Abstract
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) has been suggested as a reliable method for bacterial identification from cultures. Direct analysis of clinical samples might increase the usefulness of this method, shortening the time for microorganism identification. We compared conventional methods for the diagnosis of urinary tract infections (UTIs) and identification of the urinary tract pathogens (automated screening, plate cultures, and identification based on biochemical characteristics) and a fast method based on conventional screening and MALDI-TOF MS. For this latter method, 4 ml of urine was centrifuged at a low-revolution setting (2,000 × g) to remove leukocytes and then at high revolutions (15,500 × g) to collect bacteria. The pellet was washed and then applied directly to the MALDI-TOF MS plate. Two hundred sixty urine samples, detected as positive by the screening device (UF-1000i), were processed by culture and MALDI-TOF MS. Twenty samples were positive in the screening device but negative in culture, and all of them were also negative by MALDI-TOF MS. Two-hundred thirty-five samples displayed significant growth of a single morphological type in culture. Two-hundred twenty of them showed bacterial growth of >105 CFU/ml. Microorganism identifications in this group were coincident at the species level in 202 cases (91.8%) and at the genus level in 204 cases (92.7%). The most frequent microorganism was Escherichia coli (173 isolates). MALDI-TOF MS identified this microorganism directly from the urine sample in 163 cases (94.2%). Our results show that MALDI-TOF MS allows bacterial identification directly from infected urine in a short time, with high accuracy, and especially when Gram-negative bacteria with high bacterial counts are involved.
Urinary tract infections (UTIs) are among the most common human bacterial infections. More than 80% of uncomplicated UTIs are caused by Escherichia coli (9). Other common uropathogens include Staphylococcus saprophyticus in uncomplicated UTIs and Gram-negative rods (enterobacteria other than E. coli or Pseudomonas aeruginosa) and Gram-positive cocci in complicated UTIs (9). UTIs affect an estimated 1 out of 3 women before the age of 24 (7). Up to 40 to 50% of the female population will develop a symptomatic UTI at some time during their life, and about 33% of women with acute uncomplicated UTIs have frequent recurrences (7).
Several tests are available for screening patients for UTIs, including urine dipstick testing, urinalysis, Gram staining, and urine culture. Urine culture is the “gold standard” for defining the diagnosis of UTIs, because it allows the quantification and identification of the uropathogenic species. However, this method is time-consuming and expensive. Up to 70% of urine cultures are negative, with high costs for unnecessary testing (4). Thus, automated analyzers for UTI screening to rapidly identify negative samples that can be reported promptly to clinicians are now widely used. Samples positive in this screening are cultured to count colonies and identify the microorganism.
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) has been suggested as a fast and reliable method for bacterial identification (14), based on the characteristic protein profiles for each microorganism. Databases have been developed that include the main pathogenic microorganisms, thus allowing the use of this method in routine bacterial identification from plate cultures.
Direct analysis of clinical samples may further increase the usefulness of this method, since it can significantly shorten the time required for the identification of microorganisms. Here we compared the conventional methods for UTI diagnosis and identification of urinary tract pathogens in microbiology laboratories (automated screening and plate culture) and a fast method based on conventional screening plus MALDI-TOF MS.
MATERIALS AND METHODS
Processing of urine samples and biochemical identification.
We analyzed 260 urine samples submitted to the Microbiology Laboratory from outpatients and inpatients with symptoms suggestive of UTIs and reported as positive by the automated screening device. The samples were processed using an automated screening device based on flow cytometry (UF-1000i; bioMérieux, Marcy l'Étoile, France). The samples judged negative by the screening device were discarded and reported as negative without culture. For samples reported as positive, 10 μl was cultured on blood agar plates and MacConkey's agar plates. Plates were incubated in an aerobic atmosphere at 37°C for 18 h. Negative plates were incubated for a further 24 h and, if they remained negative, were discarded. When there was bacterial growth, colonies on blood agar were counted, and colonies from blood agar or MacConkey's agar plates were identified by conventional methods (WIDER MIC/ID Gram positives; manufactured for Francisco Soria Melguizo SA, Madrid, Spain, by MicroScan, Sacramento, CA; and Vitek-2 GNB, from bioMérieux, Marci l'Étoile, France), except when more than two colony morphologies were found, in which case the urine sample was rejected.
MALDI-TOF mass spectrometry.
Samples were first studied by MALDI-TOF MS directly, without extraction. When no reliable identification was obtained, samples were extracted and then studied again.
(i) Sample preparation for MALDI-TOF MS.
Urine (4 ml) was centrifuged at 2,000 × g for 30 s to remove leukocytes. The supernatant was centrifuged at 15,500 × g for 5 min to collect bacteria. The pellet was washed once with deionized water. A small amount of bacteria was applied to the MALDI plate in a thin film as a direct method.
Furthermore, an ethanol-formic acid extraction procedure was applied when the direct method did not yield reliable identification. First, a small amount of bacteria was resuspended in 300 μl of water. Then, 900 μl of absolute ethanol was added, the mixture was centrifuged at 15,500 × g for 2 min, and the supernatant was discarded. Subsequently, 50 μl of formic acid (70% [vol/vol]) was added to the pellet and mixed thoroughly by pipetting before the addition of 50 μl acetonitrile to the mixture. The mixture was centrifuged again at 15,500 × g for 2 min. One microliter of the supernatant was placed onto a spot of the steel target and air dried at room temperature.
Both the microbial film and the supernatant were overlaid with 1 μl of matrix solution (saturated solution of HCCA [α-cyano-4-hydroxy cinnamic acid] in organic solvent [50% acetonitrile and 2.5% trifluoroacetic acid]) and air dried.
(ii) MALDI-TOF mass spectrometry.
Measurements were performed on an Autoflex III MALDI-TOF/TOF mass spectrometer (Bruker Daltonics, Leipzig, Germany) equipped with a 200-Hz Smartbeam laser. Spectra were recorded in the linear, positive mode at a laser frequency of 200 Hz within a mass range from 2,000 to 20,000 Da. The IS1 voltage was 20 kV, the IS2 voltage was maintained at 18.6 kV, the lens voltage was 6 kV, and the extraction delay time was 40 ns.
For each spectrum, 500 laser shots were collected and analyzed (10 × 50 laser shots from different positions of the target spot). The spectra were calibrated externally using the standard calibrant mixture (Escherichia coli extracts including the additional proteins RNase A and myoglobin; Bruker Daltonics). The calibration masses were as follows: RL36, 4,364.3 Da; RS22, 5,095.8 Da; RL34, 5,380.4 Da; RL33meth, 6,254.4 Da; RL32, 6,315 Da; RL29, 7,273.5 Da; RS19, 10,299.1 Da; RNase A, 13,682.2 Da; and myoglobin, 16,952.5 Da.
(iii) Data analysis.
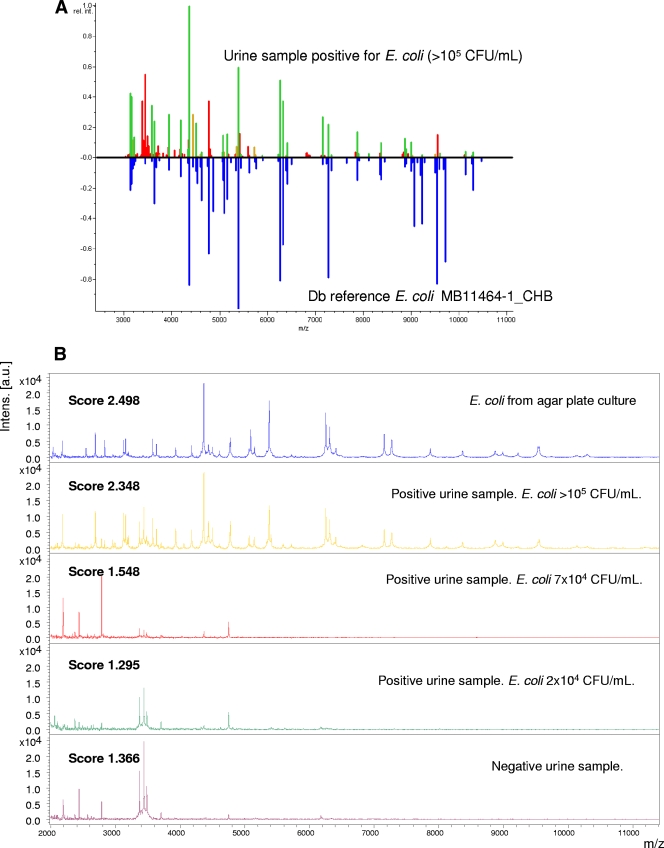
For automated data analysis, raw spectra were processed using the MALDI Biotyper 2.0 software (Bruker Daltonics, Leipzig, Germany) at default settings. The software performs normalization, smoothing, baseline subtraction, and peak picking, creating a list of the most significant peaks of the spectrum (m/z values with a given intensity, with the threshold set to a minimum of 1% of the highest peak and a maximum of 100 peaks). To identify unknown bacteria, each peak list generated was matched directly against reference libraries (3,290 species) using the integrated pattern-matching algorithm of the Biotyper 2.0 software (Bruker Daltonics, GmbH, Germany). The unknown spectra were compared with a library of reference spectra based on a pattern recognition algorithm using peak position, peak intensity distributions, and peak frequencies. A comparison between a urine sample positive for E. coli and an E. coli profile included in the Biotyper database for comparison purposes is shown in Fig. 1A. Once a spectrum has been generated and captured by the software, the whole identification process is performed automatically, without any user intervention.
FIG. 1.
(A) Mirror view of the best match of urine sample against the Biotyper database (Db). Green indicates good matching peaks, yellow indicates a moderate match, and red indicates no match. (B) Mass spectra (size, 2,000 to 11,000 Da) of Escherichia coli cells grown under different conditions.
(iv) Result scoring.
MALDI-TOF MS identifications were classified using modified score values proposed by the manufacturer: a score of ≥2 indicates species identification; a score between 1.7 and 1.9 indicates genus identification, and a score of <1.7 indicates no identification.
MALDI-TOF MS detection limits.
In order to determine the minimal bacterial concentration allowing reliable MALDI-TOF MS identifications, we inoculated 50-ml aliquots of sterile water with one strain of Escherichia coli, one strain of Enterococcus faecalis, or one strain of Pseudomonas aeruginosa at a bacterial count of 2.5 × 108 CFU/ml. We performed sequential dilutions in order to achieve aliquots of each microorganism at the following bacterial counts: 5 × 105, 2 × 105, 1.5 × 105, 1.2 × 105, 1 × 105, 8 × 104, 6 × 104, 2.5 × 104, 3 × 104, and 1.2 × 104 CFU/ml. When necessary, aliquots were furthermore diluted before spreading on plates to obtained a countable number of colonies (50 to 500 colonies) when plated. Three 100-μl aliquots of each dilution were plated onto blood agar plates. These plates were incubated at 37°C in an aerobic atmosphere, and colonies were counted manually. The mean of three aliquots of each dilution was considered to be the final count for that aliquot. Four-milliliter samples of each dilution were taken for MALDI-TOF MS, which was performed according the method previously described.
RESULTS
Two-hundred sixty urine samples reported as positive by the screening device were processed both by plate culture and MALDI-TOF MS. The results are shown in Tables 1 and 2. The screening device reported as positive 20 samples that were negative in culture. MALDI-TOF MS did not find any significant protein profile in any of these cases. Five samples positive in the screening led to significant growth of two colony morphologies in the culture. In these samples, MALDI-TOF MS reported an unreliable identification in 2 cases but reported a microorganism identification in 3 cases.
TABLE 1.
Correlation between conventional identification and MALDI-TOF MS in 260 urine samples positive in the screening for UTIs
| Conventional identification (no. of cases) | MALDI-TOF MS identification (no. of cases) |
|---|---|
| Negative (20) | Negative (20) |
| Positive, 2 morphology colony types (5) | Not reliable identification (2) |
| Microorganism identification (3)a | |
| Positive, 1 morphology colony type (235) | Positive with same identification (205)b |
| Positive with different identification (2) | |
| Negative or not reliable identification (28)c |
Two E. coli cases and one Enterobacter aerogenes case.
A total of 203 correct identifications at the species level and 2 correct identifications at the genus level.
Fourteen of these 28 cases were cultures with <105 CFU/ml.
TABLE 2.
Identification by MALDI-TOF MS and conventional identification of 220 microorganism causing UTIs with bacterial counts of >100,000 CFU/ml
| Conventional identification (no. of isolates) | Correlation (%) at: |
MALDI-TOF MS identification (no. of isolates) | |
|---|---|---|---|
| Species level | Genus level | ||
| Escherichia coli (167) | 97.6 | 97.6 | Escherichia coli (163) |
| No reliable identification (4) | |||
| Klebsiella pneumoniae (7) | 100 | 100 | Klebsiella pneumoniae (7) |
| Klebsiella oxytoca (9) | 77.8 | 77.8 | Klebsiella oxytoca (7) |
| No reliable identification (2) | |||
| Citrobacter freundii (1) | 100 | 100 | Citrobacter freundii (1) |
| Citrobacter koseri (1) | 100 | 100 | Citrobacter koseri (1) |
| Enterobacter cloacae (6) | 83.3 | 83.3 | Enterobacter cloacae (5) |
| No reliable identification (1) | |||
| Enterobacter asburiae (1) | 0 | 100 | Enterobacter sp. (1) |
| Serratia marcescens (2) | 100 | 100 | Serratia marcescens (2) |
| Proteus mirabilis (5) | 80 | 80 | Proteus mirabilis (4) |
| No reliable identification (1) | |||
| Morganella morganii (1) | 100 | 100 | Morganella morganii (1) |
| Pseudomonas aeruginosa (2) | 50 | 50 | Pseudomonas aeruginosa (1) |
| No reliable identification (1) | |||
| Raoultella planticola (2) | 0 | 50 | Raoultella ornithinolytica (1) |
| Escherichia coli (1) | |||
| Raoultella ornithinolytica (1) | 0 | 0 | Citrobacter sp. (1) |
| Enterococcus faecalis (12) | 66.7 | 66.7 | Enterococcus faecalis (8) |
| No reliable identification (4) | |||
| Staphylococcus aureus (2) | 100 | 100 | Staphylococcus aureus (2) |
| Streptococcus agalactiae (1) | 0 | 0 | No reliable identification (1) |
| Total (220) | 91.8 | 92.7 | |
Two-hundred thirty-five samples led to significant growth of one colony morphology in culture, 220 of them (93.6%) with colony counts >105 CFU/ml; 7 (3.0%) with colony counts between 5 × 104 and 105 CFU/ml, and 8 (3.4%) with colony counts lower than 5 × 104 CFU/ml. Among the 220 samples with colony counts of >105 CFU/ml, microorganism identifications coincided at the species level in 202 cases (91.8%), and at the genus level in 204 cases (92.7%). In 14 cases (6.4%), MALDI-TOF MS did not afford a reliable identification. We found major discrepancies (discrepancies at the genus level) in only 2 cases: Raoultella planticola (conventional identification) versus E. coli (MALDI-TOF MS) and Raoultella ornithinolytica (conventional identification) versus Citrobacter freundii (MALDI-TOF MS).
For 7 samples in which the bacterial count was between 5 × 104 and 9 × 104 CFU/ml (2 samples of E. coli and 1 each of P. aeruginosa, Pseudomonas putida, Acinetobacter lwoffii, Staphylococcus aureus, and E. faecalis), MALDI-TOF MS reported a reliable identification in one sample (P. putida), and for 8 samples where the bacterial count was between 5 × 103 and 4 × 104 CFU/ml (4 samples of E. coli and 1 sample each of P. aeruginosa, P. putida, Staphylococcus aureus, and E. faecalis), MALDI-TOF MS did not report a reliable identification in any of the samples.
Experimental studies of scores provided by different bacterial counts (Table 3) support these results. MALDI-TOF MS seems to require high bacterial counts to be able to provide reliable scores. Experimental inocula with at least 8 × 104 CFU/ml were required to obtain reliable scores for E. coli, and the required inocula were even higher for P. aeruginosa (1.2 × 105 CFU/ml) and E. faecalis (1.5 × 105 CFU/ml) (Table 3). This result agrees with the results for the clinical samples, since only one out of seven samples with bacterial counts between 5 × 104 and 9 × 104 CFU/ml (P. putida, 6 × 104 CFU/ml) and none of eight samples with bacterial counts of <5 × 104 CFU/ml (range, 5 × 103 to 4 × 104 CFU/ml) provided reliable MALDI-TOF profiles.
TABLE 3.
MALDI-TOF MS scores for different E. coli, P. aeruginosa, and E. faecalis bacterial counts
| Microorganism | MALDI-TOF MS score for bacterial count (CFU/ml) ofa: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 5 × 105 | 2 × 105 | 1.5 × 105 | 1.2 × 105 | 1 × 105 | 8 × 104 | 6 × 104 | 2.5 × 104 | 3 × 104 | 1.2 × 104 | |
| E. coli | 2.384 | 2.255 | 2.199 | 2.17 | 1.945 | 2.065 | 1.438 | NPF | NPF | NPF |
| P. aeruginosa | 2.267 | 1.892 | 2.121 | 2.107 | NPF | NPF | NPF | NPF | NPF | NPF |
| E. faecalis | 1.976 | 1.767 | 2.006 | 1.674 | NPF | NPF | NPF | NPF | NPF | NPF |
NPF, no peaks found.
Figure 1B shows a comparison between the MALDI spectra of colonies grown on agar plates and those of urine samples. The profiles from agar plate culture and urine samples with high bacterial counts are similar. Most peaks in the profile seem to correspond to bacterial peptides, since they appear in both positive urine samples and in E. coli cultures, and only some peaks around or under 3,500 Da seem to correspond to urinary proteins, since they do not appear in E. coli cultures and do appear in negative urine samples.
The extraction protocol did not improve the identification of microorganisms in samples with a negative culture, samples in which two colony morphologies grew, and samples with low bacterial counts. Twenty out of 220 samples with high bacterial counts and growing in the form of a single bacterial morphology on agar plates were not correctly identified by the direct method. In six cases, the extraction procedure led to a reliable bacterial identification in these samples. Thus, the extraction method improved the results of identification by 2.3%.
The most frequent microorganism isolated was Escherichia coli (173 isolates). MALDI-TOF MS identified the microorganism directly from the urine sample in 163 cases (94.2%). Failures mainly appeared when the bacterial counts were <105 CFU/ml. When the colony count was >105 CFU/ml, the correct identification rate was as high as 97.6%.
Overall, MALDI-TOF MS identified correctly the microorganisms involved in UTIs, directly from the urine sample, once it had been reported as positive by the screening system, in 79.2% of cases at the species level and 80% of cases at the genus level. Of these 260 samples, 206 samples (of which 202 samples had bacterial counts of >105 CFU/ml, 1 sample had a bacterial count of <105 CFU/ml, and 3 samples contained more than one colony morphology) resulted in bacterial species identification directly from centrifuged cells or after extraction. Two samples allowed identification only to the genus level. Twenty samples did not result in microbial identification, but these were also negative in culture (7.7%). Two samples in the MALDI-TOF MS identification were not in accordance with conventional identification (0.77%). Thirty samples (11.5%) did not result in any reliable identification either with the direct method or with extraction. Of these, 14 samples had bacterial counts of <105 CFU/ml, 6 samples had bacterial counts between 5 × 104 and 9 × 104 CFU/ml, 8 samples had bacterial counts between 5 × 103 and 4 × 104 CFU/ml, and 2 samples contained more than one colony morphology. As happened with E. coli, these failures mainly appeared with lower bacterial counts.
In 7.3% of the urine samples with bacterial counts of >105 CFU/ml and only one colony morphology type (14 samples), the MALDI-TOF MS result was considered a failure, either because the protein peaks were too faint to obtain a suitable profile (6 cases) or because the peak profiles generated did not match any profile stored in the database (8 cases).
MALDI-TOF MS seems to identify Gram-negative bacteria more reliably. Among the samples with bacterial counts of >105 CFU/ml, the coincidence rate of conventional identification and MALDI-TOF MS for Gram negatives was 93.7% at the species level and 94.6% at the genus level. The coincidence rate was lower for Gram-positive microorganisms (10/15 samples), although the number of samples was low, and a higher number of samples should be studied to corroborate these results.
DISCUSSION
Previous studies have shown that MALDI-TOF MS might be a useful resource for bacterial identification from culture, especially when a good protein profile database allows a comparison of the profiles obtained with a large number of bacterial species and strains (2, 14). Other studies have described an excellent correlation between MALDI-TOF MS identification and conventional microbiological identification in clinical bacterial and fungal isolates (1, 3, 5, 6, 8, 10, 12). In some cases, when there are discrepancies between MALDI-TOF MS and conventional identification, as sometimes happens for nonfermenting Gram-negative bacilli, 16S rRNA sequencing frequently agrees with MALDI-TOF MS identification (11).
Direct analysis of clinical samples may further increase the usefulness of MALDI-TOF MS, because it allows clinically useful results to be obtained some minutes after sample reception in the laboratory. The use of screening devices has increased steadily in the diagnosis of UTIs, since at least 60 to 70% of urine samples sent for UTI investigation are usually negative, and such devices are now a typical resource for diagnosis at UTI laboratories. The specificity of these devices is usually low, so that very high sensitivity can be obtained. When samples reported as positive by the screening device used (UF-1000i) were studied by conventional methods and with MALDI-TOF MS, all of the false-positive samples reported were discarded by both methods.
There is scant information available about the use of MALDI-TOF devices for the direct identification of microorganisms growing in positive samples. A report at the 48th ICAAC involving 37 urine samples (13) revealed that MALDI-TOF MS correctly identified the etiological microorganism in 33 cases (17 E. coli cases, 4 E. faecalis cases, 6 K. pneumoniae cases, 2 P. aeruginosa cases, 1 S. anginosus case, and 3 S. agalactiae cases), and failed in only 4 cases, all of which led to a mixed culture on conventional culture plates. The present results of MALDI-TOF MS for bacterial identification directly from urine samples, without culture, are excellent. MALDI-TOF MS provided no identification in two mixed cultures but reported identification in 3 mixed cultures. The results of MALDI-TOF MS in mixed cultures probably depend on the proportion between both populations.
The best results are obtained when there is a high bacterial count (>105 CFU/ml) and the microorganism involved is Gram negative. In this respect, the failure rate was only 5.4%. The whole study of 220 positive urine samples with high bacterial counts showed a 91.8% correlation at the species level and 92.7% correlation at the species or genus level.
The bacterial count seems to be a critical issue in obtaining good results with MALDI-TOF MS directly from clinical samples. The results obtained with clinical samples, when the bacterial count was <105 CFU/ml, were poorer than those obtained with high bacterial counts, and in vitro studies on E. coli, E. faecalis, and P. aeruginosa different inocula strengthen this impression. The bacterial count and MALDI-TOF MS show that reliable protein profiles and identifications are obtained only when the bacterial counts reach at least 8 × 104 CFU/ml. This is the diagnostic threshold for most UTIs, although some UTIs such as cystitis may emerge with quite lower bacterial counts (102 to 103 CFU/ml). Identification failures at bacterial counts of <105 CFU/ml should therefore be considered real failures only in some cases, since a number of conventional cultures with these low bacterial counts would be also reported as negative, depending on the clinical characteristics of the patient. Clearly, our results are associated with the method used, in which 4 ml of urine was centrifuged and processed by MALDI-TOF MS. Further modifications to the methodology would probably increase its sensitivity. Nevertheless, while urine samples with bacterial counts of >105 CFU/ml are almost always clinically relevant, urine samples with lower bacterial counts may be relevant or not. Thus, they should probably remain to be studied by conventional methods that allow one to know this bacterial count and manage the patient by taking this finding into account.
MALDI-TOF MS allows the identification of the etiological agent, but antibiotic susceptibility should be studied by conventional methods. Thus, culture is still necessary, since bacterial growth is still needed for susceptibility studies. This suggests that the benefits obtained from fast identification of pathogens by MALDI-TOF MS would be modest, since it does not allow other procedures to be discontinued. Nevertheless, in our opinion this would be a mistaken impression. The evolution of the susceptibility to antibiotics of some of the usual urinary tract pathogens (such as fluoroquinolone resistance in E. coli, spread of extended-spectrum β-lactamases [ESBLs] in E. coli and Klebsiella, and increasing resistance to trimethoprim-sulfamethoxazole [SXT] in some countries) has significantly reduced the number of antibiotics that can be used empirically with a high probability of success. MALDI-TOF MS allows the diagnosis of a UTI and its etiology in minutes, thus affording the opportunity of guiding empirical treatment much more efficaciously. Using the threshold proposed here (4 ml of urine, associated with MALDI-TOF MS positivity when the CFU count is >105 CFU/ml) almost guarantees that identification will correlate with the UTI, and according to our results allows the etiology of >90% of UTIs to be diagnosed in just a few minutes.
In conclusion, the data gathered in the present study demonstrate that MALDI-TOF MS is a quick and reliable method for the identification of bacteria from infected urine, whose shortened analysis time enables an earlier and more accurate selection of antibiotics for patient treatment.
Footnotes
Published ahead of print on 14 April 2010.
REFERENCES
- 1.Barbuddhe, S. B., T. Maier, G. Schwarz, M. Kostrzewa, H. Hof, E. Domann, T. Chakraborty, and T. Hain. 2008. Rapid identification and typing of Listeria species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 74:5402-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blondiaux, N., O. Gaillot, and R. J. Courcol. 2009. MALDI-TOF mass spectrometry to identify clinical bacterial isolates: evaluation in a teaching hospital. Pathol. Biol. (Paris). [Epub ahead of print.] [DOI] [PubMed]
- 3.Degand, N., E. Carbonnelle, B. Dauphin, J. L. Beretti. M. Le Bourgeois, Semet-Gaudelus, C. Segonds, P. Berche, X. Nassif, and A. Ferroni. 2008. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of nonfermenting gram-negative bacilli isolated from cystic fibrosis patients. J. Clin. Microbiol. 46:3361-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devillé, W. L., J. C. Yzermans, N. P. van Duijn, P. D. Bezemer, D. A. van der Windt, and L. M. Bouter. 2004. The urine dipstick test useful to rule out infections. A meta-analysis of the accuracy. BMC Urol. 4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dieckmann, R., R. Helmuth, M. Erhard, and B. Malorny. 2008. Rapid classification and identification of salmonellae at the species and subspecies levels by whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 74:7767-7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenselau, C., and P. A. Demirev. 2001. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom. Rev. 20:157-171. [DOI] [PubMed] [Google Scholar]
- 7.Foxman, B., R. Barlow, H. d'Arcy, B. Gillespie, and J. D. Sobe. 2000. Urinary tract infection: estimated incidence and associated costs. Ann. Epidemiol. 10:509-515. [DOI] [PubMed] [Google Scholar]
- 8.Grosse-Herrenthey, A., T. Maier, F. Gessler, R. Schaumann, H. Böhnel, M. Kostrzewa, and M. Krüger. 2008. Challenging the problems of clostridial identification with matrix-assisted laser desorption and ionization-time-of-flight mass spectrometry (MALDI-TOF MS). Anaerobe 14:242-249. [DOI] [PubMed] [Google Scholar]
- 9.Hooton, T. M., and W. E. Stamm. 1997. Diagnosis and treatment of uncomplicated urinary tract infection. Infect. Dis. Clin. North Am. 11:551-581. [DOI] [PubMed] [Google Scholar]
- 10.Marklein, G., M. Josten, U. Klanke, E. Müller, R. Horré, T. Maier, T. Wenzel, M. Kostrzewa, G. Bierbaum, A. Hoerauf, and H. G. Sahl. 2009. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 47:2912-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellmann, A., J. Cloud, T. Maier, U. Keckevoet, I. Ramminger, P. Iwen, J. Dunn, G. Hall, D. Wilson, P. Lasala, M. Kostrzewa, and D. Harmsen. 2008. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J. Clin. Microbiol. 46:1946-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagy, E., T. Maier, E. Urban, G. Terhes, M. Kostrzewa, and the ESCMID Study Group on Antimicrobial Resistance in Anaerobic Bacteria. 2009. Species identification of clinical isolates of Bacteroides by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 15:796-802. [DOI] [PubMed] [Google Scholar]
- 13.Schwarz, G., T. Maier, M. Kostrzewa, C. Boogen, and U. Weller. 2008. Rapid identification of bacteria causing urinary tract infections by MALDI-TOF, abstr. D-4025. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother. (ICAAC)-Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet. American Society for Microbiology and Infectious Diseases Society of America, Washington, DC.
- 14.Seng, P., M. Drancourt, F. Gouriet, B. La Scola, P. E. Fournier, J. M. Rolain, and D. Raoult. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49:543-551. [DOI] [PubMed] [Google Scholar]