Abstract
Mouse-human chimeric antibodies (cAbs) against hepatitis C virus (HCV) core, NS3 (nonstructural), NS4, and NS5 antigens were developed as quality control (QC) reagents to replace the use of human sera/plasma for Abbott HCV immunoassays. The cAb retains the mouse monoclonal antibody (MAb) specificity and affinity but still reacts in the existing HCV assay format, which measures human anti-HCV immunoglobulin. Mouse heavy-chain (VH) and light-chain (VL) variable regions of anti-HCV core, NS3, NS4, and NS5 antigens were PCR amplified from hybridoma lines and then cloned with human IgG1 heavy-chain (CH) and light-chain (CL) constant regions, respectively. A single mammalian expression plasmid containing both heavy-chain and light-chain immunoglobulin genes was constructed and transfected into dihydrofolate reductase (DHFR)-deficient Chinese hamster ovary (CHO) cells. The transfected CHO cells were selected using hypoxanthine- and thymidine-free medium and screened by an enzyme immunoassay (EIA). The clone secreting the highest level of antibody was isolated from the CHO transfectants and further subcloned. Each cAb-expressing CHO cell line was weaned into serum-free medium, and the cAb was purified by protein A affinity chromatography. The levels of cAb production for the various CHO cell lines varied from 10 to 20 mg/liter. Purified anti-HCV cAbs were tested with Abbott HCV immunoassays and showed reactivity. Moreover, yeast surface display combined with alanine-scanning mutagenesis was used to map the epitope at the individual amino acid level. Our results suggest that these HCV cAbs are ideal controls, calibrators, and/or QC reagents for HCV assay standardization.
Infection with hepatitis C virus (HCV) causes an inflammation of the liver and is the most common chronic blood-borne infection in the United States. According to the U.S. Centers for Disease Control and Prevention, approximately 1.8% of the U.S. population, or 3.9 million Americans, have been infected with the virus. About 35,000 new cases of HCV are estimated to occur in the United States each year. Common routes of infection include needle stick accidents, blood transfusions, and injection drug use. Most individuals acutely infected with HCV become chronically infected. Once a person is chronically infected, the virus is almost never cleared without treatment (20).
Abbott HCV immunoassays designed to detect anti-HCV antibodies in patient samples provide a fast and reliable serological diagnostic method. Typically, diagnostic kits contain one or more antibodies as calibrators/positive controls. Traditionally, these controls consist of human plasma and/or serum samples from infected individuals. The quality control (QC) reagents, such as assay sensitivity panels, are human plasma/serum samples selected for antibodies against HCV core, NS3 (nonstructural), NS4, and NS5 antigen epitopes. However, the use of human serum/plasma has several significant disadvantages, including increasing regulatory concerns about patient sample drawing, sample storage and transportation, difficulty in sourcing large volumes with high titers and specificities, lot-to-lot variability, limitations with respect to characterization (epitope and affinity), and cost. Recombinant DNA technology has made it possible to combine the heavy-chain (VH) and light-chain (VL) variable regions of a desired monoclonal antibody (MAb) with human IgG constant regions, creating a chimeric antibody (cAb) (11, 12). The chimeric antibody retains the MAb specificity and affinity and is reactive in assays using an indirect or anti-human IgG antibody as a conjugate for detection.
In this study, we first developed MAbs against HCV core, NS3, NS4, and NS5 antigens by using a conventional hybridoma fusion. The mouse variable genes from the heavy and light chains were cloned by reverse transcription-PCR (RT-PCR) using mRNA purified from hybridoma lines. The mouse heavy-chain variable gene was cloned with a human IgG1 constant region, and the mouse light-chain variable gene was cloned with a human kappa constant region to create mouse-human cAb. Finally, a stable Chinese hamster ovary (CHO) cell line was generated to express mouse-human cAb.
MATERIALS AND METHODS
Hybridoma line development.
The anti-HCV hybridoma lines were developed using the polyethylene glycol-mediated fusion technique described by Galfre et al. (8). Briefly, female BALB/c mice were immunized with a purified HCV recombinant antigen. The animal immunization regimen utilized Freund's adjuvant (Difco, Detroit, MI), and serum samples were monitored using an enzyme immunoassay (EIA) until an anti-HCV titer was identified. For the EIA, HCV core (amino acids [aa] 1 to 150), NS3 (aa 1192 to 1457), NS4 (aa 1696 to 1931), and NS5 (aa 2054 to 2481) (Abbott Laboratories, IL) recombinant antigens were coated on 96-well EIA plates, incubated for at least 1 h at room temperature, and then blocked with 2% bovine serum albumin in phosphate-buffered saline (BSA-PBS), pH 7.2, for 1 h. The mouse serum samples were added to the coated wells, and the plates were incubated for at least 1 h at room temperature. After incubation, the plates were washed with distilled water and incubated with horseradish peroxidase-labeled goat anti-mouse IgG antibody for about 1 h. The plates were developed using O-phenylenediamine-2 HCl, and the absorbance at 492 nm was measured.
Spleen-derived mouse B cells were fused with the SP2/0 myeloma and cultured at 37°C in hypoxanthine aminopterin thymidine (HAT)-supplemented Iscove's modified Dulbecco's medium (IMDM) containing 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA). The hybridoma supernatants were tested for anti-HCV reactivity 10 to 14 days later by EIA. Hybridomas secreting anti-HCV monoclonal antibodies were cloned by standard single-cell dilution techniques, and subsequent clones were tested for reactivity by EIA. Final clones were expanded in IMDM containing 10% FBS and then frozen in a cryopreservative in liquid nitrogen.
DNA constructs for chimeric antibodies.
Each anti-HCV hybridoma line obtained as described above was cultured in hybridoma serum-free medium to obtain ∼5 × 106 cells for mRNA purification. Poly(A)+ mRNA was isolated from the cells and purified using an Oligotex direct mRNA micro kit (Qiagen, Valencia, CA) following the manufacturer's instructions. The purified mRNA was then used as a template in an RT-PCR using a mouse Ig primer set (Novagen, San Diego, CA) as described previously (13). Each RT-PCR was executed using 2× reaction buffer (deoxynucleoside triphosphate [dNTP]), each 5′ and 3′ primer pair, and 2.5 U of RT-Platinum Taq HiFi mix (Invitrogen). The cDNA synthesis and predenaturation were performed at 1 cycle of 50°C for 30 min and 1 cycle of 94°C for 2 min, followed by the PCR, comprising denaturation for 1 min at 94°C, annealing for 1 min at 50°C, and extension for 2 min at 68°C, with a final extension for 6 min at 68°C. A total of 45 cycles were performed.
PCR products (∼400 bp) were gel purified, cloned into the pCR TOPO 2.1 TA vector (Invitrogen), and transformed into Escherichia coli DH5α. The plasmid DNA was prepared using a QIAprep spin miniprep kit (Qiagen) following the manufacturer's instructions, and the plasmid DNA containing the VH or VL insert was confirmed by EcoRI digestion. The final plasmid clone for each of the VH or VL inserts was selected by sequencing. With the selected VL clone plasmid DNA used as the template, a pair of PCR primers was designed to amplify the mouse VL sequence. The 5′-end primer contained a partial human kappa signal sequence and an NruI restriction site, and the 3′-end primer contained a BsiWI restriction site (primers are listed in Table 1). A similar process was used to amplify the VH sequences from the selected VH clones. Specifically, the 5′-end primer contained a partial human IgG heavy-chain signal sequence and an NruI restriction site, and the 3′-end primer contained a SalI restriction site. PCR used 2× Pfx amplification buffer with 15 pmol each of the 5′-end and 3′-end primers, 1.25 U of Pfx DNA polymerase (Invitrogen), and 100 ng of plasmid DNA. The PCR was performed for 30 cycles of 15 s at 94°C followed by 1 min at 68°C. The VL and VH PCR products were restriction enzyme digested by NruI/BsiWI and NruI/SalI digestion, respectively, and then cloned into the pBOS-hck vector (for the VL sequence) or the pBOS-hcg vector (for the VH sequence) and transformed into E. coli DH5α. The pBOS-hck vector (Abbott Laboratories, IL) comprises an ampicillin resistance gene, a pUC origin, a simian virus 40 (SV40) origin, an EF-1a promoter, a kappa signal peptide, and a human kappa gene (hck). The pBOS-hcg vector (Abbott Laboratories) comprises an ampicillin resistance gene, a pUC origin, an SV40 origin, an EF-1a promoter, a heavy-chain signal peptide, and a human constant IgG gamma 1 gene (hcg). The transformed E. coli clones were grown in LB broth overnight with shaking at 37°C. Plasmid DNA was purified from each individual clone with a QIAprep spin miniprep kit (Qiagen), followed by sequencing confirmation using a Big Dye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA).
TABLE 1.
Primers used for cloning variable genes of anti-HCV antibodies into pBos vectors
| Target | Primer | Primer sequence |
|---|---|---|
| HCV core antigen | VL 5′ end | 5′-GCTCGCGATGCGACATTGTGATGTCACAGTCT-3′ |
| VL 3′ end | 5′-CACCGTACGTTTTATTTCCAGCTTGGT-3′ | |
| VH 5′ end | 5′-TTGTCGCGATTTTAAAAGGTGTCCAGTGCCAGATCCAGTTGGTGCAGTCTGGACCT-3′ | |
| VH 3′ end | 5′-TTGGTCGACGCTGAGGAGACGGTGACTGAGGTT-3′ | |
| HCV NS3 antigen | VL 5′ end | 5′-CTGTGGTTCCCCGGCTCGCGATGCGATGTTGTGATGGCCCAAACTCCACTCTCCCC-3′ |
| VL 3′ end | 5′-GCGCATGCGTCGTACGTTTTATTTCCAGCTTGGTCCCCCC-3′ | |
| VH 5′ end | 5′-GGCTTTTTCTTGTCGCGATTTTAAAAGGTGTCCAGTGCGAAGTGAAGCTGGTGGAGTCTG GGGGAGGC-3′ | |
| VH 3′ end | 5′-GCGCATGCATGCATTGTCGACGCGAGGAGACTGTGAGAGTGGTGCCTTGGCCC-3′ | |
| HCV NS4 antigen | VL 5′ end | 5′-GCTCGCGATGCGATGTTGTGATGACCCAAAC-3′ |
| VL 3′ end | 5′-CACCGTACGTTTGATTTCCAGCTTGGTGC-3′ | |
| VH 5′ end | 5′-CTTGTCGCGATTTTAAAAGGTGTCCAGTGCCAGATCCAGTTGGTGCAGTC-3′ | |
| VH 3′ end | 5′-TGGTCGACGCTGAGGAGACTGTGAGAGTGGT-3′ | |
| HCV NS5 antigen | VL 5′ end | 5′-GCTCGCGATGCGACATTGTGATGTCACAGT-3′ |
| VL 3′ end | 5′-CACCGTACGTTTCAGCTCCAGCTTGGT-3′ | |
| VH 5′ end | 5′-CTTGTCGCGATTTTAAAAGGTGTCCAGTGCGAGGTTCAGCTGCAGCAGT-3′ | |
| VH 3′ end | 5′-TGGTCGACGCTGCAGAGACAGTGACCAG-3′ |
Transient expression of chimeric antibodies.
pBOS-hck-L and pBOS-hcg-H plasmid DNAs were prepared using an Endofree plasmid maxi kit (Qiagen) by standard techniques. The high-purity plasmid DNA obtained was then transiently transfected into COS 7L cells by electroporation (gene pulser; Bio-Rad, CA). The transfected COS 7L cells were incubated at 37°C in a 5% CO2 incubator for 3 days and harvested by centrifugation at 4,000 rpm for 20 min, and the supernatant was collected. Following filtration and standard protein A agarose (Invitrogen) affinity purification, the purified anti-HCV chimeric antibodies were assayed with the EIA described above to confirm reactivity with HCV antigens.
CHO cell line development.
Following positive confirmation of immunoreactivity of the transiently expressed chimeric antibodies, both pBOS-hck-L and pBOS-hcg-H plasmids were digested with SrfI and NotI to extract the heavy-chain or light-chain gene. These heavy- and light-chain genes were gel purified and cloned into the pBV and pJV vectors (Abbott Laboratories), respectively. Once the correct pBV or pJV clone was identified, both plasmids were digested with PacI and AscI. The heavy-chain or light-chain genes containing DNA fragments from pBV or pJV were gel purified and ligated together to form the pBJ plasmid, which contains both heavy- and light-chain genes.
A dihydrofolate reductase (DHFR)-deficient CHO cell line was used for transfection and to establish a stable cAb-expressing cell line. The CHO cells were cultured at 37°C and 8% CO2 and transfected with the pBJ HCV cAb plasmid by use of a standard calcium phosphate-mediated procedure. The transfected CHO cells were selected using hypoxanthine- and thymidine-free medium for about 2 weeks prior to single-cell cloning in 96-well plates using a BD fluorescence-activated cell sorter (FACS) Aria flow cytometer. The cells from the 96-well plates were cultured and screened in an antigen-specific EIA to rank the performances of the CHO subclones. The EIA was conducted as described above, using various concentrations of the cAb. The subclone secreting the highest level of the cAb, as determined by the EIA, was weaned into chemically defined CHO serum-free medium (Invitrogen). Once the subclone was weaned, subcloning using the flow cytometer was performed one additional time to ensure a clonal-antibody-producing cell line.
Purification of chimeric antibody.
The CHO cell line secreting the chimeric antibody was cultured in a shaker flask for 7 days at 37°C. The supernatants were collected and centrifuged at 4,000 rpm for 15 min at ambient temperature and then passed through a 0.45-μm filter. Purification of chimeric IgG was performed using standard protein A (POROS A50; Applied Biosystems) purification procedures, followed by Sephadex G-25 Superfine desalting-column purification. Analysis of a sample from each purification step and HCV cAb preparations were carried out by SDS-PAGE as described by Laemmli (15).
HCV chimeric antibody testing by Abbott immunoassay.
The four chimeric antibodies against HCV core, NS3, NS4, and NS5 antigens, prepared as described above, were tested using the Abbott HCV immunoassays on PRISM and AxSYM instruments according to package insert instructions (22).
The PRISM assay utilized microparticles coated with HCV NS3, NS4, NS5, and core antigens, and the AxSYM assay used NS3, NS4, and core antigens as the solid phase to capture anti-HCV-specific antibodies. The captured antibodies were detected with labeled anti-human IgG antibody conjugates. The positive calibrator was human plasma reactive for anti-HCV, and the negative calibrator was HCV human plasma nonreactive for anti-HCV. Both the PRISM and AxSYM HCV assays calculate a result based on the ratio of the sample rate (S) to the cutoff rate (CO) for each sample and control, S/CO, per the package inserts.
Yeast strains and epitope mapping.
Cells from Saccharomyces cerevisiae strain EBY100 (a GAL1-AGA1::URA3 ura3-52 trp1 leu2Δ1 his3Δ200 pep4::HIS2 prb1Δ1.6R can1 GAL) were used as host cells for the surface display of HCV antigen fragments (1). The pYD41 vector (Invitrogen) was used for cloning HCV antigen fragments fused to the carboxy terminus of Aga2p, and the V5 epitope tag was fused at the HCV antigen C-terminus fragment. The fusion protein tethers to the yeast cell surface by disulfide bonding to the anchoring subunit Aga1p. The overlapping HCV antigen sequence oligonucleotides were designed to cover specific regions corresponding to the immunogen used to generate the individual MAbs. These pYD41 constructs were then transfected into EBY100 (9). Yeast cells containing HCV antigen surface display plasmids were grown in minimal SD-CAA medium (20.0 g/liter dextrose, 6.7 g/liter yeast nitrogen base, 5.0 g/liter Casamino Acids, 10.19 g/liter Na2HPO4·7H2O, 8.56 g/liter NaH2PO4·H2O) overnight in a 30°C shaking incubator until an optical density at 600 nm (OD600) of 1 to 2 was reached. Yeast cells were recovered by centrifugation and resuspended in SG-CAA medium (same as SD-CAA medium, except with 20.0 g/liter galactose replacing dextrose) to induce HCV antigen display. The induction phase was carried out at 20°C for 20 h. The induced yeast cells were incubated with a specific anti-HCV cAb, followed by Alexa Fluor 633-labeled goat anti-human IgG, and analyzed with a FACs Calibur flow cytometer to identify binding. The anti-V5 MAb, Alexa Fluor 633-labeled goat anti-human IgG, and Alexa Fluor 488-labeled goat anti-human IgG were used as controls. Once the antibody binding fragment was identified, each amino acid in the fragment was further mutated to alanine by using a set of synthetic DNA oligonucleotides. These DNA oligonucleotides were cloned into pYD41, transfected into EBY100 yeast cells, and tested for loss of antibody binding, indicative of the participation of the mutated residue in antibody binding.
RESULTS
Selection of monoclonal antibodies.
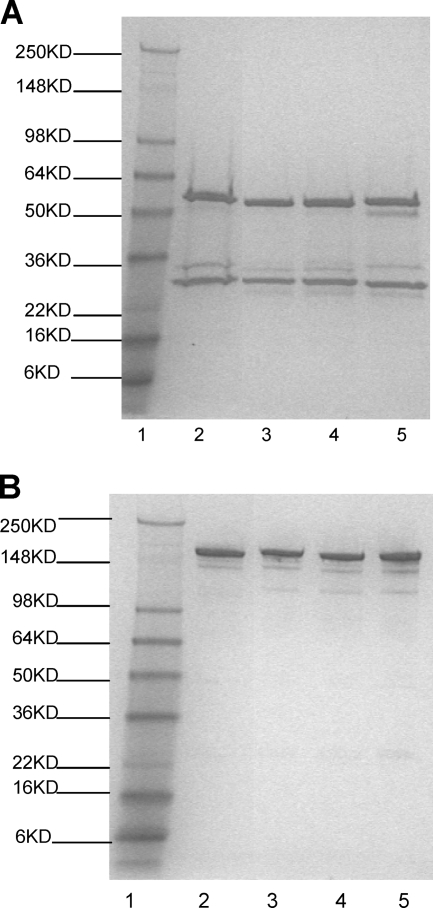
A panel of murine monoclonal antibodies specific for HCV core, NS3, NS4, and NS5 antigens was established. Four MAbs with potential utility as quality control reagents were selected based on antigen specificity and performance in the EIA (data not shown). Reducing SDS-PAGE analysis of IgG purified on a protein A column revealed a purity of approximately 90% as determined by densitometric scanning (Fig. 1).
FIG. 1.
SDS-PAGE analysis of anti-HCV MAbs. SDS-PAGE was performed with a 4 to 20% Tris-glycine gel, followed by staining with an Imperial protein stain. (A) MAbs analyzed under reduced conditions. (B) MAbs analyzed under nonreduced conditions. Lane 1, Seeblue plus2 molecular size marker; lane 2, anti-HCV core antigen MAb; lane 3, anti-HCV NS3 MAb; lane 4, anti-HCV NS4 MAb; lane 5, anti-HCV NS5 MAb.
Expression of chimeric antibodies in CHO cells.
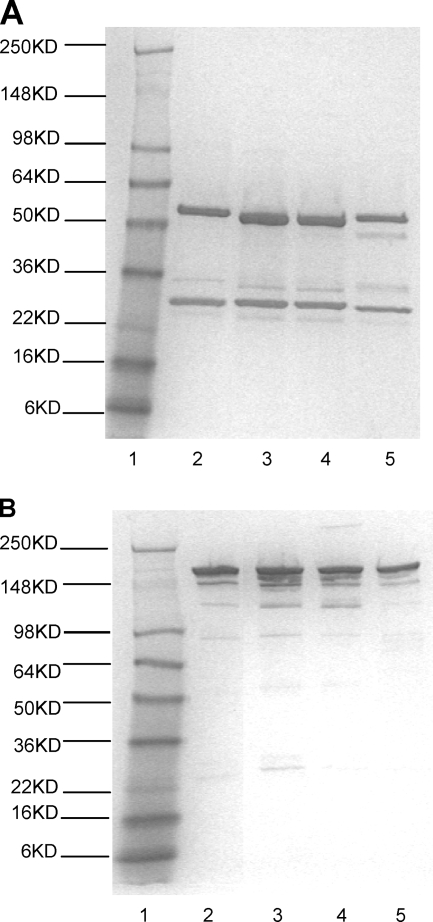
The observed production levels for cAbs against HCV core, NS3, NS4, and NS5 antigens were 10 to 20 μg/ml without methotrexate amplification. Reducing SDS-PAGE analysis of material purified on a protein A column revealed a purity of approximately 90% as determined by densitometric scanning (Fig. 2).
FIG. 2.
SDS-PAGE analysis of protein A-purified anti-HCV chimeric antibodies. SDS-PAGE was performed with a 4 to 20% Tris-glycine gel, followed by staining with an Imperial protein stain. (A) Chimeric antibodies analyzed under reduced conditions. (B) Chimeric antibodies analyzed under nonreduced conditions. Lane 1, Seeblue plus2 molecular size marker; lane 2, anti-HCV core antigen cAb; lane 3, anti-HCV NS3 cAb; lane 4, anti-HCV NS4 cAb; lane 5, anti-HCV NS5 cAb.
Evaluation of anti-HCV chimeric antibodies.
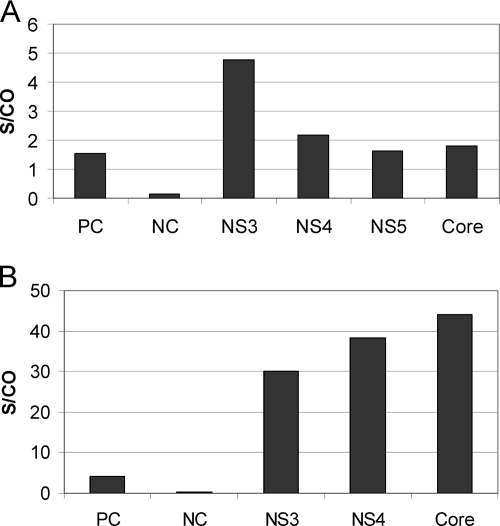
Purified chimeric antibodies were examined for immunoreactivity to HCV by use of the Abbott PRISM and AxSYM HCV assays. In the PRISM HCV assay, specimens with an S/CO value of ≥1.00 were considered reactive, per the Abbott PRISM HCV assay package insert. All four mouse-human cAbs were diluted in anti-HCV-negative human plasma for testing. All four anti-HCV cAbs showed reactivity at the tested concentration (Fig. 3 A).
FIG. 3.
Testing of anti-HCV chimeric antibodies using the Abbott PRISM and AxSYM assays. (A) Chimeric antibodies were tested with the Abbott PRISM assay at a concentration of 500 ng/ml, with the exception of the anti-HCV NS5 cAb, which was tested at a concentration of 10 μg/ml. (B) Chimeric antibodies were tested with the Abbott AxSYM assay at a concentration of 500 ng/ml. Only HCV NS3, NS4, and core antigens are utilized in this assay. The positive controls (PC) and the negative controls (NC) were from the assay kits.
The positive control in the AxSYM HCV assay is human plasma reactive for anti-HCV (S/CO, 2.50 to 7.00), and the negative control is human plasma nonreactive for anti-HCV (S/CO, 0.01 to 0.60). Three mouse-human chimeric antibodies, anti-HCV core antigen, anti-HCV NS3, and anti-HCV NS4, were diluted in anti-HCV-negative human plasma and tested using the AxSYM HCV assay. Specimens with S/CO values of ≥1.21 are considered reactive for IgG antibody to HCV. Specimens with S/CO values between 0.80 and 1.20 are considered to be in the gray-zone (indeterminate) range. Specimens with S/CO values between 0.00 and 0.79 are considered nonreactive. The three cAbs showed reactivity at the tested concentration (Fig. 3B). The anti-HCV core antigen, anti-HCV NS3, and anti-HCV NS4 cAbs were also diluted in HCV-negative human plasma and tested using the Abbott Architect HCV assay. All three anti-HCV cAbs showed reactivity (data not shown). The anti-HCV NS5 cAb was not tested using the Abbott Architect and AxSYM HCV assays, because in these assays, the HCV NS5 antigen is not present during the microparticle preparation.
Epitope mapping.
To further characterize these antibodies, three to five amino acids overlapping synthetic peptides ranging from 15 to 25 amino acids were used to map the epitope. The anti-HCV core antigen MAb was mapped to the core antigen region from aa 32 to 36, and the anti-HCV NS4 MAb was mapped to aa 1697 to 1708 (data not shown). Then, yeast surface display was used to map epitopes for anti-HCV NS3 and anti-HCV NS5.
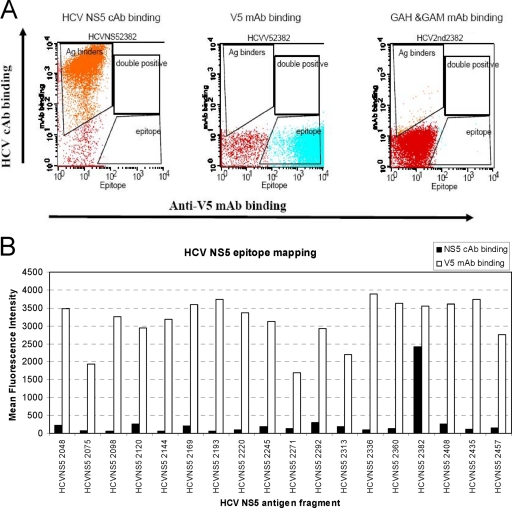
The anti-HCV NS5 MAb was raised against the HCV NS5 immunogen region from aa 2054 to 2481 (428 aa). In order to map the epitope, the 428-aa antigen was divided into 18 overlapping segments. Six extra amino acids on the N-terminal end and five extra amino acids on the C-terminal end of the 428-aa fragment were included. These yeast clones expressing HCV NS5 fragments were detected with the HCV NS5 cAb and Alexa Fluor 633-labeled goat anti-human IgG secondary antibody. FACs Calibur flow cytometry analysis showed that the HCV NS5 fragment from aa 2382 to 2408 (2382-AESYSSMPPLEGEPGDPDLSDGSWSTV-2408) was reactive to the anti-HCV NS5 cAb (Fig. 4), which indicates the presence of the HCV NS5 epitope in this fragment. All 18 yeast clones stained with the anti-V5 MAb and Alexa Fluor 488-labeled goat anti-mouse IgG reacted positively to the anti-V5 MAb.
FIG. 4.
Display of HCV NS5 antigen fragments on the yeast cell surface. (A) Flow cytometry analysis of the HCV NS5 antigen fragments. The presence of the V5 tag confirms the expression of the full-length polypeptide on the yeast cell surface. The Alexa Fluor 633-labeled goat anti-human IgG secondary antibody verifies the low background. The HCV NS5 epitope-containing fragment showed that yeast cells shifted after binding to the anti-HCV NS5 cAb. GAH, goat anti-human; GAM, goat anti-mouse. (B) The HCV NS5 fragment from aa 2382 to 2408 was the only one with a positive signal. The diagram depicts the mean fluorescence intensity (MFI) of the anti-HCV NS5 chimeric antibody, as determined by flow cytometry.
Similarly, an anti-HCV NS3 murine monoclonal antibody was raised against the HCV NS3 antigen region from aa 1192 to 1457 (265 aa). The 265-aa antigen was divided into 12 overlapping pieces and displayed on the yeast surface. Two extra amino acids on both the N- and C-terminal ends of the 265-aa fragment were included. Only the HCV NS3 fragment from aa 1190 to 1216 showed positive binding to the HCV NS3 chimeric antibody (1190-AKAVDFVPVESLETTMRSPVFTDNSSP-1216). Meanwhile, all 12 HCV NS3 yeast clones reacted positively to the anti-V5 MAb.
Fine epitope mapping using yeast display.
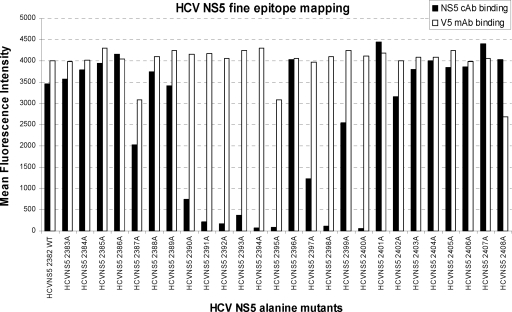
The anti-HCV core antigen MAb has been mapped to aa 32 to 36, the anti-HCV NS4 MAb to aa 1697 to 1708, the HCV NS3 cAb to aa 1190 to 1216, and the HCV NS5 cAb to aa 2382 to 2408. To fine map the epitopes, alanine scanning combined with yeast display was used to probe the epitopes at the individual amino acid level. Each amino acid of the epitope fragment was substituted with alanine. For example, HCV NS5 aa 2382 to aa 2408 had alanine substituted at each position of the epitope, with the exclusion of aa 2382, which was alanine in the wild type. These yeast cells expressing alanine mutations were cultured, induced, and analyzed by FACs Calibur to determine which alanine mutant had lost the antibody binding activity, indicating that the mutated amino acid was part of the antibody binding epitope. The data showed that HCV NS5 antigen residues Pro2390, Leu2391, Glu2392, Gly2393, Glu2394, Pro2395, Asp2397, Pro2398, and Leu2400 were the anti-HCV NS5 chimeric antibody binding sites (Fig. 5).
FIG. 5.
Residue-level epitope mapping of the mutations carried by HCV NS5 alanine mutants. The diagram depicts the mean fluorescence intensities of 26 HCV NS5 alanine mutants and the HCV NS5 wild type (WT) on the yeast cell surface, as determined by flow cytometry. Note that aa 2382 is alanine in the wild type.
We applied the same technique to the other HCV cAbs, and the results are summarized in Fig. 6. Specifically, the anti-HCV core antigen cAb mapped to residues Gln29, Val31, Gly32, Gly33, and Leu36. The anti-HCV NS3 cAb mapped to residues Asp1194, Phe1195, Val1196, Pro1197, Glu1199, Leu1201, Glu1202, and Met1205. The anti-HCV NS4 cAb mapped to residues Leu1701, Tyr1702, Gly1704, Phe1705, and Asp1706.
FIG. 6.
Summary of the residue-level epitope mapping for the four anti-HCV chimeric antibodies.
DISCUSSION
Diagnostic tests for HCV infection include serological assays for antibodies and molecular techniques for detecting virus particles. Automated enzyme immunoassays allow for processing of large numbers of samples and are used mainly for screening of blood and initial detection of anti-HCV antibodies. The immunoassay kits contain positive controls and calibrators as described above. Manufacturers use performance evaluation panels as one of the QC reagents to monitor the manufacturing process. Current HCV immunoassay QC reagents are derived from HCV-infected patient serum and/or plasma samples. For replacement of these reagents, a panel of anti-HCV chimeric antibodies were developed and characterized for use as HCV immunoassay controls and QC reagents.
The HCV chimeric antibodies were purified by protein A agarose affinity chromatography. Yields of purified antibody were 10 to 20 μg/ml of cell culture supernatant. Reducing SDS-PAGE analysis of purified MAbs and cAbs revealed a purity of approximately 90%, as determined by densitometric scanning of gels stained with an Imperial stain. The expected antibody heavy chains and light chains were seen at apparent molecular sizes of 45,000 and 25,000 Da, respectively. There were some smaller bands in the nonreducing SDS-polyacrylamide gel, which may be due to heavy-heavy-light, heavy-heavy, heavy-light, and/or light-light combinations. These kinds of heavy-chain and light-chain combinations have been seen with the production of other recombinant antibodies (16). Sizing exclusion chromatography was used to further purify intact full-length cAbs that achieved >95% purity for all four HCV cAbs. These anti-HCV chimeric antibodies were tested using Abbott immunoassay platforms and demonstrated suitability for use in HCV immunoassays as positive controls, calibrators, and QC reagents to monitor the manufacturing process, as well as for standardization of the HCV assays.
The use of chimeric antibodies in the preparation of quality controls for immunoassays offers several substantial advantages over traditional methods utilizing high-titer plasma or serum, as well as other alternatives, such as chemical coupling of human IgG Fc fragments to a murine monoclonal antibody (18). First, the chimeric antibody can be produced continuously with the same affinity and specificity (5). A reduction in lot-to-lot variability over time with respect to antibody class composition, titer, specificity, and affinity would be expected to yield a consistent QC reagent that should give the most reproducible patient results. Second, virtually unlimited quantities of the chimeric antibodies can be generated. Recombinant cell lines can stably produce chimeric antibodies at manufacturable levels and at a reasonable cost. Finally, the homogeneous nature of the chimeric antibodies allows for easier characterization, and creating the chimeric antibodies will mean eliminating the high costs and risks associated with sourcing, transporting, and standardizing serum-derived reagents. Mouse-human chimeric antibodies have previously been shown to be useful as quality control reagents and for quantitation of specific antibodies in reference standards (10, 11, 12).
For the high-level expression of recombinant antibodies, one of the most widely used mammalian expression systems is the gene amplification procedure offered by use of DHFR-deficient CHO cells (14, 21). The advantage of this system lies in its ability to amplify genes linked to DHFR, which leads to enhanced levels of protein expression. We transfected CHO cells with a single plasmid containing cAb light-chain, cAb heavy-chain, and DHFR expression cassettes. However, after careful screening of positive CHO clones, we were able to identify the clones producing quantities of chimeric antibodies significant enough to satisfy the requirements of the assay. There was no need to amplify these CHO subclones with methotrexate.
Since there are a number of HCV antigens utilized in HCV assays, the methodology for characterizing HCV-specific antibody binding epitopes is very important to identify diagnostically relevant antibodies. Previous methods for antibody epitope mapping using biosynthetic systems involved the presentation of protein or peptide fragments on the surface of E. coli and the measurement of antibody binding (3). Overlapping and/or mutated synthetic peptides have also been used to map antibody binding epitopes (19, 23). These methods have been successful in defining linear or continuous regions of antibody interactions, but they are less useful for characterizing antibodies that are reactive toward a discontinuous binding epitope. The conformational epitope has been successfully detected using phage display (24). Moreover, yeast display has been used to determine the domain of an antigen recognized by an antibody (2, 4, 25). We extended this technique by combining yeast display with scanning alanine mutagenesis to elucidate the antigen amino acid residues directly involved in antibody recognition.
Since all four anti-HCV MAbs were characterized as possible linear epitopes by Western blotting (data not shown), we decided to fine map the epitope to the residue level by using yeast cell surface display. Yeast cells displaying the targeted HCV antigen overlapping fragments were constructed and screened for binding. HCV antigen fragments were defined using a computer-assisted prediction based on a hydrophilicity algorithm. In general, yeast cells express folded, functional proteins due to the protein folding and quality control machinery in the endoplasmic reticulum, which is different from bacterial expression systems (7, 17). Tethering of HCV fragments to the yeast cell surface removed the need for soluble expression, purification, and characterization of individual protein fragments, which is laborious, time-consuming, and not guaranteed to produce usable protein fragments for analysis. Once the yeast display system of HCV fragments was established, the fragment mapping and characterization were rapid and could be carried out over the course of a few days, in comparison with biochemical and biophysical analysis using soluble protein domains.
Alanine scanning, a method in which residues of interest are mutated to alanine and subsequent changes in binding are measured, is another useful tool in dissecting protein-protein interactions (6). A yeast-displayed single alanine point mutation panel was generated in the epitope-containing HCV antigen fragment and was screened for a reduction in or a loss of binding to anti-HCV cAbs. This method was successfully used to identify nonlinear epitopes of antibodies binding to complex eukaryotic proteins without prior knowledge of potential contact residues (2, 4). A single point mutation of the HCV target antigen that resulted in reduced or lost cAb binding suggested that the residue was involved in antigen-antibody contact. Using this method, we have identified the key residues energetically important for the binding of HCV to each chimeric antibody of the anti-HCV core, NS3, NS4, and NS5 antigens. For example, the HCV NS3 cAb epitope was mapped to eight amino acids of the NS3 antigen: aa 1194, aa 1195, aa 1196, aa 1197, aa 1199, aa 1201, aa 1202, and aa 1205. Interestingly, alanine mutation of aa 1198, aa 1200, aa 1203, and aa 1204 within the epitope fragment from aa 1194 to 1205 has no impact on cAb binding. Since the chimeric antibody was engineered from the MAb by cloning the murine V genes from the hybridoma line, it should have the same epitope as the corresponding murine MAb. Using the HCV NS3 MAb, we mapped the same residues as those for the HCV NS3 cAb. (Note that positions aa 1190 and 1192 are alanine in the wild type). The findings for the anti-HCV NS3 MAb are not shown. Every alanine mutant was confirmed to have C-terminal V5 expression, which indicated correct expression.
The anti-HCV core antigen MAb was first mapped to aa 32 to 36 by use of synthetic peptides (data not shown); however, fine epitope mapping using yeast display demonstrated that it actually binds to aa 29, 31, 32, 33, and 36. Amino acids 29 and 31 were not identified by synthetic peptides, showing the limitation of this technique. For example, HCV core antigen aa 29, 30, and 31 were not identified as part of the anti-HCV core antigen epitope by peptide mapping. If the designed recombinant HCV core antigen in the assay does not consist of aa 29 to 31, then there may be a loss of or reduction in binding by the patient's anti-HCV core antibody and the immunoassay may not reflect the patient's actual condition.
In conclusion, we have cloned, expressed, and characterized four anti-HCV chimeric antibodies for use as quality control reagents in diagnostic immunoassays. By this protocol, highly pure and immunoreactive chimeric antibodies can be produced continuously on a laboratory scale. Further scale-up and streamlining of production and purification methods should be possible by standard procedures.
Acknowledgments
We thank Chung-Ming Hsieh and Gerard Carson for their support of this work, and we thank Scott Muerhoff for critical review of the manuscript.
Footnotes
Published ahead of print on 28 April 2010.
REFERENCES
- 1.Boder, E. T., and K. D. Wittrup. 1997. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15:553. [DOI] [PubMed] [Google Scholar]
- 2.Chao, G., J. R. Cochran, and K. D. Wittrup. 2004. Fine epitope mapping of anti-epidermal growth factor receptor antibodies through random mutagenesis and yeast surface display. J. Mol. Biol. 342:539-550. [DOI] [PubMed] [Google Scholar]
- 3.Christmann, A., A. Wentzel, C. Meyer, G. Meyers, and H. Kolmar. 2001. Epitope mapping and affinity purification of mono-specific antibodies by Escherichia coli cell surface display of gene-derived random peptide libraries. J. Immunol. Methods 257:163-173. [DOI] [PubMed] [Google Scholar]
- 4.Cochran, J. R., Y. S. Kim, M. J. Olsen, R. Bhandari, and K. D. Wittrup. 2004. Domain-level antibody epitope mapping through yeast surface display of epidermal growth factor receptor fragments. J. Immunol. Methods 287:147-158. [DOI] [PubMed] [Google Scholar]
- 5.Colcher, D., D. Milenic, M. Roselli, A. Raubitshek, G. Yanranton, D. King, J. Adair, N. Whittle, M. Bodmer, and J. Schlom. 1989. Characterization and biodistribution of recombinant and recombinant/chimeric constructs of monoclonal antibody B72.3. Cancer Res. 49:1738-1745. [PubMed] [Google Scholar]
- 6.Cunningham, B. C., and J. A. Wells. 1989. High resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science 244:1081-1085. [DOI] [PubMed] [Google Scholar]
- 7.Ellgaard, L., and A. Helenius. 2003. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4:181-191. [DOI] [PubMed] [Google Scholar]
- 8.Galfre, G., S. C. Howe, C. Milstein, G. W. Butcher, and J. C. Howard. 1977. Antibodies to major histocompatibility antigens produced by hybrid cell lines. Nature 266:550-552. [DOI] [PubMed] [Google Scholar]
- 9.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 10.Hackett, J., Jr., J. Hoff-Velk, A. Golden, J. Brashear, J. Robinson, M. Rapp, M. Klass, D. H. Ostrow, and W. Mandecki. 1998. Recombinant mouse-human chimeric antibodies as calibrators in immunoassays that measure antibodies to Toxoplasma gondii. J. Clin. Microbiol. 36:1277-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton, R. G. 1990. Engineered human antibodies as immunologic quality control reagents. Ann. Biol. Clin. 48:473-477. [PubMed] [Google Scholar]
- 12.Hamilton, R. G. 1991. Application of engineered chimeric antibodies to the calibration of human antibody standards. Ann. Biol. Clin. 49:242-248. [PubMed] [Google Scholar]
- 13.Jones, S. T., and M. M. Bendig. 1991. Rapid PCR-cloning of full-length mouse immunoglobulin variable regions. Biotechnology 9:579. [DOI] [PubMed] [Google Scholar]
- 14.Kaufman, R. J., P. A. Sharp, and S. A. Latt. 1983. Evolution of chromosomal regions containing transfected and amplified dihydrofolate reductase sequences. Mol. Cell. Biol. 3:699-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Mazor, Y., T. Van Blarcom, R. Mabry, B. L. Iverson, and G. Georgiou. 2007. Isolation of engineered, full length antibodies from libraries expressed in Escherichia coli. Nat. Biotechnol. 25:563-565. [DOI] [PubMed] [Google Scholar]
- 17.Mischo, A., A. Wadle, K. Wätzig, D. Jäger, E. Stockert, D. Santiago, G. Ritter, E. Regitz, E. Jäger, A. Knuth, L. Old, M. Pfreundschuh, and C. Renner. 2003. Recombinant antigen expression on yeast surface (RAYS) for the detection of serological immune responses in cancer patients. Cancer Immun. 3:5-16. [PubMed] [Google Scholar]
- 18.Miyachi, J., K. Doi, K. Kitamura, T. Jitsukawa, and H. Watanabe. 1992. Chemically humanized murine monoclonal antibody against a cell nuclear antigen: usefulness in autoimmune diagnostics. J. Clin. Lab. Anal. 6:343-350. [DOI] [PubMed] [Google Scholar]
- 19.Rosa, C., S. Osborne, F. Garetto, S. Griva, A. Rivella, G. Calabresi, R. Guaschino, and F. Bonelli. 1995. Epitope mapping of NS4 and NS5 gene products of hepatitis V virus and the use of a chimeric NS4-NS5 synthetic peptide for serodiagnosis. J. Virol. Methods 55:219-232. [DOI] [PubMed] [Google Scholar]
- 20.Shepard, C. W., L. Finelli, and M. J. Alter. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558-567. [DOI] [PubMed] [Google Scholar]
- 21.Urlaub, G., and L. Chasin. 1980. Isolation of Chinese hamster cell mutants deficient in dihydrofolate reductase activity. Proc. Natl. Acad. Sci. U. S. A. 77:4216-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wild, D. (ed.) 2005. The immunoassay handbook, 3rd ed., p. 297-303, 329-333. Elsevier, Oxford, United Kingdom.
- 23.Yip, Y. L., G. Smith, J. Koch, S. Dubel, and R. L. Ward. 2001. Identification of epitope regions recognized by tumor inhibitory and stimulatory anti-ErbB-2 monoclonal antibodies: implications for vaccine design. J. Immunol. 166:5271-5278. [DOI] [PubMed] [Google Scholar]
- 24.Yip, Y. L., J. Novotny, M. Edwards, and R. L. Ward. 2003. Structural analysis of the ErbB-2 receptor using monoclonal antibodies: implications for receptor signaling. Int. J. Cancer 104:303-309. [DOI] [PubMed] [Google Scholar]
- 25.Zhang, X., J. Wang, K. Wen, Z. Mou, L. Zou, X. Che, B. Ni, and Y. Wu. 2009. Antibody binding site mapping of SARS-CoV spike protein receptor-binding domain by a combination of yeast surface display and phage peptide library screening. Viral Immunol. 22:407-415. [DOI] [PubMed] [Google Scholar]