Abstract
Bacterial fimbriae can accept foreign peptides and display them on the cell surface. A highly efficient gene replacement method was used to generate peptide vaccines based on Salmonella enterica serovar Typhimurium SL3261. The T-cell epitopes (NY-ESO-1 p157-165 and p157-167) from NY-ESO-1, which is a promising target antigen in patients for the specific immune recognition of cancer, were incorporated into the gene encoding AgfA (the major subunit protein of thin aggregative fimbriae of Salmonella) by replacing an equal length of the DNA segment. To improve cytotoxic T-lymphocyte recognition, both termini of the peptide were flanked by double alanine (AA) residues. Immunofluorescence microscopy with AgfA-specific antiserum verified the expression of chimeric AgfA, which was also proved by a Congo red binding assay. Oral immunizations of HLA-A*0201 transgenic mice with recombinant SL3261 strains encoding NY-ESO-1 p157-165 or p157-167 induced NY-ESO-1 p157-165-specific CD8+ T cells, detected by an HLA-A*0201 pentamer, and induced a T-cell response detected by an enzyme-linked immunospot assay. The Salmonella fimbrial display system was efficient at the induction of an antitumor cellular immune response in vivo, providing a new strategy for the development of efficient cancer vaccinations.
Modulating the immune system to control cancer has been a challenge for cancer immunotherapists for many years. A number of clinical trials have indeed demonstrated that objective tumor regressions can occur in the setting of cancer vaccinations (1, 19, 31). One of the major problems, however, is that robust clinical responses or durable clinical benefits are not consistently seen, and concerted efforts are being made to optimize vaccination strategies (1, 31).
It has been clearly established that the use of attenuated Salmonella strains as live oral vaccines is a safe and effective means of inducing significant humoral and secretory antibody responses in animal species, including humans and mice (8, 16, 21, 32-34). The use of plasmid-based expression systems to elevate foreign antigen levels in Salmonella live vaccine vectors is ideal, but these systems have the serious disadvantages of instability and selective requirements in vivo (10, 14, 17, 38). The problem of plasmid instability can be overcome by integrating the genes for the foreign antigens into the Salmonella chromosome, but the level of expression of the antigens from a single locus is usually below the desired levels (33). One solution is to express the foreign antigen from the Salmonella chromosome as part of a preexisting, cellular protein carrier which is expressed at a high level. The cell surface organelles called fimbriae (or pili) are excellent candidates for use for the presentation of heterologous antigens.
Fimbriae are composed of a large number of polymerized protein subunits called fimbrins and are found on the surfaces of a wide variety of pathogenic bacteria. In addition, many are known to be involved in the colonization of host cell surfaces. They are polymers composed of large numbers of identical protein subunits called fimbrins. Thin aggregative fimbriae (called Tafi or curli in Escherichia coli) and the major subunit AgfA (CsgA or curlin in E. coli) have been found in a wide range of Salmonella enterica subsp. enterica serovars, such as S. enterica subsp. enterica serovars Typhimurium (S. Typhimurium) and Enteritidis (S. Enteritidis) (6). Tafi fimbriae are highly flexible at displaying foreign antigens and could induce a viral epitope-specific immune response, but it is unknown whether this presentation system could act as a carrier of tumor antigens to induce specific immune responses against cancer antigens (36, 37).
In the study described here, we used the Tafi display system to present T-cell epitopes of the tumor antigen NY-ESO-1 and evaluated its immune effects. We engineered the agfA gene of attenuated S. Typhimurium SL3261 (an S. Typhimurium aroA-negative mutant) by replacing a 48-bp segment with a DNA segment encoding a T-cell epitope, i.e., NY-ESO-1 p157-165 and p157-167 (4, 7, 12, 20). Oral immunization of transgenic mice carrying a human class I major histocompatibility complex (MHC) antigen, HLA-A*0201, with the engineered bacterial strains induced an epitope-specific cytotoxic T-lymphocyte (CTL) response.
MATERIALS AND METHODS
Mice, bacterial strains, media, and growth condition.
We maintained HLA-A*0201 transgenic mice in a pathogen-free facility. HLA-A2.1 cell surface expression by transgenic mice was verified by flow cytometry with a fluorescein isothiocyanate (FITC)-conjugated mouse anti-human HLA-A*0201 antibody (Biolegend) on a FACSCanto II flow cytometer (Becton Dickinson).
S. Typhimurium SL3261 was grown in Luria-Bertani (LB) broth or on LB agar for 14 to 18 h at 37°C. SL3261 or E. coli containing a recombinant plasmid was grown in LB broth or on LB agar supplied with kanamycin (50 μg/ml) for 20 to 24 h at 28°C, as described by White et al. (37). To express the AgfA fimbrin, strain SL3261 isolates were grown on T agar for 48 to 60 h at 28°C, as described by Romling et al. (32a). For Congo red (CR) binding experiments, the strains were grown on T agar supplemented with 100 μg CR/ml (TCR).
Construction of SL3261 strains containing agfA::eso9 or agfA::eso11 in the chromosome.
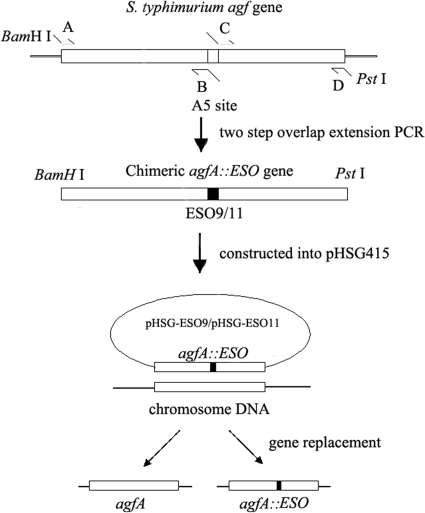
The chimeric agfA::eso9 genes were generated by two-step overlap extension PCR with primers A, B, C, and D (Fig. 1). The primers used are listed in Table 1. The chimeric genes were recombined into the chromosome of SL3261 by the procedures outlined by White et al. (35, 37) and replaced the wild-type agfA gene (Fig. 1). To facilitate the presentation of the ESO9 epitope, two alanine residues flanked both sides of the epitope. Using the same procedure, we replaced a DNA segment in the agfA gene with the eso11 gene. The chimeric agf::eso genes produced by PCR were then cloned into pHSG415.
FIG. 1.
Schematic of generation of Salmonella enterica serovar Typhimurium SL3261 strains carrying a foreign epitope in fimbrin. Salmonella enterica serovar Typhimurium SL3261 genome DNA was used as the target for amplification of the chimeric agfA genes by two-step overlap PCR. The chimeric genes were constructed in pHSG415, which is an unstable and temperature-sensitive plasmid, to generate pHSG-ESO9 and pHSG-ESO11. The recombinant plasmids were then transferred into SL3261. Finally, we obtained mutant strains through gene replacement by the use of temperature control and biotic pressure, as described in the text.
TABLE 1.
Primers used to generate agfA::eso9 and agfA::eso11
| Primera | Sequenceb (5′-3′) |
|---|---|
| eso9-A | GACTGGATCCTGTCCGTTATTTCACAAG |
| eso9-B | CTGAGTAATCCACATAAGAAGAGAAGCTGCTTTACGGGCATCGCTTTG |
| eso9-C | CTTCTTATGTGGATTACTCAGTGCGCTGCGGGCCAGGGTGCGGATAAC |
| eso9-D | GACTCTGCAGAGGGTTCGTTTAATGTGA |
| eso11-A | GACTGGATCCTGTCCGTTATTTCACAAG |
| eso11-B | GAAGCACTGAGTAATCCACATAAGAAGAGACGCTGCTTTACGGGCATCGCTTTG |
| eso11-C | CTTCTTATGTGGATTACTCAGTGCTTCCTTGCTGCGGGCCAGGGTGCGGATAAC |
| eso11-D | GACTCTGCAGAGGGTTCGTTTAATGTGA |
Primers used to generate agfA::eso, as noted in Fig. 1.
Boldface nucleotides encode the NY-ESO-1 epitopes.
The resultant recombinant plasmids, pHSG-ESO9 and pHSG-ESO11, were used to construct two bacterial strains. Using the same procedure, we replaced a DNA segment in the agfA gene with several termination codons to construct an AgfA-deficient (ΔagfA) strain. For the final confirmation of gene replacement, fragments containing agfA and the surrounding DNA region were amplified with primers agf1 and agf2 and sequenced by BGI LifeTech (Beijing, China). High-fidelity Taq enzyme, restriction enzymes, and ligase were supplied by New England Biolabs.
CR binding assay.
The CR binding assay was performed as described by White et al. (36). Strain SL3261 and the SL3261-ESO9, SL3261-ESO11, and SL3261-ΔagfA mutant strains were grown on TCR plates; the cells were then scraped off the plates and suspended in 10 mM Tris buffer (pH 7.0) at an A650 value of 1.0. The cell suspension was equilibrated at room temperature for 1 h, and 1-ml aliquots were transferred into Eppendorf tubes containing 50 μl of 30% polyethylene glycol 8000 in 100 mM Tris buffer (pH 7.0). The cells were removed from the suspension by centrifugation (15,000 × g for 5 min), before determination of the amount of CR released into the supernatant by measuring the absorbance at 480 nm with a spectrophotometer.
Immunofluorescence.
Bacterial cells were heat fixed on glass slides prior to incubation with AgfA-specific rabbit anti-AgfA antiserum (5). The secondary antibody, Alexa Fluor 488 goat anti-rabbit IgG (heavy and light chains), was purchased from Invitrogen. The cells were examined and photographed with a fluorescence microscope (BX60-32FB3-E01; Olympus, Japan) with a ×100 magnification oil immersion lens.
Animal immunization.
Female HLA-A2*0201 transgenic mice (age, 6 to 8 weeks; six mice per group) were immunized with wild-type strain SL3261, SL3261-ESO9, or SL3261-ESO11. Each mouse was administered a dose of 108 CFU bacteria orally in 100 μl saline (0.9% NaCl solution) by pipette. As a negative control, a group of mice was immunized with saline only, without bacteria. The oral immunization was repeated twice, at intervals of 14 days, for a total of three immunizations.
HLA-A*0201 pentamer staining.
One week after the last immunization, the spleens of all immunized mice were removed and monosplenocytes were prepared. Splenocytes were incubated for 15 min at room temperature with the allophycocyanin-labeled Pro5 HLA-A*0201/SLLMWITQC pentamer (ProImmune, United Kingdom) in staining buffer (phosphate-buffered saline, 0.5% bovine serum albumin, 0.05% sodium azide, pH 7.2 to 7.4), and then FITC-labeled anti-mouse CD8 antibody (ProImmune) was added for additional 20 min at 4°C. The cells were washed twice in staining buffer, fixed with fix buffer (staining buffer supplemented with 1% formaldehyde), and analyzed by flow cytometry on a FACSCanto II flow cytometer (Becton Dickinson).
Synthetic peptides and ELISPOT assay.
The HLA-A*0201-restricted CTL epitope derived from the NY-ESO-1 protein, referred to as peptide p157-165 (NH2-SLLMWITQC-COOH), was used in this study and was synthesized by BGI LifeTech. The peptides were dissolved in dimethyl sulfoxide at 1 mg/ml and stored at −20°C. A mouse gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) assay kit was purchased from U-CyTech Biosciences. The mice were euthanized 1 week after the last immunization, and their spleens were removed. Monosplenocytes were prepared and cultured for 7 days in RPMI 1640 (Gibco) supplemented with 10% calf serum (Gibco), 100 μg/ml streptomycin, 100 U/ml penicillin, and 10 U/ml murine interleukin (IL-2; PeproTech); the culture medium was refreshed every 48 h after the start of incubation. On the day before testing by the ELISPOT assay, 10 μg/ml of peptide NY-ESO-1 p157-165 was added to the cell culture clusters. Cells were collected and counted before the ELISPOT assay was performed under the conditions specified by the manufacturer. Four replicates of 5 × 105 and 1 × 105 cells per well were added to 96-well mixed polyvinylidene difluoride plates precoated with anti-mouse IFN-γ antibody, and each well was supplemented with 10 μg/ml synthesized peptides and 10 U/ml murine IL-2. No peptides were applied to the medium for the negative-control group. After incubation at 37°C for 24 h without agitation, the plates were washed and first incubated with a biotinylated secondary antibody and then with alkaline phosphatase-conjugated streptavidin, followed by addition of freshly prepared 3-amino-9-ethylcarbazole (AEC) substrate buffer. The plates were photographed and analyzed with an immunospot analyzer (Cellular Technology Ltd.).
Statistical analyses.
The experimental data were analyzed by one-way analysis of variance (ANOVA), followed by Tukey's multiple range test for significant differences. In all cases, the criterion for statistical significance was a P value of <0.05.
RESULTS
Generation of mutants and expression of AgfA in SL3261 strains.
Two individual strains of S. Typhimurium SL3261 were engineered to express chimeric AgfA proteins containing the NY-ESO-1 T-cell epitope. Strain SL3261-ESO9 contained NY-ESO-1 p157-165 and strain SL3261-ESO11 contained NY-ESO-1 p157-167, and each NY-ESO-1 T-cell epitope was flanked by two alanine residues at both ends. An AgfA-deficient (ΔAgfA) strain of SL3261 was also generated.
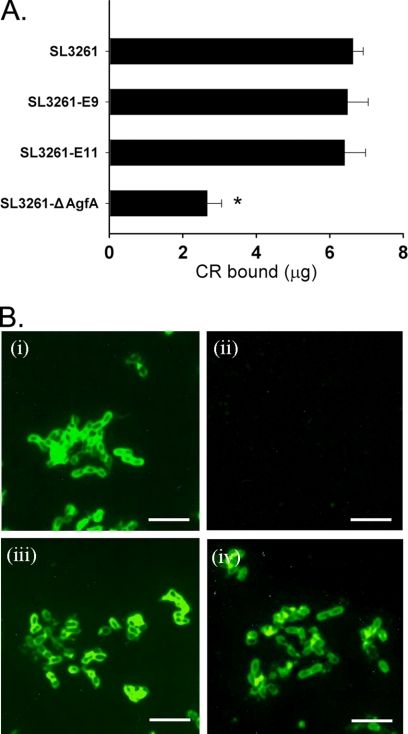
The two SL3261 mutant strains, along with the SL3261 parent strain, were analyzed for their ability to bind to the hydrophobic dye Congo red. The level of binding of Congo red to Salmonella strains is proportional to the relative amounts of thin aggregative fimbriae (Tafi, curli) produced (36). The mutated strains had a morphology similar to that of the SL3261 parent strain on TCR plates, indicating that the AgfA::ESO9 proteins and the AgfA::ESO11 proteins were expressed and assembled into functional Tafi fimbriae. The level of binding of CR by the three mutated strains with chimeric AgfA was a little less than or equal to that by the parent strain but significantly higher than that by the ΔagfA strain (P < 0.05) (Fig. 2 A). This verified the expression of AgfA in the constructed strains and demonstrated that the insertion of ESO9 and ESO11 did not cause a significant reduction in the level of AgfA expression.
FIG. 2.
Expression of chimeric fimbriae. (A) Congo red binding assay. The amount of Congo red bound by 1 A650 unit of Salmonella cells from TCR plates was measured. The SL3261 mutants can bind similar amounts of Congo red as parent strain SL3261, but the amount bound by the ΔAgfA strain was much less than that bound by the other strains (P < 0.05). (B) Immunofluorescence of SL3261 strains by rabbit anti-AgfA serum. Cells were scraped from T medium and fixed on glass slides. AgfA-specific antiserum was applied. (i) strain SL3261; (ii) strain SL3261 ΔAgfA; (iii) strain SL3261-ESO9, and (iv) strain SL3261-ESO11. Scale bars, 5 μm.
The expression of AgfA in the SL3261 strains grown on TCR plates was further examined by immunofluorescence, and the expression of thin aggregative fimbriae on the cell surface of the SL3261 parent strain (Fig. 2B, panel i) and the SL3261 mutant strains (Fig. 2B, panels iii and iv) but not the ΔagfA strain (Fig. 2B, panel ii) was shown.
HLA-A*0201 pentamer staining in mice immunized with SL3261 vaccine strains.
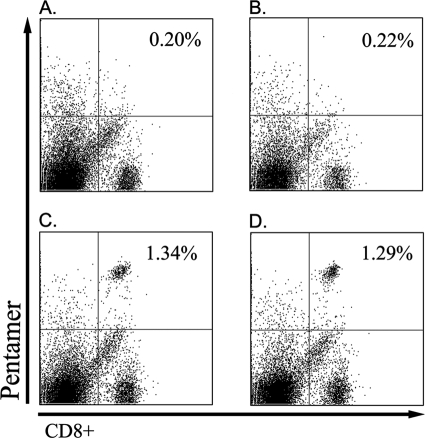
The presence of NY-ESO-1-specific CD8+ T cells in the mice immunized with the transgenic SL3261 mutant strains was detected by NY-ESO-1 p165-157/HLA-A*0201 pentamer staining. The positive cells occupied 1.34% ± 0.15% (SL3261-ESO9) and 1.29% ± 0.09% (SL3261-ESO11) of the whole splenocytes or 9.97% ± 1.54% (SL3261-ESO9) and 9.84% ± 1.43% (SL3261-ESO11) of the CD8+ T cells, whereas CD8+ T cells from the mice immunized with the SL3261 parent strain and the mice treated with saline showed negative staining (Fig. 3).
FIG. 3.
HLA-A*0201/NY-ESO-1 p157-165 pentamer staining of mouse spleen cells. One week after the last immunization, spleen cells were stained with the HLA-A*0201/NY-ESO-1 p157-165 pentamer. The mean response for a typical mouse from a group of four mice is shown. The mean response for each group is provided. (A) SL3261 parental group; (B) saline group; (C) SL3261-ESO9 group; (D) SL3261-ESO11 group.
Epitope-specific T-cell immune response in mice immunized with SL3261 vaccine strains.
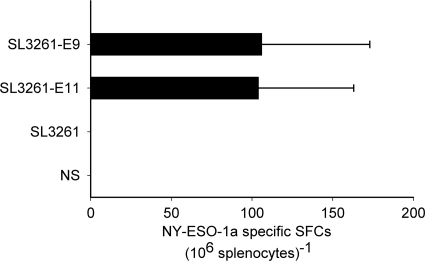
The epitope-specific T-cell immune response in HLA-A*0201 transgenic mice immunized with the SL3261 vaccine strains with the parental strain and saline as the controls was analyzed by the ELISPOT assay. The two SL3261 mutant strains both induced p157-165 peptide-specific T-cell responses in the mice, and there was no significant difference between these two groups (Fig. 4). In mice immunized with parental SL3261 strain or saline, the ELISPOT assay showed no p157-165 peptide-specific spot-forming cells (SFCs).
FIG. 4.
ELISPOT assay of IFN-γ of mouse spleen cells. HLA-A*0201 transgenic mice were euthanized 1 week after the last immunization, and their spleens were removed. Single-cell suspension of spleen cells were prepared and cultured in RPMI 1640 for 7 days with IL-2. The ELISPOT assay was performed by use of a mouse IFN-γ ELISPOT assay kit. The values on the x axis indicate the NY-ESO-1 p157-165-specific SFCs (106 splenocytes)−1 in each group. The error bars indicate the significant differences for each group. Epitope-specific SFCs were observed in the mutant strain groups but not in the parental SL3261 group or the saline group.
DISCUSSION
This is the first report of an attenuated Salmonella thin aggregative fimbria-based tumor antigen peptide vaccine that induced epitope-specific T-cell responses in transgenic mice. In this study, we used a site-specific chromosomal gene replacement system (35, 37) to introduce foreign DNA encoding a heterologous peptide into agfA, which encodes the major subunit of Salmonella thin aggregative fimbriae. The bacterial strain used in our study was S. Typhimurium SL3261, which is aroA deficient and attenuated (2, 15, 24). We have also demonstrated that the inserted chimeric ESO9 gene and ESO11 gene, which were recombined into the chromosome of S. Typhimurium SL3261, can be expressed and that an epitope-specific T-cell response was induced in transgenic mice by this recombinant organism.
Through growth on TCR plates, a Congo red binding assay, and AgfA immunofluorescence, we found that the two SL3261 mutant strains (strains SL3261-ESO9 and SL3261-ESO11) had functional and antigenic properties similar to those of the SL3261 parental strain, which indicated that the chimeric AgfA genes were expressed and assembled into functional Tafi without causing a significant reduction in AgfA expression. This means that the AgfA::ESO9 proteins and the AgfA::ESO11 proteins can be stably expressed on the Salmonella thin aggregative fimbriae.
However, the differences shown by immunofluorescence may be due to the change in the fimbrial structure caused by the insertion of foreign peptides; meanwhile, these data also confirm the expression of the various chimeric proteins in the mutated SL3261 strains. As NY-ESO-1 p157-165 and p157-167 are T-cell epitopes, we failed to detect the epitopes on the fimbriae by immunofluorescence with the NY-ESO-1 polyclonal antibody (data not shown).
In our research, the target epitopes applied were T-cell epitopes NY-ESO-1 p157-165 and p157-167. NY-ESO-1 is a promising target antigen for the specific immune recognition of cancer in patients because it has restricted expression in healthy tissue but frequently occurs on human tumors. The presence of NY-ESO-1 is seen in approximately one-third to one-fourth of all cases of melanoma and lung, ovarian, esophageal, bladder, and prostate cancers; and expression is often associated with high-grade tumors (13). NY-ESO-1 is spontaneously immunogenic in 50% of these patients, eliciting both CD8 and CD4 T-cell responses that correlate with the presence of serum antibodies to NY-ESO-1 (11, 13, 19). However, the immune responses to the antigen are downregulated by regulatory T cells (Tregs) and are not protective. It has previously been shown that stimulation of the innate immune system is essential to break immunological tolerance (18). S. Typhimurium is able to induce strong innate immune responses through Toll-like receptor signals, which not only can block the suppressive activity of CD4+ CD25+ Tregs but also can break the tolerance of CD8 cells, even in the presence of CD4+ CD25+ Tregs (28-30, 39).
To be of value, we used transgenic mice carrying a human class I major histocompatibility complex antigen, HLA-A2, to assay the efficacies of the vaccines. Due to the central role of MHC in the generation of an immune response, transgenic mice carrying human MHC products (HLA-A*0201) could provide an important model system in which to assess the induction of an HLA-associated immune response (3, 22). Antigen processing and presentation by murine cells can reveal the same set of HLA-restricted antigenic epitopes recognized by human T cells.
To determine the T-cell response in immunized mice, we conducted MHC class I pentamer staining and the ELISPOT assay. Compared to the CD8+ T cells from the mice immunized with the SL3261 parental strain and the mice treated with saline, the CD8+ T cells from the mice immunized with the SL3261 mutant strains expressing chimeric AgfA::ESO9 or AgfA::ESO11 showed positive staining with the NY-ESO-1 p157-165 peptide/HLA-A*0201 pentamer, but there were no significant differences between these two groups. Our result is similar to or better than the results of former studies with a naked DNA vaccine encoding polypeptide or a virus-based whole antigen vaccine or peptide vaccine (26, 27).
Consistent with the results of the pentamer staining assay, we found that in transgenic mice, the SL3261 mutant strains expressing chimeric AgfA::ESO9 or AgfA::ESO11 that we constructed induced epitope-specific T-cell immune responses, determined by ELISPOT assay with the NY-ESO-1 p157-165 peptide. However, there was no significant difference between the intensity of the immune responses of the two groups, which suggests that both the AgfA::ESO9 proteins and the AgfA::ESO11 proteins had the same antigen presentation in vivo and that both were efficient in supporting the recognition of epitopes by CTLs. The frequency of epitope-specific SFCs was 106 ± 67/106 splenocytes for SL3261-ESO9 or 104 ± 59/106 splenocytes for SL3261-ESO11. In a former study, a lentivirus-based NY-ESO-1 vaccine elicited a three times greater T-cell response, which may have been due to the high level of replication of lentivirus in the spleen, but this approach needed a subcutaneous injection and had a biosafety problem (9, 23). Another study employed a Salmonella serovar Typhimurium vaccine strain to deliver NY-ESO-1 through a type III protein secretion system and showed a rate of IFN-γ-secreting T cells and tumor regression in mice similar to that found in the present study (25). In this system, the recombinant salmonellae specifically secreted NY-ESO-1 into Salmonella-infected cells and the tumor antigen could be efficiently presented to T cells, but the plasmid-based expression system was instable and needed selection in vivo.
All together, from the results of the specific CD8+ T-cell responses of the mice immunized with the SL3261 mutant strains, we conclude that SL3261 mutants expressing the tumor antigen NY-ESO-1 p157-165 peptide can invite a specific T-cell immune response in vivo, which also proves that the antigen display system that we constructed is effective at delivering a foreign antigen to the mouse immune system. In contrast to other vaccine strategies for the treatment of cancer, this vaccine strategy has the following advantages. First, Salmonella replicates directly in lymphoid organs, and antigens are presented more efficiently; second, fimbriae are strong immunogens which can serve as an effective natural adjuvant; third, epitopes have stable and high levels of expression in this system, which is very advantageous, since by conventional methods recombinant proteins are either unstable or poorly expressed; and fourth, oral immunization is very convenient and economical. This study describes a good means of delivery of peptides that may be used to combat tumor diseases in the future.
Acknowledgments
This work was supported by NSFC (no. 30771995 and no. 30671962) and NFFTBS (no. J0630853/J0108).
Footnotes
Published ahead of print on 7 April 2010.
REFERENCES
- 1.Ayyoub, M., A. Zippelius, M. J. Pittet, D. Rimoldi, D. Valmori, J. C. Cerottini, P. Romero, F. Lejeune, D. Lienard, and D. E. Speiser. 2003. Activation of human melanoma reactive CD8+ T cells by vaccination with an immunogenic peptide analog derived from Melan-A/melanoma antigen recognized by T cells-1. Clin. Cancer Res. 9:669-677. [PubMed] [Google Scholar]
- 2.Benitez, A. J., N. McNair, and J. R. Mead. 2009. Oral immunization with attenuated Salmonella enterica serovar Typhimurium encoding Cryptosporidium parvum Cp23 and Cp40 antigens induces a specific immune response in mice. Clin. Vaccine Immunol. 16:1272-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernhard, E. J., A. X. Le, J. A. Barbosa, E. Lacy, and V. H. Engelhard. 1988. Cytotoxic T lymphocytes from HLA-A2 transgenic mice specific for HLA-A2 expressed on human cells. J. Exp. Med. 168:1157-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi, E. M., J. L. Chen, L. Wooldridge, M. Salio, A. Lissina, N. Lissin, I. F. Hermans, J. D. Silk, F. Mirza, M. J. Palmowski, P. R. Dunbar, B. K. Jakobsen, A. K. Sewell, and V. Cerundolo. 2003. High avidity antigen-specific CTL identified by CD8-independent tetramer staining. J. Immunol. 171:5116-5123. [DOI] [PubMed] [Google Scholar]
- 5.Collinson, S. K., L. Emody, K. H. Muller, T. J. Trust, and W. W. Kay. 1991. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 173:4773-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doran, J. L., S. K. Collinson, J. Burian, G. Sarlos, E. C. Todd, C. K. Munro, C. M. Kay, P. A. Banser, P. I. Peterkin, and W. W. Kay. 1993. DNA-based diagnostic tests for Salmonella species targeting agfA, the structural gene for thin, aggregative fimbriae. J. Clin. Microbiol. 31:2263-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebert, L. M., Y. C. Liu, C. S. Clements, N. C. Robson, H. M. Jackson, J. L. Markby, N. Dimopoulos, B. S. Tan, I. F. Luescher, I. D. Davis, J. Rossjohn, J. Cebon, A. W. Purcell, and W. Chen. 2009. A long, naturally presented immunodominant epitope from NY-ESO-1 tumor antigen: implications for cancer vaccine design. Cancer Res. 69:1046-1054. [DOI] [PubMed] [Google Scholar]
- 8.Echchannaoui, H., M. Bianchi, D. Baud, M. Bobst, J. C. Stehle, and D. Nardelli-Haefliger. 2008. Intravaginal immunization of mice with recombinant Salmonella enterica serovar Typhimurium expressing human papillomavirus type 16 antigens as a potential route of vaccination against cervical cancer. Infect. Immun. 76:1940-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia Casado, J., J. Janda, J. Wei, L. Chapatte, S. Colombetti, P. Alves, G. Ritter, M. Ayyoub, D. Valmori, W. Chen, and F. Levy. 2008. Lentivector immunization induces tumor antigen-specific B and T cell responses in vivo. Eur. J. Immunol. 38:1867-1876. [DOI] [PubMed] [Google Scholar]
- 10.Garmory, H. S., R. W. Titball, K. A. Brown, and A. M. Bennett. 2003. Construction and evaluation of a eukaryotic expression plasmid for stable delivery using attenuated Salmonella. Microb. Pathog. 34:115-119. [DOI] [PubMed] [Google Scholar]
- 11.Gnjatic, S., D. Atanackovic, E. Jager, M. Matsuo, A. Selvakumar, N. K. Altorki, R. G. Maki, B. Dupont, G. Ritter, Y. T. Chen, A. Knuth, and L. J. Old. 2003. Survey of naturally occurring CD4+ T cell responses against NY-ESO-1 in cancer patients: correlation with antibody responses. Proc. Natl. Acad. Sci. U. S. A. 100:8862-8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gnjatic, S., E. Jager, W. Chen, N. K. Altorki, M. Matsuo, S. Y. Lee, Q. Chen, Y. Nagata, D. Atanackovic, Y. T. Chen, G. Ritter, J. Cebon, A. Knuth, and L. J. Old. 2002. CD8(+) T cell responses against a dominant cryptic HLA-A2 epitope after NY-ESO-1 peptide immunization of cancer patients. Proc. Natl. Acad. Sci. U. S. A. 99:11813-11818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gnjatic, S., H. Nishikawa, A. A. Jungbluth, A. O. Gure, G. Ritter, E. Jager, A. Knuth, Y. T. Chen, and L. J. Old. 2006. NY-ESO-1: review of an immunogenic tumor antigen. Adv. Cancer Res. 95:1-30. [DOI] [PubMed] [Google Scholar]
- 14.Haga, T., S. Kumabe, A. Ikejiri, Y. Shimizu, H. Li, Y. Goto, H. Matsui, H. Miyata, and T. Miura. 2006. In vitro and in vivo stability of plasmids in attenuated Salmonella enterica serovar Typhimurium used as a carrier of DNA vaccine is associated with its replication origin. Exp. Anim. 55:405-409. [DOI] [PubMed] [Google Scholar]
- 15.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 16.Huang, H., Y. J. Wang, A. P. White, J. Z. Meng, G. R. Liu, S. L. Liu, and Y. D. Wang. 2009. Salmonella expressing a T-cell epitope from Sendai virus are able to induce anti-infection immunity. J. Med. Microbiol. 58:1236-1242. [DOI] [PubMed] [Google Scholar]
- 17.Isoda, R., S. P. Simanski, L. Pathangey, A. E. Stone, and T. A. Brown. 2007. Expression of a Porphyromonas gingivalis hemagglutinin on the surface of a Salmonella vaccine vector. Vaccine 25:117-126. [DOI] [PubMed] [Google Scholar]
- 18.Iwasaki, A., and R. Medzhitov. 2004. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5:987-995. [DOI] [PubMed] [Google Scholar]
- 19.Jager, E., Y. Nagata, S. Gnjatic, H. Wada, E. Stockert, J. Karbach, P. R. Dunbar, S. Y. Lee, A. Jungbluth, D. Jager, M. Arand, G. Ritter, V. Cerundolo, B. Dupont, Y. T. Chen, L. J. Old, and A. Knuth. 2000. Monitoring CD8 T cell responses to NY-ESO-1: correlation of humoral and cellular immune responses. Proc. Natl. Acad. Sci. U. S. A. 97:4760-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karbach, J., S. Gnjatic, C. Pauligk, A. Bender, M. Maeurer, J. L. Schultze, K. Nadler, C. Wahle, A. Knuth, L. J. Old, and E. Jager. 2007. Tumor-reactive CD8+ T-cell clones in patients after NY-ESO-1 peptide vaccination. Int. J. Cancer 121:2042-2048. [DOI] [PubMed] [Google Scholar]
- 21.Kulkarni, R. R., V. R. Parreira, S. Sharif, and J. F. Prescott. 2008. Oral immunization of broiler chickens against necrotic enteritis with an attenuated Salmonella vaccine vector expressing Clostridium perfringens antigens. Vaccine 26:4194-4203. [DOI] [PubMed] [Google Scholar]
- 22.Le, A. X., E. J. Bernhard, M. J. Holterman, S. Strub, P. Parham, E. Lacy, and V. H. Engelhard. 1989. Cytotoxic T cell responses in HLA-A2.1 transgenic mice. Recognition of HLA alloantigens and utilization of HLA-A2.1 as a restriction element. J. Immunol. 142:1366-1371. [PubMed] [Google Scholar]
- 23.Lopes, L., M. Dewannieux, U. Gileadi, R. Bailey, Y. Ikeda, C. Whittaker, M. P. Collin, V. Cerundolo, M. Tomihari, K. Ariizumi, and M. K. Collins. 2008. Immunization with a lentivector that targets tumor antigen expression to dendritic cells induces potent CD8+ and CD4+ T-cell responses. J. Virol. 82:86-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massis, L. M., C. J. Braga, M. E. Sbrogio-Almeida, C. Lauand, S. M. Newton, P. E. Klebba, and L. C. Ferreira. 2008. Anti-flagellin antibody responses elicited in mice orally immunized with attenuated Salmonella enterica serovar Typhimurium vaccine strains. Mem. Inst. Oswaldo Cruz 103:606-610. [DOI] [PubMed] [Google Scholar]
- 25.Nishikawa, H., E. Sato, G. Briones, L. M. Chen, M. Matsuo, Y. Nagata, G. Ritter, E. Jager, H. Nomura, S. Kondo, I. Tawara, T. Kato, H. Shiku, L. J. Old, J. E. Galan, and S. Gnjatic. 2006. In vivo antigen delivery by a Salmonella typhimurium type III secretion system for therapeutic cancer vaccines. J. Clin. Invest. 116:1946-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmowski, M. J., E. M. Choi, I. F. Hermans, S. C. Gilbert, J. L. Chen, U. Gileadi, M. Salio, A. Van Pel, S. Man, E. Bonin, P. Liljestrom, P. R. Dunbar, and V. Cerundolo. 2002. Competition between CTL narrows the immune response induced by prime-boost vaccination protocols. J. Immunol. 168:4391-4398. [DOI] [PubMed] [Google Scholar]
- 27.Palmowski, M. J., L. Lopes, Y. Ikeda, M. Salio, V. Cerundolo, and M. K. Collins. 2004. Intravenous injection of a lentiviral vector encoding NY-ESO-1 induces an effective CTL response. J. Immunol. 172:1582-1587. [DOI] [PubMed] [Google Scholar]
- 28.Pasare, C., and R. Medzhitov. 2004. Toll-dependent control mechanisms of CD4 T cell activation. Immunity 21:733-741. [DOI] [PubMed] [Google Scholar]
- 29.Pasare, C., and R. Medzhitov. 2003. Toll pathway-dependent blockade of CD4+CD25+ T cell-mediated suppression by dendritic cells. Science 299:1033-1036. [DOI] [PubMed] [Google Scholar]
- 30.Peng, G., Z. Guo, Y. Kiniwa, K. S. Voo, W. Peng, T. Fu, D. Y. Wang, Y. Li, H. Y. Wang, and R. F. Wang. 2005. Toll-like receptor 8-mediated reversal of CD4+ regulatory T cell function. Science 309:1380-1384. [DOI] [PubMed] [Google Scholar]
- 31.Peterson, A. C., H. Harlin, and T. F. Gajewski. 2003. Immunization with Melan-A peptide-pulsed peripheral blood mononuclear cells plus recombinant human interleukin-12 induces clinical activity and T-cell responses in advanced melanoma. J. Clin. Oncol. 21:2342-2348. [DOI] [PubMed] [Google Scholar]
- 32.Qu, D., S. Wang, W. Cai, and A. Du. 2008. Protective effect of a DNA vaccine delivered in attenuated Salmonella typhimurium against Toxoplasma gondii infection in mice. Vaccine 26:4541-4548. [DOI] [PubMed] [Google Scholar]
- 32a.Römling, U., Z. Bian, M. Hammar, W. D. Sierralta, and S. Normark. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180:722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spreng, S., G. Dietrich, and G. Weidinger. 2006. Rational design of Salmonella-based vaccination strategies. Methods 38:133-143. [DOI] [PubMed] [Google Scholar]
- 34.Wang, Y. J., Y. Hou, H. Huang, G. R. Liu, A. P. White, and S. L. Liu. 2008. Two oral HBx vaccines delivered by live attenuated Salmonella: both eliciting effective anti-tumor immunity. Cancer Lett. 263:67-76. [DOI] [PubMed] [Google Scholar]
- 35.White, A. P., E. Allen-Vercoe, B. W. Jones, R. DeVinney, W. W. Kay, and M. G. Surette. 2007. An efficient system for markerless gene replacement applicable in a wide variety of enterobacterial species. Can. J. Microbiol. 53:56-62. [DOI] [PubMed] [Google Scholar]
- 36.White, A. P., S. K. Collinson, P. A. Banser, D. J. Dolhaine, and W. W. Kay. 2000. Salmonella enteritidis fimbriae displaying a heterologous epitope reveal a uniquely flexible structure and assembly mechanism. J. Mol. Biol. 296:361-372. [DOI] [PubMed] [Google Scholar]
- 37.White, A. P., S. K. Collinson, J. Burian, S. C. Clouthier, P. A. Banser, and W. W. Kay. 1999. High efficiency gene replacement in Salmonella enteritidis: chimeric fimbrins containing a T-cell epitope from Leishmania major. Vaccine 17:2150-2161. [DOI] [PubMed] [Google Scholar]
- 38.Xu, C., Z. S. Li, Y. Q. Du, Y. F. Gong, H. Yang, B. Sun, and J. Jin. 2007. Construction of recombinant attenuated Salmonella typhimurium DNA vaccine expressing H. pylori ureB and IL-2. World J. Gastroenterol. 13:939-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang, Y., C. T. Huang, X. Huang, and D. M. Pardoll. 2004. Persistent Toll-like receptor signals are required for reversal of regulatory T cell-mediated CD8 tolerance. Nat. Immunol. 5:508-515. [DOI] [PubMed] [Google Scholar]