Further phylogenetic comparisons of species closely and more distantly related to S. pombe are likely to reveal further insights, similar to the studies of the hemiascomycetes by Butler and colleagues discussed above (32).
Mating in S. cerevisiae and other related hemiascomycetous yeasts can involve both outcrossing to the general population and several different types of inbreeding/selfing. The three different forms of inbreeding that have been detected are termed (i) intertetrad selfing, which involves mating between haploid isolates that are derived from different meiotic events (different tetrads) from the same diploid progenitor; (ii) intratetrad selfing, which involves the mating of two haploids derived from the same meiotic event and contained within the same tetrad; and (iii) haploselfing, which involves a haploid isolate mating with a clone (also commonly involving mother-daughter cell mating). In experimental population-based studies of Saccharomyces paradoxus, a closely related sibling species of S. cerevisiae, Tsai et al. found that this species is asexually reproducing ∼99.9% of the time and is sexually reproducing 0.1% of the time (288). Those isolates that are derived from sexual reproduction could be attributed to outcrossing 1% of the time, intratetrad mating 94% of the time, and self-mating with a clone following mating type switching 5% of the time. Thus, a variety of patterns of selfing can occur, including examples in which diploids are produced with two seemingly identical copies of the genome, similar to same-sex mating in Candida albicans and Cryptococcus neoformans (as discussed further in the section on same-sex mating below).
Sex in the Candida Clade
The hemiascomycetes also contain the Candida clade, which is closely related to but diverged from S. cerevisiae ∼200 million years ago (mya). The Candida clade is characterized by a unique CUG codon, encoding serine rather than leucine, and ∼150 mya, the CUG codon capture event punctuated the evolution of this clade of species (263). The clade consists of both human-pathogenic and nonpathogenic species (50). C. albicans, C. tropicalis, C. parapsilosis, C. lusitaniae, and C. guilliermondii are human pathogens causing candidiasis, of which C. albicans is the most prevalent human-pathogenic fungus among not only Candida species but also fungi in general (234). Lodderomyces elongisporus has recently been identified as a bloodstream infectious agent related to C. parapsilosis, C. orthopsilosis, and C. metapsilosis (170).
Fungi within the Candida clade have evolved strikingly divergent patterns of sexual development (33, 254). C. albicans undergoes a parasexual cycle wherein two diploid cells mate, resulting in cell fusion and a ploidy increase (2N to 4N), and the tetraploid cells then undergo mitosis and random chromosome loss to return to the diploid state with no recognized meiosis (220). L. elongisporus and Debaryomyces hansenii are diploid and haploid fungi, respectively, that are self-fertile (homothallic) and, thus, do not need an opposite-mating-type partner for sexual reproduction (293, 294). The haploid species C. lusitaniae and C. guilliermondii are known to mate with a partner of a compatible mating type (heterothallic) (255, 324). In these species, the a/α diploid produced by mating proceeds to meiosis and completes the sexual cycle (254). No sexual cycle has yet been described for C. tropicalis or C. parapsilosis (294). Hence, it is intriguing to study how sexual development and determination have evolved within the Candida clade. Here, we discuss the mating-type-like locus (MTL) and parasexual/meiotic cycles in the Candida clade.
The MTL locus and parasexual cycle of C. albicans.
For more than a century, C. albicans was thought to be strictly asexual. This idea was supported by decades of observations of no apparent sexual development (e.g., no cell-cell fusion, meiosis, asci, or spores). Furthermore, the population structure exhibits a high degree of clonality, providing corroborative evidence that the fungus might lack a sexual cycle (244, 273). However, a low level of recombination was observed by population studies, indicating the possibility for sexual development in C. albicans (83).
The discovery of a genetic locus (mating-type-like [MTL] locus) related to the MAT locus of S. cerevisiae by Hull and Johnson revolutionized our understanding of sexual reproduction in C. albicans (116). Both MAT and the MTL locus encode idiomorphic a and α genes. As discussed above, S. cerevisiae MATa encodes a1 (homeodomain), and MATα encodes α1 (α box) and α2 (homeodomain) (Fig. 2A). The C. albicans MTLα idiomorph encodes α1 and α2, whereas the C. albicans MTLa idiomorph encodes a2 (a high-mobility-group [HMG] protein) in addition to a1. Unlike the S. cerevisiae MAT locus, the C. albicans MTL locus encodes three additional genes: PAP [poly(A) polymerase], OBP (oxysterol binding protein), and PIK (phosphatidylinositol 4-kinase), whose functions remain to be elucidated (Fig. 2A) (116).
Based on the known features of the S. cerevisiae mating system, the C. albicans a/α cell type would be predicted to be unable to mate (i.e., sterile) (117). Hull and colleagues then isolated genetically modified strains, termed the “a” type and “α” type, which resulted from the deletion of either the a1 or α1 and α2 genes, resulting in a/Δ and α/Δ cells. Following coinfection of mice with “a”-type and “α”-type cells, they recovered progeny with karyotypes higher than 2N resulting from cell-cell fusion (mating) in the host. Magee and Magee conducted a similar experiment. They constructed a/a and α/α homozygous strains by selecting for a loss of one copy of chromosome 5 (2N − 1) (on which MTL resides) by selection on sorbose as a sole carbon source followed by reduplication on yeast extract-peptone-dextrose (YPD) medium (a/α→a/a or α/α). Mating between genetically marked opposite-mating-type cells was found to occur under in vitro laboratory conditions (185), further supporting the existence of mating in C. albicans. However, in both cases, mating occurred with a very low efficiency.
Subsequent analyses by Miller and Johnson (199) revealed that the a/α stage is unable to mate, as a1 and α2 form a heterodimer, and the resulting a1-α2 complex inhibits mating. The a1-α2 complex inhibits the expression of a- and α-specific genes and confers a unique cell type identity to the a/α cells, resulting in a loss of the mating ability of the cell (Fig. 2A). Those researchers found that only a/a and α/α diploid or a/Δ and α/Δ strains can undergo mating (199). Thus, cells lacking the a1-α2 complex can mate.
In addition to the homozygosis of the MTL locus, a phenotypic switch from a white to an opaque colony morphology is critical for the fungus to mate. a/α diploids do not undergo the white-to-opaque switch (see below), whereas a/a and α/α diploids can transition to the opaque form. Opaque cells are specialized for mating, in which mating is a millionfold more efficient between opaque cells than between white cells (199). To undergo sexual development, a/α strains must first undergo homozygosis to yield a/a or α/α cell types, and the phenotypic switch from the white to the opaque mating-specialized cell type is then required for high-efficiency mating (171, 172). Finally, bona fide sex of C. albicans has been observed both under laboratory conditions and in the mammalian host (51).
The key element regulating white-to-opaque switching in C. albicans is the master regulator Wor1, a transcriptional regulator that is repressed by the action of the a1-α2 complex. WOR1 was identified as one of a subset of genes repressed by the a1-α2 complex and subsequently found to be both necessary and sufficient for white-to-opaque switching based on gene deletion and enforced-expression studies (113, 276, 328). Furthermore, Wor1 has been found to control its own expression via a direct positive-feedback loop and thus represents a bistable genetic switch with two states: white/infertile and opaque/fertile. Subsequent studies have revealed in elegant detail a complex cellular circuitry that governs the white-to-opaque transition involving not only Wor1 but also several other regulatory elements, including Czf1, Wor2, and Efg1 (329). Thus, it is now understood at a molecular level why a/α cells are infertile in C. albicans: this cell type expresses a1/α2, which represses Wor1 and thereby blocks white-to-opaque switching and mating. In contrast, in MTL homozygous or hemizygous strains, either a1 or α2 is missing, Wor1 expression is therefore permissible, and in some cells in the population, the bistable state switches from white (low level of or no Wor1 expressed) to opaque (high level of Wor1 expressed).
It is quite striking that mating in C. albicans involves a cell type morphology switch that does not occur in S. cerevisiae. Why might the white-to-opaque transition have been interposed into the mating cycle of C. albicans? Given that the environmental niches occupied by S. cerevisiae and C. albicans are remarkably different, involving fruit or arboreal niches versus mucosal surfaces of humans as part of the microbiota, mating is likely to occur in different environments. Given that C. albicans must run the gauntlet to avoid recognition, or at least survive, assault by the human immune system, it may be that aspects of mating result from this selective pressure. This could involve white-to-opaque transitions and also the apparent absence of the formation of an ascus or ascospores (which might be antigenic). The ways in which white and opaque cells interact with human immune cells differ, and this may be a mechanism to enable mating to occur in this harsh environment. White cells release chemoattractants for neutrophils, whereas opaque cells do not, and thus, in some cell culture systems, neutrophils readily phagocytose white cells but ignore opaque cells (75, 174). Thus, this may be a mechanism to shield or protect mating events from the immune system.
Because C. albicans mating involves diploid partners, their fusion generates a tetraploid cell. One might expect subsequent meiosis to generate diploid progeny in a complete sexual cycle. However, interestingly, no recognized meiosis has been observed, and instead, the tetraploid breaks down to diploid or near-diploid stages through chromosome loss via a parasexual cycle (18). A recent study revealed that genetic recombination during the parasexual cycle does occur within some progeny and that some progeny are also aneuploid (e.g., 2N + 1 and 2N + 2) (64). SPO11 encodes a recombinase that generates double-strand DNA breaks that stimulate meiotic recombination (reviewed in reference 267). Interestingly, SPO11 is required for this recombination during the parasexual cycle of C. albicans, indicating that either Spo11 plays a novel mitotic role or cryptic meiosis (parameiosis) occurs in C. albicans. In summary, C. albicans has evolved a unique sexual cycle involving the MTL locus and at least a parasexual cycle to produce recombinant progeny.
Evolution of the MTL locus and paradoxical signs of sex in the Candida clade.
Recent genome surveys to elucidate pathogenicity and sexuality for eight Candida species revealed that the MTL loci within the Candida clade are surprisingly different between species (Fig. 2B) (33, 254). The differences in the MTL locus components are illustrated in Fig. 2B (33, 173, 33, 254, 33).
Each Candida species possesses different patterns of sexuality and differences in the genetic compositions of the MTL loci, suggesting that during speciation within the Candida clade, novel regulatory systems have evolved for sexual development. Soll et al. suggested a possible evolutionary trajectory during speciation within the Candida clade based on the patterns of sexuality and differences in MTL loci (274). The common ancestral genome contained a MAT locus with all of the a1, a2, α1, and α2 genes. S. cerevisiae lost the a2 gene during an early divergence. The genomes of the sexual haploid Candida species, including C. guilliermondii, C. lusitaniae, and D. hansenii, all lack the α2 gene, and furthermore, C. guilliermondii has also lost the a1 gene (33, 254). This is striking, as these species have extant, complete sexual cycles, including meiosis. How meiosis occurs in C. lusitaniae and C. guilliermondii without α2 or a1/α2 is a mystery given that a1 is known to function only with α2 in S. cerevisiae and a1/α2 is required for meiosis in S. cerevisiae. Two diploid Candida species, C. albicans and C. tropicalis, parasexual and asexual diploid species, respectively, retain all of the ancestral MAT locus components (33); however, in the two other diploid species C. parapsilosis and L. elongisporus, C. parapsilosis, which has no known sexual cycle, lost a1 (nonfunctional a1 pseudogene), α1, and α2 (173), and L. elongisporus lost all four of the MAT locus components even though it is classified as a sexual homothallic (self-fertile) fungus (Fig. 2B).
Butler et al. and Reedy et al. analyzed the genomes of Candida spp. and found that a large number of genes known to be involved in sexual development and meiosis in S. cerevisiae are missing throughout Candida species whether they are sexual or asexual species (33, 254). The examples include the IME1 gene, which is a master regulator of meiosis (130); genes for meiotic recombination (RAD55, REC104, and REC114); genes for synaptonemal complexes (RED1 and ZIP1); and genes for chromosomal segregation (UBC11, MAM1, and SPI13) (33). These examples might explain the apparent absence of meiosis in C. albicans and several asexual Candida species.
However, one confronts a paradox in the sexuality of a species in this aspect in searching the genomes of the sexual Candida species C. lusitaniae and C. guilliermondii. In addition to also lacking all of the missing meiotic genes described above, both species also lack additional gene sets known to play critical roles in sexual development. For example, genes involved in meiotic recombination (DMC1, MEI5, and SAE3), synaptonemal complex formation (ZIP4 and HOP1), and crossover/interference (MSH4, MSH5, and MER3) are missing in both species (254). Detailed genetic and molecular studies revealed that meiosis occurs in C. lusitaniae despite the loss of additional key meiotic genes. First, a restriction fragment length polymorphism (RFLP) map for three chromosomes revealed that genetic recombination occurs frequently during C. lusitaniae sexual reproduction, with a recombination frequency of 4 to 17 kb/cM (where 1 centimorgan [cM] equals 1% recombination), on par with known meiotic recombination rates for other fungi (254). Second, the conserved meiotic recombinase Spo11 was shown to be required for this recombination. Third, analyses by fluorescence-activated cell sorting (FACS) and comparative genome hybridization (CGH) revealed that two-thirds of the progeny were haploid and euploid (254). Thus, by these three criteria, meiosis occurs during C. lusitaniae sexual reproduction, as was inferred based on classical mating studies in which asci and spores were observed. Interestingly, one-third of the meiotic progeny were observed to be aneuploid or diploid, which results from nondisjunction in meiosis I or II or precocious sister chromatid segregation. Thus, in this species, meiosis results in a high degree of aneuploid progeny.
There are interesting implications for modes of sexual reproduction that result in a high level of aneuploidy. With respect to C. lusitaniae, about one-third of the meiotic progeny produced are aneuploid or diploid (254), which is a much higher level than those reported by previous studies of S. cerevisiae. It may be that this is the natural consequence of an organism undergoing sexual reproduction in the absence of many conserved meiotic components. What are the consequences of aneuploidy, and does this represent an additional route by which meiosis can contribute to genetic diversity? We know and appreciate that aneuploidy is often deleterious, and a notable example is Down's syndrome, which results from trisomy for chromosome 21 in humans, and Turner's (XO) and Kleinfelter's (XXY) syndromes in humans. Aneuploidy in fungi could be phenotypically advantageous, deleterious, or neutral. There are several recent examples in which aneuploidy has been found to confer a variety of phenotypes, some of which can be considered detrimental, such as temperature-sensitive growth (285). However, in other cases, aneuploidy results in antifungal drug resistance associated with an isochromosome that frequently forms in fluconazole-resistant C. albicans isolates or in which aneuploidy has been found to promote adaptive evolution (250, 268). Thus, there may be either short-term or longer-term advantages conferred by modes of sexual reproduction that produce aneuploid isolates.
It is notable that many studies of S. cerevisiae meiosis have focused on strains that efficiently sporulate, germinate, and produce a high level of euploid progeny. However, many natural isolates sporulate or germinate poorly and may result in a higher level of aneuploidy. Thus, sex in nature may be considerably messier than sex in the laboratory. Crosses between isogenic or congenic strains may result in a higher level of euploidy than crosses between nonisogenic strains, or those with differing karyotypes, and this would be interesting and useful to study in the laboratory.
Studies of meiosis and its consequences in fungi may also prove to be informative models to provide insights into the relatively high rates of aneuploidy that occur during gametogenesis in humans, involving both oocytes and sperm. We know that the frequency of chromosomal abnormalities increases precipitously with advanced maternal age, spontaneous miscarriage in humans can occur in up to one-third of pregnancies, the frequency of spontaneous miscarriage is independently associated with both increased maternal and paternal age, and up to 50% of spontaneous miscarriages are attributed to chromosomal abnormalities and aneuploidy. Thus, up to one-sixth of human fertilization events may involve the production of aneuploid zygotes, few of which are carried to term birth.
With the loss of a large set of meiotic genes, how do these Candida species still undergo a complex sexual cycle, including meiosis? These findings suggest that a different meiotic paradigm could have evolved in the Candida clade; for example, they may have evolved a reduced meiotic tool kit or a completely different meiotic machinery (33). As noted by Soll et al., the S. cerevisiae meiotic gene set may not be sufficient to explain sexuality in Candida species. Yarrowia lipolytica, the earlier common ancestor of S. cerevisiae and the Candida clade, also lacks a large set of meiotic genes present in the S. cerevisiae genome, indicating that S. cerevisiae might have acquired additional meiotic genes or that they could be rapidly evolving (274).
A central paradox has been when and where C. albicans mating might occur, given that the opaque mating-specialized cell type is unstable at 37°C under many in vitro growth conditions. If mating occurs in the body, this might require environments in which the temperature is lower, opaque cells are stabilized, or opaque cells are favored in some way. Classical studies by Lachke and colleagues demonstrated that mating of C. albicans can occur on the skin of baby mice, where the temperature was shown to be 31.5°C, similar to the temperature on the surface of human skin (32°C) (150). Thus, C. albicans, which is commonly resident on the mucosa of the mouth and oropharynx, gastrointestinal (GI) tract, and vagina might transiently be present in environments involving skin adjacent to these areas, undergo mating, and then either return to other niches in the same individual or be transmitted to another host. Similar studies have provided evidence that both anaerobic conditions and the elevated CO2 levels found in the host can promote white-to-opaque switching and, thereby, mating (51, 112, 249). C. albicans mating might also occur and be promoted in the context of biofilms present on mucosal surfaces. Daniels and colleagues found that a and α cells can signal one another over considerable distances in the context of a biofilm, and those authors hypothesized that the matrix of the biofilm serves to facilitate pheromone communication by serving as a diffusion gradient between rare mating partners that arise in the population (41). In turn, their studies revealed that pheromone signaling promotes cell cohesiveness and promotes biofilm formation; thus, one physiological role for C. albicans biofilms may be to serve as a forum that promotes mating. Finally, a series of recent studies revealed that not only opaque but also white cells respond to pheromones. While many of the components that mediate pheromone sensing are shared between the two cell types, a unique role for a novel domain of the α pheromone receptor and differences in the transcription factors involved show that the two pathways likely play different physiological roles (260, 261, 322, 323).
Current models posit that rare MTL homozygosis events occur, yielding distant a/a and α/α potential mating partners in the context of a biofilm under anaerobic/high-CO2 conditions (or at lower temperatures). Next, one of the two partners switches to opaque. Signaling can then occur from a solo opaque cell to potential white MTL homozygous mating partners, which can sense and respond to pheromone via this novel white cell response pathway. This may then promote, in some fashion, white-to-opaque switching such that both partners are then in the mating-specialized cell type and are competent to produce long conjugation tubes that lead to cell and then nuclear fusion. The very recent discovery that C. albicans can also undergo same-sex mating in addition to opposite-sex mating (2) broadens the conditions under which mating might occur (98) and will be discussed in further detail below. The divergent sexuality, differentiated MTL loci, and different sets of meiotic genes in the sibling species of the Candida complex provide insights into the plasticity of sex determination and sexuality during evolution.
Sex in Aspergilli
The genus Aspergillus represents some of the most common fungi in the environment and contains ∼250 species comprising diverse groups (76). This includes (i) species popular in industrial processes, such as Aspergillus oryzae, which is used in the fermentation of products containing starchy ingredients, and A. niger, which is the major world source of citric acid and valuable enzyme products; (ii) human pathogens, such as A. fumigatus, which causes respiratory tract diseases, and A. clavatus, which causes pneumonitis, such as malt worker's lung; (iii) plant pathogens, such as A. flavus and A. parasiticus, which produce the potent carcinogen aflatoxin; and (iv) a classical genetic model, A. nidulans, studied to address myriad biological questions. The genomes of these Aspergillus species have been sequenced, and the genome sequences can be accessed at the NCBI Entrez Genome Project Database (http://www.ncbi.nlm.nih.gov/sites/entrez?DB=genomeprj), the Aspergillus Comparative Database at the Broad Institute (http://www.broadinstitute.org/annotation/genome/aspergillus_group/MultiHome.html), and the Aspergillus Genome Database (http://www.aspgd.org/).
Aspergilli, which include both known asexual and sexual species, provide an excellent system to study the genetic basis of reproductive modes in fungi. All Aspergillus species reproduce asexually by forming a dandelion seed head-like structure, called a conidiophore, which has a vesicle head bearing long chains of asexual spores (conidia) (Fig. 3). Approximately 70 named species can also propagate through sexual development, exhibiting either homothallic or heterothallic patterns of mating (77). Phylogenetic analysis of both the mitochondrial and nuclear genomes suggests that most of these species have arisen from a common ancestor. In particular, asexual species arise frequently from sexual ones (77). Therefore, it is of particular interest to study sexual reproduction and the functions of mating type (MAT) locus genes in the aspergilli.
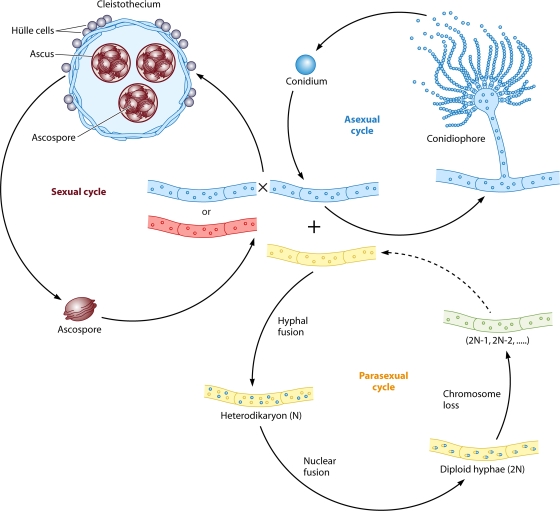
FIG. 3.
Life cycle of A. nidulans. A. nidulans can undergo three life cycles. First, it produces conidia robustly during an asexual cycle. A conidium germinates to form hyphae, from which conidiophores develop to produce more conidia. Second, it can undergo a homothallic sexual cycle involving selfing or out-crossing to generate fruiting bodies (called cleistothecia) containing thousands of ascospores, which germinate to form hyphae. Third, it can undergo a parasexual cycle. Heterohyphae fuse to form a heterodikaryon, followed by nuclear fusion to generate diploid hyphae, in which random chromosome loss occurs to restore the haploid chromosome number.
Asexual, sexual, and parasexual cycles in the aspergilli.
All Aspergillus species robustly produce conidia, the asexual spores. Conidia are generated through an asexual reproductive cycle, which is divided into two main phases: vegetative growth and development. The vegetative growth phase starts with the germination of a conidium and is followed by the formation of mycelia composed of hyphal cells. After a period of vegetative growth, under appropriate conditions, certain hyphal cells cease normal growth and initiate asexual development, which includes conidiophore formation and conidial maturation (Fig. 3) (1, 212). Conidia are infectious propagules of most Aspergillus pathogens.
About one-third of aspergilli, including A. nidulans, can also reproduce sexually. Sexual development starts with the formation of a cleistothecial initial (CI), in which ascogonial coils form by hyphal branching. Ascogonial coils from two hyphal partners then fuse to produce a dikaryon. A CI is wrapped in a nest-like structure comprised of thick-walled Hülle cells, which serve as nurse cells. Under these Hülle cells, each dikaryon undergoes nuclear fusion followed by meiosis and then a postmeiotic mitosis, resulting in the formation of an ascus containing eight haploid ascospores. Finally, a fruiting body called a cleistothecium is formed and serves to house tens of thousands of ascospores (Fig. 3) (272). Most sexual Aspergillus species are self-fertile (homothallic) but are also capable of outcrossing. The first identified heterothallic Aspergillus species is A. heterothallicus (141). Recently, several other well-known “asexual” Aspergillus species, including A. fumigatus and A. parasiticus, were discovered to have extant heterothallic sexual cycles (see below). Compared to asexual development, sexual development, especially outcrossing, has many potential benefits. These benefits include the generation of new genotypes that may be better adapted to novel environments, purging the genome from the otherwise relentless accumulation of deleterious mutations, and enabling the formation of thick-walled fruiting bodies that are resistant to harsh conditions.
Besides the asexual and sexual cycles, many Aspergillus species can undergo a parasexual cycle that enables recombination during mitosis. The parasexual cycle starts with the formation of a dikaryon by hyphal fusion followed by haploid (N) nuclear fusion, which results in the formation of diploid (2N) hyphae. Instead of undergoing meiosis, the vegetative cells continue dividing mitotically, and the haploid chromosome number (N) is restored by random chromosome loss (Fig. 3) (36, 240). As no known sexual cycle had been found for several species (including A. fumigatus, A. niger, A. flavus, and A. parasiticus) for many years, the parasexual cycle has been applied widely for gene identification, the generation of new strains, and linkage mapping for these species (42, 59, 128, 159, 201, 229, 230, 278, 295).
Although a parasexual cycle has been defined for many Aspergillus species, relatively less was known about its evolutionary importance until recently. Schoustra et al. examined the growth fitnesses of both haploid and diploid A. nidulans strains, and they found that diploid strains attained higher fitnesses than isogenic haploid strains (i.e., the diploid strains' progenitors) after ∼3,000 mitotic generations, and invariably, these faster-growing isolates evolved from a diploid progenitor had undergone a parasexual reduction to return to the haploid state. Thus, mitotic recombination occurring during the parasexual cycle can accelerate adaptation under laboratory conditions (266). The higher fitness obtained is due to “sign epistasis” effects, where mutations occurring in diploid nuclei could be neutral or deleterious on their own in a haploid but are instead advantageous when combined. After mitotic recombination through the parasexual cycle, the advantageous mutations were selected for based on faster growth. This study revealed that the parasexual cycle can serve as a capacitor for evolution and might generate genotypic diversity de novo rather than admixing genetic differences from two divergent parental isolates.
Genomic evidence of mating abilities in “asexual” Aspergillus species.
Unlike A. nidulans, which has a defined sexual cycle (discussed above), certain Aspergillus species such as A. fumigatus, A. niger, A. oryzae, and A. parasiticus were long considered to have only asexual development. However, several lines of evidence have revealed or suggested that a sexual stage for these organisms is or may be extant.
First, whole genome sequences of several well-known “asexual” species, such as A. fumigatus, A. oryzae, and A. niger, have shown that they contain a suite of genes associated with different stages of the sexual cycle in ascomycete fungi, including the mating process, pheromone response, meiosis, and fruiting-body development (71, 179, 219, 233, 236). Two different MAT locus genes function in establishing sexual compatibility in these fungi: one encodes a protein with a high-mobility-group (HMG) domain, and the other encodes a protein with an alpha box domain. Homothallic fungi, such as A. nidulans, have both MAT genes, which are located on different chromosomes, and as a consequence, these species are self-fertile (139, 228). A. nidulans cells need to be able to express both MAT1-1 and MAT1-2 to be self-fertile. According to Paoletti et al., mutation in either the MAT1-1 or MAT1-2 gene led to a failure of the formation of fertile cleistothecia (228), indicating that both the MAT1-1 and MAT1-2 alleles are required for homothallic sexual development. However, it is not understood in detail how the presence of both MAT alleles enables self-fertility. In one model, genetic noise results in different subpopulations of cells randomly expressing MAT1-1 or MAT1-2 and two different cell types undergoing mating as if these were heterothallic. In other models, single cells expressing both MAT alleles are fertile.
Heterothallic fungi contain only one MAT locus idiomorph and require a partner with the opposite MAT locus. Whole-genome sequencing discovered a single HMG mating type gene and a single alpha box mating type gene located in the MAT locus of A. fumigatus and A. oryzae, respectively (71). In addition, 215 genes with potential roles in regulating sexual development were found in the genome of A. fumigatus (71, 219). These findings, combined with previous results showing that A. fumigatus or A. oryzae isolates contain either HMG or alpha box mating type (MAT1-1 or MAT1-2) genes, but never intact copies of both, suggest that they are heterothallic species (227).
Second, population genetic analyses revealed that the two mating types are equally distributed globally and provided evidence of recombination. Paoletti et al. (227) screened a worldwide collection of 290 A. fumigatus isolates from both the environment and clinic for mating type and showed a nearly 1:1 ratio of MAT1-1 to MAT1-2, in accordance with extant sexuality. Those researchers also showed the expression of both MAT1-1 and MAT1-2 and of genes encoding sex pheromones and pheromone receptors. Another study also presented a ∼1:1 ratio of the two mating types in 91 isolates from five locations in Dublin, Ireland (221). Examination of the genetic variability of A. fumigatus by multiple phenotypic and genotypic techniques, including multilocus enzyme electrophoresis (MLEE), RFLP, randomly amplified polymorphic DNA (RAPD), sequence-specific DNA polymorphism (SSDP), microsatellite length polymorphism (MLP), and DNA fingerprinting, revealed that recombination occurs in A. fumigatus natural populations (227, 243, 296). In A. flavus and A. parasiticus, the existence of both MAT idiomorphs (MAT1-1 and MAT1-2) in equal proportions in the populations, coupled with their expression at the mRNA level, also indicated the potential functional role of these genes and the possible existence of an extant sexual state for these plant pathogens (248).
Third, mating type proteins from “asexual” Aspergillus species were shown to play potential roles in regulating sexual development. The evidence that HMG domain and alpha box mating type genes are required for the formation of sexual spores in A. nidulans (228) and that MAT1-1 and MAT1-2 are expressed during the mycelial growth of A. fumigatus suggested that they might be functional proteins in A. fumigatus (227). As no known sexual stage was identified at that time, two groups studied the functions of A. fumigatus mating type proteins during the sexual development of A. nidulans. Pyrzak et al. (245) showed that the ectopic integration of the A. fumigatus MAT1-2 gene driven by the A. nidulans MAT2 (matA) promoter was able to restore partial fertility in a sterile A. nidulans strain with a disrupted HMG MAT gene. Grosse and Krappmann (90) found that the overexpression of the sexual developmental regulator nsdD resulted in the formation of coiled hyphae reminiscent of those seen in the early stages of cleistothecium formation in A. nidulans. Those authors also reported that replacing the A. nidulans MAT1 (matB) open reading frame (ORF) with the A. fumigatus MAT1-1 ORF resulted in normal sexual development. These findings demonstrate the potential functions of A. fumigatus mating type genes in controlling sexual development, thereby suggesting cryptic sexuality in this “asexual” Aspergillus species.
Discovery of an extant sexual cycle for A. fumigatus and A. parasiticus.
The accumulating evidence (discussed above) for the existence of sexual stages in several Aspergillus species known to reproduce only asexually culminated with the discovery of extant sexual cycles (109, 221). O'Gorman et al. showed that A. fumigatus generates sexual fruiting bodies (cleistothecia) with viable sexual spores (ascospores) by accomplishing a sexual cycle (Fig. 4) and named its teleomorph Neosartorya fumigata (note that a teleomorph is the sexual stage of a fungus) (221). Different from other Aspergillus species, mature cleistothecia of A. fumigatus were found in cultures grown on parafilm-sealed oatmeal agar plates at 30°C in the dark after an extremely long incubation period—6 months. Furthermore, RAPD analysis of five genetic markers in 15 ascospore progeny provided clear evidence of recombination occurring during the sexual cycle.
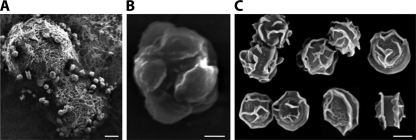
FIG. 4.
Sexual structures of A. fumigatus. Shown are scanning electron micrographs of cleistothecia (A), an ascus with eight ascospores (B), and ascospores (C). Scale bars represent 100 μm (A), 2 μm (B), and 2 μm (C). (Courtesy of Celine O'Gorman.)
Horn et al. (109) reported that A. parasiticus also undergoes a complete sexual cycle, which results in the development of ascospore-bearing ascocarps embedded within stromata. Strains with opposite MAT loci (MAT1-1 and MAT1-2) from different vegetative compatibility groups were crossed by inoculating mixed conidial suspensions on mixed cereal agar, and cultures were incubated at 30°C in the dark for 6 to 9 months. Multilocus sequence typing (MLST) analysis of three genetic markers in 57 progeny from three crosses demonstrated that recombination occurred during the sexual cycle. The discovery of sexual cycles of both A. fumigatus and A. parasiticus provides an invaluable tool for genetic analyses to better understand the biology and evolution of these species. At the same time, it raises the question of why the completion of the sexual cycle is so protracted and when and where sexual development occurs in nature. One possibility is that the prolonged sexual cycle is a novel type of overwintering strategy.
Model for evolution of the MAT locus in aspergilli.
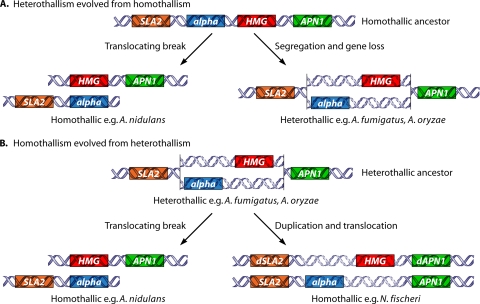
Two different models have been proposed for the evolution of the MAT locus in aspergilli (Fig. 5). The first model (Fig. 5A) is that heterothallism evolved from homothallism in the aspergilli. The alpha MAT (MAT1-2) locus of A. fumigatus is flanked by a fragmented HMG gene that can reflect an ancestral linkage of alpha box and HMG genes (227). In addition, few Aspergillus species are heterothallic, and one of them (A. heterothallicus) evolved closely aligned with known homothallic species (77), suggesting that homothallism could be ancestral. The MAT locus idiomorphs of A. nidulans were proposed to have arisen from a translocating break from an ancestral homothallic state, in which the linked MAT genes were contained in a single MAT locus (71). The MAT loci of either A. oryzae or A. fumigatus were proposed to have been generated by a segregation event followed by speciation from the ancestral homothallic state.
FIG. 5.
Model of evolution of MAT loci within aspergilli. (A) In the model of homothallism as ancestral, the last common ancestor contained both alpha box and HMG domain genes adjacent to each other flanked by the SLA2 and APN1 genes. Next, in one lineage, a chromosomal break and translocation occurred to rearrange the alpha box and HMG domain genes to different chromosomes flanked by either the SLA2 or APN1 gene, giving rise to extant homothallic species. In addition, in an alternative lineage, chromosome segregation and gene loss occurred to maintain either the alpha box or HMG domain genes at the original locus, giving rise to extant heterothallic species. (B) In the model of heterothallism as ancestral, the last common ancestor contained either the alpha box or the HMG domain gene at the same locus flanked by the SLA2 and APN1 genes. This species underwent a chromosomal break and translocation to rearrange the alpha box and HMG genes to different chromosomes flanked by the SLA2 and APN1 genes, respectively, to evolve into extant homothallic species (A. nidulans). In addition, this ancestor also underwent gene duplication and chromosomal translocation to maintain either the alpha box or HMG domain gene at both the original locus and an unlinked locus with modified flanking genes to evolve into other homothallic species (N. fischeri).
The second model is that homothallism evolved from heterothallism within the aspergilli. A comparison of the MAT loci in the genomes of multiple aspergilli, including A. nidulans, Neosartorya fischeri, A. fumigatus, A. oryzae, and A. terreus (Fig. 5B) (259), provides evidence to support this model. A. fumigatus has either a MAT1-1 (alpha box) or a MAT1-2 (HMG) idiomorph located adjacent to a single genomic copy of the APN1 (or, in some cases, indicated as APN2; both genes are paralogs of a S. cerevisiae gene encoding a DNA lyase) or SLA2 (cytoskeleton assembly control) gene (71). In this case, chromosomal translocation severed the MAT locus into two unlinked loci in the genome. N. fischeri, a homothallic fungus, contains two MAT locus regions, MAT1 and MAT2, in unlinked regions of the genome. Intact and functional alleles of the APN1 and SLA2 genes flank the MAT1 locus, whereas partial copies of the APN1 and SLA2 alleles flank the MAT2 locus that was generated by gene duplication (259). It is hypothesized that the transposition of MAT to an unlinked genome location and a loss or decay of the functional syntenic flanking genes (Fig. 5B) might have led to the current N. fischeri MAT system. A distinct pattern is observed in the A. nidulans genome, where MATA (alpha box) and MATB (HMG) are located on different chromosomes, but both genes are flanked by only one copy of the APN1 or SLA2 gene where no gene duplication event is hypothesized (Fig. 5B) (71). These data suggest that N. fischeri and A. fumigatus may share an ancestral heterothallic MAT locus with either an HMG or an alpha domain MAT gene flanked by the APN1 and SLA2 genes. In addition, a similar heterothallic MAT locus arrangement was also found for A. oryzae (71). Since A. fumigatus and A. oryzae are taxonomically divergent within the genus Aspergillus, their MAT sequence arrangement may reflect the ancestral state. Although both N. fischeri and A. nidulans are homothallic, the configurations of their MAT loci are different, and thus, both appear to be derived character states. The MAT locus of A. nidulans possibly arose from an ancestral heterothallic state through chromosome breakage and segregation.
As additional Aspergillus genomes become available, comparative genomics will further enhance our understanding of genome evolution to refine these evolving models for transitions between heterothallic and homothallic sexual reproduction, which commonly occur throughout the fungi.
Conclusions and perspective.
Aspergilli, which include both known asexual and sexual species, provide robust systems to study the evolution of sex in fungi. The discovery that A. fumigatus and A. parasiticus retain extant sexual cycles yields insight into the potential for sexual reproduction in other purportedly “asexual” fungi. A central question is whether there are any truly asexual fungi in nature. These results will also facilitate research into the genetic basis of pathogenicity and fungicide resistance of A. fumigatus and A. parasiticus with the aim of improving methods for the control of aspergillosis and mycotoxin contamination. Another interesting question is whether there is an association between mating type and virulence. One recent study showed a 4-fold-higher frequency of MAT1-1 than MAT1-2 genotypes among A. fumigatus isolates from an invasive origin (5). Therefore, further investigations into the possible link between mating type and other relevant traits, including virulence, are warranted.
Sex in Dimorphic Fungi and Dermatophytes
In this section, we discuss sex and the MAT locus in two groups of human-pathogenic ascomycetes: dimorphic fungi and dermatophytes. These two groups are found to be phylogenetically closely related to each other (22, 104, 152).
Dimorphic fungi comprise a group of important human-pathogenic fungi capable of converting between two morphologies: mold and yeastlike forms. At room temperature in vitro, they grow as molds with numerous filaments. They grow as pathogenic yeastlike forms at body temperature when invading mammalian hosts or in vitro. While many fungi can undergo morphological transitions between mold and yeast, the medically important pathogens generally recognized as dimorphic fungi are Histoplasma capsulatum, Coccidioides immitis, Coccidioides posadasii, Paracoccidioides brasiliensis, Blastomyces dermatitidis, Sporothrix schenckii, and Penicillium marneffei (252). Except for S. schenckii and H. capsulatum, which have a worldwide distribution, dimorphic fungi are geographically restricted. P. marneffei is the most important thermally dimorphic fungus causing respiratory, skin, and systemic mycosis in Southeast Asia (44). P. brasiliensis, the causative agent of paracoccidioidomycosis, is widespread in Latin America (262). C. immitis and C. posadasii are causative agents of coccidioidomycosis, or valley fever, which is endemic to the Southwestern United States and Central and South America (13). B. dermatitidis is the causative agent of blastomycosis and is endemic to the Americas (26).
Dermatophytes are the most common fungal infection worldwide and cause infections of the nails, hair, and skin via their ability to degrade keratin in these tissues (303, 307). Three anamorphic (asexual) genera, Microsporum, Epidermophyton, and Trichophyton, of dermatophytes are grouped into the sexual family Arthrodermataceae, the order Onygenales, and the class Eurotiomycetes of the Ascomycota in the fungal kingdom (303). Based on host preference and natural habitat, the dermatophytes are divided into three ecological groups: anthropophiles, zoophiles, and geophiles (78). Geophiles are primarily soil-dwelling organisms, but some taxa are pathogenic to humans. Zoophiles are essentially animal pathogens, although they may also cause human infections in some circumstances. Anthropophiles are restricted largely to humans, but some species can also infect other animals in some cases. So far, a correlation between phylogeny and ecological niches has not been found. Most dermatophytes are thought to be capable of reproducing sexually based on direct observations of mating structures or indirect observations derived from population genetic studies (307). By direct mating assays and indirect population genetic studies, it was suggested that geophilic dermatophytes typically have an extant sexual cycle, and the zoophilic dermatophytes also frequently retain the ability to reproduce sexually, whereas anthropophilic dermatophytes are frequently infertile (307).
The dimorphic fungi and dermatophytes are closely related members of the Ascomycota based on rRNA and chitin synthase 1 gene (CHS1) sequences (22, 104, 152). On the basis of the substitution rate of the nucleotides of the small-subunit rRNA genes, it is estimated that the dermatophytes diverged within the last 50 million years, which is comparable to mammalian evolution (95). Although closely related, dimorphic fungi and dermatophytes are recognized as distinct groups of species that have broadly differentiated.
Sexual cycle and mating ability of dimorphic fungi and the dermatophytes.
Sexual reproduction has been described for H. capsulatum (142, 146), B. dermatitidis (194), and most dermatophyte species (307). While successful mating has not yet been observed for P. marneffei, C. immitis, or C. posadasii, genes related to sexual reproduction have been identified in the genomes of these dimorphic fungal species (70, 189, 311). Data from population genetic studies also support extant sexual reproduction (30, 61, 62, 138, 189). No sexual cycle or MAT locus has been described for S. schenckii, but it has been speculated that sexual reproduction may be extant in this fungal species on the basis of population genetic studies (191, 196). Even though the sexual cycle has been well defined for H. capsulatum, a loss of mating ability has also been observed with repeated passage, but the mechanism has not yet been elucidated (28, 146). Since the MAT locus genes are intact and major meiotic genes are conserved in the completed genomic sequences from four different H. capsulatum isolates, this loss of mating ability could involve epigenetic factors. In Microsporum gypseum, cleistothecium formation and the production of asci and ascospores were observed by mating assays (160, 302). By scanning electron microscopy, the cleistothecium was found to be enveloped by coiled and spiral hyphae. Inside the cleistothecium, each ascus contains eight ascospores. Ascospores germinate to produce hyphae when spread onto Sabouraud dextrose agar at room temperature. Sexual structures have not been observed for some pairs of strains with opposite mating types, even following coincubation for more than 2 months (160). This finding suggests that sexual reproduction may occur only under special circumstances to generate recombinant progeny. An understanding of how and why human pathogenic fungi maintain a sexual cycle is important to elucidate their pathogenicity.
Dimorphic fungi and dermatophytes are important human fungal pathogens. Most dimorphic fungi are pathogenic to humans by causing pulmonary infection and can also disseminate to cause systemic infections (241). The dermatophytes utilize superficial keratin as a nutrient source and cause the most common fungal infections in the world. However, like other human-pathogenic fungi, the dermatophytes and dimorphic fungi are thought to undergo predominantly asexual reproduction in the human host.
Investigation of the sexual cycles of these human-pathogenic fungi will further enhance our understanding of their pathogenicity because the mating process has the potential to play a role in the virulence of human pathogens by generating progeny with altered virulence via genetic recombination (216). Different mating type strains may also have distinct virulence properties. An unequal prevalence of the two opposite mating types has been observed for clinical isolates of H. capsulatum (149). In contrast, a 1:1 ratio of two mating types was recently observed for 71 clinical isolates of P. brasiliensis in South America, and the coincubation of some isolates results in coiled hyphae that may represent an early stage of sexual reproduction (286). Similarly, in Microsporum gypseum, the two mating types have an almost equal prevalence among clinical isolates (106, 302). A virulence test of M. gypseum in the Galleria mellonella larva heterologous host further supports that the two mating type strains of M. gypseum may have similar virulences (160). Characterization of the MAT locus of these pathogenic fungi now enables specific PCR-based tests to scan a large strain population to correlate mating types with virulence.
General structure of the MAT locus of dermatophytes and dimorphic fungi.
The sexual reproduction of dimorphic fungi and dermatophytes is governed by a specialized genomic region, the mating type (MAT) locus, as described for other fungi (160). The recent completion of a larger number of fungal genome sequencing projects, including dermatophytes and dimorphic fungi, enables a detailed analysis of the MAT locus and its evolution and the correlation of sexual reproduction with a defined genetic basis.
As for other euscomycetes, the MAT locus in dimorphic fungi and the dermatophytes is a relatively small genomic region compared to that of basidiomycetes such as Cryptococcus, in which the MAT locus is as large as ∼100 kb and includes more than 20 genes (66, 157) (see the section on basidiomycetes below) (40, 289). Typically, the plus mating type contains an alpha box gene and is termed the MAT1-1 locus. The minus mating type contains an HMG gene and is termed MAT1-2 (40, 289). The MAT locus is closely linked to the SLA2, APN2 (paralogous with APN1), and COX13 genes (40, 289). While both homothallic and heterothallic mating systems have been identified in the Ascomycota, no homothallic species in the dermatophyte or dimorphic fungi have been described. This suggests that the ancestral mating system of the dimorphic fungi and dermatophytes was heterothallic.
MAT locus size of dimorphic fungi and dermatophytes.
The MAT locus size is determined by the sequence diversity between the two MAT alleles. High sequence identity (>99%) and gene synteny reflect the boundaries of the MAT locus (160). While there is no direct association between the size of the MAT locus and mating ability, a larger MAT locus may facilitate the suppression of recombination of this unique region. In humans, animals, and plants, the genomic region responsible for sexual differentiation independently evolved as sex chromosomes, with suppressed homologous recombination during meiosis (66). The sex-determining region in the fungal kingdom is typically more restricted but shares the general feature of suppressed recombination in many species.
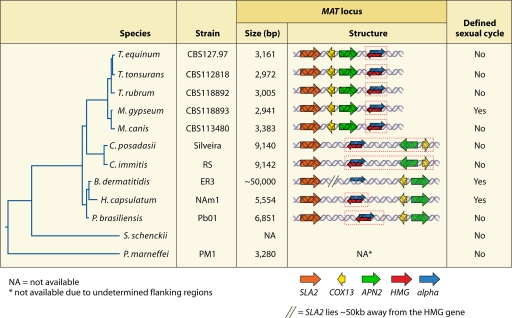
The MAT locus size for the dermatophytes ranges from 2.9 to 3.5 kb, which is relatively small compared to those for dimorphic fungi and other euascomycetes (160) (Fig. 6). In M. gypseum, the first dermatophyte fungus with a defined MAT locus, the locus is only ∼3 kb (2,941 bp for MAT1-1 and 3,184 bp for MAT1-2). However, in mating assays, this small MAT locus is fully sufficient to control the cell identity and sexual reproduction of M. gypseum (160). According to comparative analyses of several species, dermatophytes have similar MAT sizes, ranging from 2.9 to 3.5 kb.
FIG. 6.
MAT loci of the dermatophytes and dimorphic fungi. The phylogenetic organization of dermatophytes and dimorphic fungi was deduced from partial 18S rRNA gene and internal transcribed spacer (ITS) sequences. The MAT locus of Coccidioides species expanded (broken red line) by the capture of the APN2 and COX13 genes into the MAT locus, which typically flank the MAT locus in other fungal species. This analysis also revealed a unique gene arrangement of the MAT locus and flanking region of the dermatophytes in which the APN2 and COX13 genes lie on the same side as the SLA2 gene. P. marneffei also has a small MAT locus (3.3 kb) similar in size to that of the dermatophytes and much smaller than those of the dimorphic fungi. In B. dermatitidis, the SLA2 gene is located more than 50 kb away from the HMG domain gene in the MAT1-2 locus, but the size of MAT is as yet unknown, as the MAT1-1 idiomorph has not yet been defined. T. equinum, Trichophyton equinum; T. tonsurans, Trichophyton tonsurans; T. rubrum, Trichophyton rubrum; M. canis, Microsporum canis.
In dimorphic fungal pathogens, by sequence comparison between two MAT alleles, the MAT locus size is ∼3.3 kb in P. marneffei, ∼5.5 kb in H. capsulatum, ∼6.8 kb in P. brasiliensis, and ∼9.1 kb in C. immitis and C. posadasii, thus reflecting a gradual increase of the MAT locus sizes (Fig. 6). Further analysis revealed that the significantly larger MAT locus in Coccidioides species results from the capture of the flanking APN2 and COX13 genes, which now exhibit higher sequence divergence (70, 189). In the genomic sequences of three B. dermatitidis strains (ATCC 26199, sequenced at Washington University in St. Louis, MO [http://genome.wustl.edu/genomes/view/blastomyces_dermatitidis/], and strains ER3 and SLH-14081, sequenced at the Broad Institute [http://www.broadinstitute.org/annotation/genome/blastomyces_dermatitidis/MultiHome.html]), a MAT1-2 HMG gene flanked by SLA2, APN2, and COX13 was identified by using Blastn and tBlastx searches (W. Li and J. Heitman, unpublished data). The orientation of these genes is the same as that of the most closely related species, H. capsulatum. A larger distance (∼30 kb in ER3, 56 kb in SLH-14081, and 58 kb in ATCC 26199) is apparent in these B. dermatitidis strains between the SLA2 and APN2 genes, both of which often flank the MAT locus, but it is not currently possible to determine the size of MAT until the MAT1-1 sequence is determined (Fig. 6). The sequence alignment of three B. dermatitidis strains revealed a dramatically high sequence diversity. The MAT locus in S. schenckii has not been studied. Taken together, an expansion model of the MAT locus from the dermatophytes to the dimorphic fungi H. capsulatum, P. brasiliensis, C. immitis, C. posadasii has been proposed (160). Surprisingly, the MAT locus size of P. marneffei is as small (3.2 kb) as those in the dermatophytes, but P. marneffei is relatively distinct phylogenetically from the dermatophytes.
Gene organization in the MAT locus of dermatophytes and dimorphic fungi.
The MAT locus and flanking genes are a dynamic genomic region. Typically, the MAT locus of the euscomycetes is flanked by the SLA2 gene on one side and the APN2 and COX13 genes on the other (40, 289). The SLA2 and APN2 genes have the same orientation with the alpha box gene in MAT1-1 but a reverse orientation compared to those of the HMG and COX13 genes. This gene organization is useful for identifying the MAT locus in some species by degenerate PCR approaches using universal primers complementary to the SLA2 and APN2 genes (259). However, the SLA2, APN2, and COX13 genes are all linked on one side of the MAT locus of dermatophytes (Fig. 6). This uncommon gene organization of the MAT locus has been observed only for dermatophytes. This may be an evolutionary facet of the MAT locus within the dermatophytes that can serve as a marker to discriminate dermatophytes from dimorphic fungi.
In dimorphic fungi, the gene organization of the MAT locus and flanking regions resembles those of other ascomycetes. However, an exception has been found in the Coccidioides species, in which the direction of the COX13 and APN2 genes is reversed (Fig. 6) (70, 189, 259). Given that these two genes have been incorporated into the MAT locus, the change in their orientation may be associated with the capture event.
Further analysis of the B. dermatitidis MAT locus revealed a number of transposable elements (TEs) inserted between the SLA2 and HMG genes in strain SLH-14081. TEs are widespread in fungal genomes and have potential effects on genomic diversity (312). A TE has also been found in the MAT locus of C. neoformans (157). A Pogo TE has also been found inserted into MAT1-2 of P. brasiliensis (160). Asymmetrical insertions of TE into one MAT idiomorph contribute to increased sequence diversity and suppress the recombination of this important genomic region, but the influence on mating ability, if any, remains to be addressed.
SEX IN BASIDIOMYCETES
Basidiomycetes are also derived fungi that descended from a last common ancestor with the ascomycetes as the dikarya. However, unlike ascomycetes, sexual spores (basidiospores) of basidiomycetes are not enveloped in an ascus but instead are exposed to the air on the surface of the basidia (Fig. 1). Also, even though they grow vegetatively, no mechanism of asexual spore production is known. Another characteristic of sex in basidiomycetes is found in the karyotype of the sexual hyphae, in which two parental nuclei are stably maintained separately without fusion. Karyogamy occurs in the basidia, specialized cells at the apex of sexual hyphae, followed by meiosis and budding to produce long chains of haploid progeny spores. Basidiomycete sexual reproduction is also genetically governed by a MAT locus. However, compared to the ascomycetes, the basidiomycetes evolved a tetrapolar MAT locus with pheromone/pheromone receptors and transcription factors encoded by two unlinked loci. In some cases, the MAT loci have expanded, and in other species, they have even fused to evoke a tetrapolar-to-bipolar transition. In this section, we discuss the evolutionary trajectory of the MAT locus in the basidiomycetes, including how the basidiomycete MAT locus evolved as two unlinked loci; how the two unlinked loci underwent fusion, resulting in an expansion of the MAT locus; and how the evolution of the MAT locus can contribute to an understanding of sex chromosome evolution.
More than Two Mating Types in Ustilago maydis and Other Basidiomycetes
In the 1930s, Fisher argued that “the sexes are, in fact always two” (63). His observation was contradicted by “a more than two sexes system” found in caste determination systems in the harvest ants Pogonomyrmex rugosus and P. barbatus (reviewed in reference 231). Both ant species have two female queens (AA and BB genotypes) and two males (A and B) to maintain three genotypically diverged populations in the colony consisting of a queen (AA or BB), a male (A or B), or workers (AB) (reviewed in reference 7). Mating systems in basidiomycete fungi provide further insight into the evolutionary transitions to more than two mating types or sexes.
Most sexual fungi are bipolar (two mating types), in which a single locus with two alleles or idiomorphs encodes transcription factors that define cell fate. However, more than 50% of basidiomycete fungi are known to be tetrapolar, in which two unlinked loci (A and B) must differ between the two cells for sexual reproduction to occur (reviewed in reference 207). In this section, we discuss how tetrapolarity is genetically orchestrated in basidiomycetous fungi based on the Ustilago maydis MAT locus paradigm.
U. maydis belongs to the smut fungi, which are a group of plant-pathogenic fungi that infect mainly cereal crops and transform the infected tissues into galls. Interestingly, only the sexual stage is virulent to the plant host, and mating occurs preferentially in association with the plant host. U. maydis exists as a budding yeast, which is nonpathogenic. When two compatible mating type cells meet, conjugation tubes form, cell fusion occurs, and the resulting dikaryotic hyphae are infectious (reviewed in references 11 and 207). Unlike other smut fungi, U. maydis has two unlinked sex determinants, designated the a and b loci (94, 257). The a locus has two known alleles (a1 and a2), whereas the b locus is multiallelic, comprising at least 25 alleles (reviewed in reference 127). Mating occurs only between partner combinations compatible at both loci. For example, a strain with the a1 allele is compatible only with isolates with the a2 allele, and one with the b1 allele is compatible with ones with b alleles other than with b1 (Table 1). Both alleles must differ to complete sexual reproduction; thus, a1b1 mates with a2b2 but not with a1b2 or a2b1. Because any productive cross yields meiotic progeny with four different a b combinations (i.e., a1b1 × a2b2 yields a1b1, a1b2, a2b1, and a2b2), this type of mating system is termed tetrapolar.
TABLE 1.
Mating compatibility in U. maydisa
| Mating type | Compatibility with allele |
|||
|---|---|---|---|---|
| a1b1 | a1b2 | a2b1 | a2b2 | |
| a1b1 | − | − | − | + |
| a1b2 | − | − | + | − |
| a2b1 | − | + | − | − |
| a2b2 | + | − | − | − |
−, no mating; +, mating.
The b locus of U. maydis carries two homeodomain transcription factors (bW and bE) of different functional classes (HD2 and HD1, respectively) that are divergently transcribed (Fig. 7A). With only an ∼200-bp intergenic space between them, the bW and bE genes are not readily separated by meiotic recombination (reviewed in reference 127). For example, the bW1 allele is linked to bE1, and thus, it is not expected that a cross between b1- and b2-type cells will generate haploid progeny with a b locus with the bW1 and bE2 alleles (thus avoiding the generation of progeny with self-compatible bE-bW combinations). After the fusion of two compatible cells, bW from one mating type binds to the compatible bE from its partner to form a heterodimer. The bW-bE complex (for example, bW1-bE2 or bW2-bE1) then activates subsequent developmental steps, including invasive hyphal growth and tumor formation in plant hosts (300). The bE/bW heterodimeric complex governs the dikaryotic state and orchestrates the postfusion stages of sexual reproduction by controlling the expression of key target genes, both directly and indirectly, via other induced transcription factors. Expression analysis by Bakkeren and colleagues resulting from the enforced expression of bE/bW identified 350 differentially regulated genes, including genes that are directly and indirectly regulated (8). Key targets include the zinc finger transcription factors Biz1 and Rbf1 and the homeodomain protein Hdp2. Notably, Rbf1 was subsequently found to play a key role in promoting the expression of genes indirectly controlled by bE/bW. These findings reveal that a homeodomain-zinc finger transcription factor relay network underpins gene regulatory events that are central to the dimorphic transition from yeast to hyphal growth and other genetic, cell biological, and morphological events that occur in the pathogenic dikaryotic state.
FIG. 7.
MAT loci of the tetrapolar basidiomycete U. maydis and bipolar basidiomycetes U. hordei, M. globosa, and C. neoformans. (A) The pheromone/pheromone receptor and transcription factor loci are unlinked in U. maydis. There are two known a loci, containing the mfa and pra genes. Two representative b loci (of an estimated ∼25 loci) are presented, which encode two divergently transcribed homeodomain proteins (HD1 and HD2). (B) However, the pheromone/pheromone receptor and transcription factor genes are linked in the U. hordei, M. globosa, and C. neoformans genomes. Many additional genes have been incorporated into the C. neoformans MAT locus. (The MAT locus alleles of the serotype D strains JEC21 for the α mating type and JEC20 for the a mating type are presented.)
The U. maydis a locus encodes elements of the pheromone response pathway (pheromones and their receptors) mediating cell-to-cell recognition. The a locus regulates cell fusion between two compatible haploid cells (257) and the maintenance of hyphal growth (12). Two distinct a1 and a2 alleles share an architecture of genes encoding the pheromone (mfa1) and pheromone receptor (pra1) (Fig. 7A), with the sequences of the genes being diverged between alleles (24). The a1 locus spans ∼4.5 kb containing mfa1 and pra1. The a2 allele harbors mfa2 and pra2 and three additional genes, lga2, rga2, and a pheromone pseudogene (Fig. 7A) (292). The lga2 and rga2 genes have been implicated in mitochondrial inheritance and function during sexual reproduction and infection (25, 56, 186).
Other basidiomycetes also harbor a tetrapolar mating system. In the mushroom fungus Coprinopsis cinerea, the A locus carries two divergently transcribed homeodomain genes, and the B locus encodes pheromones/pheromone receptors (38). Another mushroom, Schizophyllum commune, also contains A and B loci encoding homeodomain proteins and pheromones/pheromone receptors, respectively (251). U. maydis is closely related to another smut fungus, Sporisorium reilianum, both of which infect maize. Recent studies have characterized the mating type loci of S. reilianum, providing further insights into MAT evolutionary trajectories. Like U. maydis, S. reilianum harbors a tetrapolar mating system in which the b locus encodes two divergently transcribed homeodomain proteins (bE and bW), and at least five alleles are extant in the population (265). Most interestingly, the a locus encoding pheromones/pheromone receptors has three extant alleles, in contrast to the biallelic system present in U. maydis.
Thus, two models could apply. In one model, an ancestral a locus was multiallelic and collapsed to the triallelic form extant in S. reilianum and even further to the biallelic state extant in U. maydis. An alternative model is that the ancestral state was biallelic, and this has expanded by the incorporation of an additional novel allele to a triallelic form in S. reilianum and even further to the dramatic multiallelic state in other fungi, such as S. commune and C. cinerea. Analysis of other closely and more divergently related fungi will allow these models to be tested further.
Notably, each a allele in S. reilianum encodes two pheromones and one receptor, enabling each of the three a mating types to signal to and respond to each of the other two possible mating partners but not themselves. One feature of the U. maydis a2 allele involving the presence of one active variant and one pseudogene variant of the mating pheromone suggests that the ancestral state of the two species might have involved at least a triallelic a locus system, with the loss of one allele (a3) and the loss of one of the two pheromone genes from the remaining two extant alleles (a1 and a2) in U. maydis. If so, this represents transitions from a multiallelic, ancestral tetrapolar mating system to first a multiallelic, triallelic tetrapolar system and then to a multiallelic, biallelic tetrapolar form. Notably, these transitions serve to reduce outcrossing potential (from >99% to 50%) and retain the restriction to inbreeding (from 50% to 25%) exhibited by tetrapolar mating systems (111). For some features, these transitions mirror the transitions that have occurred in the emergence of the pathogenic Cryptococcus species complex and suggest that pathogenic microbes might in general transition from outcrossing to inbreeding/self-fertile modes of reproduction concomitant with their emergence as successful pathogens of plants or animals.
In tetrapolar mating systems, there is a 25% chance of mating compatibility among any given progeny from a sexual cross, whereas there is a 50% chance in a bipolar system. Therefore, the tetrapolar mating system may contribute to reduce inbreeding that could be detrimental to the fitness of a species under certain selective pressures. Furthermore, the multiallelic mating locus confers a greater level of outcrossing (99.9% versus 50%). For example, C. cinerea has more than 200 different alleles resulting in thousands of potential mating types (37, 251). Thus, basidiomycetes established a genetic basis to achieve highly efficient outcrossing through mating locus evolution.
Transition from Tetrapolar to Bipolar and Expansion of the MAT Locus in U. hordei and Cryptococcus Species
The bipolar mating system is also found in the basidiomycetes, in which pheromones/pheromone receptors and homeodomain loci are linked, whereas in tetrapolar species, they are unlinked and segregated independently. Two evolutionary scenarios might be hypothesized: (i) a transition from a bipolar system to a tetrapolar one or (ii) a transition to a bipolar system from a tetrapolar one (251). Because ascomycetes and zygomycetes (see the section on zygomycetes below) have bipolar mating systems and groups of basidiomycetes retain bipolarity, the most parsimonious model is one in which bipolarity is the ancestral sexual form (68). Subsequent tetrapolar systems evolved from bipolar species, and in some examples, transitions back to bipolar mating may have also occurred. Thus, within the basidiomycetes, bipolarity could be both an ancestral and a derived state (68, 207, 251). Tetrapolarity is known only for basidiomycetes and thus may have arisen only once. A detailed model of how tetrapolar sex determination might have evolved was presented by Fraser et al. (69). Given the expansion of the MAT locus and the linkage of previously unlinked sex determinant genes, the secondary transition to a bipolar system (see the next two paragraphs and Fig. 8) serves as a model for sex chromosome evolution in multicellular eukaryotes. The MAT loci of Ustilago hordei and C. neoformans, basidiomycetes with bipolar mating systems, support this view.
FIG. 8.
Model of transition to extant bipolar systems from an ancestral tetrapolar system in basidiomycete MAT loci. Linkage of the a and b loci generates an extended MAT locus in the U. hordei and M. globosa genomes. In the C. neoformans genome, gene acquisition occurred into the MAT locus, and the two loci were then linked, generating an intermediate tripolar system in which the a and b loci were linked in one mating type MAT allele with the other mating alleles unlinked, followed by gene conversion and chromosome rearrangement to generate the extant extended bipolar MAT locus.
U. hordei is a dimorphic basidiomycete that infects mainly barley, causing covered smut. The role of its sexual cycle in pathogenesis is similar to that of U. maydis as outlined above. However, unlike U. maydis, which has a tetrapolar mating system, U. hordei has a bipolar system despite the close pathogenetic relationship between the two species. In the genome of U. hordei, the two mating type loci, a and b, are linked and segregate together (9, 153). Surprisingly, the MAT locus of U. hordei is expanded up to ∼500 kb, with the a locus at one end and the b locus at the other, whereas the U. maydis MAT loci consist of ∼4 kb for the b locus and ∼4.5 to 8 kb for the a locus, and the a and b loci lie on different chromosomes (Fig. 7B). This expansion of the bipolar U. hordei MAT locus resulted in the incorporation of additional DNA occupying a significant portion of the chromosome. Recently, the whole genome sequence of another basidiomycetous fungus, Malassezia globosa, causing dandruff, provides an independent example in which the tetrapolar MAT loci have become linked in what may represent a bipolar mating system (Fig. 7B) (319).
Another tetrapolar-to-bipolar transition of the MAT locus occurred in the human-pathogenic fungi Cryptococcus neoformans and C. gattii. C. neoformans infects immunocompromised and, sometimes, immunocompetent individuals and causes meningoencephalitis. C. neoformans is a dimorphic fungus; it produces exclusively budding yeast cells during vegetative growth and infection, whereas hyphal growth is observed during sexual development. The sexual cycle of C. neoformans was first defined by Kwon-Chung (143-145). C. neoformans has two mating types, a and α, and similar to S. cerevisiae, when two compatible mating type cells meet, mating initiates. During the response to pheromone, yeast cells of both mating types undergo morphological changes (conjugation tube formation or enlargement via isotropic growth), undergo cell-cell fusion, and then produce dikaryotic hyphae. Dikaryotic hyphae are characteristic of basidiomycetes, as discussed above in reference to the U. maydis sexual cycle. In response to unknown signals, the hyphae grow aerially and form basidia at the apex of the aerial hyphae. Inside a basidium, the opposite-mating-type nuclei fuse (karyogamy) to form a diploid, followed by meiosis. Four chains of haploid spores are produced by mitosis and basipetal budding from the basidial surface (Fig. 1).
The MATα locus of C. neoformans was identified by a differential cloning approach and initially found to contain a pheromone gene (MFα1) (204). Further study revealed that the C. neoformans MAT locus spanned at least ∼50 kb and contained additional genes (Fig. 7) (129). Subsequent work discovered that the MAT locus harbors a homeodomain gene and that the pheromone/pheromone receptor cluster is linked to this homeodomain gene spanning ∼100 to 120 kb (Fig. 7B) (115, 157). Interestingly, compared to other basidiomycetes, the C. neoformans MAT locus alleles have a different paradigm for the homeodomain genes, in which the MATa locus encodes only an HD2 factor (Sxi2a), while the MATα locus encodes only an HD1 factor (Sxi1α), in contrast to other basidiomycete MAT loci with two divergently transcribed homeodomain genes (HD1 and HD2) (114). Another significant characteristic is that the Cryptococcus MAT locus contains many additional genes linked to the pheromone/pheromone receptor and homeodomain genes. These genes include those encoding components of the mitogen-activated protein (MAP) kinase pathway and meiosis and sporulation (sexual spore) genes (Fig. 7B) (66, 157). Taken together, the evolutionary trajectory of the Cryptococcus MAT locus is as follows: (i) new genes were acquired into ancestral tetrapolar loci; (ii) a chromosomal translocation linked the two loci in one mating type, yielding an intermediate tripolar state; and (iii) recombination between the two loci completed the formation of a bipolar-mating-type system (Fig. 8). The formation of an extended MAT locus (>100 kb) in the C. neoformans genome led to a transition from tetrapolar to bipolar, forming a gene cluster that functions in both a-α opposite-sex and α-α same-sex mating (see the section on same-sex mating below) and virulence.
Notably, the Cryptococcus MAT locus is unique among fungi in that it contains five essential genes, including two ribosomal genes. The presence of these embedded essential genes likely punctuated MAT evolution by constraining gene deletion to only those events that would not result in inviability. The transitions from a multiallelic to a biallelic MAT locus and tetrapolar to bipolar and the loss of one paired homeodomain gene or the other all reduce outcrossing, promote inbreeding/selfing, and may therefore have contributed to the success of this pathogen species complex.
Features of the MAT locus expansions in basidiomycete fungi mimic the evolutionary path of the sex chromosomes of plants and animals. Genetic recombination between homeodomain and pheromone/pheromone receptor loci of U. hordei and C. neoformans is suppressed, conferring a stringent bipolar mating system. In the anther smut fungus, Microbotryum violaceum, the emergence of sex chromosomes (chromosomes containing the MAT locus) further extends this phenomenon, where the nonrecombining region across the MAT locus spans 25% (∼1 Mb) of the chromosome (81, 299). Karyotype analyses and mating assays identified that the fungus's sex chromosomes exhibit size dimorphism, ranging from 2.2 to 4.2 Mb, and furthermore, no recombination was found across the MAT locus in these chromosomes (107, 108). A chromosome containing the mating type locus of the ascomycete fungus Neurospora tetrasperma also displays an absence of recombination (74, 195). This is a common characteristic of mammalian sex chromosomes, in which recombination between the X and Y chromosomes is suppressed and restricted to only the pseudoautosomal regions (84). Thus, the evolution of MAT loci in fungi provides insight into how mammalian sex chromosomes might have evolved (68).
There is a series of conserved features of the evolution of mating type and sex chromosome evolution observed based on related studies of fungi, plants, and animals (67). These features involve a common set of properties. First, the sex determinants first emerged on an autosome and then served as the starting point upon which a larger nonrecombining region of the genome originated. Second, the mating type locus and sex chromosomes are comprised of strata of genes of distinct evolutionary ages based on when they were acquired on the locus or chromosome. Third, there is a coherence of gene function in these sex-specific regions of the genome, involving pheromone production and sensing in fungi and spermatogenesis in mammalian genes located on the Y chromosome. Fourth, sex-specific genes in both fungi (the pheromone genes) and the mammalian Y chromosome (genes involved in the production and function of sperm) are arranged in inverted palindromic repeats, enabling repair via intrachromosomal recombination for these genes that do not have a partner for repair on the other allele or chromosome. Fifth, these regions of the genome are sheltered from recombination, and as a consequence, repetitive sequences and transposons accumulate, leading to a marked propensity to undergo rearrangements, deletions, inversions, and gene conversion. These examples of convergent evolution to similar patterns of sex determination in divergent lineages, including fungi, birds, insects, and plants, illustrate the key underlying principles applicable to all eukaryotes.
Sex and the MAT Locus in Sibling Species of Cryptococcus
The model proposed for the evolution of the Cryptococcus pathogenic species complex involving an ancestral tetrapolar mating system, the acquisition of additional genes related to mating and meiosis, and collapse to a tripolar intermediate and then a bipolar system has been addressed by two experimental approaches. First, to model the proposed tripolar intermediate and ancestral tetrapolar state, the SXI1α and SXI2a genes were deleted from MAT and relocated to an unlinked genomic location (110). The resulting strains were fertile in genetic crosses and able to undergo sexual reproduction in patterns reflecting how the tripolar and tetrapolar ancestors would have mated. Moreover, these studies provided experimental evidence that the tripolar intermediate might have led to the production of a higher level of self-sterile, self-fertile, or inviable progeny and therefore might have been at a selective disadvantage, perhaps facilitating the transition from a tetrapolar to a bipolar system.
The second approach to address the proposed MAT evolutionary model involves comparative genomics of other related but diverged species in both the sensu stricto and sensu lato complexes related to the pathogenic species complex. The species analyzed include those most closely related, Cryptococcus amylolentus, Tsuchiyaea wingfieldii, and Filobasidiella depauperata, and those more distantly related, including Cryptococcus heveanensis and Tremella mesenterica (57, 255a). None of these related species are pathogens, but they can serve as evolutionary windows to provide insights into how MAT might have been configured in the last common ancestral nodal points (Fig. 9). The most distantly related of these, T. mesenterica, has a well-defined tetrapolar mating system, supporting this assignment as the ancestral state. This line of investigation has resulted in two key findings. First, extant sexual cycles have been discovered for both C. amylolentus and C. heveanensis (196a; K. Findley and J. Heitman, unpublished results). Second, analysis of the MAT locus of both species revealed that two large gene clusters have been assembled, corresponding to the homeodomain and pheromone/pheromone receptor loci, into which many additional genes have been recruited, similar to the pathogenic species complex MAT locus. Moreover, in both cases, these MAT-related gene clusters are not fused. Thus, both species represent extant examples of species that closely mirror proposed evolutionary intermediates in the MAT evolution model. In other words, they represent fungal genomic equivalents of “transitional fossils.”
FIG. 9.
Sex recognition systems and evolutionary trajectory in C. neoformans sibling species. The clade of pathogenic Cryptococcus species is characterized as having bipolar mating systems. Only one of the genes for HD1 or HD2 transcription factors is found in the a and α MAT alleles. However, the C. amylolentus, T. wingfieldii, and C. heveanensis MAT loci contain both HD1 and HD2 genes, suggesting that two paired, divergently oriented HD1 and HD2 genes in the MAT locus are the ancestral arrangement. Interestingly, F. depauperata grows only as a filamentous form resembling the Cryptococcus sexual state, indicating that this fungus may be an obligate sexual species. Fungi in the Tremellales group retain the tetrapolar mating system. One species in the Kwoniella clade is sexual and bipolar or tetrapolar. Thus, bipolarity may have emerged during the origin of the Kwoniella and Filobasidella clades from a common tetrapolar ancestor. The tree was drawn based on six-gene MLST as described previously by Findley et al. (57). C. dejecticola, Cryptococcus dejecticola; B. dendrophila, Bullera dendrophila; K. mangroviensis, Kwoniella mangroviensis. C. bestiolae, Cryptococcus bestiolae; T. globispora, Tremella globispora; C. humicola, Cryptococcus humicola.
LESSONS FROM BASAL FUNGI: ZYGOMYCETES AND MICROSPORIDIA
Sex in Zygomycetes
Zygomycetes are one of the early-diverged groups within the fungal kingdom and, as such, are known as the basal fungi, together with the chytridiomycetes. The zygomycetes are characterized by forming coenocystic (aseptated) hyphae, nonmotile cells, and nonenveloped zygospores for sexual reproduction. The phylum Zygomycota includes nine orders (20), eight of which are known to sexually reproduce (119). The order Mucorales is the most well-studied group. These fungi propagate through both asexual and sexual life cycles. In the asexual life cycle, spores germinate to form complex mycelia, and aerial hyphae then emerge, followed by the formation of sporangia at the apex. The sporangia harbor asexual spores called sporangiospores.
In the sexual life cycle, hyphae from two different mating types, (+) and (−), fuse to form zygospores, which later send a hypha to form a sporangium containing spores at the apex. Sexual development is mediated by a zygomycete-specific pheromone: trisporic acid. As illustrated in Fig. 10, precursors of trisporic acid are produced from β-carotene in each (−) and (+) mating type, and delivery to the opposite mating type is necessary to complete trisporic acid synthesis. In brief, (+) strains produce 4-dihydromethyl trisporate, which is delivered into (−) strains. Interestingly, the key enzyme for this reaction, 4-dihydrotrisporin-dehydrogenase, exists in both mating types, and only enzyme activity is differentially regulated in the Mucorales fungus Mucor mucedo (305). The (−) strains then produce methyltrisporate from 4-dihydromethyl trisporate and finally form trisporic acid. On the other hand, (−) strains produce trisporin and trisporol, both of which are exported to (+) cells to produce trisporic acid (Fig. 10) (313). Trisporic acid triggers the formation of specialized hyphae supporting the zygospores, called the zygophore, followed by the formation of a zygote through hyphal fusion between two opposite mating types. Meiosis occurs in the zygote, and finally, zygospores are formed.
FIG. 10.
Pathway of trisporic acid synthesis in zygomycetes. In the (+) strains, 4-dihydromethyl is produced from β-carotene, secreted, and taken up by the (−) strains, where it is finally converted to trisporic acid. Trisporin and trisporol are produced in the (−) strains, exported, and then imported by the (+) strains and converted to trisporic acid.
sex Locus of Zygomycetes
More than a century ago, Blakeslee described two mating types in zygomycetes (23); however, subsequently, the genetics of sex in zygomycetes were not as extensively studied as the two more derived fungal groups, the ascomycetes and basidiomycetes (119). A recent study identified the sex locus, orchestrating sexual development in Phycomyces blakesleeanus, a zygomycetous fungus (120). The sex locus gene cluster includes a putative triose phosphate transporter (TPT) homolog, SexP/M, and RNA helicase genes. The sexP and sexM genes encode HMG proteins for the plus (+) and minus (−) mating types, respectively, which may serve differentially as transcription factors during sexual reproduction (Fig. 11).
FIG. 11.
sex loci in the genomes of three zygomycetes. The sex loci in the three zygomycetes M. circinelloides, P. blakesleeanus, and R. oryzae are within a gene cluster encompassing the TPT, HMG, and RNA helicase genes. The sex loci encode idiomorphic HMG transcription factors, SexP and SexM, for (+) and (−) mating types, respectively. The P. blakesleeanus (+) sex locus contains a repetitive element, and the R. oryzae (+) sex locus carries an additional ORF in the gene cluster. The orientation of the sex genes is divergent in P. blakesleeanus and the TPT genes in the R. oryzae sex locus alleles are inverted compared to those of M. circinelloides and P. blakesleeanus. Gray boxes indicate the extent of the sex locus. Note that the promoter of the TPT gene in M. circinelloides is within the border of the sex locus. BTB, bric-a-brac, tramtrack, and broad complex.
The evidence that the sex locus governs the establishment of sexual identity and orchestrates sexual reproduction of zygomycetes is based on several converging lines of evidence (120, 154). First, the sexM and sexP genes are expressed during mating, and only one is found in the genome of any given haploid isolate from the population. Second, isolates with the sexP gene mate as the (+) mating type, those harboring the sexM gene mate as (−) isolates, and no mating was observed between two (+) or two (−) isolates. Third, rare strains that are disomic for the sexM and sexP genes (aneuploids that harbor two copies of the sex chromosome), isolated following genetic crosses, are self-fertile, produce spiraling zygophore-like structures, and produce an unusual “fluffy” colony morphology. These findings are analogous to data from previous studies of other fungi in which the introduction of the opposite-mating-type MAT allele induces homothallic self-fertile behavior. One notable example involves C. neoformans haploid α strains expressing the a sex determinant Sxi2a or haploid a strains expressing the α sex determinant Sxi1α, both of which are self-fertile (114, 115). Fourth, genetic crosses and meiotic mapping provide additional evidence linking the sex locus to sex determination and sexual reproduction. Following genetic crosses, the sexM gene is linked to the (−) mating type and sexP is linked to the (+) mating type in meiotic progeny. Moreover, an analysis of recombinant progeny with crossovers near the sex locus identified based on RFLP mapping delimits the sex-specific region of the genome to a 34-kb interval encompassing the sex locus in the ∼65-Mb genome of P. blakesleeanus. Finally, the finding that a similar region was identified to be sex specific and linked to sexual identity in two related but diverged species, Mucor circinelloides and Rhizopus oryzae (120, 154), provides compelling evidence for the identity and function of sex in cell identity and sexual reproduction. The disruption of the sexM gene in M. circinelloides results in a failure of zygospore formation in a cross with (+) strains (S. C. Lee, C. H. Li, M. Cervantes, R. M. Ruiz-Vazquez, S. Torres-Martinez, and J. Heitman, unpublished data), further supporting that the sex genes are key regulators of zygomycete mating.
How do SexM and SexP function? The two are diverged paralogs and members of the HMG domain family of transcription factors. Three models have been considered for their action, all of which center on the regulation of key target genes (Fig. 12). First, SexM and SexP might converge to regulate the same target genes but via distinct promoter regulatory elements in independent DNA-protein complexes (model I). Second, these proteins may serve a role in defining cell type identity prior to cell fusion and also in the zygote formed after cell fusion. Analogous to the homeodomain determinants of the dikarya, SexM and SexP might form a heterodimeric complex to regulate key target genes (model II). Third, SexM and SexP might regulate complementary target genes that themselves interact, directly or indirectly, to define diploid fate (model III). Approaches to address these and other models will involve gene deletion and heterologous complementation studies, protein-protein and protein-DNA interaction studies, and expression analysis.
FIG. 12.
Models for the mode of action of SexP and SexM. First, SexP and SexM may function in distinct promoter areas with different regulatory complexes to converge on common target genes (model I). Second, SexP and SexM may interact to form a heterodimer to function to transcribe mating genes (model II). Third, SexP and SexM may regulate different subsets of mating genes (model III). Note that SexP and SexM likely function both pre- and postfusion.
The fact that the zygomycetes utilize solely HMG transcription factors for sex determination is coincident with multicellular eukaryotes, including humans, for which the HMG factor Sry governs sexual identity (269). This fungal group diverged from a common ancestor shared with the metazoans at an early point (10, 301) and, accordingly, could share evolutionary relationships with the metazoans not shared with the dikarya (124, 178) (Fig. 1). Therefore, zygomycetes might have retained an ancestral sex recognition system that could reveal early patterns of sex evolution. Interestingly, although the architecture of sex synteny is similar, there are several differences among the sex loci within the three zygomycetes: (i) the orientations of the genes differ in the three species, especially in P. blakesleeanus, in which sexM and sexP are oppositely oriented; (ii) the R. oryzae genome harbors an additional ORF between the TPT and SexM/P genes (A. Gryganskyi, A. P. Litvintseva, S. C. Lee, G. Bonito, I. Anishchenko, R. Vilgalys, and J. Heitman, unpublished results); and (iii) the P. blakesleeanus sex locus harbors a repetitive element (Fig. 11). These differences indicate that the zygomycete sex locus might be a hot spot for recombination and gene conversion.
The presence of a repetitive element, recombination, and gene conversion in the zygomycete sex loci contributes to elucidate early stages in sex chromosome evolution (see Fig. 14), in which (i) a gene for sex determination arises on an autosomal chromosome as proposed by Ohno (222); (ii) a gene inversion of one allele blocks recombination, enabling the inverted gene pair to diverge to become idiomorphic; (iii) a repetitive element (blue box) emerges on the primitive sex chromosome; and (iv) a transposition of the repetitive element occurs into one sex allele, followed by additional inversions caused by the presence of the repetitive elements at different loci, resulting in the expansion of the sex locus throughout a significant portion of the chromosome. This hypothesis is supported by the sexP and sexM gene orientations and the presence of a repetitive element in the P. blakesleeanus genomes. However, the fact that the M. circinelloides and R. oryzae sexP and sexM genes lie in the same orientation could be interpreted by a different hypothesis in which the order of the divergence and inversion steps is reversed with respect to each other (divergence-inversion model) (Fig. 13) (120, 154).
FIG. 14.
The microsporidian sex-related locus. (A) Differential interference contrast (DIC) and transmission electron microscopy (TEM) images of an E. cuniculi spore. A polar tube is everted from the spore. The spore contains a coiled polar tube. (B) sex-related loci observed for four different microsporidian species. The E. cuniculi and A. locustae genomes contain a syntenic region encompassing TPT (green), HMG (red), and RNA helicase (blue) genes similar to that observed at the zygomycete sex loci (Fig. 11). The E. cuniculi sex-related locus has an additional weak homology HMG gene (purple). All sex-related loci from the four different microsporidian species have a conserved hypothetical protein gene (yellow). Two other microsporidians, E. bieneusi and N. ceranae, have an unlinked RNA helicase gene. Only microsporidia and zygomycetes share the synteny of the TPT, HMG, and RNA helicase genes compared to other fungi for which complete genome sequences are available.
FIG. 13.
Early steps in sex chromosome evolution. The P. blakesleeanus sex locus supports the hypothesis posited by Ohno (222): a sex determinant gene arises on an autosome and undergoes gene inversion followed by gene divergence in the two alleles (top). A repetitive element is inserted at the locus and elsewhere on the proto-sex chromosome. The sex locus expands to form an early sex chromosome. However, the sex loci in M. circinelloides and R. oryzae present an alternative scenario in which sex genes in the same orientation could be an ancestral character, and therefore, gene inversion might follow rather than precede gene divergence (bottom).
The zygomycete genomes provide insights into early stages of sex chromosome evolution. However, further studies are needed to understand sexual reproduction more broadly within this understudied fungal lineage. For example, a connection between the sex locus and trisporic acid production is not yet known. The possible roles of the linked TPT and RNA helicase genes in sexual reproduction, if any, are also not yet known. Finally, the precise roles of SexM and SexP in establishing cell type identity (prefusion) and in orchestrating sexual reproduction (postfusion) remain to be elucidated.
Microsporidia, Obligate Intracellular Pathogens
Microsporidia are single-celled obligate intracellular pathogens that infect a wide range of animals, from nematodes to humans and some protists (134, 287). The infection cycle of these pathogens includes several steps that are distinguished from other infectious agents (320). Microsporidian dormant spores contain a cylinder-like tube, called the polar tube, which originates from an anterior part of the spores and is coiled within the spore to serve as a “tunnel” for moving infectious material into the hosts. When stimulated upon encountering and recognizing the host, the spores rapidly uncoil and evert the polar tube, which penetrates the host cell membrane and serves as a conduit to deliver the sporoplasm while the spore wall remains outside (Fig. 14A). Inside the host, microsporidian cells lacking a cell wall undergo replication, producing cell wall-deficient progeny called meronts. Cell walls are eventually built surrounding the meronts to form mature spores, which are released from the host cells.
The main site of infection by microsporidia involves intestinal cells. Insects are well-known microsporidian hosts (16). Nosema apis and Nosema ceranae infect honeybees, causing nosemosis, resulting in a significant loss of honey production. Interestingly, another insect microsporidian, Antonospora locustae, is used as an insecticide to control harmful locusts and grasshoppers (175). A dozen microsporidian species are known to infect humans, mainly immunocompromised hosts, causing severe and long-lasting diarrhea; in some cases, infections of immunocompetent individuals have been observed (48). Microsporidian infections of humans were recently recognized as an emerging infectious disease, and both the NIH and the Environmental Protection Agency (EPA) have issued a caution on these pathogens (reviewed in reference 49).
Among microsporidia, 1,300 species in 150 genera have been reported (310). However, the phylogenic placement and evolutionary origin of microsporidia have been ambiguous since the first discovery of microsporidian species in 1857. The first identified microsporidian species, Nosema bombycis, which infects silkworms, was placed within the schizomycetes, an artificially defined group based on microscopic morphology (reviewed in reference 132). Later, their obligate parasitic life cycle brought microsporidia aligned with the cnidosporidia within protists. One interesting characteristic of microsporidian species is that they were thought to lack mitochondria, which led to microsporidia being considered an ancient and early-diverged eukaryote before the origin of mitochondria (298). However, the presence of the mitosome, a remnant of mitochondria, challenged their position as a primitive eukaryote (309). Instead, phylogenic analyses revealed that they are related to fungi (53, 124, 133). Recent findings that microsporidia share a synteny for two rRNA genes (RPS9 and RPL21) that is conserved exclusively within fungal lineages also strongly suggest that microsporidia are fungi (154, 155).
Molecular phylogenetic analyses based on the concatenation of eight genes suggested that microsporidia are closely related to ascomycetes and basidiomycetes (79, 284). In contrast, Keeling analyzed α- and β-tubulin genes and positioned microsporidia within the zygomycetes (131). Microsporidia are highly diverged organisms with a high level of sequence variation and a loss of many genes; thus, sequence-based phylogenetic reconstruction is problematic and might be insufficient to unambiguously resolve their origin. Free from being dependent upon sequence-based phylogeny, Lee et al. reported that microsporidia share gene order exclusively with zygomycetes, further supporting that microsporidia and zygomycetes are diverged from a common ancestor (154). Another striking feature that supports the zygomycete origin hypothesis is that microsporidian genomes harbor a similar genomic locus as that seen in the sex locus of zygomycetes (Fig. 11 and 14B), where genes for a putative TPT, HMG protein (SexP/M), and RNA helicase form a cluster. Genomes of two human-pathogenic microsporidia, Encephalitozoon cuniculi and Enterocytozoon bieneusi, carry genes for TPT, HMG, and RNA helicase in a single locus. The genomes of the other two insect-pathogenic microsporidia, Antonospora locustae and Nosema carenae (155a), also harbor syntenic TPT and HMG genes with an unlinked RNA helicase gene (Fig. 14B). Another ORF for a hypothetical protein is common to the four microsporidia in the putative sex-related locus. The syntenic gene cluster for TPT, HMG, and RNA helicase is unique, thus far, to the zygomycetes and microsporidia (154), further supporting the zygomycete origin hypothesis. These findings support models in which the microsporidia evolved from ancestral basal fungi.
sex-Related Locus and Meiotic Genes in Microsporidian Genomes
A few previous reports suggested that some microsporidian species might undergo sexual reproduction according to morphological inferences (97, 279). However, a bona fide sexual cycle has not been verified molecularly or genetically. Thus, it is unclear whether microsporidia have an extant sexual cycle.
Microsporidia have a syntenic sex-related locus that mimics the zygomycete sex locus (see above). During evolution, zygomycetes retained an HMG protein as a major sex determinant (120) that had been adapted in the common ancestor of opisthokonts, including the kingdoms Fungi and Animali (also see Fig. 19). The syntenic sex locus containing an HMG gene with TPT and RNA helicase genes is found exclusively in the genomes of zygomycetes and microsporidia, where the overall architecture of the locus is conserved within the two lineages despite minor divergence, for example, an unlinked RNA helicase gene in some microsporidian genomes (Fig. 11 and 14). This observation could be a strong harbinger of microsporidian sex.
FIG. 19.
Evolution of transcription factors encoded by the fungal MAT locus. Zygomycetes, representative basal fungi, utilize an HMG transcription factor for sexual recognition, which parallels the human sex determination system with the Sry HMG protein. The common ancestor to the opisthokont clade may have had an HMG protein as a sex determinant (52). Alternatively, given the ubiquity of both HMG and homeodomain (HD) factors, there may have been two ancestral sex-determining systems that competed for preeminence, or in some cases, such as C. albicans, both remain resident at the MAT locus. There were multiple losses of HMG as a sex determinant, and in some lineages, α domain and/or HD transcription factors were adopted. Basidiomycetes lost the MAT-encoded HMG but acquired HD factors as sex determinants. In addition, they adapted multiallelism (e.g., U. maydis and C. cinerea). However, some basidiomycetes have returned to the biallelic bipolar state (e.g., C. neoformans, U. hordei, and M. globosa). The α domain proteins were likely incorporated into MAT during the evolution of the ascomycete MAT loci but also share sequence identity with HMG domains and may therefore be distantly related. In Saccharomyces spp., the HMG protein no longer functions as a sex determinant, and the mating type switching system evoked by the Ho endonuclease was recently acquired within this group. In contrast, several species of the Candida CTG clade retained the HMG gene in the MTL locus, and thus, three classes of transcription factors are present (HMG, HD, and alpha box), and no silent cassette or mating type switching occurs.
It was suggested that genomics can inform us about the possible existence of sex in organisms (267). There is a set of genes that specifically function during meiosis, especially genes encoding Spo11, Rad50/Mre11, Dmc1, Msh4/5, and Mlh1, that were identified as “core meiotic recombination machinery” (see also the section on Candida above) (247, 297). Therefore, it was hypothesized that the presence of these genes identifies an organism as a sexual species or at least that its immediate ancestors were sexual species. Microsporidian genomes contain this meiosis-specific machinery (Table 2) (154, 187). Genes for Rad50 and Mre11 are present in all three microsporidian species, including E. cuniculi, A. locustae, and E. bieneusi. MSH4 is present only in E. bieneusi, whereas no ortholog of MSH5 was found. In addition to MSH5, DMC1 is absent in the three microsporidian genomes. Dmc1 is also absent in the Drosophila melanogaster and Caenorhabditis elegans genomes; either functional homologs with less sequence similarity could be present in their genomes, and this might also apply to microsporidia, or Dmc1 is simply dispensable for meiosis in these species. Other meiotic genes are also present in microsporidian genomes (Table 2). Thus, it is hypothesized that microsporidia have an extant sexual cycle.
TABLE 2.
Meiosis genes found in three microsporidians, E. cuniculi, A. locustae, and E. bieneusia
| Gene | Presence of gene in: |
||
|---|---|---|---|
| E. cuniculi | A. locustae | E. bieneusi | |
| Spo11 | + | + | − |
| Rad50 | + | + | + |
| Mre11 | + | + | + |
| Dmc1 | − | − | − |
| Rad51 | + | + | + |
| Msh4 | − | + | + |
| Msh5 | − | − | − |
| Hop1 | + | + | − |
| Hop2 | − | − | − |
| Mnd1 | + | + | + |
| Rad52/22 | + | + | − |
| Msh2 | + | + | + |
| Msh6 | + | + | + |
| Mlh1 | + | + | + |
| Mlh2 | − | + | − |
| Mlh3 | − | − | − |
| Pms1 | + | + | + |
| Mer3 | − | − | − |
| Smc1 | + | + | + |
| Smc2 | + | + | + |
| Smc3 | + | + | + |
| Smc4 | + | + | + |
| Smc5 | + | + | + |
| Rad18 | + | + | + |
| Rad21 | + | + | + |
| Rec8 | + | + | + |
| Pds5 | − | − | − |
| Scc3 | − | − | − |
A recent study has revealed a novel sexual cycle of Giardia intestinalis, a human-parasitic protozoan, which occurs during infection of host cells (242). This modified sexual cycle involves nuclear fusion (karyogamy), the expression of key meiotic homologs, and genetic exchange and segregation between nuclei. Microsporidia are also obligate parasites, so their unique chance to have sex would be during host infection. It is possible that two genetically distinct isolates produce recombinant progeny. There have been reports showing that there is some limited genetic diversity among E. cuniculi isolates (155, 316), suggesting that the population structure of E. cuniculi is not clonal and might result from genetic recombination during meiosis via an as-yet-unknown sexual cycle.
Evolutionary Trajectory of the MAT Locus within Zygomycetes and Microsporidia
The similar architectures of the sex and the sex-related loci in zygomycetes and microsporidia may imply that the synteny of TPT-HMG-RNA helicase genes is an ancestral genetic trait. However, several interesting variations were observed. First, the TPT gene of P. blakesleeanus, including the promoter and terminator, is not the part of the sex locus, whereas the promoter of the TPT genes in M. circinelloides lies within the sex locus. Second, in a reciprocal blast analysis, the microsporidian TPT gene failed to hit the TPT gene in the sex synteny of the zygomycetes (155a). Third, in both R. oryzae and microsporidians, the TPT gene is inverted relative to other zygomycetes, and an additional gene has also been acquired within the sex-related locus (Fig. 11 and 14A). It is possible that independent inversion events introduced these new genes into the extended sex-related locus.
These observations raise a hypothesis that in the genome of a last common ancestor, the entire TPT gene may have been a part of the sex locus (Fig. 15). It is clear for the fungal kingdom that additional genes have been acquired into the mating type locus and/or evicted from the mating type locus (70). Thus, the last common ancestral MAT locus could have harbored the three genes for TPT/HMG/RNA helicase and that these genes were retained within the MAT locus in the microsporidian lineage but are no longer within MAT in the zygomycete lineage (Fig. 15). This assumption leads to a hypothesis in which the TPT/RNA helicase gene was evicted by a gene conversion(s) from the ancestral sex locus and the TPT/RNA helicase gene was replaced by the one in the opposite-mating-type locus through recombination in the zygomycete lineage, whereas in the microsporidian lineage, both genes with the sequence of the ancestral MAT locus are retained in the sex-related locus. In this model, the genes flanking the HMG genes in the sex locus of zygomycetes compared to microsporidia are paralogs rather than orthologs. Repeated tests of these hypotheses will be possible when the genomes of more zygomycete, microsporidian, and other basal fungal species become available.
FIG. 15.
Evolutionary trajectory of the sex locus within zygomycetes and microsporidia. The ancestral sex loci are hypothesized to have contained diverged alleles of the TPT, HMG, and RNA helicase genes. By gene eviction from the sex locus, only the TPT or the RNA helicase gene was retained flanking the sex locus. In this evolutionary model, the microsporidian TPT and RNA helicase genes are paralogs of those that flank the zygomycete sex locus.
LESSONS FROM BASAL FUNGI: CHYTRIDIOMYCETES
Sex in Chytridiomycetes
Sexual cycles have been molecularly defined for species in several phyla within the fungal kingdom, but this area of research is largely an unchartered frontier for the Chytridiomycota. Cytological evidence supporting the occurrence of sex has been found for several species, including the water mold Allomyces macrogynus (54, 223) and the freshwater fungus Chytriomyces hyalinus (136, 197, 202). A role for sex may be implicated in the dispersal and persistence of the amphibian pathogen Batrachochytrium dendrobatidis, a species responsible for recent global amphibian declines and extinctions, although there is currently no morphological evidence for a sexual cycle (205). Overall, there is a paucity of knowledge in the field of chytrid sexual reproduction, but anticipated releases of several chytrid genome sequences will provide the foundation to investigate the occurrence and roles of sex in chytrid life cycles.
Chytrids represent basal fungal species that share the flagellar machinery with metazoan and choanoflagellates that has been lost in other more-derived fungal lineages (124, 177). Study of the molecular sexual cycle of representative chytrid species may provide insights into the origins of sex complementary to studies of zygomycetes. As evidence for sex exists for all major fungal branches, as well as for many unicellular and multicellular eukaryotic organisms outside the fungal kingdom, sex likely arose prior to or concomitant with the last common ancestor of all eukaryotes and clearly prior to that of the metazoan and fungal kingdoms early in the opisthokont lineage. Investigations into chytrid sexual reproduction may therefore provide insights into early steps in the evolution of sex.
Species within the Chytridiomycota reproduce both sexually and asexually. Asexual reproduction typically occurs in thin-walled zoosporangia in which several rounds of mitosis occur, followed by cytoplasmic divisions, to produce small, motile zoospores (54). Sexual reproduction results in the production of thick-walled, stress-tolerant, resting sporangia (RS), although RS may also be produced asexually (198, 203). Meiosis may occur within the RS or during germination, which results in a mature sporangium (reviewed in reference 119). Several variations of this cycle exist, and the majority of chytrid species have no identified sexual cycle.
We do not know whether the first sexual organism or fungus was homothallic, where a single individual has the capacity to reproduce sexually and is self-fertile, or if it was heterothallic, in which two individuals of opposite mating types must interact to produce recombinant progeny. There are examples of both homothallism and heterothallism in chytrid lineages (reviewed in reference 119).
Several chytrid species are homothallic, and a paradigmatic example is A. macrogynus. In this species, β-carotene-producing male gametes are clearly evident distal on the same hyphal branch to the larger colorless, hyaline female gametes, each contained in separate gametangia (206). This arrangement of gametangia is referred to as epigynous, whereas other species can have a different order. For example, the closely related species Allomyces arbuscula is hypogynous, where the female gametangia are terminal to the male gametangia (223). A. macrogynus is self-fertile, and a single meiospore can give rise to the sexual, gametophyte generation, a hallmark of homothallism (reviewed in reference 119).
In contrast, heterothallism is present in several chytrid species, including the mosquito/copepod parasite Coelomomyces psorophorae, which has an obligatory sexual cycle occurring in the copepod host (306). Whisler et al. discovered that C. psorophorae is heterothallic based on the observation that uniflagellar gametes liberated from infected copepods do not undergo self-fusion to form a biflagellate zygote (306). When these unfused gametes were mixed with other unfused gametes, fusion occurred to form the biflagellar zygote in approximately 50% of crosses, presumably when opposite mating types were mixed (306). Nothing is known about the molecular basis of heterothallism in this species. The mating type locus has not yet been identified, and the genome has not been sequenced. The sexual cycles of most chytridiomycete species remain undiscovered, and thus, data on whether they are homothallic or heterothallic are not available.
Gamete size and shape differ between chytridiomycete lineages in three paradigms: isogamy, anisogamy, and oogamy. Isogamy is the presence of two similar-sized gametes with similar or identical morphologies. In C. psorophorae, a heterothallic parasite, isogametes are derived from gametangia, which form in the copepod host (306). Alternatively, anisogamy is the presence of two differently sized gametes, typically with the female being larger than but morphologically similar to the male, as is the case for A. macrogynus. Outside the fungal kingdom, there have been multiple transitions from anisogamy to isogamy in ciliates, a diverse class of eukaryotic microbes. In ciliates, most genetic exchange involves the exchange of equally sized gametes, but at least two transitions to anisogamy in which the gametes are of unequal sizes and play different roles in the process of genetic exchange have occurred (58, 200, 235, 275).
Interestingly, the Monoblepharidales are unique in the fungi in that they are the only known fungal species whose sexual reproduction involves oogamy. The order Monoblepharidales is a small order consisting of only six genera and is the only known example of oogamy in the fungal kingdom. In Monoblepharis polymorpha, the same thallus can produce both zoosporangia (asexual structures) and gametangia (sexual structures) in a temperature-dependent fashion. In oogamy, large, nonmotile female gametes and smaller, motile male gametes fuse to form a zygote, very similar to the process in humans and other animals. In M. polymorpha, the motile male antherozoid fertilizes the sessile female egg, and after the zygote is formed, an oospore develops, which matures into a thallus (124). The Chytridiomycota represent a diversity of fungal mating characteristics, such as heterothallism, homothallism, isogamy, anisogamy, and oogamy. Studies delving into the origins of these characteristics will undoubtedly provide insights into the characteristics of the last common sexual ancestor of the eukaryotes and of the opisthokonts. In the following sections, we discuss both what is known and what remains to be discovered about the sexual cycles of three representative chytrid species: Allomyces macrogynus, Chytriomyces hyalinus, and Batrachochytrium dendrobatidis.
Allomyces macrogynus: a Model Homothallic Fungus
A. macrogynus is found primarily in humid tropical soil but has also been isolated from other regions globally. This zoosporic fungus was a popular model organism for teaching from the 1950s into the 1980s but has fallen out of favor for several reasons, including its slow growth, easy contamination of cultures, and complex life cycle and ploidy and the rise to prominence of other more molecularly amenable model systems (223). The release of the A. macrogynus genome sequence should renew interest in this fascinating zoosporic aquatic fungus.
The last common ancestor between the metazoans and the fungi was an aquatic unicellular flagellar organism similar to the extant choanoflagellate lineage (301). A. macrogynus shares these characteristics in that it has aquatic, unicellular, uniflagellate zoospores. Thus, A. macrogynus represents a basal lineage of the fungal kingdom that shares common features such as flagella with the last common ancestor between the metazoans and the fungi to the exclusion of other derived fungal lineages (124, 177). Study of the molecular sexual cycle of A. macrogynus gives us valuable insight into sexual mechanisms that might have operated in the last common ancestor.
The life cycle of A. macrogynus is a true alternation of generations between the free-living gametophytic phase and the sporophytic phase (Fig. 16). The sporophyte generation results from the fusion of motile uniflagellated male and female gametes. The resulting zoospores, released from zoosporangia, can maintain this diploid/tetraploid stage, but the sporophyte can also give rise to the RS in which meiosis occurs. Haploid meiospores are released from the RS (also known as the meiosporangium) and produce the haploid gametophytic generation, which enables the completion of the sexual cycle in the release and subsequent fusion of uniflagellate male and female gametes to form a diploid zygote (223).
FIG. 16.
The Allomyces macrogynus life cycle. The sporophyte generation is maintained asexually through the production of diploid zoospores. Meiosporangia that develop on the mature sporophyte produce meiospores, haploid uniflagellate spores that produce mature gametophytes capable of forming male and female gametangia. Gametes are released and undergo cell fusion (plasmogamy), followed by nuclear fusion (karyogamy), to form a biflagellate uninucleate zygote that develops into a sporophytic plant.
The sexual structures of A. macrogynus are large and readily discernible by light microscopy. A. macrogynus is homothallic, and both male and female gametangia are derived from mitotic divisions of the same haploid meiospore nucleus (206). The male gametangium is typically terminal to the female gametangium on the same hypha, although several repeats of the gametangium pattern may be present on a single hypha. The female gametangium is large, elongated, and colorless, whereas the male gametangium tends to be smaller, rounder, and orange due to β-carotene production. Female gametes are less motile than male gametes, although both are flagellated. Gametes are released into a gelatinous plug known as the exit papillae (270). This structure on both male and female gametangia fills with gametes and eventually breaks, releasing gametes ready for the fertilization process. Female gametes emerge slowly, without much apparent flagellar motion, whereas males burst out and immediately become active. Their fusion results in the formation of a biflagellate zygote, which matures to a sporophytic plant (223). The male gametangia often rupture first, releasing male gametes and enabling outcrossing (if foreign female gametes are available) until the female gametangia burst to enable selfing. Several movies detailing the Allomyces macrogynus life cycle are available from the Institut für den Wissenschaftlichen (http://www.iwf.de/iwf/).
Sexual differentiation in Allomyces macrogynus requires more energy to produce females than males. Female gametangia are typically two to three times the size of male gametangia, and the individual gametes are larger, thus requiring more energy to grow and develop. Supporting a role for an energy requirement in sexual differentiation, there is a differential distribution of mitochondria in the gametangia in which more mitochondria are allocated to female gametangia (96). A. macrogynus all-male and all-female mutants have been isolated through a variety of mutagenesis techniques. Non-Mendelian mutants producing nearly 100% male gametangia have been isolated by means of acridine dye treatment, resulting in mitochondrial DNA damage (218, 224, 256). No all-female A. macrogynus non-Mendelian mutants have been isolated, although both all-male and all-female strains have been generated through mutagenesis of the nuclear genome (218). The all-male mutants resulting from acridine dye treatment are likely the result of mitochondrial DNA mutations affecting the electron transport and ultimate energy output of the individual rather than a mutation in a sex-determining factor. Likely, the larger female gametes and gametangia require more energy to develop; thus, mutations lowering the ultimate energy available for sexual differentiation result in an all-male phenotype.
A. macrogynus males and females secrete nonpeptide sexual pheromones that enable the gametes to locate the opposite sex. Female gametangia and gametes secrete a low-molecular-mass (236 Da) pheromone called sirenin, named after the Sirens, the seductive bird-women of Greek mythology (239). This pheromone has been purified (183), and its structure has been elucidated (Fig. 17A) (182, 188). Sirenin is structurally unrelated to the pheromones of zygomycetes (trisporic acid [see the section on zygomycetes above]), basidiomycetes (farnesylated peptides), or ascomycetes (peptides and farnesylated peptides). Thus, the molecular nature of the pheromone signals that elicit mate recognition exhibits considerable plasticity. Sirenin is potent, and concentrations as low as 10−10 M are effective in in vitro assays (180). Interestingly, sirenin is secreted by female gametangia at a concentration of 10−6 M, the concentration determined to be the most effective in in vitro studies, attracting males to intact gametangia (34). This effectively attracts a swarm of male gametes waiting for the release of female gametes, resulting in high fertilization rates (181). The pheromone from unreleased female gametangia may attract recently released male gametes from nearby but genetically distinct individuals, thus increasing genetic diversity in the resulting zygote. While A. macrogynus is homothallic, it is also capable of outcrossing, as is the case with many other known homothallic fungi.
FIG. 17.
Chytrids produce a novel mating pheromone. (A) Chemical structure of sirenin. (B) Bioassay design for sirenin chemotaxis. Female gametangia are placed into the center well of a thin agar plate. Experimental groups are placed into outlying wells. If chemotaxis toward sirenin occurs, clusters toward the center well are observed. The orange circles represent male gametangia, white circles represent female gametangia, and gray circles represent zoosporangia.
How the pheromone signal is transduced to elicit chemotaxis is unknown, although it likely involves Ca2+ regulation (239) and may also lead to the inactivation of sirenin (34). Female gametes are unaffected by sirenin but are attracted to a lesser-studied pheromone secreted by male gametes, called parisin. While the structure of parisin is not known, it is thought to be similar to sirenin and stable in response to a variety of chemical and environmental stresses (238, 239). These highly effective nonpeptide pheromones promote high rates of gamete fusion.
Bioassays for sirenin attractivity involve using a cork borer to produce small holes placed close to one another in a thin agar plate. Typically, female gametangia are placed into one well, and male gametangia or another experimental group is placed into other nearby wells. After male gametangia release the male gametes, males are attracted to the edge of their well closest to the female gametangia, as visualized by light microscopy. Zoospores or female gametes, however, show no attraction to sirenin-containing wells (Fig. 17B) (237). This bioassay is a fast and efficient means of studying pheromone behavior in this species.
Interestingly, some Allomyces species readily form hybrids, both naturally and in the laboratory. Allomyces javanicus was collected in India and determined to be a natural hybrid of the closely related species A. macrogynus and A. arbuscula (223) that is able to undergo meiosis. The ability of these species to hybridize has been useful in the laboratory for creating strains producing nearly exclusively male or female gametes, as opposed to their hermaphroditic parents (181), which at the time was useful for studies requiring gamete isolation or sirenin production.
In addition to sexual reproduction through the production of gametes, A. macrogynus can also proliferate through asexual reproduction. A. macrogynus produces zoospores contained in zoosporangia but, interestingly, can proliferate asexually through parthenogenesis. Female gametes that can develop into new sporophytic or gametophytic plants producing both male and female gametangia without the presence of a male gamete have been isolated (54). This apomictic pathway is hypothesized to initiate in two different ways: (i) through the doubling of chromosome number in unfertilized gametes (54) or (ii) by the fusion of two nuclei within a single gamete (290). Male gametes have not been observed to undergo this process (54), although androgenetic development into a sporophyte can occur through the fusion of multiple nuclei within a single gamete. The resulting sporophyte shows morphological abnormalities at some frequency (225). A. macrogynus is an interesting and well-established model of sexual reproduction and several forms of asexual proliferation involving haploid male and female gametes and diploid zoospores.
A. macrogynus reproduces both sexually and asexually, but the genetic mechanisms underlying sexual reproduction are largely unknown. The Allomyces macrogynus strain Burma 3-35 genome sequence has recently been made available (258), and efforts are under way to identify candidate mating type loci with a focus on genes encoding HMG and homeodomain transcription factors (119, 154). The identification and characterization of proteins involved in sexual reproduction in other basal fungal species with defined sexual cycles will provide insight into the evolution and molecular basis of sex both within and beyond the fungal kingdom.
Chytriomyces hyalinus: Second Homothallic Example of Chytrid Sex
Chytriomyces hyalinus, a member of the order Chytridiales, is a homothallic chytrid fungus that grows saprobically in freshwater, typically on the molted shells of May flies (4). C. hyalinus has a morphologically described sexual phase studied mostly by microscopy. The sexual cycle is induced by the depletion of nutrients in medium (198), as has been seen for other fungal species such as the basidiomycete C. neoformans and the ascomycete S. pombe. The depletion of nutrients as a cue for sexual reproduction has interesting implications in natural environments. A shift to low-nutrient conditions is a signal to the fungus that there are challenging environmental conditions. Sexual reproduction usually results in the production of spores capable of withstanding environmental stress, and these resistant spores allow the organism to persist in a dormant form until conditions are favorable to resume growth.
In C. hyalinus, sexual reproduction involves two protoplasts from different thalli. These protoplasts have a common rhizoidal system, and nuclei migrate to an anastomosis of the rhizoids, which swells. This swelling is known as the fusion or resting body, and the nuclei from the two thalli fuse. Koch initially attributed this process to both sexual and asexual reproduction (136), but Moore and Miller later determined that the fusion of nuclei from two thalli, as Koch described, was always observed to result from a sexual process, resulting in a thick-walled resistant meiosporangium (197, 202, 203). Nuclear fusion occurs soon after the nuclei enter the resting body, and the resting body increases in size, producing a thick-walled, resistant structure (198). Although sexual reproduction in this species has not been studied in detail, meiosis is assumed to occur in the sporangium produced after the germination of the meiosporangium rather than in the resting body itself.
Batrachochytrium dendrobatidis: an Emerging Amphibian Pathogen
Batrachochytrium dendrobatidis is a diploid amphibian pathogen implicated in the global amphibian decline and extinction. Like C. hyalinus, B. dendrobatidis is a member of the order Chytridiales and was first described in 1999 (176). Since then, amphibian populations on every continent except Antarctica have tested positive for B. dendrobatidis (205, 246). The general life cycle of B. dendrobatidis includes a motile, waterborne zoospore that is able to disperse and a sessile thallus, which develops into a zoosporangium that undergoes asexual clonal proliferation on a wide variety of amphibian hosts (21). Unlike most fungal organisms that spend the majority of their life cycle in a haploid state, B. dendrobatidis is diploid throughout its life cycle, with no morphological evidence as yet of a sexual cycle or of RS that are commonly produced via sexual reproduction. The presence of an extant sexual cycle could have profound implications for the dispersal and persistence of the organism in amphibian populations or in areas where amphibian populations have gone extinct and in the evolution of the recent emergence of B. dendrobatidis (205).
Sex may be present in some B. dendrobatidis populations, but most evidence supports clonal reproduction. Recent studies have uncovered limited allelic diversity (i.e., number of alleles per locus) in B. dendrobatidis isolates involved in the amphibian epidemic, although there is a large amount of genotype diversity (number of genotypes per sample) as well as gene diversity (measures of Hardy-Weinberg equilibrium heterozygosity) (125, 205). Morgan et al. identified two populations in California in which genetic diversity and evidence of recombination were present, suggesting that sexual reproduction could be occurring (205). A later study attributed this genetic diversity to a loss of heterozygosity due to mitotic gene conversion rather than meiotic recombination, but this still does not provide evidence against sex in some isolates (125).
B. dendrobatidis may have recently emerged as a pathogen from a single isolate that underwent a genome rearrangement or diploidization/hybridization event to acquire virulence, although there is debate on the origin of the B. dendrobatidis epidemic. B. dendrobatidis has been hypothesized to have been dispersed throughout the world through the distribution of infected frogs for museums, science, and pregnancy tests (125, 304). A genomic rearrangement event in a nonpathogenic isolate could have led to increased pathogenicity yet an inability to undergo sexual recombination with its opposite mating type, assuming that the sexual ancestor was heterothallic. Alternatively, the opposite mating type to this clonal global population of B. dendrobatidis could be rare or extinct, resulting in clonal amplification in its absence and low overall genetic diversity. James et al. hypothesized that the current clonal epidemic may be a result of strong selection on a single genotype, essentially a single mating type resulting in the elimination of the opposite mating type (125). Several examples of a specific mating type being both more common and more virulent than its partner exist for other fungal species, such as C. neoformans (213, 214, 217).
Efforts are currently under way to investigate the sexual cycle, or lack thereof, in B. dendrobatidis. Approaches include assays for morphological evidence of a sexual cycle as well as utilizing available genomic information. This includes searching for HMG domain genes as well as other genes with products typically involved in sexual reproduction, such as homeodomain or alpha box genes in the genomes of two isolates, JEL423 (Batrachochytrium dendrobatidis Sequencing Project, Broad Institute of Harvard and MIT) and JAM81 (Joint Genome Institute), and characterizing these genes to determine if they could be involved in sexual reproduction. The presence of an extensive meiotic tool kit (data not shown) suggests that sexual recombination may occur in B. dendrobatidis. Because of the mass amphibian extinctions resulting from B. dendrobatidis and the implications in persistence and dispersal as a result of sexual recombination, it will be critical to investigate this topic further.
SAME-SEX MATING (HOMOTHALLISM) IN FUNGI
Homothallism is found throughout the fungal kingdom. For example, the A. nidulans genome contains HMG and alpha box mating genes enabling selfing (see the section on aspergilli above). Further examples can be found in mating between mother and daughter cells by the mating type switch in S. cerevisiae utilizing recombination between MAT loci and silenced HML/HMR loci evoked by the HO endonuclease (see “as the Mating Paradigm” above S. cerevisiae). A zygomycete fungus, Mycotypha africana, undergoes homothallic sexual development by the fusion of hyphae of cells of one mating type (313). A. macrogynus is an example of a homothallic chytrid fungus, where both male and female gametangia are produced through the mitosis of the same haploid meiospore nucleus (see the section on chytrids above). In this section, we discuss recent findings on same-sex mating in two pathogenic fungi, C. neoformans and C. albicans, and discuss the implications for the evolution of same-sex mating in the fungal kingdom.
Same-Sex Mating in Cryptococcus Species
The Cryptococcus opposite-sex mating cycle was defined in the laboratory more than 30 years ago (143, 144); however, whether mating occurs in nature is an open question, because hyphae and spores produced by mating are microscopic. One finding that provides evidence that mating occurs in nature is that unusual hybrid AD isolates are diploid strains produced by the mating of a and α parental isolates to yield aADα and αADa strains (39, 156). These hybrid strains result from mating between two related cryptic species and, as such, fail to undergo normal meiosis and remain trapped in the diploid state. The successful induction of sexual development under several laboratory conditions mimicking natural C. neoformans niches suggests that sexual reproduction may occur in specialized environmental niches (215, 321). Natural niches of C. neoformans include trees and pigeon excreta (35). Xue et al. showed that coculturing with Arabidopsis enhances Cryptococcus mating (321). Another study showed that C. neoformans undergoes mating on pigeon guano medium (215). These findings provide evidence that opposite-sex mating occurs in nature. If a-α mating occurs in nature, a 1:1 ratio of a and α mating type cells might exist. Instead, in clinical and environmental samples, C. neoformans isolates exhibit a largely clonal population structure in which the α mating type predominates, in some cases exclusively (27, 35, 65, 93, 147, 167-169). Only a few examples of more balanced proportions of a and α mating type cells have been observed for certain geographical regions of Australia and Botswana (93, 167). Given that the a mating type is rare, the opportunities for opposite-sex mating to occur are limited. The discovery that C. neoformans can undergo unisexual mating under defined laboratory conditions, especially between α isolates, provides an alternative means of maintaining diversity in a unisexual population (162, 163).
C. neoformans is known to undergo monokaryotic fruiting (308), in which α cells complete a fruiting cycle in the absence of a cells (Fig. 18). For a long time, this process was thought to be strictly mitotic, leading to a clonal population. However, a recent study provides evidence that monokaryotic fruiting is a sexual cycle governed by key meiotic genes, DMC1 and SPO11, in which genetic recombination occurs between two nonisogenic α cells at a frequency on par with a-α mating (Fig. 18) (163). Moreover, complete ploidy transitions from haploid to diploid to haploid occur during this process. Several recent studies provide evidence that unisexual reproduction also occurs in nature. First, the discovery and characterization of αADα hybrids produced by the mating of two α isolates of different serotypes (A and D) provide definitive evidence that unisexual mating has occurred in nature (164). Second, recent studies of C. gattii populations from tree hollows in Australia provided evidence that recombination is occurring within populations that are exclusively of the α mating type (264). Third, population genetic studies of C. neoformans isolates from tree hollows in India provide evidence for recombination in otherwise α unisexual populations, and one parsimonious hypothesis is that this involves unisexual reproduction (105). Fourth, C. neoformans serotype A veterinary isolates from infected animals in Sydney, Australia, were found to define a recombining population that is exclusively of the α mating type (29). Moreover, diploid αAAα isolates that represent intermediates produced via same-sex mating were identified within this population. Notably, these isolates undergo limited hyphal growth and basidial production when grown in confrontation with a cells as a source of mating pheromone (which stimulates same-sex mating), but no sporulation of these isolates has been observed as yet. Finally, a recent large-scale survey of ∼500 C. neoformans serotype A isolates revealed that ∼8% of the naturally occurring population of environmental and clinical isolates are diploid, and the majority of these are αAAα strains produced via same-sex mating, either by mating between genetically distinct individuals in the population or from self-mating (165).
FIG. 18.
Illustration of the same-sex mating process in the two human-pathogenic fungi C. neoformans and C. albicans. Same-sex mating in C. neoformans occurs between two α cells, whereas conventional mating involves two opposite-mating-type cells. C. albicans a/a diploid cells also need to be opaque to undergo same-sex mating. In the absence of Bar1 protease activity or in the presence of α cells, the a/a diploids undergo homothallic mating. The 4N cells then undergo a parasexual cycle to return to a diploid state as seen in opposite-sex mating.
Taken together, these studies provide robust support that same-sex mating occurs both in the laboratory and in nature and occurs in all major lineages of Cryptococcus, including both serotype A and D strains and the sibling species C. gattii. A future challenge will be to define conditions that support this alternative unisexual cycle in the laboratory for C. neoformans serotype A and C. gattii isolates of this fungal pathogen.
Same-Sex Mating in C. albicans
A recent study found that C. albicans also has the capacity to undergo same-sex mating (2). As described above in the section on ascomycetes, the conventional mating of C. albicans includes several steps: (i) a/α diploids of C. albicans undergo homozygosis, resulting in a/a or α/α diploids; (ii) a phenotypic switch from white to opaque cells and the presence of opposite-mating-type cells trigger mating to produce an a/a/α/α tetraploid; and (iii) a parasexual chromosome loss process finally produces diploid progeny (Fig. 18). Despite the presence of a well-defined mating cycle involving two compatible mating type cells, Alby et al. found that same-sex mating also occurs in C. albicans. Two observations led to their discovery (Fig. 18). First, they were puzzled why a cells of C. albicans express both the a and α pheromone genes (in contrast to S. cerevisiae). Second, they discovered that bar1 mutants produce and respond to α pheromone. Two mechanisms in C. albicans enable same-sex mating: (i) a disruption of the BAR1 gene, encoding a protease that cleaves α pheromone, stimulates a cells to undergo same-sex mating, and (ii) the presence of limiting α cells as a source of α pheromone induces same-sex mating between a cells similar to ménage à trois matings in C. neoformans (Fig. 18) (114, 115).
Bar1 protease produced by a cells degrades α pheromone from α cells, resulting in the inactivation of mating in both C. albicans and S. cerevisiae (103, 184, 190). In C. albicans, a cells produce both a and α mating pheromones, and the disruption of BAR1 causes a cells to be activated to mate by autocrine α pheromone signaling (2, 19). It is striking that same-sex mating evolved independently in two divergent fungi, C. albicans and C. neoformans, via distinct molecular mechanisms. Given that both are successful human pathogens, same-sex mating may contribute to generate highly clonal populations well adapted to virulence.
How does C. albicans mate in nature? There are two possibilities for C. albicans mating in nature. First, two opposite mating types can signal to each other and form biofilms that promote signaling and conjugation tube formation for opposite-sex mating. Alternatively, it could also be the case that MTL homozygous isolates can undergo mitosis and then mate a/a with a/a via same-sex mating. However, it is infrequent that both a/a and α/α cells are close enough to each other to mate given that they arise rarely from the dominant natural a/α starting strains. There could be a more general mechanism to generate both a/a and α/α from a single cell and its daughter progeny, akin to mating type switching in S. cerevisiae. As one possible mechanism, chromosome 5 containing the MTL locus could be replicated late, resulting in the generation of 2N − 1 progeny, one which inherits MTLa and the other which inherits MTLα, and the subsequent reduplication to drive the 2N − 1 aneuploid stage back to 2N euploidy to generate MTL homozygous rather than heterozygous isolates. This model provides a starting point for future study.
Why Self-Mating?
Homothallism is a fascinating form of sexual reproduction in the fungal kingdom. The presence of self-fertility in species with heterothallic mating systems raises an interesting question, why did self-mating arise during evolution? Self-mating is a mechanism to rediploidize rapidly and could serve to shelter the genome. An analogy is the fission yeast S. pombe, which undergoes G2 arrest with two copies of the genome that provides a greater buffer for DNA damage and repair than the haploid stage, like the diploid stage. Homothallism might also be a way to ensure that the organism survives to the next chance to mate. In this model, homothallism can be viewed as a form of practice, ensuring that the machinery for mating and meiosis is maintained, functional, and ready to play the concert when a-α mating becomes possible. Thus, self-sex would be a way to protect fertility for the longer term and promote or increase the chances to engage in cross-mating.
Homothallism might have evolved to allow any individual to mate with any other individual and not to enable self-mating. However, for a species such as S. cerevisiae, there are clear examples of isolates from nature that have completely selfed (a mother cell has switched and mated with its daughter cell such that the two genomes are identical) compared to other diploid isolates that are heterozygous. It is also the case that the natural population consists of both HO+ and ho− isolates, and the latter cannot switch mating type. A prevailing model is that the two mating types are arranged in the tetrad such that following germination and cell division, each cell is adjacent to a partner with which it can then immediately mate to return to the diploid stage. The fact that essentially all natural isolates of yeast are diploid further indicates that mating is highly efficient. This necessarily would then appear to promote both direct selfing and mating with siblings present within the ascus. Outcrossing can and does occur, but the extent to which homothallism promotes inbreeding within the ascus, or enables outcrossing with the more general population, remains to be addressed and determined.
It is also worth considering a thought experiment in which a single haploid yeast cell is present in nature with no suitable mating partner available. In this scenario, a cell that is heterothallic can never mate, whereas one that is homothallic can divide, switch mating type, mate, and be restored to the diploid state. Given that this is the preferred ploidy of this species, it will allow enhanced DNA repair and resistance to damage, enable the production of spores in response to adverse conditions, and maintain both mating types for future mating events. Cells that cannot switch mating type would remain haploid, unable to undergo pseudohyphal growth or sporulate, and would be less fit under a wide variety of conditions. Thus, it seems that there are selective pressures that promote homothallism with respect to both outcrossing potential and the benefits of direct selfing.
If same-sex mating can occur in nature, why has the opposite mating type not disappeared? First, opposite-sex mating appears more efficient than same-sex mating. Second, opposite-sex mating could lead to meiosis under certain conditions, whereas same-sex mating might not, or alternatively, the frequency or efficiency of meiosis may be higher for opposite-sex than for same-sex mating. Third, in the case of C. albicans following opposite-sex mating, the a/a/α/α products convert back to white cells, precluding further mating, whereas the a/a/a/a products of same-sex mating remain opaque/mating competent and can mate, again yielding less-fit, higher-ploidy cells (2, 98). Finally, a/α cells may exhibit hybrid fitness, such as during infection.
EXPERIMENTAL EVOLUTION OF SEX IN FUNGI
There are two main facets in studying the evolution of sex: the emergence of sexual reproduction and its maintenance. While the origin of sex remains elusive, the fungal kingdom provides insights into the genetic basis of sexual reproduction from distinct evolutionary vantage points. The fungi also provide an excellent system for studying the origin, evolution, and maintenance of sex.
On a theoretical level, many issues must be addressed, particularly why sex is beneficial although there are many possible negative outcomes of sex. One common theory as to why sex is beneficial is described by the Fisher-Müller hypothesis, which posits that an advantage of sex is that the beneficial adaptations produced by rare mutational events can be brought together into the same genome through recombination. This effectively pairs adaptive mutations together, whereas in an asexual organism, only one rare mutation may be selected for at any given time, so a beneficial mutation that is not concurrent is not available for selection to act upon. Recombination also serves to purge the genome of deleterious mutations, including those caused by transposons. Alternatively, genetic recombination can be detrimental in cases where two or more beneficial mutations are separated.
While most of the experimental evolution studies of sex focused on the benefits of increasing diversity resulting from the sexual process, Heng presented an interesting contrast (100). At an individual-gene level, sexual reproduction does introduce diversity, but at a genomic or species level, sexual reproduction functions to reduce the diversity between individuals in the same population. Because recombination depends on the pairing of homologous chromosomes, chromosomal aberrations, such as a translocation or rearrangement events, aneuploidy, and large duplications or deletions, result in decreased crossing-over and chromosomal nondisjunction. Thus, at a chromosomal level, sex functions to filter an altered karyotype from the population and decrease diversity, whereas at a genic level, sex functions to increase the genetic diversity of the population (100).
In a diploid sexual organism, a new mutation that confers a beneficial adaptation can occur. Originally, the organism will be heterozygous for this allele, but following sexual reproduction and genetic recombination with another individual heterozygous for the allele, the resulting sexual progeny, as well as their progeny, may have a significant advantage over others in the population since they are homozygous for the beneficial allele. Asexual diploid organisms must acquire the advantageous mutation through a rare mutation event and must have another rare event in order to homozygose the locus.
There is also a diploid advantage in that diploid individuals are not immediately susceptible to deleterious recessive mutations. Diploids have two copies of each gene and, thus, a higher adaptive ability. However, there may be a long-term effect of tolerating harmful, though recessive, mutations. Over time, diploids will have a larger mutation load than haploid individuals and may suffer from reduced fitness.
Saccharomyces cerevisiae: a Model for the Experimental Evolution of Sex
The budding yeast Saccharomyces cerevisiae is a paradigm to study sex and its evolution. S. cerevisiae has a short generation time, can be stably maintained as either a haploid or a diploid, and can reproduce asexually or sexually. Yeast is isogamous, meaning that the two gametes are morphologically identical. Moreover, both mating types, a and α, contribute energy equally to form a diploid cell and ultimately produce four haploid spores in a single ascus. While there is no “cost of sex” in many facultatively sexual microbes, including the majority of fungi (317), with regard to how much energy is expended by either mating type, sexual reproduction does involve more time and energy than the alternative: asexual mitotic proliferation.
In S. cerevisiae, fitness can be readily assayed. Zeyl and Bell grew asexual and sexual yeast populations on standard medium with glucose as the carbon source as well as on medium with galactose, which cannot be as readily metabolized by wild-type yeast, as the only carbon source to determine if the sexual populations developed a growth advantage on galactose medium over asexual populations (326). Interestingly, both the asexual and sexual populations grew equally well on galactose medium, but a growth advantage in the sexual population on standard glucose-containing medium was observed. Those authors attributed this growth advantage to the purging of deleterious mutations in the sexual populations. One acknowledged limitation of that study was that the asexual and sexual populations faced different selective pressures when the sexual ones were periodically grown on nitrogen-limiting medium to promote sporulation (326).
Goddard and colleagues devised a method to compare isogenic asexual and sexual populations of S. cerevisiae (82), overcoming the additional selective pressure placed on the sexual populations of the above-mentioned study by Zeyl and Bell. By deleting SPO11 and SPO13 from S. cerevisiae, the resulting double-mutant strain can respond to standard sporulation medium but does not undergo a reductional meiotic division. Rather, two diploid spores are produced instead of four haploid spores in a single ascus. Isogenic wild-type and spo11Δ spo13Δ strains were compared on harsh medium with high osmolarity and a stressful temperature. Interestingly, those researchers observed different results from those from the experiments reported by Zeyl and Bell. There was no growth difference on standard medium, whereas on the harsh medium, the sexual isolates adapted more quickly. Zeyl later attributed the difference to the nature of the mutations required for increased growth in the different environments, as galactose utilization requires fewer adaptations than does a harsh osmotic environment with high temperatures (325).
Desai et al. used S. cerevisiae to challenge the model of mutation fixation, in which one individual mutation is fixed at any given time before another mutation can begin the process of fixation (45). In that study, they used a 500-generation evolution experiment. The starting population was assumed to have a background level of mutations, with each individual containing different mutations. New mutations occur in the background of the original mutations, and if these new mutations are advantageous, they may act synergistically with the original mutation such that an individual can outcompete other individuals in the population to eventually have the two or more synergistic mutations fixed. This can allow a mutation that is not strong enough by itself to compete with other more beneficial mutations to persist and eventually thrive due to piggybacking with another synergistic beneficial mutation (45).
Interestingly, S. cerevisiae can evolve to have mating discrimination. Leu and Murray derived two different yeast strains from the same ancestor through the introduction of auxotrophic requirements and drug resistance markers (158). One population, the evolving strain (E), was passaged through several rounds of selection, whereas the reference strain (R) was newly reintroduced from a frozen stock before each round of selection. The strains were mixed and allowed to mate. Initially, the E strain did not discriminate and mated with E or R cells with similar efficiencies. After mating, a suicide gene was induced in the R cells, and 95% of the E × R diploids and R × R diploids were eliminated, creating a nearly pure population of E × E diploids with small amounts of cells expressing some genetic markers from the original R strains.
After 36 rounds of selection, the majority of the 13 E populations studied developed a preference for mating with individuals from their own E population, although different populations showed different levels of discrimination. Reciprocally, R cells also developed a mating preference for R cells after several generations. Many mutations likely accumulated in each population contributing to mating preference, explaining the phenomenon of different levels of discrimination in different populations. Furthermore, the mechanism of discrimination for many E populations involved the mating kinetics, mating either more rapidly or more slowly than the R populations. This information suggests that unique features, such as the kinetics of the mating process, play an important role in the development of reproductive isolation (158) not only in fungi but possibly also in animals and other eukaryotic species as well.
Greig and colleagues examined whether sex was more advantageous in homozygous or heterozygous yeast populations (86). They produced homozygous and heterozygous diploid strains by backcrossing them to either the same or different background strains, respectively. Initially, homozygous sexual populations fared better than their heterozygous counterparts, likely because sex initially disrupts beneficial allelic combinations, and homozygous individuals will not experience this, regardless of the level of recombination. However, heterozygous populations ultimately prevailed. This phenomenon is likely because while most combinations between genes produced by recombination are deleterious, some are advantageous, and individuals with these combinations are able to outcompete the rest of the population, including the parents (86).
Yeast has several proteins capable of forming prions, and at least one may play a role in maintaining sex in a population. Sup35 normally functions in the translation termination complex; its loss results in stop codon readthrough, ultimately revealing cryptic genetic variation (232, 327). The [PSI+] prion results from a misfolding of the Sup35 protein and occurs spontaneously in laboratory populations of S. cerevisiae (291). Once established, the prion can be vertically transmitted from mother to daughter cells. While the cryptic genetic variation that is revealed upon [PSI+] formation is typically detrimental, it may rarely but rapidly reveal beneficial adaptations, since the variation is preexisting. This prion system can be viewed as an evolutionary capacitor, since it is able to store genetic variance when the prion is absent and express variants upon the expression of the [PSI+] prion (89).
Mutator alleles are gene variants that increase the intrinsic mutation rate in an organism. Such variants are thought to be eliminated by sex, because recombination can break up the linkage between these mutator alleles and the beneficial adaptations that they create, abolishing the ability of the potentially detrimental mutator allele to “piggyback” and therefore persist with the advantageous alleles that it generates (283). The advantage of the [PSI+] system has long been debated. While the majority of the allelic variation that it generates is detrimental, the prion has been hypothesized to also contribute to the overall evolvability and adaptability of the organism.
Griswold and Masel investigated whether sex prevents the formation of [PSI+], analogous to how sex limits the spread of mutator alleles (89). In natural populations of Saccharomyces paradoxus, a species closely related to S. cerevisiae, sexual reproduction occurs rarely. Although sex is rare, it seems to actually drive the formation of [PSI+], although synergistic interactions must be present between the modifier and the adaptation acquired. Obligate sex, rather than facultative sex as observed for these yeast species, would likely function to disrupt the [PSI+] system and prevent its presence (89). Thus, the pressure for increased genetic variation during stressful situations as well as obligate synergistic interactions between two loci with effects on the fitness of the organism are sufficient to drive the evolution and persistence of the prion system. Furthermore, this experiment provides evidence that, at least in species closely related to S. paradoxus, capacitor systems sustaining cryptic genetic variation in the presence of sex are beneficial and not selected against.
Saccharomyces cerevisiae: a Model To Study Speciation
Sex, or the lack thereof, plays an important role in speciation. The biological species concept (BSC) provides one method to discriminate species: if the two populations cannot mate due to either prezygotic (e.g., differences between gametes) or postzygotic (e.g., sterile progeny) mechanisms, then the populations are considered different species. Other means of determining whether populations are different species include the genotypic cluster species concept, in which some gene flow can occur between the populations but they must be genotypically distinct, and the phylogenetic species concept (PSC), in which a species is a monophyletic group based on evolutionary history. Under the BSC, chromosomal rearrangements are one means of reproductive isolation, and thus speciation, acting as a postzygotic reproductive barrier. Other methods of genetic speciation under the BSC include the accumulation of major genic differences and sequence divergence, with both mostly contributing to prezygotic separation. Species within the model filamentous fungal genus Neurospora have been phylogenetically defined using both the BSC and PSC. Using two species initially, Neurospora crassa and N. intermedia, Dettman and colleagues determined that these two species were actually four cryptic species based on mating-incompatibility testing and phylogenetic studies (46, 47).
S. cerevisiae and related species in the sensu stricto group of Saccharomyces have been instrumental in experimental evolution with regard to speciation. Species within the sensu stricto Saccharomyces group can mate with a low frequency and generate viable hybrids (211). Fischer and colleagues investigated the chromosomal evolution in Saccharomyces sensu stricto species and the extent to which it has contributed to speciation within the clade (60). That group characterized chromosomal translocations in six species within the sensu stricto group. Interestingly, those researchers found that closely related species had chromosomal rearrangements likely resulting from nonhomologous crossing-over between Ty elements and highly repetitive sequences, and more distantly related species had colinear genomes. While chromosomal translocations do serve to reproductively isolate some closely related species, it is not a prerequisite for speciation (43, 60), and most speciation occurs due to genic incompatibilities (87, 166).
In matings between the sensu stricto species S. cerevisiae and S. paradoxus, which have ∼16% sequence divergence (118), recombination is significantly decreased, resulting in high levels of chromosome nondisjunction and spore inviability. However, mutations in PMS1 and MSH2, genes coding for proteins participating in the mismatch repair system, can restore spore viability in these crosses. Pms1 and Msh2 normally function to repair mispaired bases during DNA replication and recombination, and Hunter et al. determined that mutations in either of these genes result in increased recombination between hybrid crosses and increased spore viability (118). These results suggest that the mismatch repair system plays a positive role in speciation in that the mismatch repair pathway prevents recombination between sequences that are diverged from one another and thereby enforces species boundaries (88, 118).
Cryptococcus neoformans: an Emerging Model for the Evolution of Sex
Xu investigated the spontaneous mutation rate resulting in a loss of sex in the human fungal pathogen C. neoformans (318). The C. neoformans mating process is divided into two steps. The first involves the fusion of the two different mating types (a and α) to form a dikaryon, and the second is the formation of hyphae, with meiosis occurring at the hyphal tips in the basidium to produce haploid spores. Xu allowed mutations to accumulate in C. neoformans under laboratory conditions, resulting in the loss of sexual fecundity. After ∼600 mitotic divisions on standard laboratory media promoting asexual growth conditions, the ability of opposite mating types to fuse and the resulting diploid to filament decreased significantly. This study raises an important point in the maintenance of laboratory cultures of facultative sexual organisms: in order to maintain their ability to sexually reproduce, the organism must be periodically transferred into mating/sporulation-promoting media (89, 318).
Furthermore, Xu showed in a later study that interactions with a sexual partner, particularly in the sexual facultative pathogen C. neoformans, can be deleterious for fitness (317). The cost of sexual reproduction is composed of at least two components: the cost of producing mating signals (pheromones) to attract partners and the cost of associating with and fusing with a mating partner. These costs can be avoided through asexual mitotic division, but benefits arising from sexual reproduction, namely, recombination to produce novel genotypes, likely compensate for these costs, allowing the continued maintenance and evolution of the sexual process not only in C. neoformans but also in other facultative sexual organisms.
CONCLUDING REMARKS AND FUTURE DIRECTIONS
Sexual development is common in eukaryotic lineages, including animals, plants, and fungi. It is an intriguing question how organisms have evolved partner recognition systems and sexual development. Fungi serve as an excellent system to understand the genetics of sex in several aspects: (i) each fungal group displays a wide variety of sexual development paradigms; (ii) many fungal genomes are currently available, enabling studies of MAT loci and sex evolution and revealing that many fungi classified as “asexual” retain all the machinery required for sex; and (iii) fungi are evolutionarily close to metazoans and therefore provide insights into general principles. In this review, we discussed how fungi evolved partner recognition systems, how sexual reproduction is genetically regulated, and how genetic systems have diverged among phyla and among more closely related clades.
The HMG proteins may be ancestral to both metazoans and fungi given that in basal zygomycetes and microsporidia, they are integral components of the sex and sex-related locus and the key sex-determining factors (52, 85) (Fig. 19). This is conserved in the a2 gene in MAT that is extant in certain hemiascomycete yeast species, such as Candida albicans, and in Neurospora crassa and other euascomycetes as the MAT1-1 gene in Aspergillus species. However, strikingly, in the basidiomycetes, the role of the HMG genes has been supplanted by homeodomain (HD) proteins, which also play key roles as the a1 and α2 factors in hemiascomycetes and the matPi gene in S. pombe in the archiascomycetes. It may be that there were two ancestral sex-determining systems based on either HMG or homeodomain factors and that one or the other has been conscripted in different lineages to play the central role encoded by MAT, and the other factors contribute a supporting role encoded by other genomic regions. Alternatively, it may be that both classes of factors were present in an ancestral organism and that one or the other was either lost or transposed elsewhere in the genome, similar to the recent loss of a2 from the S. cerevisiae MAT locus and other examples of MAT gene loss in the Candida pathogenic species complex.
The evolutionary trajectory of sex determination systems in fungi implies a plasticity of sexual reproduction in biological systems, suggesting that sex may be at the forefront of evolution. The evolution of fungal MAT loci serves as a foundation to understand the evolution of sex chromosomes in higher eukaryotes; for example, zygomycete sex loci hint at the earliest steps of sex chromosome occurrence (Fig. 13), and the expansion of MAT loci in basidiomycetes gives a possible explanation as to how divergent pairs of sex chromosomes formed in higher eukaryotes (Fig. 7 and 8). Self-fertility is common in fungi, and each lineage of fungi may have evolved homothallism independently (for example, see Fig. 5 and 18). This phenomenon can contribute to explain reproduction through self-pollination in plants and the parthenogenesis occasionally observed for vertebrates. Further investigations into a wide range of fungal lineages will advance our understanding of the evolution of sex. Our knowledge about sex in basal fungal lineages, including zygomycetes, chytridiomycetes, and microsporidia, is currently limited but already revealing considerable insights.
Sexual reproduction remains mysterious in myriad ways. How did sex first evolve? How has sex been maintained throughout all eukaryotic lineages for billions of years? How is sex beneficial in the face of associated costs? The study of the evolution of sex in eukaryotic microbes such as the fungi will continue to prove informative, especially since sex likely first arose in our shared last common ancestor, a single-celled aquatic eukaryotic microbe, from which we, the fungi, and other eukaryotes descended.
Acknowledgments
We thank Yen-Ping Hsueh, Keisha Findley, John Logsdon, and Tatiana Giraud for discussions.
S.C.L. is supported by the Molecular Mycology and Pathogenesis Training Program (AI52080). C.S. is supported by a UPGG Marcy Speer Memorial fellowship. This work was supported by NIH/NIAID grants AI39115 and AI50113 to J.H.
Biography
 Soo Chan Lee, Ph.D., received B.S. and M.S. degrees in biology and microbiology from Kyung Hee University, Seoul, South Korea, and earned his Ph.D. degree in December 2007 from Texas A&M University. As a Ph.D. student, he trained with Brian Shaw and was primarily interested in how Aspergillus nidulans, a filamentous fungus, establishes and maintains hyphal polarity. He is currently a postdoctoral scholar in the Heitman Laboratory at the Department of Molecular Genetics and Microbiology in the Duke University Medical Center. His research is focused on understanding the sexual development of three different groups of human-pathogenic fungi: Cryptococcus neoformans, a basidiomycete; Mucor circinelloides, a zygomycete; and Encephalitozoon cuniculi, a microsporidian.
Soo Chan Lee, Ph.D., received B.S. and M.S. degrees in biology and microbiology from Kyung Hee University, Seoul, South Korea, and earned his Ph.D. degree in December 2007 from Texas A&M University. As a Ph.D. student, he trained with Brian Shaw and was primarily interested in how Aspergillus nidulans, a filamentous fungus, establishes and maintains hyphal polarity. He is currently a postdoctoral scholar in the Heitman Laboratory at the Department of Molecular Genetics and Microbiology in the Duke University Medical Center. His research is focused on understanding the sexual development of three different groups of human-pathogenic fungi: Cryptococcus neoformans, a basidiomycete; Mucor circinelloides, a zygomycete; and Encephalitozoon cuniculi, a microsporidian.
 Min Ni, Ph.D., received a B.S. degree in biological sciences from the University of Science and Technology of China and an M.S. degree in Molecular Engineering of Biological and Chemical Systems from the Singapore-MIT Alliance. She then obtained her Ph.D. in Genetics at the University of Wisconsin—Madison in the laboratory of Jaehyuk Yu, where she investigated the regulatory mechanisms underlying sporogenesis in the model filamentous fungus Aspergillus nidulans. She is currently a postdoctoral fellow in the laboratory of Dr. Heitman at the Duke University Medical Center, where she is studying the phenotypic and genotypic changes during same-sex mating and the functions of certain mating-type locus genes during opposite- and same-sex mating in the human pathogen Cryptococcus neoformans.
Min Ni, Ph.D., received a B.S. degree in biological sciences from the University of Science and Technology of China and an M.S. degree in Molecular Engineering of Biological and Chemical Systems from the Singapore-MIT Alliance. She then obtained her Ph.D. in Genetics at the University of Wisconsin—Madison in the laboratory of Jaehyuk Yu, where she investigated the regulatory mechanisms underlying sporogenesis in the model filamentous fungus Aspergillus nidulans. She is currently a postdoctoral fellow in the laboratory of Dr. Heitman at the Duke University Medical Center, where she is studying the phenotypic and genotypic changes during same-sex mating and the functions of certain mating-type locus genes during opposite- and same-sex mating in the human pathogen Cryptococcus neoformans.
 Wenjun Li, M.D., Ph.D., received his M.D. degree in 1998 from Luoyang Medical School University, Henan Province, China, and served as a residency surgeon in the Jianghe Hospital, Henan Province, China, for three years. He received his Ph.D. degree in Microbiology in March 2008 at the Medical School of Marseille, University of the Mediterranean. His research in Marseille, France, was mainly focused on the molecular identification and characterization of some fastidious bacteria and human lice, the vectors of Rickettsia prowazekii, Borrelia recurrentis, and Bartonella quintana. He also participated in a comparative genomics study of Francisella tularensis. He joined the Heitman laboratory as a postdoctoral fellow in July 2008 to study the population genetics of C. neoformans and C. gattii as well as to characterize the mating type (MAT) locus in model and pathogenic fungi.
Wenjun Li, M.D., Ph.D., received his M.D. degree in 1998 from Luoyang Medical School University, Henan Province, China, and served as a residency surgeon in the Jianghe Hospital, Henan Province, China, for three years. He received his Ph.D. degree in Microbiology in March 2008 at the Medical School of Marseille, University of the Mediterranean. His research in Marseille, France, was mainly focused on the molecular identification and characterization of some fastidious bacteria and human lice, the vectors of Rickettsia prowazekii, Borrelia recurrentis, and Bartonella quintana. He also participated in a comparative genomics study of Francisella tularensis. He joined the Heitman laboratory as a postdoctoral fellow in July 2008 to study the population genetics of C. neoformans and C. gattii as well as to characterize the mating type (MAT) locus in model and pathogenic fungi.
 Cecelia Shertz graduated with a B.S. in biology from Juniata College in Huntingdon, PA, in 2007. At Juniata College, she studied the function of the immunoglobulin-degrading enzyme of Streptococcus (IdeS) with Michael Boyle. She is currently attending graduate school at Duke University, investigating the evolution, maintenance, and loss of sex in basal fungi and novel mechanisms of phenotypic variation in basal fungi.
Cecelia Shertz graduated with a B.S. in biology from Juniata College in Huntingdon, PA, in 2007. At Juniata College, she studied the function of the immunoglobulin-degrading enzyme of Streptococcus (IdeS) with Michael Boyle. She is currently attending graduate school at Duke University, investigating the evolution, maintenance, and loss of sex in basal fungi and novel mechanisms of phenotypic variation in basal fungi.
 Joseph Heitman, M.D., Ph.D., is the Chair and James B. Duke Professor of the Department of Molecular Genetics and Microbiology at Duke University. He received B.S. and M.S. degrees in chemistry and biochemistry with general and special honors from the University of Chicago and M.D. and Ph.D. degrees from the Medical Scientist Training Program of Cornell University Medical College and the Rockefeller University and was an EMBO long-term postdoctoral fellow at the Biocenter in Basel, Switzerland. He joined the faculty at Duke in 1992. His research focuses on the evolution of sex in fungi and the roles of sexual reproduction in microbial pathogens, how cells sense and respond to nutrients and the environment, the targets and mechanisms of action of immunosuppressive and antimicrobial drugs, and the genetic and molecular basis of microbial pathogenesis and development.
Joseph Heitman, M.D., Ph.D., is the Chair and James B. Duke Professor of the Department of Molecular Genetics and Microbiology at Duke University. He received B.S. and M.S. degrees in chemistry and biochemistry with general and special honors from the University of Chicago and M.D. and Ph.D. degrees from the Medical Scientist Training Program of Cornell University Medical College and the Rockefeller University and was an EMBO long-term postdoctoral fellow at the Biocenter in Basel, Switzerland. He joined the faculty at Duke in 1992. His research focuses on the evolution of sex in fungi and the roles of sexual reproduction in microbial pathogens, how cells sense and respond to nutrients and the environment, the targets and mechanisms of action of immunosuppressive and antimicrobial drugs, and the genetic and molecular basis of microbial pathogenesis and development.
REFERENCES
- 1.Adams, T. H., J. K. Wieser, and J. H. Yu. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alby, K., D. Schaefer, and R. J. Bennett. 2009. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature 460:890-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexopoulos, C. J. 1962. Introductory mycology, 2nd ed. John Wiley & Sons, New York, NY.
- 4.Alexopoulos, C. J., C. W. Mims, and M. Blackwell. 1996. Introductory mycology, 4th ed. John Wiley & Sons, New York, NY.
- 5.Alvarez-Perez, S., J. L. Blanco, P. Alba, and M. E. Garcia. 2009. Mating type and invasiveness are significantly associated in Aspergillus fumigatus. Med. Mycol. 26:1-6. [DOI] [PubMed] [Google Scholar]
- 6.Arcangioli, B., and G. Thon. 2004. Mating-type cassettes: structure, switching and silencing. In R. Egel (ed.), The molecular biology of Schizosaccharomyces pombe. Springer-Verlag, Berlin, Germany.
- 7.Ashe, A., and B. Oldroyd. 2002. Genetic determination of caste in harvester ants. Trends Ecol. Evol. 17:448-449. [Google Scholar]
- 8.Bakkeren, G., J. Kamper, and J. Schirawski. 2008. Sex in smut fungi: structure, function and evolution of mating-type complexes. Fungal Genet. Biol. 45:S15-S21. [DOI] [PubMed] [Google Scholar]
- 9.Bakkeren, G., and J. W. Kronstad. 1994. Linkage of mating-type loci distinguishes bipolar from tetrapolar mating in basidiomycetous smut fungi. Proc. Natl. Acad. Sci. U. S. A. 91:7085-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldauf, S., and J. D. Palmer. 1993. Animals and fungi are each other's closest relatives: congruent evidence from multiple proteins. Proc. Natl. Acad. Sci. U. S. A. 90:11558-11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banuett, F. 2007. History of the mating types in Ustilago maydis, p. 351-375. In J. Heitman, J. W. Kronstad, J. W. Taylor, and L. A. Casselton (ed.), Sex in fungi. ASM Press, Washington, DC.
- 12.Banuett, F., and I. Herskowitz. 1989. Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. U. S. A. 86:5878-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barker, B. M., K. A. Jewell, S. Kroken, and M. J. Orbach. 2007. The population biology of Coccidioides: epidemiologic implications for disease outbreaks. Ann. N. Y. Acad. Sci. 1111:147-163. [DOI] [PubMed] [Google Scholar]
- 14.Barr, D. J. S. 1992. Evolution and kingdoms of organisms from the perspective of a mycologist. Mycologia 84:1-11. [Google Scholar]
- 15.Barsoum, E., P. Martinez, and S. U. Astrom. 2010. α3, a transposable element that promotes host sexual reproduction. Genes Dev. 24:10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becnel, J. J., and T. G. Andreadis. 1999. Microsporidia in insects, p. 1-6. In M. Wittner (ed.), The microsporidia and microsporidiosis. ASM Press, Washington, DC.
- 17.Bell, G. 1982. The masterpiece of nature: the evolution and genetics of sexuality. Croom Helm, London, United Kingdom.
- 18.Bennett, R. J., and A. D. Johnson. 2003. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 22:2505-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett, R. J., and A. D. Johnson. 2006. The role of nutrient regulation and the Gpa2 protein in the mating pheromone response of C. albicans. Mol. Microbiol. 62:100-119. [DOI] [PubMed] [Google Scholar]
- 20.Benny, G. L., R. A. Humber, and J. B. Morton. 2001. Zygomycota: zygomycetes, p. 113-146. In D. J. McLaughlin, E. G. McLaughlin, and P. A. Lemke (ed.), The mycota VIIA, systematics and evolution. Springer-Verlag, Berlin, Germany.
- 21.Berger, L., A. D. Hyatt, R. Speare, and J. E. Longcore. 2005. Life cycle stages of the amphibian chytrid Batrachochytrium dendrobatidis. Dis. Aquat. Organ. 68:51-63. [DOI] [PubMed] [Google Scholar]
- 22.Bialek, R., A. Ibricevic, A. Fothergill, and D. Begerow. 2000. Small subunit ribosomal DNA sequence shows Paracoccidioides brasiliensis closely related to Blastomyces dermatitidis. J. Clin. Microbiol. 38:3190-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blakeslee, A. F. 1904. Sexual reproduction in the Mucorineae. Proc. Am. Acad. Arts Sci. 40:205-319. [Google Scholar]
- 24.Bölker, M., M. Urban, and R. Kahmann. 1992. The a mating type locus of U. maydis specifies cell signaling components. Cell 68:441-450. [DOI] [PubMed] [Google Scholar]
- 25.Bortfeld, M., K. Auffarth, R. Kahmann, and C. W. Basse. 2004. The Ustilago maydis a2 mating-type locus genes lga2 and rga2 compromise pathogenicity in the absence of the mitochondrial p32 family protein Mrb1. Plant Cell 16:2233-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradsher, R. W. 1988. Systemic fungal infections: diagnosis and treatment. I. Blastomycosis. Infect. Dis. Clin. North Am. 2:877-898. [PubMed] [Google Scholar]
- 27.Brandt, M. E., L. C. Hutwagner, L. A. Klug, W. S. Baughman, D. Rimland, E. A. Graviss, R. J. Hamill, C. Thomas, P. G. Pappas, A. L. Reingold, and R. W. Pinner. 1996. Molecular subtype distribution of Cryptococcus neoformans in four areas of the United States. Cryptococcal disease active surveillance group. J. Clin. Microbiol. 34:912-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bubnick, M., and A. G. Smulian. 2007. The MAT1 locus of Histoplasma capsulatum is responsive in a mating type-specific manner. Eukaryot. Cell 6:616-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bui, T., X. Lin, R. Malik, J. Heitman, and D. Carter. 2008. Isolates of Cryptococcus neoformans from infected animals reveal genetic exchange in unisexual, alpha mating type populations. Eukaryot. Cell 7:1771-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burt, A., D. A. Carter, G. L. Koenig, T. J. White, and J. W. Taylor. 1996. Molecular markers reveal cryptic sex in the human pathogen Coccidioides immitis. Proc. Natl. Acad. Sci. U. S. A. 93:770-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butler, G. 2010. Fungal sex and pathogenesis. Clin. Microbiol. Rev. 23:140-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butler, G., C. Kenny, A. Fagan, C. Kurischko, C. Gaillardin, and K. H. Wolfe. 2004. Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc. Natl. Acad. Sci. U. S. A. 101:1632-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butler, G., M. D. Rasmussen, M. F. Lin, M. A. S. Santos, S. Sakthikumar, C. A. Munro, E. Rheinbay, M. Grabherr, A. Forche, J. L. Reedy, I. Agrafioti, M. B. Arnaud, S. Bates, A. J. P. Brown, S. Brunke, M. C. Costanzo, D. A. Fitzpatrick, P. W. J. de Groot, D. Harris, L. L. Hoyer, B. Hube, F. M. Klis, C. Kodira, N. Lennard, M. E. Logue, R. Martin, A. M. Neiman, E. Nikolaou, M. A. Quail, J. Quinn, M. C. Santos, F. F. Schmitzberger, G. Sherlock, P. Shah, K. A. T. Silverstein, M. S. Skrzypek, D. Soll, R. Staggs, I. Stansfield, M. P. H. Stumpf, P. E. Sudbery, T. Srikantha, Q. Zeng, J. Berman, M. Berriman, J. Heitman, N. A. R. Gow, M. C. Lorenz, B. W. Birren, M. Kellis, and C. A. Cuomo. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlile, M. J., and L. Machlis. 1965. The response of male gametes of Allomyces to the sexual hormone sirenin. Am. J. Bot. 52:478-483. [Google Scholar]
- 34a.Carroll, L. 1871. Through the looking-glass. Macmillan Publishers Ltd., London, United Kingdom.
- 35.Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans. ASM Press, Washington, DC.
- 36.Casselton, L., and M. Zolan. 2002. The art and design of genetic screens: filamentous fungi. Nat. Rev. Genet. 3:683-697. [DOI] [PubMed] [Google Scholar]
- 37.Casselton, L. A. 2002. Mate recognition in fungi. Heredity 88:142-147. [DOI] [PubMed] [Google Scholar]
- 38.Casselton, L. A., and N. S. Olesnicky. 1998. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol. Mol. Biol. Rev. 62:55-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cogliati, M., M. C. Esposto, D. L. Clarke, B. L. Wickes, and M. A. Viviani. 2001. Origin of Cryptococcus neoformans var. neoformans diploid strains. J. Clin. Microbiol. 39:3889-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coppin, E., R. Debuchy, S. Arnaise, and M. Picard. 1997. Mating types and sexual development in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 61:411-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daniels, K. J., T. Srikantha, S. R. Lockhart, C. Pujol, and D. R. Soll. 2006. Opaque cells signal white cells to form biofilms in Candida albicans. EMBO J. 25:2240-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Debets, F., K. Swart, R. F. Hoekstra, and C. J. Bos. 1993. Genetic maps of eight linkage groups of Aspergillus niger based on mitotic mapping. Curr. Genet. 23:47-53. [DOI] [PubMed] [Google Scholar]
- 43.Delneri, D., I. Colson, S. Grammenoudi, I. N. Roberts, E. J. Louis, and S. G. Oliver. 2003. Engineering evolution to study speciation in yeasts. Nature 422:68-72. [DOI] [PubMed] [Google Scholar]
- 44.Deng, Z., J. L. Ribas, D. W. Gibson, and D. H. Connor. 1988. Infections caused by Penicillium marneffei in China and Southeast Asia: review of eighteen published cases and report of four more Chinese cases. Rev. Infect. Dis. 10:640-652. [DOI] [PubMed] [Google Scholar]
- 45.Desai, M. M., D. S. Fisher, and A. W. Murray. 2007. The speed of evolution and maintenance of variation in asexual populations. Curr. Biol. 17:385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dettman, J. R., D. J. Jacobson, and J. W. Taylor. 2003. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57:2703-2720. [DOI] [PubMed] [Google Scholar]
- 47.Dettman, J. R., D. J. Jacobson, E. Turner, A. Pringle, and J. W. Taylor. 2003. Reproductive isolation and phylogenetic divergence in Neurospora: comparing methods of species recognition in a model eukaryote. Evolution 57:2721-2741. [DOI] [PubMed] [Google Scholar]
- 48.Didier, E. S., J. A. Maddry, P. J. Brindley, M. E. Stovall, and P. J. Didier. 2005. Therapeutic strategies for human microsporidia infections. Expert Rev. Anti Infect. Ther. 3:419-434. [DOI] [PubMed] [Google Scholar]
- 49.Didier, E. S., and L. M. Weiss. 2006. Microsporidiosis: current status. Curr. Opin. Infect. Dis. 19:485-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diezmann, S., C. J. Cox, G. Schonian, R. J. Vilgalys, and T. G. Mitchell. 2004. Phylogeny and evolution of medical species of Candida and related taxa: a multigenic analysis. J. Clin. Microbiol. 42:5624-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dumitru, R., D. H. M. L. P. Navarathna, C. P. Semighini, C. G. Elowsky, R. V. Dumitru, D. Dignard, M. Whiteway, A. L. Atkin, and K. W. Nickerson. 2007. In vivo and in vitro anaerobic mating in Candida albicans. Eukaryot. Cell 6:465-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dyer, P. S. 2008. Evolutionary biology: genomic clues to original sex in fungi. Curr. Biol. 18:R207-R209. [DOI] [PubMed] [Google Scholar]
- 53.Edlind, T. D., J. Li, G. S. Visvesvara, M. H. Vodkin, G. L. McLaughlin, and S. K. Katiyar. 1996. Phylogenetic analysis of beta-tubulin sequences from amitochondrial protozoa. Mol. Phylogenet. Evol. 5:359-367. [DOI] [PubMed] [Google Scholar]
- 54.Emerson, R. 1941. An experimental study of the life cycles and taxonomy of Allomyces. Lloydia 4:77-144. [Google Scholar]
- 55.Ezov, T. K., S.-L. Chang, Z. E. Frenkel, A. V. Segre, M. Bahalul, A. W. Murray, J.-Y. Leu, A. Korol, and Y. Kashi. 2010. Heterothallism in Saccharomyces cerevisiae isolates from nature: effect of HO locus on the mode of reproduction. Mol. Ecol. 19:121-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fedler, M., K.-S. Luh, K. Stelter, F. Nieto-Jacobo, and C. W. Basse. 2009. The a2 mating-type locus genes lga2 and rga2 direct uniparental mitochondrial DNA (mtDNA) inheritance and constrain mtDNA recombination during sexual development of Ustilago maydis. Genetics 181:847-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Findley, K., M. Rodriguez-Carres, B. Metin, J. Kroiss, A. Fonseca, R. Vilgalys, and J. Heitman. 2009. Phylogeny and phenotypic characterization of pathogenic Cryptococcus species and closely related saprobic taxa in the Tremellales. Eukaryot. Cell 8:353-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finley, H. E. 1943. The conjugation of Vorticella microstoma. Trans. Am. Microsc. Soc. 62:97-121. [Google Scholar]
- 59.Firon, A., A. Beauvais, J. P. Latge, E. Couve, M. C. Grosjean-Cournoyer, and C. D'Enfert. 2002. Characterization of essential genes by parasexual genetics in the human fungal pathogen Aspergillus fumigatus: impact of genomic rearrangements associated with electroporation of DNA. Genetics 161:1077-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fischer, G., S. A. James, I. N. Roberts, S. G. Oliver, and E. J. Louis. 2000. Chromosomal evolution in Saccharomyces. Nature 405:451-454. [DOI] [PubMed] [Google Scholar]
- 61.Fisher, M. C., D. Aanensen, S. de Hoog, and N. Vanittanakom. 2004. Multilocus microsatellite typing system for Penicillium marneffei reveals spatially structured populations. J. Clin. Microbiol. 42:5065-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fisher, M. C., W. P. Hanage, S. de Hoog, E. Johnson, M. D. Smith, N. J. White, and N. Vanittanakom. 2005. Low effective dispersal of asexual genotypes in heterogeneous landscapes by the endemic pathogen Penicillium marneffei. PLoS Pathog. 1:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fisher, R. A. 1930. The genetical theory of natural selection. Oxford University Press, London, United Kingdom.
- 64.Forche, A., K. Alby, D. Schaefer, A. D. Johnson, J. Berman, and R. J. Bennett. 2008. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 6:e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Franzot, S. P., J. S. Hamdan, B. P. Currie, and A. Casadevall. 1997. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for both local genetic differences and a global clonal population structure. J. Clin. Microbiol. 35:2243-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fraser, J. A., S. Diezmann, R. L. Subaran, A. Allen, K. B. Lengeler, F. S. Dietrich, and J. Heitman. 2004. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2:e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fraser, J. A., and J. Heitman. 2005. Chromosomal sex-determining regions in animals, plants and fungi. Curr. Opin. Genet. Dev. 15:645-651. [DOI] [PubMed] [Google Scholar]
- 68.Fraser, J. A., and J. Heitman. 2004. Evolution of fungal sex chromosomes. Mol. Microbiol. 51:299-306. [DOI] [PubMed] [Google Scholar]
- 69.Fraser, J. A., Y.-P. Hsueh, K. Findley, and J. Heitman. 2007. Evolution of the mating-type locus: the basidiomycetes, p. 19-34. In J. Heitman, J. W. Kronstad, J. W. Taylor, and L. A. Casselton (ed.), Sex in fungi. ASM Press, Washington, DC.
- 70.Fraser, J. A., J. E. Stajich, E. J. Tarcha, G. T. Cole, D. O. Inglis, A. Sil, and J. Heitman. 2007. Evolution of the mating type locus: insights gained from the dimorphic primary fungal pathogens Histoplasma capsulatum, Coccidioides immitis, and Coccidioides posadasii. Eukaryot. Cell 6:622-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Galagan, J. E., S. E. Calvo, C. Cuomo, L.-J. Ma, J. R. Wortman, S. Batzoglou, S.-I. Lee, M. Basturkmen, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. D'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. A. Penalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. W. Birren. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105-1115. [DOI] [PubMed] [Google Scholar]
- 72.Galgoczy, D. J., A. Cassidy-Stone, M. Llinas, S. M. O'Rourke, I. Herskowitz, J. L. DeRisi, and A. D. Johnson. 2004. Genomic dissection of the cell-type-specification circuit in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 101:18069-18074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Galitski, T., A. J. Saldanha, C. A. Styles, E. S. Lander, and G. R. Fink. 1999. Ploidy regulation of gene expression. Science 285:251-254. [DOI] [PubMed] [Google Scholar]
- 74.Gallegos, A., D. J. Jacobson, N. B. Raju, M. P. Skupski, and D. O. Natvig. 2000. Suppressed recombination and a pairing anomaly on the mating-type chromosome of Neurospora tetrasperma. Genetics 154:623-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Geiger, J., D. Wessels, S. R. Lockhart, and D. R. Soll. 2004. Release of a potent polymorphonuclear leukocyte chemoattractant is regulated by white-opaque switching in Candida albicans. Infect. Immun. 72:667-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Geiser, D. M., M. A. Klich, J. C. Frisvad, S. W. Peterson, J. Varga, and R. A. Samson. 2007. The current status of species recognition and identification in Aspergillus. Stud. Mycol. 59:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Geiser, D. M., W. E. Timberlake, and M. L. Arnold. 1996. Loss of meiosis in Aspergillus. Mol. Biol. Evol. 13:809-817. [DOI] [PubMed] [Google Scholar]
- 78.Georg, L. K. 1960. Epidemiology of the dermatophytoses sources of infection, modes of transmission and epidemicity. Ann. N. Y. Acad. Sci. 89:69-77. [DOI] [PubMed] [Google Scholar]
- 79.Gill, E. E., and N. M. Fast. 2006. Assessing the microsporidia-fungi relationship: combined phylogenetic analysis of eight genes. Gene 375:103-109. [DOI] [PubMed] [Google Scholar]
- 80.Giraud, T., G. Refregier, M. Le Gac, D. M. de Vienne, and M. E. Hood. 2008. Speciation in fungi. Fungal Genet. Biol. 45:791-802. [DOI] [PubMed] [Google Scholar]
- 81.Giraud, T., R. Yockteng, M. Lopez-Villavicencio, G. Refregier, and M. E. Hood. 2008. Mating system of the anther smut fungus Microbotryum violaceum: selfing under heterothallism. Eukaryot. Cell 7:765-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goddard, M. R., H. C. J. Godfray, and A. Burt. 2005. Sex increases the efficacy of natural selection in experimental yeast populations. Nature 434:636-640. [DOI] [PubMed] [Google Scholar]
- 83.Graser, Y., M. Volovsek, J. Arrington, G. Schonian, W. Presber, T. G. Mitchell, and R. Vilgalys. 1996. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc. Natl. Acad. Sci. U. S. A. 93:12473-12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Graves, J. A. M. 2006. Sex chromosome specialization and degeneration in mammals. Cell 124:901-914. [DOI] [PubMed] [Google Scholar]
- 85.Graves, J. A. M. 2008. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu. Rev. Genet. 42:565-586. [DOI] [PubMed] [Google Scholar]
- 86.Greig, D., R. H. Borts, and E. J. Louis. 1998. The effect of sex on adaptation to high temperature in heterozygous and homozygous yeast. Proc. Biol. Sci. 265:1017-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Greig, D., E. J. Louis, R. H. Borts, and M. Travisano. 2002. Hybrid speciation in experimental populations of yeast. Science 298:1773-1775. [DOI] [PubMed] [Google Scholar]
- 88.Greig, D., M. Travisano, E. J. Louis, and R. H. Borts. 2003. A role for the mismatch repair system during incipient speciation in Saccharomyces. J. Evol. Biol. 16:429-437. [DOI] [PubMed] [Google Scholar]
- 89.Griswold, C. K., and J. Masel. 2009. Complex adaptations can drive the evolution of the capacitor [PSI], even with realistic rates of yeast sex. PLoS Genet. 5:e1000517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grosse, V., and S. Krappmann. 2008. The asexual pathogen Aspergillus fumigatus expresses functional determinants of Aspergillus nidulans sexual development. Eukaryot. Cell 7:1724-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Haber, J. E. 2007. Decisions, decisions: donor preference during budding yeast mating-type switching, p. 159-170. In J. Heitman, J. W. Kronstad, J. W. Taylor, and L. A. Casselton (ed.), Sex in fungi. ASM Press, Washington, DC.
- 92.Haber, J. E. 2003. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32:561-599. [DOI] [PubMed] [Google Scholar]
- 93.Halliday, C. L., and D. A. Carter. 2003. Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var. gattii isolates from Australia. J. Clin. Microbiol. 41:703-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hanna, W. F. 1929. Studies in the physiology and cytology of Ustilago zeae and Sorosporium reilianum. Phytophathology 19:415-443. [Google Scholar]
- 95.Harmsen, D., A. Schwinn, M. Weig, E. B. Brocker, and J. Heesemann. 1995. Phylogeny and dating of some pathogenic keratinophilic fungi using small subunit ribosomal RNA. J. Med. Vet. Mycol. 33:299-303. [DOI] [PubMed] [Google Scholar]
- 96.Hatch, W. R. 1935. Gametogenesis in Allomyces arbuscula. Ann. Bot. 49:623-649. [Google Scholar]
- 97.Hazard, E. I., and J. W. Brookbank. 1984. Karyogamy and meiosis in an Amblyospora sp. (Microspora) in the mosquito Culex salinarius. J. Invertebr. Pathol. 44:3-11. [Google Scholar]
- 98.Heitman, J. 2009. Microbial genetics: love the one you're with. Nature 460:807-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Heitman, J., J. W. Kronstad, J. W. Taylor, and L. A. Casselton (ed.). 2007. Sex in fungi. ASM Press, Washington, DC.
- 100.Heng, H. H. 2007. Elimination of altered karyotypes by sexual reproduction preserves species identity. Genome 50:517-524. [DOI] [PubMed] [Google Scholar]
- 101.Herman, A., and H. Roman. 1966. Allele specific determinations of homothallism in Saccharomyces lactis. Genetics 53:727-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Herskowitz, I., J. Rine, and J. Strathern. 1992. Mating-type determination and mating type interconversion in Saccharomyces cerevisiae, p. 583-656. In J. R. Broach, J. R. Pringle, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 103.Hicks, J. B., and I. Herskowitz. 1976. Evidence for a new diffusible element of mating pheromone in yeast. Nature 260:246-248. [DOI] [PubMed] [Google Scholar]
- 104.Hirai, A., R. Kano, Y. Nakamura, S. Watanabe, and A. Hasegawa. 2003. Molecular taxonomy of dermatophytes and related fungi by chitin synthase 1 (CHS1) gene sequences. Antonie Van Leeuwenhoek 83:11-20. [DOI] [PubMed] [Google Scholar]
- 105.Hiremath, S. S., A. Chowdhary, T. Kowshik, H. S. Randhawa, S. Sun, and J. Xu. 2008. Long-distance dispersal and recombination in environmental populations of Cryptococcus neoformans var. grubii from India. Microbiology 154:1513-1524. [DOI] [PubMed] [Google Scholar]
- 106.Hironaga, M., S. Tanaka, and S. Watanabe. 1982. Distribution of mating types among clinical isolates of the Microsporum gypseum complex. Mycopathologia 77:31-35. [DOI] [PubMed] [Google Scholar]
- 107.Hood, M. E. 2002. Dimorphic mating-type chromosomes in the fungus Microbotryum violaceum. Genetics 160:457-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hood, M. E., J. Antonovics, and B. Koskella. 2004. Shared forces of sex chromosome evolution in haploid-mating and diploid-mating organisms: Microbotryum violaceum and other model organisms. Genetics 168:141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Horn, B. W., J. H. Ramirez-Prado, and I. Carbone. 2009. Sexual reproduction and recombination in the aflatoxin-producing fungus Aspergillus parasiticus. Fungal Genet. Biol. 46:169-175. [DOI] [PubMed] [Google Scholar]
- 110.Hsueh, Y.-P., J. A. Fraser, and J. Heitman. 2008. Transitions in sexuality: recapitulation of an ancestral tri- and tetrapolar mating system in Cryptococcus neoformans. Eukaryot. Cell 7:1847-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hsueh, Y.-P., and J. Heitman. 2008. Orchestration of sexual reproduction and virulence by the fungal mating-type locus. Curr. Opin. Microbiol. 11:517-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang, G., T. Srikantha, N. Sahni, S. Yi, and D. R. Soll. 2009. CO2 regulates white-to-opaque switching in Candida albicans. Curr. Biol. 19:330-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Huang, G., H. Wang, S. Chou, X. Nie, J. Chen, and H. Liu. 2006. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 103:12813-12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hull, C. M., M. J. Boily, and J. Heitman. 2005. Sex-specific homeodomain proteins Sxi1α and Sxi2a coordinately regulate sexual development in Cryptococcus neoformans. Eukaryot. Cell 4:526-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hull, C. M., R. C. Davidson, and J. Heitman. 2002. Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1α. Genes Dev. 16:3046-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hull, C. M., and A. D. Johnson. 1999. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science 285:1271-1275. [DOI] [PubMed] [Google Scholar]
- 117.Hull, C. M., R. M. Raisner, and A. D. Johnson. 2000. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289:307-310. [DOI] [PubMed] [Google Scholar]
- 118.Hunter, N., S. R. Chambers, E. J. Louis, and R. H. Borts. 1996. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 15:1726-1733. [PMC free article] [PubMed] [Google Scholar]
- 119.Idnurm, A., T. Y. James, and R. Vilgalys. 2007. Sex in the rest: mysterious mating in the Chytridiomycota and Zygomycota, p. 407-418. In J. Heitman, J. W. Kronstad, J. W. Taylor, and L. A. Casselton (ed.), Sex in fungi. ASM Press, Washington, DC.
- 120.Idnurm, A., F. J. Walton, A. Floyd, and J. Heitman. 2008. Identification of the sex genes in an early diverged fungus. Nature 451:193-196. [DOI] [PubMed] [Google Scholar]
- 121.Reference deleted.
- 122.James, T., D. Porter, C. Leander, R. Vilgalys, and J. Longcore. 2000. Molecular phylogenetics of the Chytridiomycota supports the utility of ultrastructural data in chytrid systematics. Can. J. Bot. 78:226-350. [Google Scholar]
- 123.Reference deleted.
- 124.James, T. Y., F. Kauff, C. L. Schoch, P. B. Matheny, V. Hofstetter, C. J. Cox, G. Celio, C. Gueidan, E. Fraker, J. Miadlikowska, H. T. Lumbsch, A. Rauhut, V. Reeb, A. E. Arnold, A. Amtoft, J. E. Stajich, K. Hosaka, G.-H. Sung, D. Johnson, B. O'Rourke, M. Crockett, M. Binder, J. M. Curtis, J. C. Slot, Z. Wang, A. W. Wilson, A. Schuszler, J. E. Longcore, K. O'Donnell, S. Mozley-Standridge, D. Porter, P. M. Letcher, M. J. Powell, J. W. Taylor, M. M. White, G. W. Griffith, D. R. Davies, R. A. Humber, J. B. Morton, J. Sugiyama, A. Y. Rossman, J. D. Rogers, D. H. Pfister, D. Hewitt, K. Hansen, S. Hambleton, R. A. Shoemaker, J. Kohlmeyer, B. Volkmann-Kohlmeyer, R. A. Spotts, M. Serdani, P. W. Crous, K. W. Hughes, K. Matsuura, E. Langer, G. Langer, W. A. Untereiner, R. Lucking, B. Budel, D. M. Geiser, A. Aptroot, P. Diederich, I. Schmitt, M. Schultz, R. Yahr, D. S. Hibbett, F. Lutzoni, D. J. McLaughlin, J. W. Spatafora, and R. Vilgalys. 2006. Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 443:818-822. [DOI] [PubMed] [Google Scholar]
- 125.James, T. Y., A. P. Litvintseva, R. Vilgalys, J. A. Morgan, J. W. Taylor, M. C. Fisher, L. Berger, C. Weldon, L. du Preez, and J. E. Longcore. 2009. Rapid global expansion of the fungal disease chytridiomycosis into declining and healthy amphibian populations. PLoS Pathog. 5:e1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Johnson, A. D. 1995. Molecular mechanisms of cell-type determination in budding yeast. Curr. Opin. Genet. Dev. 5:552-558. [DOI] [PubMed] [Google Scholar]
- 127.Kahmann, R., and J. Schirawski. 2007. Mating in the smut fungi: from a to b to the downstream cascades, p. 377-387. In J. Heitman, J. W. Kronstad, J. W. Taylor, and L. A. Casselton (ed.), Sex in fungi. ASM Press, Washington, DC.
- 128.Kale, S. P., J. W. Cary, C. Baker, D. Walker, D. Bhatnagar, and J. W. Bennett. 2003. Genetic analysis of morphological variants of Aspergillus parasiticus deficient in secondary metabolite production. Mycol. Res. 107:831-840. [DOI] [PubMed] [Google Scholar]
- 129.Karos, M., Y. C. Chang, C. M. McClelland, D. L. Clarke, J. Fu, B. L. Wickes, and K. J. Kwon-Chung. 2000. Mapping of the Cryptococcus neoformans MATα locus: presence of mating type-specific mitogen-activated protein kinase cascade homologs. J. Bacteriol. 182:6222-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kassir, Y., D. Granot, and G. Simchen. 1988. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell 52:853-862. [DOI] [PubMed] [Google Scholar]
- 131.Keeling, P. 2003. Congruent evidence from alpha-tubulin and beta-tubulin gene phylogenies for a zygomycete origin of microsporidia. Fungal Genet. Biol. 38:298-309. [DOI] [PubMed] [Google Scholar]
- 132.Keeling, P. 2009. Five questions about microsporidia. PLoS Pathog. 5:e1000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Keeling, P. J., and W. F. Doolittle. 1996. Alpha-tubulin from early-diverging eukaryotic lineages and the evolution of the tubulin family. Mol. Biol. Evol. 13:1297-1305. [DOI] [PubMed] [Google Scholar]
- 134.Keeling, P. J., and N. M. Fast. 2002. Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu. Rev. Microbiol. 56:93-116. [DOI] [PubMed] [Google Scholar]
- 135.King, K. C., L. F. Delph, J. Jokela, and C. M. Lively. 2009. The geographic mosaic of sex and the Red Queen. Curr. Biol. 19:1438-1441. [DOI] [PubMed] [Google Scholar]
- 136.Koch, W. J. 1959. The sexual stage of Chytriomyces. J. Elisha Mitchell Sci. Soc. 75:66. [Google Scholar]
- 137.Kohn, L. M. 2005. Mechanisms of fungal speciation. Annu. Rev. Phytopathol. 43:279-308. [DOI] [PubMed] [Google Scholar]
- 138.Koufopanou, V., A. Burt, T. Szaro, and J. W. Taylor. 2001. Gene genealogies, cryptic species, and molecular evolution in the human pathogen Coccidioides immitis and relatives (Ascomycota, Onygenales). Mol. Biol. Evol. 18:1246-1258. [DOI] [PubMed] [Google Scholar]
- 139.Kronstad, J. W. 2007. Self-fertility: the genetics of sex in lonely fungi. Curr. Biol. 17:R843-R845. [DOI] [PubMed] [Google Scholar]
- 140.Kronstad, J. W., and C. Staben. 2003. Mating type in filamentous fungi. Annu. Rev. Genet. 31:245-276. [DOI] [PubMed] [Google Scholar]
- 141.Kwon, K. J., and K. B. Raper. 1967. Sexuality and cultural characteristics of Aspergillus heterothallicus. Am. J. Bot. 54:36. [PubMed] [Google Scholar]
- 142.Kwon-Chung, K. J. 1972. Emmonsiella capsulata: perfect state of Histoplasma capsulatum. Science 177:368-369. [DOI] [PubMed] [Google Scholar]
- 143.Kwon-Chung, K. J. 1976. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68:821-833. [PubMed] [Google Scholar]
- 144.Kwon-Chung, K. J. 1975. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia 67:1197-1200. [PubMed] [Google Scholar]
- 145.Kwon-Chung, K. J. 1976. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia 68:943-946. [PubMed] [Google Scholar]
- 146.Kwon-Chung, K. J. 1972. Sexual stage of Histoplasma capsulatum. Science 175:326. [DOI] [PubMed] [Google Scholar]
- 147.Kwon-Chung, K. J., and J. E. Bennett. 1978. Distribution of α and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 108:337-340. [DOI] [PubMed] [Google Scholar]
- 148.Kwon-Chung, K. J., and J. A. Sugui. 2009. Sexual reproduction in Aspergillus species of medical or economical importance: why so fastidious? Trends Microbiol. 17:481-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kwon-Chung, K. J., R. J. Weeks, and H. W. Larsh. 1974. Studies on Emmonsiella capsulata (Histoplasma capsulatum). II. Distribution of the two mating types in 13 endemic states of the United States. Am. J. Epidemiol. 99:44-49. [DOI] [PubMed] [Google Scholar]
- 150.Lachke, S. A., S. R. Lockhart, K. J. Daniels, and D. R. Soll. 2003. Skin facilitates Candida albicans mating. Infect. Immun. 71:4970-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lang, B., C. O'Kelly, T. Nerad, M. Gray, and G. Burger. 2002. The closest unicellular relatives of animals. Curr. Biol. 12:1773-1778. [DOI] [PubMed] [Google Scholar]
- 152.Leclerc, M. C., H. Philippe, and E. Gueho. 1994. Phylogeny of dermatophytes and dimorphic fungi based on large subunit ribosomal RNA sequence comparisons. J. Med. Vet. Mycol. 32:331-341. [DOI] [PubMed] [Google Scholar]
- 153.Lee, N., G. Bakkeren, K. Wong, J. E. Sherwood, and J. W. Kronstad. 1999. The mating-type and pathogenicity locus of the fungus Ustilago hordei spans a 500-kb region. Proc. Natl. Acad. Sci. U. S. A. 96:15026-15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lee, S. C., N. Corradi, E. J. Byrnes, S. Torres-Martinez, F. S. Dietrich, P. J. Keeling, and J. Heitman. 2008. Microsporidia evolved from ancestral sexual fungi. Curr. Biol. 18:1675-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lee, S. C., L. M. Weiss, and J. Heitman. 2009. Generation of genetic diversity in microsporidia via sexual reproduction and horizontal gene transfer. Commun. Integr. Biol. 2:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155a.Lee, S. C., N. Corradi, S. Doan, F. S. Dietrich, P. J. Keeling, and J. Heitman. Evolution of the sex-related locus and genomic features shared in microsporidia and fungi. PLoS One, in press. [DOI] [PMC free article] [PubMed]
- 156.Lengeler, K. B., G. M. Cox, and J. Heitman. 2001. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect. Immun. 69:115-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lengeler, K. B., D. S. Fox, J. A. Fraser, A. Allen, K. Forrester, F. S. Dietrich, and J. Heitman. 2002. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell 1:704-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Leu, J.-Y., and A. W. Murray. 2006. Experimental evolution of mating discrimination in budding yeast. Curr. Biol. 16:280-286. [DOI] [PubMed] [Google Scholar]
- 159.Levadoux, W. L., K. F. Gregory, and A. Taylor. 1981. Sequential cold-sensitive mutations in Aspergillus fumigatus. II. Analysis by the parasexual cycle. Can. J. Microbiol. 27:295-303. [DOI] [PubMed] [Google Scholar]
- 160.Li, W., B. Metin, T. C. White, and J. Heitman. 2010. Organization and evolutionary trajectory of the mating type (MAT) locus in the dermatophyte and dimorphic fungal pathogens. Eukaryot. Cell 9:46-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lin, X. 2009. Cryptococcus neoformans: morphogenesis, infection, and evolution. Infect. Genet. Evol. 9:401-416. [DOI] [PubMed] [Google Scholar]
- 162.Lin, X., J. C. Huang, T. G. Mitchell, and J. Heitman. 2006. Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MATalpha allele enhances filamentation. PLoS Genet. 2:e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lin, X., C. M. Hull, and J. Heitman. 2005. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434:1017-1021. [DOI] [PubMed] [Google Scholar]
- 164.Lin, X., A. P. Litvintseva, K. Nielsen, S. Patel, A. Floyd, T. G. Mitchell, and J. Heitman. 2007. αADα hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet. 3:e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lin, X., S. Patel, A. P. Litvintseva, A. Floyd, T. G. Mitchell, and J. Heitman. 2009. Diploids in the Cryptococcus neoformans serotype A population homozygous for the α mating type originate via unisexual mating. PLoS Pathog. 5:e1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liti, G., D. B. Barton, and E. J. Louis. 2006. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics 174:839-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Litvintseva, A. P., R. E. Marra, K. Nielsen, J. Heitman, R. Vilgalys, and T. G. Mitchell. 2003. Evidence of sexual recombination among Cryptococcus neoformans serotype A isolates in sub-Saharan Africa. Eukaryot. Cell 2:1162-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Litvintseva, A. P., R. Thakur, L. B. Reller, and T. G. Mitchell. 2005. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in sub-Saharan Africa. J. Infect. Dis. 192:888-892. [DOI] [PubMed] [Google Scholar]
- 169.Litvintseva, A. P., R. Thakur, R. Vilgalys, and T. G. Mitchell. 2006. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics 172:2223-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lockhart, S. R., S. A. Messer, M. A. Pfaller, and D. J. Diekema. 2008. Lodderomyces elongisporus masquerading as Candida parapsilosis as a cause of bloodstream infections. J. Clin. Microbiol. 46:374-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lockhart, S. R., C. Pujol, K. J. Daniels, M. G. Miller, A. D. Johnson, M. A. Pfaller, and D. R. Soll. 2002. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics 162:737-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lockhart, S. R., R. Zhao, K. J. Daniels, and D. R. Soll. 2003. Alpha-pheromone-induced “shmooing” and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot. Cell 2:847-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Logue, M. E., S. Wong, K. H. Wolfe, and G. Butler. 2005. A genome sequence survey shows that the pathogenic yeast Candida parapsilosis has a defective MTLa1 allele at its mating type locus. Eukaryot. Cell 4:1009-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lohse, M. B., and A. D. Johnson. 2008. Differential phagocytosis of white versus opaque Candida albicans by Drosophila and mouse phagocytes. PLoS One 3:e1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lomer, C. J., R. P. Bateman, D. L. Johnson, J. Langewald, and M. Thomas. 2003. Biological control of locusts and grasshoppers. Annu. Rev. Entomol. 46:667-702. [DOI] [PubMed] [Google Scholar]
- 176.Longcore, J. E., A. P. Pessier, and D. K. Nichols. 1999. Batrachochytrium dendrobatidis gen. et sp. nov., a chytrid pathogenic to amphibians. Mycologia 91:219-227. [Google Scholar]
- 177.Lutzoni, F., F. Kauff, C. J. Cox, D. McLaughlin, G. Celio, B. Dentinger, M. Padamsee, D. Hibbett, T. Y. James, E. Baloch, M. Grube, V. Reeb, V. Hofstetter, C. Schoch, A. E. Arnold, J. Miadlikowska, J. Spatafora, D. Johnson, S. Hambleton, M. Crockett, R. Shoemaker, G.-H. Sung, R. Lucking, T. Lumbsch, K. O'Donnell, M. Binder, P. Diederich, D. Ertz, C. Gueidan, K. Hansen, R. C. Harris, K. Hosaka, Y.-W. Lim, B. Matheny, H. Nishida, D. Pfister, J. Rogers, A. Rossman, I. Schmitt, H. Sipman, J. Stone, J. Sugiyama, R. Yahr, and R. Vilgalys. 2004. Assembling the fungal tree of life: progress, classification, and evolution of subcellular traits. Am. J. Bot. 91:1446-1480. [DOI] [PubMed] [Google Scholar]
- 178.Ma, L.-J., A. S. Ibrahim, C. Skory, M. G. Grabherr, G. Burger, M. Butler, M. Elias, A. Idnurm, B. F. Lang, T. Sone, A. Abe, S. E. Calvo, L. M. Corrochano, R. Engels, J. Fu, W. Hansberg, J.-M. Kim, C. D. Kodira, M. J. Koehrsen, B. Liu, D. Miranda-Saavedra, S. O'Leary, L. Ortiz-Castellanos, R. Poulter, J. Rodriguez-Romero, J. Ruiz-Herrera, Y.-Q. Shen, Q. Zeng, J. Galagan, B. W. Birren, C. A. Cuomo, and B. L. Wickes. 2009. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet. 5:e1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Machida, M., K. Asai, M. Sano, T. Tanaka, T. Kumagai, G. Terai, K.-I. Kusumoto, T. Arima, O. Akita, Y. Kashiwagi, K. Abe, K. Gomi, H. Horiuchi, K. Kitamoto, T. Kobayashi, M. Takeuchi, D. W. Denning, J. E. Galagan, W. C. Nierman, J. Yu, D. B. Archer, J. W. Bennett, D. Bhatnagar, T. E. Cleveland, N. D. Fedorova, O. Gotoh, H. Horikawa, A. Hosoyama, M. Ichinomiya, R. Igarashi, K. Iwashita, P. R. Juvvadi, M. Kato, Y. Kato, T. Kin, A. Kokubun, H. Maeda, N. Maeyama, J.-I. Maruyama, H. Nagasaki, T. Nakajima, K. Oda, K. Okada, I. Paulsen, K. Sakamoto, T. Sawano, M. Takahashi, K. Takase, Y. Terabayashi, J. R. Wortman, O. Yamada, Y. Yamagata, H. Anazawa, Y. Hata, Y. Koide, T. Komori, Y. Koyama, T. Minetoki, S. Suharnan, A. Tanaka, K. Isono, S. Kuhara, N. Ogasawara, and H. Kikuchi. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157-1161. [DOI] [PubMed] [Google Scholar]
- 180.Machlis, L. 1973. The chemotactic activity of various sirenins and analogues and the uptake of sirenin by the sperm of Allomyces. Plant Physiol. 52:527-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Machlis, L. 1958. Evidence for a sexual hormone in Allomyces. Physiol. Plant. 11:181-192. [Google Scholar]
- 182.Machlis, L., W. H. Nutting, and H. Rapoport. 1968. The structure of sirenin. J. Am. Chem. Soc. 90:1674-1676. [Google Scholar]
- 183.Machlis, L., W. H. Nutting, M. W. Williams, and H. Rapoport. 1966. Production, isolation, and characterization of sirenin. Biochemistry 5:2147-2152. [DOI] [PubMed] [Google Scholar]
- 184.MacKay, V. L., S. K. Welch, M. Y. Insley, T. R. Manney, J. Holly, G. C. Saari, and M. L. Parker. 1988. The Saccharomyces cerevisiae BAR1 gene encodes an exported protein with homology to pepsin. Proc. Natl. Acad. Sci. U. S. A. 85:55-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Magee, B. B., and P. T. Magee. 2000. Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science 289:310-313. [DOI] [PubMed] [Google Scholar]
- 186.Mahlert, M., C. Vogler, K. Stelter, G. Hause, and C. W. Basse. 2009. The a2 mating-type-locus gene lga2 of Ustilago maydis interferes with mitochondrial dynamics and fusion, partially in dependence on a Dnm1-like fission component. J. Cell Sci. 122:2402-2412. [DOI] [PubMed] [Google Scholar]
- 187.Malik, S.-B., A. W. Pightling, L. M. Stefaniak, A. M. Schurko, and J. M. Logsdon, Jr. 2008. An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis. PLoS One 3:e2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Mandai, T., K. Hara, M. Kawada, and J. Nokami. 1983. A new total synthesis of dl-sirenin. Tetrahedron Lett. 24:1517-1518. [Google Scholar]
- 189.Mandel, M. A., B. M. Barker, S. Kroken, S. D. Rounsley, and M. J. Orbach. 2007. Genomic and population analyses of the mating type loci in Coccidioides species reveal evidence for sexual reproduction and gene acquisition. Eukaryot. Cell 6:1189-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Manney, T. R. 1983. Expression of the BAR1 gene in Saccharomyces cerevisiae: induction by the alpha mating pheromone of an activity associated with a secreted protein. J. Bacteriol. 155:291-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Marimon, R., J. Gene, J. Cano, L. Trilles, M. Dos Santos Lazera, and J. Guarro. 2006. Molecular phylogeny of Sporothrix schenckii. J. Clin. Microbiol. 44:3251-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.McClelland, C. M., Y. C. Chang, A. Varma, and K. J. Kwon-Chung. 2004. Uniqueness of the mating system in Cryptococcus neoformans. Trends Microbiol. 12:208-212. [DOI] [PubMed] [Google Scholar]
- 193.McCusker, J. H. 2006. Saccharomyces cerevisiae: an emerging and model pathogenic fungus, p. 245-259. In J. Heitman, S. G. Filler, J. E. Edwards, and A. P. Mitchell (ed.), Molecular principles of fungal pathogenesis. ASM Press, Washington, DC.
- 194.McDonough, E. S., and A. L. Lewis. 1967. Blastomyces dermatitidis: production of the sexual stage. Science 156:528-529. [DOI] [PubMed] [Google Scholar]
- 195.Merino, S. T., M. A. Nelson, D. J. Jacobson, and D. O. Natvig. 1996. Pseudohomothallism and evolution of the mating-type chromosome in Neurospora tetrasperma. Genetics 143:789-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mesa-Arango, A. C., M. Del Rocio Reyes-Montes, A. Perez-Mejia, H. Navarro-Barranco, V. Souza, G. Zuniga, and C. Toriello. 2002. Phenotyping and genotyping of Sporothrix schenckii isolates according to geographic origin and clinical form of sporotrichosis. J. Clin. Microbiol. 40:3004-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196a.Metin, B., K. Findley, and J. Heitman. The mating type locus (MAT) and sexual reproduction of Cryptococcus heveanensis: insight into the evolution of sex and sex-determining chromosomal regions in fungi. PLoS Genet., in press. [DOI] [PMC free article] [PubMed]
- 197.Miller, C. E. 1977. A developmental study with the SEM of sexual reproduction in Chytriomyes hyalinus. Bull. Soc. Bot. Fr. 124:281-289. [Google Scholar]
- 198.Miller, C. E., and D. P. Dylewski. 1981. Syngamy and resting body development in Chytriomyces hyalinus (Chytridiales). Am. J. Bot. 68:342-349. [Google Scholar]
- 199.Miller, M. G., and A. D. Johnson. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110:293-302. [DOI] [PubMed] [Google Scholar]
- 200.Miyake, A. 1996. Fertilization and sexuality in ciliates, p. 243-290. In H. K. and P. Bradbury (ed.), Ciliates: cells as organisms. Gustav Fischer, Stuttgart, Germany.
- 201.Montiel-Gonzalez, A. M., F. J. Fernandez, G. Viniegra-Gonzalez, and O. Loera. 2002. Invertase production on solid-state fermentation by Aspergillus niger strains improved by parasexual recombination. Appl. Biochem. Biotechnol. 102-103:63-70. [DOI] [PubMed] [Google Scholar]
- 202.Moore, E. D., and C. E. Miller. 1971. Observations on sexual fusions in Chytriomyces. Am. J. Bot. 48:474-475. [Google Scholar]
- 203.Moore, E. D., and C. E. Miller. 1973. Resting body formation by rhizoidal fusion in Chytriomyces hyalinus. Mycologia 65:145-154. [Google Scholar]
- 204.Moore, T. D. E., and J. C. Edman. 1993. The α-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol. Cell. Biol. 13:1962-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Morgan, J. A., V. T. Vredenburg, L. J. Rachowicz, R. A. Knapp, M. J. Stice, T. Tunstall, R. E. Bingham, J. M. Parker, J. E. Longcore, C. Moritz, C. J. Briggs, and J. W. Taylor. 2007. Population genetics of the frog-killing fungus Batrachochytrium dendrobatidis. Proc. Natl. Acad. Sci. U. S. A. 104:13845-13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Morrison, P. J. 1977. Gametangial development in Allomyces macrogynus. Arch. Microbiol. 113:173-179. [Google Scholar]
- 207.Morrow, C. A., and J. A. Fraser. 2009. Sexual reproduction and dimorphism in the pathogenic basidiomycetes. FEMS Yeast Res. 9:161-177. [DOI] [PubMed] [Google Scholar]
- 208.Mortimer, R. K. 1993. Founder of yeast genetics, p. 3-16. In M. N. Hall (ed.), The early days of yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 209.Mortimer, R. K. 1993. Iconoclastic father of Neurospora and yeast genetics, p. 17-38. In M. N. Hall (ed.), The early days of yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 210.Nasmyth, K. 1983. Molecular analysis of a cell lineage. Nature 302:670-676. [DOI] [PubMed] [Google Scholar]
- 211.Naumov, G. I. 1996. Genetic identification of biological species in the Saccharomyces sensu stricto complex. J. Ind. Microbiol. 17:295-302. [Google Scholar]
- 212.Ni, M., and J.-H. Yu. 2007. A novel regulator couples sporogenesis and trehalose biogenesis in Aspergillus nidulans. PLoS One 2:e970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Nielsen, K., G. M. Cox, A. P. Litvintseva, E. Mylonakis, S. D. Malliaris, D. K. Benjamin, Jr., S. S. Giles, T. G. Mitchell, A. Casadevall, J. R. Perfect, and J. Heitman. 2005. Cryptococcus neoformans α strains preferentially disseminate to the central nervous system during coinfection. Infect. Immun. 73:4922-4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Nielsen, K., G. M. Cox, P. Wang, D. L. Toffaletti, J. R. Perfect, and J. Heitman. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 71:4831-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Nielsen, K., A. L. De Obaldia, and J. Heitman. 2007. Cryptococcus neoformans mates on pigeon guano: implications for the realized ecological niche and globalization. Eukaryot. Cell 6:949-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Nielsen, K., and J. Heitman. 2007. Sex and virulence of human pathogenic fungi. Adv. Genet. 57:143-173. [DOI] [PubMed] [Google Scholar]
- 217.Nielsen, K., R. E. Marra, F. Hagen, T. Boekhout, T. G. Mitchell, G. M. Cox, and J. Heitman. 2005. Interaction between genetic background and the mating-type locus in Cryptococcus neoformans virulence potential. Genetics 171:975-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Nielsen, T. A. B., and L. W. Olson. 1982. Nuclear control of sexual differentiation in Allomyces macrogynus. Mycologia 74:303-312. [Google Scholar]
- 219.Nierman, W. C., A. Pain, M. J. Anderson, J. R. Wortman, H. S. Kim, J. Arroyo, M. Berriman, K. Abe, D. B. Archer, C. Bermejo, J. Bennett, P. Bowyer, D. Chen, M. Collins, R. Coulsen, R. Davies, P. S. Dyer, M. Farman, N. Fedorova, N. Fedorova, T. V. Feldblyum, R. Fischer, N. Fosker, A. Fraser, J. L. Garcia, M. J. Garcia, A. Goble, G. H. Goldman, K. Gomi, S. Griffith-Jones, R. Gwilliam, B. Haas, H. Haas, D. Harris, H. Horiuchi, J. Huang, S. Humphray, J. Jimenez, N. Keller, H. Khouri, K. Kitamoto, T. Kobayashi, S. Konzack, R. Kulkarni, T. Kumagai, A. Lafon, J. P. Latge, W. Li, A. Lord, C. Lu, W. H. Majoros, G. S. May, B. L. Miller, Y. Mohamoud, M. Molina, M. Monod, I. Mouyna, S. Mulligan, L. Murphy, S. O'Neil, I. Paulsen, M. A. Penalva, M. Pertea, C. Price, B. L. Pritchard, M. A. Quail, E. Rabbinowitsch, N. Rawlins, M. A. Rajandream, U. Reichard, H. Renauld, G. D. Robson, S. Rodriguez de Cordoba, J. M. Rodriguez-Pena, C. M. Ronning, S. Rutter, S. L. Salzberg, M. Sanchez, J. C. Sanchez-Ferrero, D. Saunders, K. Seeger, R. Squares, S. Squares, M. Takeuchi, F. Tekaia, G. Turner, C. R. Vazquez de Aldana, J. Weidman, O. White, J. Woodward, J. H. Yu, C. Fraser, J. E. Galagan, K. Asai, M. Machida, N. Hall, B. Barrell, and D. W. Denning. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151-1156. [DOI] [PubMed] [Google Scholar]
- 220.Noble, S. M., and A. D. Johnson. 2007. Genetics of Candida albicans, a diploid human fungal pathogen. Annu. Rev. Genet. 41:193-211. [DOI] [PubMed] [Google Scholar]
- 221.O'Gorman, C. M., H. T. Fuller, and P. S. Dyer. 2009. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature 457:471-474. [DOI] [PubMed] [Google Scholar]
- 222.Ohno, S. 1967. Sex chromosomes and sex-linked genes. Springer-Verlag, New York, NY.
- 223.Olson, L. W. 1984. Allomyces—a different fungus. Oper. Bot. 73:1-96. [Google Scholar]
- 224.Olson, L. W., T. A. B. Nielsen, H. P. Heldt-Hansen, and N. Grant. 1982. Maleness, its inheritance and control in the aquatic phycomycete Allomyces macrogynus. Trans. Br. Mycol. Soc. 78:331-336. [Google Scholar]
- 225.Olson, L. W., and M. Ronne. 1975. Induction of abnormal gametes and androgenesis in the aquatic phycomycete Allomyces. Protoplasma 84:327-344. [DOI] [PubMed] [Google Scholar]
- 226.Otto, S. P., and T. Lenormand. 2002. Resolving the paradox of sex and recombination. Nat. Rev. Genet. 3:252-261. [DOI] [PubMed] [Google Scholar]
- 227.Paoletti, M., C. Rydholm, E. U. Schwier, M. J. Anderson, G. Szakacs, F. Lutzoni, J. P. Debeaupuis, J. P. Latge, D. W. Denning, and P. S. Dyer. 2005. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Biol. 15:1242-1248. [DOI] [PubMed] [Google Scholar]
- 228.Paoletti, M., F. A. Seymour, M. J. C. Alcocer, N. Kaur, A. M. Calvo, D. B. Archer, and P. S. Dyer. 2007. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr. Biol. 17:1384-1389. [DOI] [PubMed] [Google Scholar]
- 229.Papa, K. E. 1973. The parasexual cycle in Aspergillus flavus. Mycologia 65:1201-1205. [PubMed] [Google Scholar]
- 230.Papa, K. E. 1978. The parasexual cycle in Aspergillus parasiticus. Mycologia 70:766-773. [PubMed] [Google Scholar]
- 231.Parker, J. D. 2004. A major evolutionary transition to more than two sexes? Trends Ecol. Evol. 19:83-86. [DOI] [PubMed] [Google Scholar]
- 232.Paushkin, S. V., V. V. Kushnirov, V. N. Smirnov, and M. D. Ter-Avanesyan. 1996. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 15:3127-3134. [PMC free article] [PubMed] [Google Scholar]
- 233.Pel, H. J., J. H. de Winde, D. B. Archer, P. S. Dyer, G. Hofmann, P. J. Schaap, G. Turner, R. P. de Vries, R. Albang, K. Albermann, M. R. Andersen, J. D. Bendtsen, J. A. E. Benen, M. van den Berg, S. Breestraat, M. X. Caddick, R. Contreras, M. Cornell, P. M. Coutinho, E. G. J. Danchin, A. J. M. Debets, P. Dekker, P. W. M. van Dijck, A. van Dijk, L. Dijkhuizen, A. J. M. Driessen, C. d'Enfert, S. Geysens, C. Goosen, G. S. P. Groot, P. W. J. de Groot, T. Guillemette, B. Henrissat, M. Herweijer, J. P. T. W. van den Hombergh, C. A. M. J. J. van den Hondel, R. T. J. M. van der Heijden, R. M. van der Kaaij, F. M. Klis, H. J. Kools, C. P. Kubicek, P. A. van Kuyk, J. Lauber, X. Lu, M. J. E. C. van der Maarel, R. Meulenberg, H. Menke, M. A. Mortimer, J. Nielsen, S. G. Oliver, M. Olsthoorn, K. Pal, N. N. M. E. van Peij, A. F. J. Ram, U. Rinas, J. A. Roubos, C. M. J. Sagt, M. Schmoll, J. Sun, D. Ussery, J. Varga, W. Vervecken, P. J. J. van de Vondervoort, H. Wedler, H. A. B. Wosten, A.-P. Zeng, A. J. J. van Ooyen, J. Visser, and H. Stam. 2007. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25:221-231. [DOI] [PubMed] [Google Scholar]
- 234.Pfaller, M. A., and D. J. Diekema. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Phadke, S., and R. A. Zufall. 2009. Rapid diversification of mating systems in ciliates. Biol. J. Linn. Soc. Lond. 98:187-197. [Google Scholar]
- 236.Poggeler, S. 2002. Genomic evidence for mating abilities in the asexual pathogen Aspergillus fumigatus. Curr. Genet. 42:153-160. [DOI] [PubMed] [Google Scholar]
- 237.Pommerville, J. 1977. Chemotaxis of Allomyces gametes. Exp. Cell Res. 109:43-51. [DOI] [PubMed] [Google Scholar]
- 238.Pommerville, J., and L. W. Olson. 1987. Evidence for a male-produced pheromone in Allomyces macrogynus. Exp. Mycol. 11:245-248. [Google Scholar]
- 239.Pommerville, J. C., J. B. Strickland, and K. E. Harding. 1990. Pheromone interactions and ionic communication in gametes of aquatic fungus Allomyces macrogynus. J. Chem. Ecol. 16:121-131. [DOI] [PubMed] [Google Scholar]
- 240.Pontecorvo, G. 1956. The parasexual cycle in fungi. Annu. Rev. Microbiol. 10:393-400. [DOI] [PubMed] [Google Scholar]
- 241.Pound, M. W., R. H. Drew, and J. R. Perfect. 2002. Recent advances in the epidemiology, prevention, diagnosis, and treatment of fungal pneumonia. Curr. Opin. Infect. Dis. 15:183-194. [DOI] [PubMed] [Google Scholar]
- 242.Poxleitner, M. K., M. L. Carpenter, J. J. Mancuso, C.-J. R. Wang, S. C. Dawson, and W. Z. Cande. 2008. Evidence for karyogamy and exchange of genetic material in the binucleate intestinal parasite Giardia intestinalis. Science 319:1530-1533. [DOI] [PubMed] [Google Scholar]
- 243.Pringle, A., D. M. Baker, J. L. Platt, J. P. Wares, J. P. Latge, and J. W. Taylor. 2005. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution 59:1886-1899. [PubMed] [Google Scholar]
- 244.Pujol, C., J. Reynes, F. Renaud, M. Raymond, M. Tibayrenc, F. J. Ayala, F. Janbon, M. Mallie, and J. M. Bastide. 1993. The yeast Candida albicans has a clonal mode of reproduction in a population of infected human immunodeficiency virus-positive patients. Proc. Natl. Acad. Sci. U. S. A. 90:9456-9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Pyrzak, W., K. Y. Miller, and B. L. Miller. 2008. Mating type protein Mat1-2 from asexual Aspergillus fumigatus drives sexual reproduction in fertile Aspergillus nidulans. Eukaryot. Cell 7:1029-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Rachowicz, L. J., J. M. Hero, R. A. Alford, J. W. Taylor, J. A. T. Morgan, V. T. Vredenburg, J. P. Collins, and C. J. Briggs. 2005. The novel and endemic pathogen hypothesis: competing explanations for the origin of emerging infectious diseases of wildlife. Curr. Biol. 19:1441-1448. [Google Scholar]
- 247.Ramesh, M. A., S.-B. Malik, and J. M. Logsdon, Jr. 2005. A phylogenomic inventory of meiotic genes: evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr. Biol. 15:185-191. [DOI] [PubMed] [Google Scholar]
- 248.Ramirez-Prado, J. H., G. G. Moore, B. W. Horn, and I. Carbone. 2008. Characterization and population analysis of the mating-type genes in Aspergillus flavus and Aspergillus parasiticus. Fungal Genet. Biol. 45:1292-1299. [DOI] [PubMed] [Google Scholar]
- 249.Ramirez-Zavala, B., O. Reuss, Y.-N. Park, K. Ohlsen, and J. Morschhauser. 2008. Environmental induction of white-opaque switching in Candida albicans. PLoS Pathog. 4:e1000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Rancati, G., N. Pavelka, B. Fleharty, A. Noll, R. Trimble, K. Walton, A. Perera, K. Staehling-Hampton, C. W. Seidel, and R. Li. 2008. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell 135:879-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Raper, J. R. 1966. Genetics of sexuality in higher fungi. Ronald Press Company, New York, NY.
- 252.Rappleye, C. A., and W. E. Goldman. 2006. Defining virulence genes in the dimorphic fungi. Annu. Rev. Microbiol. 60:281-303. [DOI] [PubMed] [Google Scholar]
- 253.Raudaskoski, M., and E. Kothe. 26 February 2010. Basidiomycete mating type genes and pheromone signaling. Eukaryot. Cell doi: 10.1128/EC.00319-09. [DOI] [PMC free article] [PubMed]
- 254.Reedy, J. L., A. M. Floyd, and J. Heitman. 2009. Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr. Biol. 19:891-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Rodrigues de Miranda, L. 1979. Clavispora, a new yeast genus of the Saccharomycetales. Antonie Van Leeuwenhoek 45:479-483. [DOI] [PubMed] [Google Scholar]
- 255a.Rodriguez-Carres, M., K. Findley, S. Sun, F. S. Dietrich, and J. Heitman. 2010. Morphological and genomic characterization of Filobasidiella depauperata: a homothallic sibling species of the pathogenic Cryptococcus species complex. PLoS One 5:e9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Ronne, M., and L. W. Olson. 1976. The isolation of male strains of Allomyces. Hereditas 83:191-202. [Google Scholar]
- 257.Rowell, J. B., and J. F. DeVay. 1954. Genetics of Ustilago zeae in relation to basic problems of its pathogenecity. Phytopathology 45:370-374. [Google Scholar]
- 258.Ruiz-Trillo, I., G. Burger, P. W. H. Holland, N. King, B. F. Lang, A. J. Roger, and M. W. Gray. 2007. The origins of multicellularity: a multi-taxon genome initiative. Trends Genet. 23:113-118. [DOI] [PubMed] [Google Scholar]
- 259.Rydholm, C., P. S. Dyer, and F. Lutzoni. 2007. DNA sequence characterization and molecular evolution of MAT1 and MAT2 mating-type loci of the self-compatible ascomycete mold Neosartorya fischeri. Eukaryot. Cell 6:868-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Sahni, N., S. Yi, K. J. Daniels, T. Srikantha, C. Pujol, and D. R. Soll. 2009. Genes selectively up-regulated by pheromone in white cells are involved in biofilm formation in Candida albicans. PLoS Pathog. 5:e1000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Sahni, N., S. Yi, C. Pujol, and D. R. Soll. 2009. The white cell response to pheromone is a general characteristic of Candida albicans strains. Eukaryot. Cell 8:251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.San-Blas, G. 1993. Paracoccidioidomycosis and its etiologic agent Paracoccidioides brasiliensis. J. Med. Vet. Mycol. 31:99-113. [PubMed] [Google Scholar]
- 263.Santos, M. A. S., and M. F. Tuite. 1995. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 23:1481-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Saul, N., M. Krockenberger, and D. Carter. 2008. Evidence of recombination in mixed-mating-type and alpha-only populations of Cryptococcus gattii sourced from single eucalyptus tree hollows. Eukaryot. Cell 7:727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Schirawski, J., B. Heinze, M. Wagenknecht, and R. Kahmann. 2005. Mating type loci of Sporisorium reilianum: novel pattern with three a and multiple b specificities. Eukaryot. Cell 4:1317-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Schoustra, S. E., A. J. M. Debets, M. Slakhorst, and R. F. Hoekstra. 2007. Mitotic recombination accelerates adaptation in the fungus Aspergillus nidulans. PLoS Genet. 3:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Schurko, A. M., and J. M. J. Logsdon. 2008. Using a meiosis detection toolkit to investigate ancient asexual “scandals” and the evolution of sex. BioEssays 30:579-589. [DOI] [PubMed] [Google Scholar]
- 268.Selmecki, A., A. Forche, and J. Berman. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313:367-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Sinclair, A. H., P. Berta, M. S. Palmer, J. R. Hawkins, B. L. Griffiths, M. J. Smith, J. W. Foster, A. M. Frischauf, R. Lovell-Badge, and P. N. Goodfellow. 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346:240-244. [DOI] [PubMed] [Google Scholar]
- 270.Skucas, G. P. 1966. Structure and composition of zoosporangial discharge papillae in the fungus Allomyces. Am. J. Bot. 53:1006-1011. [Google Scholar]
- 271.Smith, J. M. 1978. The evolution of sex. Cambridge University Press, Cambridge, United Kingdom.
- 272.Sohn, K. T., and K. S. Yoon. 2002. Ultrastructural study on the cleistothecium development in Aspergillus nidulans. Mycobiology 30:117-127. [Google Scholar]
- 273.Soll, D., and K. J. Daniels. 2007. MAT, mating, switching, and pathogenesis in Candida albicans, Candida dubliniensis, and Candida glabrata, p. 215-234. In J. Heitman, J. W. Kronstad, J. W. Taylor, and L. A. Casselton (ed.), Sex in fungi. ASM Press, Washington, DC.
- 274.Soll, D. R., C. Pujol, and T. Srikantha. 2009. Sex: deviant mating in yeast. Curr. Biol. 19:R509-R511. [DOI] [PubMed] [Google Scholar]
- 275.Sonneborn, T. M. 1978. Genetics of cell-cell interaction in ciliates, p. 417-427. In R. A. Lerner and D. Bergsma (ed.), The molecular basis of cell-cell interaction: proceedings of first international conference, La Jolla, California, 1977. Alan R. Liss, Inc., New York, NY.
- 276.Srikantha, T., A. R. Borneman, K. J. Daniels, C. Pujol, W. Wu, M. R. Seringhaus, M. Gerstein, S. Yi, M. Snyder, and D. R. Soll. 2006. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot. Cell 5:1674-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Stearn, S. C. 1987. The evolution of sex and its consequence. Birkhaeuser Verlag, Basel, Switzerland.
- 278.Stromnaes, O., and E. D. Garber. 1963. Heterocaryosis and the parasexual cycle in Aspergillus fumigatus. Genetics 148:653-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Sweeney, A. W., M. F. Graham, and E. I. Hazard. 1988. Life cycle of Amblyospora dyxenoides sp. nov. in the mosquito Culex annulirostris and the copepod Mesocyclops albians. J. Invertebr. Pathol. 51:46-57. [DOI] [PubMed] [Google Scholar]
- 280.Takahashi, T. 1958. Complementary genes controlling homothallism in Saccharomyces. Genetics 43:705-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Takano, I., and Y. Oshima. 1967. An allele specific and a complementary determinant controlling homothallism in Saccharomyces oviformis. Genetics 57:875-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Tanabe, Y., M. M. Watanabe, and J. Sugiyama. 2005. Evolutionary relationships among basal fungi (Chytridiomycota and Zygomycota): insights from molecular phylogenetics. J. Gen. Appl. Microbiol. 51:267-276. [DOI] [PubMed] [Google Scholar]
- 283.Tenaillon, O., H. Le Nagard, B. Godelle, and F. Taddei. 2000. Mutators and sex in bacteria: conflict between adaptive strategies. Proc. Natl. Acad. Sci. U. S. A. 97:10465-10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Thomarat, F., C. P. Vivarès, and M. Gouy. 2004. Phylogenetic analysis of the complete genome sequence of Encephalitozoon cuniculi supports the fungal origin of microsporidia and reveals a high frequency of fast-evolving genes. J. Mol. Evol. 59:780-791. [DOI] [PubMed] [Google Scholar]
- 285.Torres, E. M., T. Sokolsky, C. M. Tucker, L. Y. Chan, M. Boselli, M. J. Dunham, and A. Amon. 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317:916-924. [DOI] [PubMed] [Google Scholar]
- 286.Torres, I., A. M. Garcia, O. Hernandez, A. Gonzalez, J. G. McEwen, A. Restrepo, and M. Arango. 2010. Presence and expression of the mating type locus in Paracoccidioides brasiliensis isolates. Fungal Genet. Biol. 47:373-380. [DOI] [PubMed] [Google Scholar]
- 287.Troemel, E. R., M.-A. Félix, N. K. Whiteman, A. Barriere, and F. M. Ausubel. 2008. Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans. PLoS Biol. 6:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Tsai, I. J., D. Bensasson, A. Burt, and V. Koufopanou. 2008. Population genomics of the wild yeast Saccharomyces paradoxus: quantifying the life cycle. Proc. Natl. Acad. Sci. U. S. A. 105:4957-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Turgeon, B. G., and O. C. Yoder. 2000. Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genet. Biol. 31:1-5. [DOI] [PubMed] [Google Scholar]
- 290.Turian, G. 1958. Recherches sur les bases cytochimiques et cytophysiologiques de la morphogenese chez le champignon aquatique Allomyces. Rev. Cytol. Biol. 19:241-272. [Google Scholar]
- 291.Tyedmers, J., M. L. Madariaga, and S. Lindquist. 2008. Prion switching in response to environmental stress. PLoS Biol. 6:e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Urban, M., R. Kahmann, and M. Bolker. 1996. Identification of the pheromone response element in Ustilago maydis. Mol. Gen. Genet. 251:31-37. [DOI] [PubMed] [Google Scholar]
- 293.van der Walf, J. P. 1966. Lodderomyces, a new genus of the Saccharomycetaceae. Antonie Van Leeuwenhoek 32:1-5. [DOI] [PubMed] [Google Scholar]
- 294.van der Walf, J. P., M. B. Taylor, and N. V. Liebenberg. 1977. Ploidy, ascus formation and recombination in Torulaspora (Debaryomyces) hansenii. Antonie Van Leeuwenhoek 43:205-218. [DOI] [PubMed] [Google Scholar]
- 295.van de Vondervoort, P. J. I., B. R. Poulsen, G. J. G. Ruijter, T. Schuleit, J. Visser, and J. J. L. Iversen. 2004. Isolation of a fluffy mutant of Aspergillus niger from chemostat culture and its potential use as a morphologically stable host for protein production. Biotechnol. Bioeng. 86:301-307. [DOI] [PubMed] [Google Scholar]
- 296.Varga, J., and B. Toth. 2003. Genetic variability and reproductive mode of Aspergillus fumigatus. Infect. Genet. Evol. 3:3-17. [DOI] [PubMed] [Google Scholar]
- 297.Villeneuve, A. M., and K. J. Hillers. 2001. Whence meiosis? Cell 106:647-650. [DOI] [PubMed] [Google Scholar]
- 298.Vossbrinck, C. R., J. V. Maddox, S. Friedman, B. A. Debrunner-Vossbrinck, and C. R. Woese. 1987. Ribosomal-RNA sequence suggests microsporidia are extremely ancient eukaryotes. Nature 326:411-414. [DOI] [PubMed] [Google Scholar]
- 299.Votintseva, A. A., and D. A. Filatov. 2009. Evolutionary strata in a small mating-type-specific region of the smut fungus Microbotryum violaceum. Genetics 182:1391-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Wahl, R., A. Zahiri, and J. Kamper. 2010. The Ustilago maydis b mating type locus controls hyphal proliferation and expression of secreted virulence factors in planta. Mol. Microbiol. 75:208-220. [DOI] [PubMed] [Google Scholar]
- 301.Wainright, P. O., G. Hinkle, M. L. Sogin, and S. K. Stickel. 1993. Monophyletic origins of the metazoa: an evolutionary link with fungi. Science 260:340-342. [DOI] [PubMed] [Google Scholar]
- 302.Weitzman, I., M. A. Gordon, and S. A. Rosenthal. 1971. Determination of the perfect state, mating type and elastase activity in clinical isolates of the Microsporum gypseum complex. J. Invest. Dermatol. 57:278-282. [DOI] [PubMed] [Google Scholar]
- 303.Weitzman, I., and R. C. Summerbell. 1995. The dermatophytes. Clin. Microbiol. Rev. 8:240-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Weldon, C., L. H. du Preez, A. D. Hyatt, R. Muller, and R. Spears. 2004. Origin of the amphibian chytrid fungus. Emerg. Infect. Dis. 10:2100-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Wetzel, J., O. Scheibner, A. Burmester, C. Schimek, and J. Wostemeyer. 2009. 4-Dihydrotrisporin-dehydrogenase, an enzyme of the sex hormone pathway of Mucor mucedo: purification, cloning of the corresponding gene, and developmental expression. Eukaryot. Cell 8:88-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Whisler, H. C., S. L. Zebold, and J. A. Shemanchuk. 1975. Life history of Coelomomyces psorophorae. Proc. Natl. Acad. Sci. U. S. A. 72:693-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.White, T. C., B. G. Oliver, Y. Graser, and M. R. Henn. 2008. Generating and testing molecular hypotheses in the dermatophytes. Eukaryot. Cell 7:1238-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Wickes, B. L., M. E. Mayorga, U. Edman, and J. C. Edman. 1996. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the α-mating type. Proc. Natl. Acad. Sci. U. S. A. 93:7327-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Williams, B. A. P., R. P. Hirt, J. M. Lucocq, and T. M. Embley. 2002. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature 418:865-869. [DOI] [PubMed] [Google Scholar]
- 310.Wittner, M., and L. M. Weiss. 1999. The microsporidia and microsporidiosis. ASM Press, Washington, DC.
- 311.Woo, P. C., K. T. Chong, H. Tse, J. J. Cai, C. C. Lau, A. C. Zhou, S. K. Lau, and K. Y. Yuen. 2006. Genomic and experimental evidence for a potential sexual cycle in the pathogenic thermal dimorphic fungus Penicillium marneffei. FEBS Lett. 580:3409-3416. [DOI] [PubMed] [Google Scholar]
- 312.Wostemeyer, J., and A. Kreibich. 2002. Repetitive DNA elements in fungi (Mycota): impact on genomic architecture and evolution. Curr. Genet. 41:189-198. [DOI] [PubMed] [Google Scholar]
- 313.Wostemeyer, J., and C. Schimek. 2007. Trisporic acid and mating in zygomycetes, p. 431-444. In J. Heitman, J. W. Kronstad, J. W. Taylor, and L. A. Casselton (ed.), Sex in fungi. ASM Press, Washington, DC.
- 314.Wu, C., K. Weiss, C. Yang, M. A. Harris, B.-K. Tye, C. S. Newlon, R. T. Simpson, and J. E. Haber. 1998. Mcm1 regulates donor preference controlled by the recombination enhancer in Saccharomyces mating-type switching. Genes Dev. 12:1726-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Wu, X., and J. E. Haber. 1996. A 700 bp cis-acting region controls mating-type dependent recombination along the entire left arm of yeast chromosome III. Cell 87:277-285. [DOI] [PubMed] [Google Scholar]
- 316.Xiao, L., L. Li, G. S. Visvesvara, H. Moura, E. S. Didier, and A. A. Lal. 2001. Genotyping Encephalitozoon cuniculi by multilocus analyses of genes with repetitive sequences. J. Clin. Microbiol. 39:2248-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Xu, J. 2005. Cost of interacting with sexual partners in a facultative sexual microbe. Genetics 171:1597-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Xu, J. 2002. Estimating the spontaneous mutation rate of loss of sex in the human pathogenic fungus Cryptococcus neoformans. Genetics 162:1157-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Xu, J., C. W. Saunders, P. Hu, R. A. Grant, T. Boekhout, E. E. Kuramae, J. W. Kronstad, Y. M. DeAngelis, N. L. Reeder, K. R. Johnstone, M. Leland, A. M. Fieno, W. M. Begley, Y. Sun, M. P. Lacey, T. Chaudhary, T. Keough, L. Chu, R. Sears, B. Yuan, and T. L. Dawson. 2007. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc. Natl. Acad. Sci. U. S. A. 104:18730-18735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Xu, Y., and L. M. Weiss. 2005. The microsporidian polar tube: a highly specialised invasion organelle. Int. J. Parasitol. 35:941-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Xue, C., Y. Tada, X. Dong, and J. Heitman. 2007. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe 1:263-273. [DOI] [PubMed] [Google Scholar]
- 322.Yi, S., N. Sahni, K. J. Daniels, C. Pujol, T. Srikantha, and D. R. Soll. 2008. The same receptor, G protein, and mitogen-activated protein kinase pathway activate different downstream regulators in the alternative white and opaque pheromone responses of Candida albicans. Mol. Biol. Cell 19:957-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Yi, S., N. Sahni, C. Pujol, K. J. Daniels, T. Srikantha, N. Ma, and D. R. Soll. 2009. A Candida albicans-specific region of the alpha-pheromone receptor plays a selective role in the white cell pheromone response. Mol. Microbiol. 71:925-947. [DOI] [PubMed] [Google Scholar]
- 324.Young, L. Y., M. C. Lorenz, and J. Heitman. 2000. A STE12 homolog is required for mating but dispensable for filamentation in Candida lusitaniae. Genetics 155:17-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Zeyl, C. 2006. Experimental evolution with yeast. FEMS Yeast Res. 6:685-691. [DOI] [PubMed] [Google Scholar]
- 326.Zeyl, C., and G. Bell. 1997. The advantage of sex in evolving yeast populations. Nature 388:465-468. [DOI] [PubMed] [Google Scholar]
- 327.Zhouravleva, G., L. Frolova, X. Le Goff, R. Le Guellec, S. Inge-Vechtomov, L. Kisselev, and M. Philippe. 1995. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 14:4065-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Zordan, R. E., D. J. Galgoczy, and A. D. Johnson. 2006. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc. Natl. Acad. Sci. U. S. A. 103:12807-12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Zordan, R. E., M. G. Miller, D. J. Galgoczy, B. B. Tuch, and A. D. Johnson. 2007. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 5:e256. [DOI] [PMC free article] [PubMed] [Google Scholar]