Abstract
Invasive disease caused by meningococcal capsular groups A, C, W-135, and Y is now preventable by means of glycoconjugate vaccines that target their respective polysaccharide capsules. The capsule of group B meningococci (MenB) is poorly immunogenic and may induce autoimmunity. Vaccines based on the major immunodominant surface porin, PorA, are effective against clonal epidemics but, thus far, have a limited scope of coverage against the wider MenB population at large. In an alternative approach, the first-generation, investigational, recombinant MenB (rMenB) plus outer membrane vesicle (OMV) (rMenB-OMV) vaccine contains a number of relatively conserved surface proteins, fHBP, NHBA (previously GNA2132), and NadA, alongside PorA P1.4-containing OMVs from the New Zealand MeNZB vaccine. MenB currently accounts for approximately 90% of cases of meningococcal disease in England and Wales. To assess potential rMenB-OMV vaccine coverage of pathogenic MenB isolates within this region, all English and Welsh MenB case isolates from January 2008 (n = 87) were genetically characterized with respect to fHBP, NHBA, NadA, and PorA. Alleles for fHbp, nhba, and porA were identified in all of the isolates, of which 22% were also found to harbor nadA alleles. On the basis of genotypic data and predicted immunological cross-reactivity, the potential level of rMenB-OMV vaccine coverage in England and Wales ranges from 66% to 100%.
Invasive meningococcal disease (IMD) typically manifests as meningitis and/or septicemia, has a mortality rate approaching 10%, and causes severe physical and neurological sequelae in approximately 20% of survivors (31). Each of the major pathogenic capsular groups displays a characteristic global tendency, ranging from large epidemics in sub-Saharan Africa (capsular group A [MenA]) and MenW-135) to sporadic cases, outbreaks, and occasional epidemics in developed countries (MenB, MenC, and MenY) (13). The MenC glycoconjugate vaccine, introduced in the United Kingdom in 1999, is immunogenic from 2 months of age, induces immunological memory, and promotes herd immunity by interrupting the acquisition of carriage and reducing transmission (4). Glycoconjugate vaccines against MenA, MenW-135, and MenY have subsequently been developed (26), whereas the MenB capsular polysaccharide is poorly immunogenic in humans and may induce autoimmunity (41). A number of clonal MenB epidemics have been curtailed by the use of outer membrane vesicle (OMV) preparations in which the meningococcal subtype determinant, PorA, constitutes the immunodominant antigen (2, 3, 15, 30, 34). Immunity against PorA tends to be highly subtype specific (specifically, for variable region 2 [VR2]), however, and so a single PorA vaccine component would achieve limited coverage against the more diverse MenB populations endemic to many countries and regions (8, 19, 20, 37).
Efforts to broaden the OMV coverage of MenB are ongoing, e.g., by the incorporation of several PorA subtypes (39). In an alternative approach, the availability of the MenB strain MC58 genome in 2000 (38) led to the discovery of a number of relatively well conserved and cross-reactive surface proteins (28), of which factor H-binding protein (fHBP), neisserial adhesin A (NadA), and neisserial heparin-binding antigen (NHBA; formerly genome-derived neisserial antigen 2132 [GNA2132]), in particular, warranted further investigation as potential components of a broadly cross-protective MenB vaccine.
The lipoprotein fHBP is ubiquitous among meningococci and is a virulence factor that binds to human factor H, thereby downregulating complement activation on the bacterial surface (17, 33). Three major fHBP subgroups (variants) exist, termed variant 1 (or subfamily B) and variants 2 and 3 (or, collectively, subfamily A) (9, 21). Intravariant immunological cross-reactivity is good; however, while some intervariant cross-reactivity exists between variants 2 and 3, these are found to cross-react poorly with variant 1 (21). NHBA is also a lipoprotein and is ubiquitous among meningococci. It recruits heparin to the bacterial surface, putatively enhancing serum resistance (Novartis Vaccines, unpublished data). NHBA subvariants are highly cross-reactive (28), and antibody-mediated protection is believed to act through opsonophagocytosis (29, 40). NadA is a pathogenicity factor with a role in host cell adhesion and invasion. It is reported to be present in about 50% of case isolates, in which its distribution ranges from 100% in some clonal complexes (CCs), such as CC11 and CC32, to 0% in others, e.g., CC41/44. NadA is poorly represented among carriage isolates (5, 6). There are currently five NadA variants that have been described, of which NadA-1, NadA-2, and NadA-3 are highly cross-reactive (6). NadA-4 is poorly cross-reactive with NadA-1, -2, and -3 and is associated with carriage isolates (6). NadA-5 is closely related to NadA-4 (16) and is associated with CC213 isolates (1).
Collectively, these antigens, specifically, recombinant fHBP (rfHBP) subvariant 1.1 (fused with a further surface protein, GNA2091), recombinant NHBA (rNHBA) subvariant 2 (fused with a further surface protein, GNA1030), and recombinant NadA (rNadA) variant 3, alongside PorA P1.4 OMVs (as used in the MeNZB vaccine campaign), constitute Novartis Vaccines' first-generation recombinant MenB (rMenB) plus OMV (rMenB-OMV) investigational vaccine (10), currently undergoing phase III clinical trials. In a separate venture, Wyeth Vaccine Research (now part of Pfizer) is developing a vaccine comprising a recombinant lipidated fHBP from each of the two fHBP subfamilies, subfamilies A and B (27).
MenB currently causes approximately 90% of cases of IMD in England and Wales, where the incidence of IMD increases during the winter months, typically peaking in January. The aim of the present study was to determine the presence and genetic diversity of the rMenB-OMV vaccine antigens in all English and Welsh MenB case isolates received by the Health Protection Agency (HPA) Meningococcal Reference Unit (MRU) in January 2008. This recent cross-section of local fHbp, nadA, nhba, and porA diversity will contribute to estimates of potential vaccine coverage within the two countries while informing future vaccine design.
MATERIALS AND METHODS
Isolates.
All isolates used in the study (n = 87) were English and Welsh MenB case isolates received by the HPA MRU during January 2008. Grouping, typing, subtyping, and multilocus sequence typing (MLST) were performed by the HPA MRU (11). Isolates were preserved at −80°C on Microbank cryovials containing glycerol broth (ProLab Diagnostics, Ontario, Canada). Cultures were prepared on Columbia agar supplemented with 5% (vol/vol) horse blood (Oxoid, Basingstoke, United Kingdom) and were incubated overnight at 37°C in an atmosphere containing 5% CO2.
Genomic DNA extraction.
Genomic DNA was extracted by a previously published protocol (16), in which 1-ml bacterial suspensions (optical density at 650 nm = 0.1) in physiological saline were heat killed at 60°C for 70 min and then pelleted at 6,000 × g for 10 min. Following aspiration of the supernatant, the extraction was completed with a DNeasy blood and tissue kit (Qiagen, Crawley, United Kingdom), according to the manufacturer's protocol for Gram-negative bacteria. The DNA extracts were stored at 4°C.
PCR and sequencing.
The primers used for PCR and the sequencing reactions can be viewed in Table 1 . PCR for porA was carried out in 50-μl reaction volumes consisting of 0.2 μl (1.5 U) Taq DNA polymerase (Invitrogen, Paisley, Scotland), 5 μl 10× PCR buffer (Invitrogen), 1.5 μl MgCl2 (50 mM stock) (Invitrogen), 1 μl W1 (polyethylene glycol ether) (1% stock) (Invitrogen), 1.25 μl primer 210 (20 μM stock), 1.25 μl primer 211 (20 μM stock), 10 μl deoxynucleoside triphosphate mixture (1 mM stock) (Roche), and 28.8 μl ultrapure distilled water (Invitrogen). Thermocycling was performed on a GeneAmp PCR system 9700 thermocycler (Applied Biosystems, CA) with an initial step of 94°C for 2 min and a final step of 72°C for 2 min. The intervening PCR conditions can be viewed in Table 2. Before the porA PCR products were sequenced, they were cleaned by using a Multiscreen PCR cleanup plate (Millipore, MA), in accordance with the manufacturer's protocol. The sequencing of porA was carried out with a CEQ Dye Terminator cycle sequencing kit (Beckman Coulter, CA) in one-quarter-volume reaction mixtures, according to the manufacturer's protocol. Thermocycling for porA sequencing was performed on a GeneAmp PCR system 9700 thermocycler (Applied Biosystems); the thermocycle conditions can be viewed in Table 2. The porA sequencing products were cleaned by an ethanol cleanup procedure, according to the manufacturer's instructions (Beckman Coulter), and were analyzed on a CEQ 8000 automated DNA sequencer (Beckman Coulter). The porA variable-region database (http://neisseria.org) was consulted to assign VR1 and VR2 subtypes. PCR and sequencing of fHbp, nhba, and nadA were performed by previously published methods (6, 14, 16). PCRs were performed with HotStar Taq DNA polymerase (Qiagen) and an initial activation step of 95°C for 15 min. The conditions used for the remaining thermocycle steps can be viewed in Table 2. The PCR products were cleaned with ExoSAP-IT (USB Corporation, OH), according to the manufacturers' instructions, and then diluted 1 in 3 in nuclease-free water. Sequencing was performed with a BigDye (version 3.1) cycle sequencing kit (Applied Biosystems) at 1/16 strength in 10-μl reaction volumes, according to the manufacturer's protocol. The sequencing products were cleaned by ethanol-sodium acetate precipitation and were resuspended in 15 μl of HiDi formamide (Applied Biosystems), prior to electrophoresis on a 3130xl sequence analyzer (Applied Biosystems). Contig assembly and manual adjustment of the bases were performed with the Sequencher (version 4.8) program (Gene Codes Corporation, MI).
TABLE 1.
PCR and sequencing primers used in the study
| Target | Primer use (direction)h | Primer identifier | Sequence (5′ to 3′) | Reference |
|---|---|---|---|---|
| porA | PCR (fwd) | 210 | ATGCGAAAAAAACTTACCGCCCTC | 7 |
| PCR (rev) | 211 | AATGAAGGCAAGCCGTCAAAAACA | 7 | |
| porA (VR1) | Seq (fwd) | P3 | CAAAGCCGGCGTGGAAG | 32 |
| Seq (rev) | 103L | AACGGATACGTCTTGCTC | 35 | |
| porA (VR2) | Seq (fwd) | 8U | TCCGTACGCTACGATTCTCC | 35 |
| Seq (rev) | 122L | GGCGAGATTCAAGCCGCC | 35 | |
| fhbp | PCR/Seq (fwd) | gna1870F | TGACCTGCCTCATTGATGC | 14 |
| PCR/Seq (rev) | gna1870R | CGGTAAATTATCGTGTTCGGACGGC | 14 | |
| PCR/Seq (rev) | gna1870v3Ra | CGTGCCGTCGTGTCCTAG | 16 | |
| Seq (fwd) | gna1870S2 | CAAATCGAAGTGGACGGGCAG | 14 | |
| Seq (fwd) | gna1870S3 | TGTTCGATTTTGCCGTTTCCCTG | 14 | |
| gna2132 | PCR/Seq (fwd) | gna2132F | GGCGTTCAGACGGCATATTTTTACA | 14 |
| PCR/Seq (rev) | gna2132R | GGTTTATCAACTGATGCGGACTTGA | 14 | |
| Seq (fwd) | gna2132S2 | GCGGACACGCTGTCAAAACC | 14 | |
| Seq (fwd) | gna2132S4 | GGCGTTCTGCACGGTCGAGG | 14 | |
| Seq (fwd) | gna2132S5 | ATGGGTACGCAAAAATTCAA | 14 | |
| Seq (rev) | gna2132S7 | AATGCAGTACTTCGCCGTTGT | 14 | |
| Seq (rev) | gna2132S8 | CCTCGACCGTGCAGAACGCC | 14 | |
| Seq (rev) | gna2132S9 | CCGCACCGCCATTGCCTGTA | 14 | |
| Seq (fwd) | gna2132s3new | AGAAAATACAGGCAATGGCGGTGC | NAi | |
| Seq (fwd) | gna2132s3s4b | ACGGAAT(G/A)CAGGG(G/T)GACGATCC | NA | |
| Seq (rev) | gna2132s8s9b | GGATCGTC(A/C)CCCTG(C/T)ATTCCGT | NA | |
| nadA | PCR/Seq (fwd) | nadAFc,d,e,f | GTGGACGTACTCGACTACGAAGG | 6 |
| PCR/Seq (rev) | nadARc,d,e,f | CGAGGCGATTGTCAAACCGTTC | 6 | |
| PCR/Seq (fwd) | nadAintFc,d,e,f,g | TATGTAAACAAACTTGGTGGGG | 16 | |
| PCR/Seq (rev) | nadAintRc,d,e,f,g | GAAATAGAAAAGTTAACAACCAAGTT | 16 | |
| Seq (fwd) | nadAS1c,d | TATGTAAACAAACTTGGTGGGG | 6 | |
| Seq (fwd) | nadAS2c | GAAATAGAAAAGTTAACAACCAAGTT | 6 | |
| Seq (fwd) | nadAS3c,d,e | GACATCAAAGCTGATATCGCTAC | 6 | |
| Seq (rev) | nadAS4c,d,e,f | TTTCGAGGTGGCGCGTTCGGG | 6 | |
| Seq (rev) | nadAS5c | GTAGCGATATCAGCTTTGATGTC | 6 | |
| Seq (rev) | nadAS6c | CTTGGTTGTTAACTTTTCTATTTC | 6 | |
| Seq (fwd) | NadA4fd,e,f | TCAACGGATTCACAGTCGGAGACA | NA | |
| Seq (rev) | NadA4rd,e,f | CCAATCCATTGGCAGTAGTGTTCAG | NA | |
| Seq (fwd) | 328-Ff | GCCATCCTTGCCTCCTTCTG | NA | |
| Seq (rev) | 328-Re | TTTACCGATAGCAGTCTCGT | NA | |
| Seq (fwd) | E030-Af | GTCGTGGCTCAACACGACC | NA | |
| Seq (rev) | E030C-Re | CCGGACACTTCATCCAGTTTG | NA |
Alternative primer for PCR failures due to deletion in gna1870R primer binding site.
Bridges chromatograms for primers gna2132S3 and gna2132S4 or primers gna2132S8 and gna2132S9 (where required).
Use for nadA-1, nadA-2, and nadA-3.
Use for nadA-4.
Use for nadA-5.
Use for nadA-4/nadA-5-related variant.
Internally directed PCR primers; provides additional confirmation of the genome-wide absence of nadA.
Seq, sequencing; for, forward; rev, reverse.
NA, not applicable.
TABLE 2.
Thermocycle conditions used in PCR and sequencing reactions
| Target | Reaction | Conditions |
No. of cycles | ||
|---|---|---|---|---|---|
| Denaturation | Annealing | Extension | |||
| porA | PCR | 94°C, 1 min | 68°C, 1 min | 72°C, 2 min | 30 |
| Seqa | 96°C, 40 s | 50°C, 40 s | 60°C, 4 min | 40 | |
| fHbp | PCR | 96°C, 30 s | 63°C, 30 s | 72°C, 1 min | 30 |
| Seq | 96°C, 10 s | 53°C, 5 s | 60°C, 4 min | 25 | |
| gna2132 | PCR | 96°C, 30 s | 57°C, 30 s | 72°C, 1 min | 30 |
| Seq | 96°C, 10 s | 57°C, 5 s | 60°C, 4 min | 25 | |
| nadA | PCR | 96°C, 30 s | 56°C, 30 s | 68°C, 1 min 20 s | 30 |
| PCRb | 96°C, 30 s | 70°C, 30 s | 72°C, 1 min 20 s | 33 | |
| Seq | 96°C, 10 s | 52°C, 5 s | 60°C, 4 min | 25 | |
Seq, sequencing.
Conditions for PCR using internally directed PCR primers.
Sequence alignments and phylogenetic analyses.
Nucleotide and amino acid sequences were aligned by the use of BioEdit (version 6.0.8.0) software (12) and manual adjustment. P-distance neighbor-joining dendrograms were created by use of the MEGA (version 4.0) software package (36), in which positions containing alignment gaps and missing data were eliminated in pairwise sequence comparisons (pairwise deletion option).
Nucleotide sequence accession numbers.
The sequences identified in the present study have been deposited in the GenBank database and may be found under the following accession numbers (the corresponding subvariant identifiers are displayed in parentheses): for fHbp, GQ144336 (subvariant 1.1), FJ750977 (subvariant 1.4), EU541903 (subvariant 1.10), EU541890 (subvariant 1.13), GQ144338 (subvariant 1.14), FJ750976 (subvariant 1.15), FJ615433 (subvariant 1.86), FJ615438 (subvariant 1.87), GQ144337 (subvariant 2.16), EU541889 and EU541899 (subvariant 2.19), FJ615436 (subvariant 2.21), FJ615437 (subvariant 2.24), FJ615430 (subvariant 2.25), FJ153809 (subvariant 2.68), FJ615427 (subvariant 3.30), FJ153808(subvariant 3.45), FJ615431 (subvariant 3.47), FJ615422 (subvariant 3.59), FJ615428 (subvariant 3.84), and FJ615429 (subvariant 3.85); for nhba, FJ615439, FJ615440, and FJ750982 (subvariant 2), GQ144339 (subvariant 3), FJ615445 (subvariant 17), FJ717480 (subvariant 18), GQ144340 (subvariant 20), FJ615442 and FJ615444 (subvariant 21), FJ615446 (subvariant 24), FJ615458 (subvariant 25), FJ750980 (subvariant 29), FJ615455 (subvariant 30), FJ615459 (subvariant 47), FJ615452 (subvariant 93), FJ615456 (subvariant 115), FJ615453 (subvariant 116), FJ615460 (subvariant 117), FJ615462 (subvariant 118), FJ615454 (subvariant 119), and FJ615463 (subvariant 120); and for nadA, FJ619641 (variant 1), FJ619647 (variant 1 [339-bp deletion]), FJ619644 (variant 4), FJ619645 (variant 5), and FJ619648 (nadA4/nadA5-related variant).
RESULTS
fHbp and nhba subvariants are named in terms of their translated (protein) counterparts, in accordance with the public fHBP and NHBA databases (http://neisseria.org), in which new allelic subvariants are assigned a sequentially allocated numerical identifier and a preexisting or new (sequentially allocated) numerical protein identifier, alongside the corresponding Novartis variant designation (variant 1, 2, or 3); e.g., fHbp 1.15 refers to Novartis variant 1, Neisseria.org protein subvariant 15. nadA subvariants are classified in terms of their main variant group, i.e., variant 1, 2, 3, 4, or 5 (1, 5, 6). Some protein subvariants are encoded by several alleles either within or between clonal complexes.
MLST.
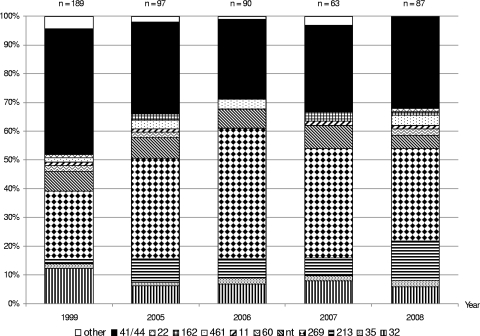
There were 10 clonal complexes present among the isolates. Two-thirds of the isolates belonged to CC41/44 or CC269, each of which represented 28 (32%) isolates. The remaining isolates belonged to CC213 (n = 12; 14%), CC32 (n = 5; 6%), CC461 (n = 3; 3%), CC35 (n = 2; 2%), CC60 (n = 2; 2%), CC22 (n = 1; 1%), CC11 (n = 1; 1%), and CC162 (n = 1; 1%), while 4 isolates (5%) belonged to sequence types (STs) not currently assigned to any clonal complexes (ST1162, ST1167, ST6790, and ST6798) (Fig. 1).
FIG. 1.
Clonal complex profile versus year among all English/Welsh MenB case isolates received by the HPA MRU in the Januaries of 1999, 2005, 2006, 2007, and 2008. “Other” indicates clonal complexes not present among January 2008 MenB isolates. Isolates belonging to STs that are unassigned to any CC are labeled “nt.”
porA.
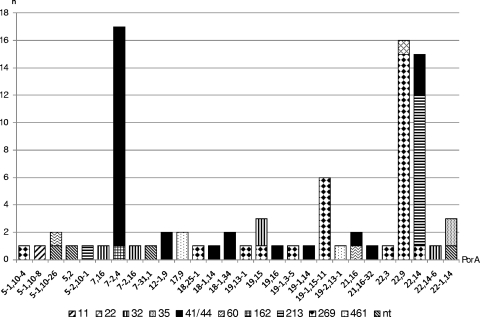
The OMV component of the rMenB-OMV investigational vaccine contains the PorA protein of subtype P1.7-2,4. The immune response elicited by this antigen has been shown to specifically target the VR2 (P1.4) epitope (19). There were 28 porA subtypes (VR1 and VR2 combinations) present among the isolates, 24 of which were identified in just one (17 subtypes), two (5 subtypes), or three (2 subtypes) isolates each. The four major (n ≥ 5 isolates) subtypes (representing 62% of the isolates in total) were P1.7-2,4 (20%), P1.22,9 (18%), P1.22,14 (17%), and P1.19-1,15-11 (7%). With the exception of the two CC35 isolates (PorA P1.22-1,14), all CCs with more than one isolate were found to be heterogeneous with respect to porA. Of the major (n ≥ 5 isolates) CCs, CC213 was the least diverse, representing just two subtypes, of which P1.22,14 predominated (92%). The remaining major CCs were relatively heterogeneous, with four subtypes representing just five CC32 isolates and nine subtypes each representing CC41/44 and CC269, in which P1.7-2,4 and P1.22,9 constituted the predominant respective subvariants (57% and 54%, respectively). Also notable within CC269 was subtype P1.19-1,15-11, which represented a further 21% of CC269 isolates. There was considerable subtype crossover between CCs, with P1.7-2,4 being present among CC41/44 and CC162; P1.22,9 being present among CC269 and CC22; P1.22,14 being present among CC41/44, CC269, and CC213; P1.19,15 being present among CC269 and CC32; P1.22-1,14 being present among CC35 and in the unassigned ST1167 isolate; and P1.21,16 being present among CC41/44 and CC60; and P1.5-1,10-26 being present among CC60 and in the unassigned ST6798 isolate. The porA composition of each CC can be viewed in Fig. 2.
FIG. 2.
porA subtype versus clonal complex for all English/Welsh MenB case isolates received by the HPA MRU in January 2008. Isolates belonging to STs that are unassigned to any CC are labeled “nt.”
nadA.
The rMenB-OMV investigational vaccine contains a recombinant NadA-3 component that is highly immunologically cross-reactive with NadA-1, NadA-2, and other NadA-3 subvariants but poorly cross-reactive with NadA-4 (6, 10). In total, 20 of 87 (23%) isolates harbored a nadA allele, comprising 1 (of 28) CC269 isolates; 1 (of 4) unassigned isolates (ST6790); and all CC213 (n = 12), CC32 (n = 5), and CC11 (n = 1) isolates. The remaining isolates were devoid of nadA alleles. The five CC32 isolates (ST32, ST34, ST259, ST749, and ST33) each harbored nadA-1 alleles; however, the ST33 isolate harbored an in-frame 339-bp deletion. All of the CC213 isolates and the single nadA-positive CC269 isolate (ST6791) harbored nadA-5 alleles, each apparently frame shifted at the internal poly(C) tract (8, 10, 11, or 13 C residues). The CC11 isolate harbored a previously undescribed allele which shares significant homology with nadA-4 and nadA-5 (Table 3; Fig. 3) and which was found to be frame shifted. The ST6790 isolate (not assigned to a clonal complex) harbored a nadA-4 allele.
TABLE 3.
Pairwise amino acid sequence identity matrix for nadA-1, nadA-2, nadA-3, nadA-4, and nadA-5 and the previously undescribed nadA-4/nadA-5-related varianta
| Allele | Pairwise identity |
|||||
|---|---|---|---|---|---|---|
| nadA-3 | nadA-1 | nadA-2 | nadA-4 | nadA-5 | nadA-4/5 | |
| nadA-3b | IDc | 0.861 | 0.972 | 0.48 | 0.47 | 0.426 |
| nadA-1 | ID | 0.841 | 0.44 | 0.432 | 0.42 | |
| nadA-2 | ID | 0.486 | 0.476 | 0.427 | ||
| nadA-4 | ID | 0.907 | 0.703 | |||
| nadA-5 | ID | 0.703 | ||||
| nadA-4/5 | ID | |||||
The previously undescribed nadA-4/nadA-5-related variant is labeled nadA-4/5.
rMenB-OMV vaccine variant.
ID, identical.
FIG. 3.
Pairwise amino acid alignment of nadA-1, nadA-2, nadA-3, nadA-4, nadA-5, and the previously undescribed nadA-4/nadA-5-related variant (labeled nadA-4/5 for the purpose of the alignment). Identical pairwise amino acids are shaded in black, and similar pairwise amino acids are shaded in gray. The nadA-4/nadA-5-related variant has been un-frame shifted for the purpose of the alignment.
fHbp.
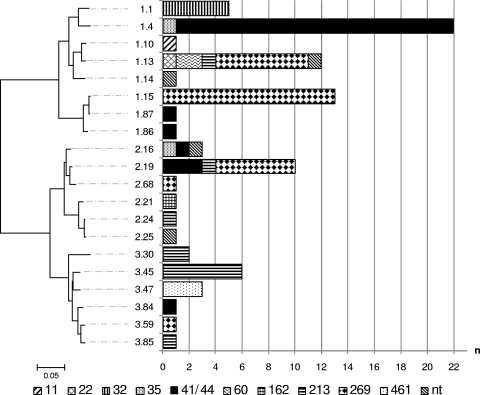
The fHBP component within rMenB-OMV is subvariant 1.1, which is highly immunologically cross-reactive with other variant 1 (subfamily B) subvariants but which is poorly cross-reactive with variant 2 and 3 (subfamily B) subvariants. All of the isolates harbored fHbp alleles, representing 20 subvariants in total (8 variant 1, 6 variant 2, and 6 variant 3 subvariants in 64% [n = 56], 20% [n = 17], and 16% [n = 14] of isolates, respectively) (Fig. 4). Of the major CCs, only CC32 (n = 5 isolates) was represented by just a single subvariant (subvariant 1.1). The remaining major CCs (CC41/44, CC269, and CC213) were each represented by all three variant groups in various proportions, with variant 1 predominating in CC41/44 (82%) and CC269 (71%) and variants 2 and 3 predominating in CC213 (17 and 75%, respectively). At the subvariant level, the major CCs were fairly heterogeneous, with CCs 41/44 and 213 each representing six subvariants and CC269 representing five subvariants. Some subvariant crossover was observed, with 4 of the 20 subvariants representing multiple CCs; i.e., subvariant 1.4 was present among CC41/44 and CC35 isolates; subvariant 2.16 was present among CCs 35 and 41/44 and in the unassigned ST1167 isolate; subvariant 2.19 was present among CCs 41/44, 269, and 213; and subvariant 1.13 was present among CCs 22, 60, 213, and 269 and in the unassigned ST6798 isolate. Despite this subvariant crossover, the predominant subvariant(s) was found to differ between each of the major CCs, with subvariants 1.4 (75%) and 3.45 (50%) predominating in CC41/44 and CC213, respectively, and subvariants 1.15 (46%), 1.13 (25%), and 2.19 (21%) predominating within CC269. The subvariant composition of individual CCs can be viewed in Fig. 4. All of the isolates harboring subvariant 1.4 (21/28 CC41/44 isolates and 1/2 CC35 isolates) and a single isolate harboring subvariant 2.19 (CC41/44) had a 181-bp insertion within the promoter region.
FIG. 4.
P-distance, neighbor-joining dendrogram of translated fHbp subvariants present among all English/Welsh MenB case isolates received by the HPA MRU in January 2008. The distribution of the subvariants among the constituent clonal complexes is represented in the adjoining bar chart. The dendrogram is drawn to scale, with the sum of the branch lengths between two subvariants representing the proportion of amino acid differences between those subvariants within the pairwise alignment. Isolates belonging to STs that are unassigned to any CC are labeled “nt.”
nhba.
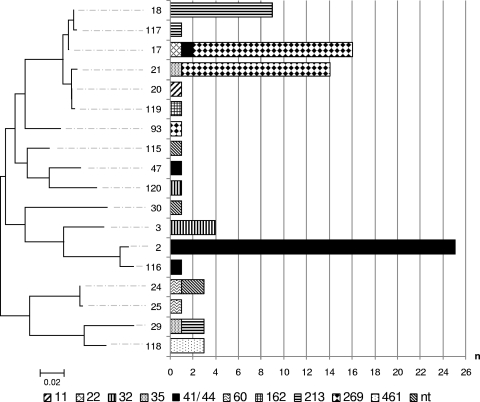
The NHBA component of the rMenB-OMV investigational vaccine is subvariant 2; however, NHBA has been demonstrated to elicit antibodies that are cross-reactive with heterologous subvariants and that are cross-protective in the infant rat challenge model of bacteremia (40). All of the isolates harbored nhba alleles, representing 18 subvariants in total. Among the major CCs, CC32 represented two nhba subvariants, CC269 and CC213 represented three nhba subvariants each, and CC41/44 represented four nhba subvariants. A different subvariant(s) predominated within each of the major CCs: subvariant 2 (89%) in CC41/44, subvariant 18 (75%) in CC213, subvariant 3 (80%) in CC32, and subvariants 17 (50%) and 21 (46%) in CC269. Of the CCs with smaller numbers (n < 5) of isolates, CC35 and CC60 each represented two subvariants, even though they represented just two isolates each. The remaining lesser CCs (CC11 [n = 1], CC22 [n = 1], CC162 [n = 1], and CC461 [n = 3]) each represented one subvariant, while the nonassigned isolates (n = 4; ST1162, ST1167, ST6790, and ST6798) represented three nhba subvariants between them. A low level of subvariant crossover was observed, with subvariant 17 being present in CCs 22, 41/44, and 269; subvariant 21 being present in CCs 35 and 269, subvariant 24 being present in CC60 and STs 1162 and 6798, and subvariant 29 being present in CCs 35 and 213. The subvariant composition of individual CCs can be viewed in Fig. 5.
FIG. 5.
P-distance, neighbor-joining dendrogram of translated nhba subvariants present among all English/Welsh MenB case isolates received by the HPA MRU in January 2008. The distribution of the subvariants among the constituent clonal complexes is represented in the adjoining bar chart. The dendrogram is drawn to scale, with the sum of the branch lengths between two subvariants representing the proportion of amino acid differences between those subvariants within the pairwise alignment. Isolates belonging to STs that are unassigned to any CC are labeled “nt.”
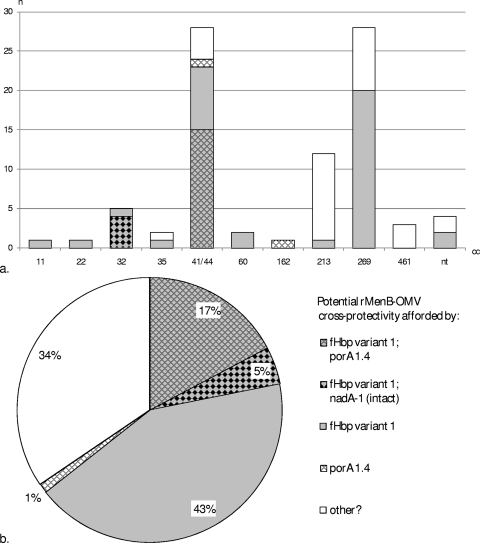
A summary of the CC distribution of the fHbp, nadA, and porA alleles encoding (sub)variants with potential rMenB-OMV cross-protectivity (i.e., variant 1 fHbp, nadA-1 [intact], and porA 1.4) can be viewed in Fig. 6a. The overall distribution of potentially covered (sub)variants is illustrated in Fig. 6b.
FIG. 6.
Potential rMenB-OMV coverage specifically due to the fHBP and/or the NadA/PorA component among all English/Welsh MenB case isolates received by the HPA MRU in January 2008. (a) Potential coverage versus clonal complex; (b) overall potential coverage afforded by the three antigens. “Other” signifies isolates for which coverage is not predicted to be afforded by any of the three antigens. Isolates belonging to STs that are unassigned to any CC are labeled “nt.” The pattern key represents the charts in both panels.
DISCUSSION
The poor immunogenicity of the group B capsular polysaccharide and its potential to elicit autoimmune antibodies have precluded the development of a potentially universal MenB glycoconjugate vaccine. Existing PorA-based vaccines have been highly effective in combating clonal epidemics but would have a limited impact against the high levels of diversity experienced in many regions. The rMenB-OMV vaccine consists of several recently discovered, relatively conserved surface antigens, NadA, fHBP, and NHBA, and a PorA-containing OMV component (10). This study aims to genetically characterize these antigens in a recent cross-section of MenB isolates from England and Wales to aid with the prediction of the level of rMenB-OMV vaccine coverage within the region.
The global presence of nadA alleles among meningococci has been estimated to be approximately 50% (5). Among the group B isolates collected by the HPA MRU in January 2008, that value was somewhat lower at 23%. This is largely due to the high proportion of CC41/44 and CC269 isolates (66%), both of which are typically devoid of the gene (6, 16). Of the highly cross-protective NadA variants (NadA-1, NadA-2, and NadA-3), only NadA-1 was represented among the isolates in which the gene occurred in all of the CC32 isolates (n = 5; 6% overall), one of which (ST33) harbored an allele containing an in-frame 339-bp deletion. A single isolate was found to harbor the carriage-associated variant, nadA-4, which cross-reacts poorly with variants 1, 2, and 3 (6). This isolate was found to belong to ST6790, which is currently unassigned to and, in terms of its allelic profile, is relatively distant from any of the established hypervirulent CCs/lineages. Therefore, while a recent horizontal transfer event involving nadA-4 is possible, it seems likely that this is an opportunistic case (meningitis in a 35-week-old child) caused by an isolate that would typically be associated with carriage. The remaining nadA-positive isolates harbored nadA-5 (12/12 CC213 isolates and 1/28 CC269 isolates) or a previously undescribed nadA-4/nadA5-related variant (1/2 CC11 isolates). The cross-reactivity of these two variants with the highly cross-protective variants, NadA-1, NadA-2, and NadA-3, has not been reported; however, their relatively high level of amino acid identity with NadA-4 (Table 3) suggests that this is likely to be poor. Therefore, before taking into account the apparent truncation of the nad-5 and nadA4/nadA-5-related variant alleles, it is unlikely that these would be covered by a NadA-1, NadA-2, or NadA-3 vaccine component. The presence of the NadA-5 protein in cultures apparently harboring truncated nadA-5 alleles (data not presented) would suggest that these cultures consist of a heterogeneous population containing both on and off genes. Such a frequency of phase variation would presumably, however, result in the immunological escape of this antigen within the host's bloodstream.
PorA is known to afford poor cross-protection between subtypes (19, 37) and between different family members of subtypes (20). Therefore, while it has proved efficacious as a vaccine component against clonal epidemics, a single PorA vaccine component would achieve limited coverage among more heterogeneous populations, such as the population that exists in England and Wales. Studies have shown that immune protection against PorA is predominantly directed at the VR2 epitope (8, 19, 20). Among all of the group B isolates collected by the MRU in January 2008 and even within the constituent CCs, porA was relatively heterogeneous, spanning 28 VR1-VR2 combinations representing 19 different VR2 epitopes. The predominant subtype was P1.7-2,4 (20%), which almost exclusively consisted of CC41/44 isolates; however, a significant proportion (47%) of the CC41/44 isolates belonged to one of eight further subtypes, none of which contained the P1.4 VR2 region. The potential levels of coverage for PorA vaccine components with VR2 regions corresponding to each of the four major subtypes (P1.7-2,4, P1.22,9, P1.22,14, and P1.19-1,15-11) are 20%, 23%, 23%, and 7%, respectively, or 73% collectively. The addition of further PorA components would increase the rate of potential coverage by between 1 and 6% per component.
In accordance with the findings of previous studies (14, 21), all of the isolates harbored fHbp and nhba alleles. Each of the major CCs was found to represent several subvariants of each antigen, and a single subvariant was often found to be present within several CCs, which, as with porA (although to a lesser extent), precludes their use in predicting the CC. With the exception of CC32, the major CCs each had fHbp subvariants spanning all three variant groups, groups 1, 2, and 3; however, each was characterized as having just one or two predominant subvariants, which were largely in agreement with the geographically and temporally diverse data from other studies (1, 24). NHBA has been implicated in opsonophagocytic protection (29, 40) and is reported to elicit antibody that cross-reacts with heterologous NHBA subvariants (28, 40). Up to 100% of the isolates in the study may potentially, therefore, be covered by an NHBA vaccine component. The intravariant cross-reactivity between fHBP subvariants is also reported to be excellent; however, the intervariant cross-reactivity between variant 1 subvariants and variant 2 and 3 subvariants is poor. The predominant variant among the 2008 isolates was variant 1 (65% overall), which favors the use of a variant 1 vaccine component within the region. Of the major CCs, however, only CC32 would potentially have been fully covered by such a component. The rates of potential coverage of a variant 1 vaccine component among the remaining major CCs, CC41/44, CC269, and CC213, would be 82%, 71%, and 8%, respectively. Caution must be exercised when inferences relating to the potential coverage of the minor CCs covered in this study are made. If the major CCs are typical, then it would be reasonable to expect that, in greater numbers, the minor CCs are also heterogeneous for fHbp. Over the period studied, a variant 1 fHBP vaccine component would potentially have prevented one of one CC11, one of one CC22, one of two CC32, two of two CC60, zero of one CC162, and zero of three CC461 cases among the minor CCs and two of four cases among the nonassigned STs.
The investigational rMenB-OMV vaccine of Novartis Vaccines contains components corresponding to each of the antigens mentioned above, specifically, recombinant NadA-3, recombinant fHBP 1.1 (presented as a fusion protein with GNA2091), recombinant NHBA 2 (presented as a fusion protein with GNA1030), and PorA P1.7-2,4 OMVs (10). On the basis of genotypic data, the rates of potential coverage of the individual rMenB-OMV vaccine components against the MenB isolates received by the MRU in January 2008 are up to 100% for NHBA, 65% for fHBP, 20% for PorA P1.7-2,4, and at least 5% for NadA (those isolates with an intact nadA-1). Alongside NHBA, approximately 66% of the isolates are potentially covered by at least one further antigen (fHBP, NadA, or PorA), while 22% are potentially covered by at least two further antigens (either fHBP and NadA or fHBP and PorA). Further coverage may be afforded by the other OMV antigens and the two surface proteins (GNA1030 and GNA2091) that are presented in fusion with fHBP and NHBA. From a genotypic point of view, the Wyeth (now part of Pfizer) investigational MenB vaccine (comprising a single recombinant, lipidated fHBP from each of the two fHBP subfamilies, subfamilies A and B [27]) also has a level of potential coverage against these isolates of up to 100%. The emergence of escape mutants may present a greater risk, however, against a vaccine effectively comprising a single antigen. In the case of the rMenB-OMV vaccine, isolates that are potentially covered by more than one antigen may present less of a risk in terms of vaccine escape through, e.g., mutation or recombination. Those isolates that are potentially covered by just a single vaccine component (i.e., NHBA) consist of 4/28 (14%) CC41/44, 8/28 (29%) CC269, 11/12 (92%) CC213, 1/2 CC32, 3/3 CC461, and 2/4 nonassigned isolates. Of these, the minor CCs have remained at low levels over the past decade; however, CC213 and a distinct lineage of CC269 harboring fHbp 2.19 (16) have recently expanded, and it is these, along with the 14% of CC41/44 isolates and perhaps isolates of CC461, that are worthy of careful surveillance as potential escape mutants, should such a vaccine be introduced.
The incidence of invasive meningococcal disease in England and Wales increases during the winter months, typically peaking in January. The genotypic data presented here are indicative of high levels of rMenB-OMV vaccine coverage (66% to 100%) against all English and Welsh MenB case isolates received by the MRU during January 2008. In terms of MLST, these isolates were found to be broadly representative of all English and Welsh MenB case isolates received during the corresponding epidemiological year (July 2007 to June 2008) (unpublished data). These putatively high levels of coverage are supported, in part, by the antigenic cross-protectivity data recently reported for adults (25). The breadth of coverage may be reduced in infants, however, in whom the levels of cross-protection afforded by heterologous subvariants are likely to be reduced (23). The apparent (albeit low level) intra-CC heterogeneity of fHbp, nhba, and porA and the potential for antigenically distinct lineages to fluctuate within a defined CC (16) mean that one must be cautious if these findings are generalized (on the basis of the CC) to other countries where capsular group B is endemic. Phenotypic expression levels are also likely to affect the susceptibility of a given isolate to vaccine-induced immunity (22), and so further work in this area will include phenotypic expression analysis of each of the antigens across the full set of January 2008 MenB isolates. While this will serve to highlight the significance of the 181-bp insertion identified in the fHbp promoter region of a number of the isolates, it will also facilitate the selection of a panel of genotypic/phenotypic representatives that may then be subjected to serum bactericidal antibody assays that will compare the bactericidal activities of pre- and postvaccination sera from clinical trials. Ultimately, these data will aid with informing decisions on potential vaccine usage and future vaccine design.
Acknowledgments
We thank Lynne Newbold, Andrew Birtles, Jayne Blake, Tony Carr, and Stefanie Gilchrist for their contribution to the MLST and PorA subtyping of the isolates used in the study.
This work was sponsored by Novartis Vaccines.
Footnotes
Published ahead of print on 7 April 2010.
REFERENCES
- 1.Bambini, S., A. Muzzi, P. Olcen, R. Rappuoli, M. Pizza, and M. Comanducci. 2009. Distribution and genetic variability of three vaccine components in a panel of strains representative of the diversity of serogroup B meningococcus. Vaccine 27:2794-2803. [DOI] [PubMed] [Google Scholar]
- 2.Bjune, G., E. A. Høiby, J. K. Grønnesby, O. Arnesen, J. H. Fredriksen, A. Halstensen, E. Holten, A. K. Lindbak, H. Nøkleby, E. Rosenqvist, L. K. Solberg, O. Closs, J. Eng, L. O. Froholm, A. Lystad, L. S. Bakketeig, and B. Hareide. 1991. Effect of outer membrane vesicle vaccine against group B meningococcal disease in Norway. Lancet 338:1093-1096. [DOI] [PubMed] [Google Scholar]
- 3.Boslego, J., J. Garcia, C. Cruz, W. Zollinger, B. Brandt, S. Ruiz, M. Martinez, J. Arthur, P. Underwood, W. Silva, E. Moran, W. Hankins, J. Gilly, and J. Mays. 1995. Efficacy, safety, and immunogenicity of a meningococcal group B (15:P1.3) outermembrane protein vaccine in Iquique, Chile. Chilean National Committee for Meningococcal Disease. Vaccine 13:821-829. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, H., R. Borrow, D. Salisbury, and E. Miller. 2009. Meningococcal C conjugate vaccine: the experience in England and Wales. Vaccine 27(Suppl. 2):B20-B29. [DOI] [PubMed] [Google Scholar]
- 5.Comanducci, M., S. Bambini, B. Brunelli, J. Adu-Bobie, B. Aricò, B. Capecchi, M. M. Giuliani, V. Masignani, L. Santini, S. Savino, D. M. Granoff, D. A. Caugant, M. Pizza, R. Rappuoli, and M. Mora. 2002. NadA, a novel vaccine candidate of Neisseria meningitidis. J. Exp. Med. 195:1445-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comanducci, M., S. Bambini, D. A. Caugant, M. Mora, B. Brunelli, B. Capecchi, L. Ciucchi, R. Rappuoli, and M. Pizza. 2004. NadA diversity and carriage in Neisseria meningitidis. Infect. Immun. 72:4217-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feavers, I. M., and M. C. Maiden. 1998. A gonococcal porA pseudogene: implications for understanding the evolution and pathogenicity of Neisseria gonorrhoeae. Mol. Microbiol. 30:647-656. [DOI] [PubMed] [Google Scholar]
- 8.Findlow, J., A. Lowe, S. Deane, P. Balmer, G. van den Dobbelsteen, M. Dawson, N. Andrews, and R. Borrow. 2005. Effect of sequence variation in meningococcal PorA outer membrane protein on the effectiveness of a hexavalent PorA outer membrane vesicle vaccine in toddlers and school children. Vaccine 23:2623-2627. [DOI] [PubMed] [Google Scholar]
- 9.Fletcher, L. D., L. Bernfield, V. Barniak, J. E. Farley, A. Howell, M. Knauf, P. Ooi, R. P. Smith, P. Weise, M. Wetherell, X. Xie, R. Zagursky, Y. Zhang, and G. W. Zlotnick. 2004. Vaccine potential of the Neisseria meningitidis 2086 lipoprotein. Infect. Immun. 72:2088-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giuliani, M. M., J. Adu-Bobie, M. Comanducci, B. Aricò, S. Savino, L. Santini, B. Brunelli, S. Bambini, A. Biolchi, B. Capecchi, E. Cartocci, L. Ciucchi, F. Di Marcello, F. Ferlicca, B. Galli, E. Luzzi, V. Masignani, D. Serruto, D. Veggi, M. Contorni, M. Morandi, A. Bartalesi, V. Cinotti, D. Mannucci, F. Titta, E. Ovidi, J. A. Welsch, D. Granoff, R. Rappuoli, and M. Pizza. 2006. A universal vaccine for serogroup B meningococcus. Proc. Natl. Acad. Sci. U. S. A. 103:10834-10839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray, S. J., C. L. Trotter, M. E. Ramsay, M. Guiver, A. J. Fox, R. Borrow, R. H. Mallard, and E. B. Kaczmarski. 2006. Epidemiology of meningococcal disease in England and Wales 1993/94 to 2003/04: contribution and experiences of the Meningococcal Reference Unit. J. Med. Microbiol. 55:887-896. [DOI] [PubMed] [Google Scholar]
- 12.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. 41:95-98. [Google Scholar]
- 13.Harrison, L. H., C. L. Trotter, and M. E. Ramsay. 2009. Global Epidemiology of meningococcal disease. Vaccine 27(Suppl. 2):B51-B63. [DOI] [PubMed] [Google Scholar]
- 14.Jacobsson, S., S. Thulin, P. Mölling, M. Unemo, M. Comanducci, R. Rappuoli, and P. Olcén. 2006. Sequence constancies and variations in genes encoding three new meningococcal vaccine candidate antigens. Vaccine 24:2161-2168. [DOI] [PubMed] [Google Scholar]
- 15.Kelly, C., R. Arnold, Y. Galloway, and J. O'Hallahan. 2007. A prospective study of the effectiveness of the New Zealand meningococcal B vaccine. Am. J. Epidemiol. 166:817-823. [DOI] [PubMed] [Google Scholar]
- 16.Lucidarme, J., M. Comanducci, J. Findlow, S. J. Gray, E. B. Kaczmarski, M. Guiver, E. Kugelberg, P. J. Vallely, P. Oster, M. Pizza, S. Bambini, A. Muzzi, C. M. Tang, and R. Borrow. 2009. Characterization of fHbp, nhba (gna2132), nadA, porA, sequence type (ST), and genomic presence of IS1301 in group B meningococcal ST269 clonal complex isolates from England and Wales. J. Clin. Microbiol. 47:3577-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madico, G., J. A. Welsch, L. A. Lewis, A. McNaughton, D. H. Perlman, C. E. Costello, J. Ngampasutadol, U. Vogel, D. M. Granoff, and S. Ram. 2006. The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J. Immunol. 177:501-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reference deleted.
- 19.Martin, D. R., N. Ruijne, L. McCallum, J. O'Hallahan, and P. Oster. 2006. The VR2 epitope on the PorA P1.7-2,4 protein is the major target for the immune response elicited by the strain-specific group B meningococcal vaccine MeNZB. Clin. Vaccine Immunol. 13:486-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin, S. L., R. Borrow, P. van der Ley, M. Dawson, A. J. Fox, and K. A. Cartwright. 2000. Effect of sequence variation in meningococcal PorA outer membrane protein on the effectiveness of a hexavalent PorA outer membrane vesicle vaccine. Vaccine 18:2476-2481. [DOI] [PubMed] [Google Scholar]
- 21.Masignani, V., M. Comanducci, M. M. Giuliani, S. Bambini, J. Adu-Bobie, B. Arico, B. Brunelli, A. Pieri, L. Santini, S. Savino, D. Serruto, D. Litt, S. Kroll, J. A. Welsch, D. M. Granoff, R. Rappuoli, and M. Pizza. 2003. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J. Exp. Med. 197:789-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNeil, L. K., E. Murphy, X. J. Zhao, S. Guttmann, S. L. Harris, A. A. Scott, C. Tan, M. Mack, I. DaSilva, K. Alexander, K. Mason, H. Q. Jiang, D. Zhu, T. L. Mininni, G. W. Zlotnick, S. K. Hoiseth, T. R. Jones, M. W. Pride, K. U. Jansen, and A. S. Anderson. 2009. Detection of LP2086 on the cell surface of Neisseria meningitidis and its accessibility in the presence of serogroup B capsular polysaccharide. Vaccine 27:3417-3421. [DOI] [PubMed] [Google Scholar]
- 23.Miller, E., A. J. Pollard, R. Borrow, J. Findlow, T. Dawson, A. Morat, T. Waterhouse, M. Snape, J. Southern, R. Morris, K. Cartwright, and P. Oster. 2008. Safety and immunogenicity of Novartis meningococcal serogroup B vaccine after three doses administered in infancy, abstr. P-16. Abstr. 26th Annu. Meet. Eur. Soc. Paediatr. Infect. Dis.
- 24.Murphy, E., L. Andrew, K. L. Lee, D. A. Dilts, L. Nunez, P. S. Fink, K. Ambrose, R. Borrow, J. Findlow, M. K. Taha, A. E. Deghmane, P. Kriz, M. Musilek, J. Kalmusova, D. A. Caugant, T. Alvestad, L. W. Mayer, C. T. Sacchi, X. Wang, D. Martin, A. von Gottberg, M. du Plessis, K. P. Klugman, A. S. Anderson, K. U. Jansen, G. W. Zlotnick, and S. K. Hoiseth. 2009. Sequence diversity of the factor H binding protein vaccine candidate in epidemiologically relevant strains of serogroup B Neisseria meningitidis. J. Infect. Dis. 200:379-389. [DOI] [PubMed] [Google Scholar]
- 25.Oster, P., D. Toneatta, P. Dull, K. Vienken, E. Ympa, L. DeTora, C. Webster, and L. Danzig. 2009. First use of an investigational meningococcal serogroup B vaccine administered in combination with New Zealand (NZ) strain (NZ98/254) outer membrane vesicles (OMV) in healthy adults, p. 52-53. Abstr. Meningitidis Res. Found. Int. Conf. Meningitis Septicaemia Children Adults.
- 26.Pace, D., and A. J. Pollard. 2007. Meningococcal A, C, Y and W-135 polysaccharide-protein conjugate vaccines. Arch. Dis. Child. 92:909-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pillai, S., A. Howell, K. Alexander, B. E. Bentley, H. Q. Jiang, K. Ambrose, D. Zhu, and G. Zlotnick. 2005. Outer membrane protein (OMP) based vaccine for Neisseria meningitidis serogroup B. Vaccine 23:2206-2209. [DOI] [PubMed] [Google Scholar]
- 28.Pizza, M., V. Scarlato, V. Masignani, M. M. Giuliani, B. Aricò, M. Comanducci, G. T. Jennings, L. Baldi, E. Bartolini, B. Capecchi, C. L. Galeotti, E. Luzzi, R. Manetti, E. Marchetti, M. Mora, S. Nuti, G. Ratti, L. Santini, S. Savino, M. Scarselli, E. Storni, P. Zuo, M. Broeker, E. Hundt, B. Knapp, E. Blair, T. Mason, H. Tettelin, D. W. Hood, A. C. Jeffries, N. J. Saunders, D. M. Granoff, J. C. Venter, E. R. Moxon, G. Grandi, and R. Rappuoli. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287:1816-1820. [DOI] [PubMed] [Google Scholar]
- 29.Plested, J. S., and D. M. Granoff. 2008. Vaccine-induced opsonophagocytic immunity to Neisseria meningitidis group B. Clin. Vaccine Immunol. 15:799-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenqvist, E., E. A. Høiby, E. Wedege, K. Bryn, J. Kolberg, A. Klem, E. Rønnild, G. Bjune, and H. Nøkleby. 1995. Human antibody responses to meningococcal outer membrane antigens after three doses of the Norwegian group B meningococcal vaccine. Infect. Immun. 63:4642-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenstein, N. E., B. A. Perkins, D. S. Stephens, T. Popovic, and J. M. Hughes. 2001. Meningococcal disease. N. Engl. J. Med. 344:1378-1388. [DOI] [PubMed] [Google Scholar]
- 32.Saunders, N. B., W. D. Zollinger, and V. B. Rao. 1993. A rapid and sensitive PCR strategy employed for amplification and sequencing of porA from a single colony-forming unit of Neisseria meningitidis. Gene 137:153-162. [DOI] [PubMed] [Google Scholar]
- 33.Schneider, M. C., R. M. Exley, H. Chan, I. Feavers, Y. H. Kang, R. B. Sim, and C. M. Tang. 2006. Functional significance of factor H binding to Neisseria meningitidis. J. Immunol. 176:7566-7575. [DOI] [PubMed] [Google Scholar]
- 34.Sierra, G. V., H. C. Campa, N. M. Varcacel, I. L. Garcia, P. L. Izquierdo, P. F. Sotolongo, G. V. Casanueva, C. O. Rico, C. R. Rodriguez, and M. H. Terry. 1991. Vaccine against group B Neisseria meningitidis: protection trial and mass vaccination results in Cuba. NIPH Ann. 14:195-207. [PubMed] [Google Scholar]
- 35.Suker, J., I. M. Feavers, M. Achtman, G. Morelli, J. F. Wang, and M. C. Maiden. 1994. The porA gene in serogroup A meningococci: evolutionary stability and mechanism of genetic variation. Mol. Microbiol. 12:253-265. [DOI] [PubMed] [Google Scholar]
- 36.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 37.Tappero, J. W., R. Lagos, A. M. Ballesteros, B. Plikaytis, D. Williams, J. Dykes, L. L. Gheesling, G. M. Carlone, E. A. Høiby, J. Holst, H. Nøkleby, E. Rosenqvist, G. Sierra, C. Campa, F. Sotolongo, J. Vega, J. Garcia, P. Herrera, J. T. Poolman, and B. A. Perkins. 1999. Immunogenicity of 2 serogroup B outer-membrane protein meningococcal vaccines: a randomized controlled trial in Chile. JAMA 281:1520-1527. [DOI] [PubMed] [Google Scholar]
- 38.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 39.van den Dobbelsteen, G. P., H. H. van Dijken, S. Pillai, and L. van Alphen. 2007. Immunogenicity of a combination vaccine containing pneumococcal conjugates and meningococcal PorA OMVs. Vaccine 25:2491-2496. [DOI] [PubMed] [Google Scholar]
- 40.Welsch, J. A., G. R. Moe, R. Rossi, J. Adu-Bobie, R. Rappuoli, and D. M. Granoff. 2003. Antibody to genome-derived neisserial antigen 2132, a Neisseria meningitidis candidate vaccine, confers protection against bacteremia in the absence of complement-mediated bactericidal activity. J. Infect. Dis. 188:1730-1740. [DOI] [PubMed] [Google Scholar]
- 41.Wyle, F. A., M. S. Artenstein, B. L. Brandt, E. C. Tramont, D. L. Kasper, P. L. Altieri, S. L. Berman, and J. P. Lowenthal. 1972. Immunologic response of man to group B meningococcal polysaccharide vaccines. J. Infect. Dis. 126:514-521. [DOI] [PubMed] [Google Scholar]