Abstract
Sapoviruses (SaVs) belong to the Caliciviridae family and can cause gastroenteritis in humans and swine. Despite extensive testing, human sapoviruses have been found only in sporadic cases and in one mixed outbreak in children between 1994 and 2007 in the Netherlands. Here we describe a change in sapovirus epidemiology in the Netherlands resulting in sapovirus outbreaks and infections in adults. From November 2007 to January 2009, 478 outbreaks of acute gastroenteritis were reported to the National Institute for Public Health and the Environment in the Netherlands as a part of ongoing surveillance. Sapoviruses were found to be the most likely cause of 19 outbreaks (4%). During the same 2-year period, sapovirus infections were reported in Sweden, Slovenia, and Hungary. In the Netherlands, further characterization of outbreak strains showed that 12 (63%) sapovirus outbreaks were caused by genotype I.2 viruses. Most patients were adults older than 60 years (range, 1 to 100 years). Phylogenetic analysis using all presently available SaV sequences showed high homology between genotype I.2 strains detected in different geographical regions (Sweden, Slovenia, Taiwan, Japan, and Russia) since 2007. These first reported outbreaks of sapovirus infections in adults in the Netherlands were remarkable. Detection of identical genotypes in many samples might suggest that these viruses have the same origin, and since the infection is spreading fast, the prevalence of sapovirus infection may be increasing. The incidence of sapovirus infections in these countries suggests that a substantial part of Europe is affected by this virus.
Sapoviruses (SaVs) and noroviruses (NoVs) are human caliciviruses that are recognized as a major cause of acute gastroenteritis (GE) worldwide (7, 19, 33). Human NoV and SaV strains cannot be reliably cultivated in vitro, and currently, reverse transcription-PCR (RT-PCR) is the most widely used method for their detection. NoVs and SaVs differ in their epidemiologies and host ranges. NoV infections are common in all age groups and are responsible for about 80% of all acute-GE outbreaks (29). On the other hand, SaV infections are less common and known to cause disease primarily in children, usually under the age of 5 years (10, 11, 13, 24). However, SaVs have recently been reported as an occasional cause of outbreaks in hospitals and other health care facilities. The age groups affected in these facilities have ranged from young adults to the elderly, suggesting less age restriction for illness than previously thought (8, 14, 15, 19, 22, 34). Although SaV-associated diarrhea is generally mild, severe cases can occur (25).
SaV has a single-stranded positive-sense RNA genome of approximately 7.3 to 7.5 kb, containing two to three open reading frames (ORF) that encode nonstructural proteins, a capsid protein (ORF 1), and a putative protein with an unknown function (ORF 2 and 3) (9). Based on complete capsid gene sequences, SaV strains are divided in five genogroups (GI to GV), of which genogroup III has been found to infect porcine species, whereas GI, GII, GIV, and GV are known to infect humans (4, 26).
In the Netherlands, a GE outbreak surveillance program has been implemented since 1994 to monitor outbreaks and their potential causative agents, including SaV (29).
In this study, we describe the emergence of SaV as a cause of outbreaks of acute GE in the Netherlands. In addition, we analyzed SaV-positive samples from GE outbreaks in Sweden, Slovenia, and Hungary (23). The emergence of sapoviruses may be explained by the genetic variability of SaV strains. The evolutionary dynamics of SaV strains were investigated using the BEAST program package (2) to assess whether the increased number of SaV infections is a result of an increased incidence or an artifact of new, more sensitive detection methods.
MATERIALS AND METHODS
Reporting of outbreaks of acute gastroenteritis in the Netherlands.
All the stool samples used for this study were obtained from outbreak cases reported to the National Institute for Public Health and the Environment (RIVM) through the ongoing surveillance of viral GE outbreaks. Outbreak surveillance has been operational in the Netherlands since 1994 and provides a systematic overview of all notified outbreaks (29). From 1 November 2007 to 1 January 2009, 478 GE outbreaks (3,233 samples) were reported to the surveillance program, and samples were tested for the presence of noroviruses, rotaviruses, enteric adenoviruses, astroviruses, and SaVs by a combination of multiplex RT-PCR assays (3, 28-30). Outbreaks were attributed to one of the pathogens mentioned above, depending on the diagnostic yield as a proportion of samples tested (3). To evaluate if changes observed in SaV incidence were not due to modifications in the detection protocols used, outbreak samples from 2006 and 2007 were retested.
Sequencing of the sapovirus-positive samples.
Detection of sapoviruses was preformed as described by Svraka et al. (30). A multiplex RT-PCR that uses random primers and is sapovirus specific was used; it proved to be more sensitive than previously used sapovirus PCR assays, as described by Svraka et al. (30). Sequencing of partial capsid genes of SaVs was performed using primers previously described by Yan et al. (35). The RT reaction step was slightly modified and performed using random primers and Superscript III (30). PCR amplification was performed as described by Svraka et al. (30) and by adding 0.1 μl forward and reverse primers (100 pmol/μl) to the PCR mixture (Table 1) (35).
TABLE 1.
Primers and probes, with corresponding concentrations, and label used for detection and typing of sapoviruses
| SaV analysis (reference) | Primer | Primer direction | Sequence (5′-3′) | Primer positionsa | Concn (pmol) used; probe label |
|---|---|---|---|---|---|
| Detection | SVLCF | Forward | GAYCWGGCYCTCGCCACCT | 5074-5092 | 10 |
| SVLCR | Reverse | GCCCTCCATYTCAAACACTA | 5159-5178 | 10 | |
| SVLCP | TGYACCACCTATRAACCAVG | 5101-5118 | 5; Texas Red-BHQ2 (610 nm)b | ||
| Capsid typing (35) | SLV5749 | Forward | CGGRCYTCAAAVSTACCBCCCCA | 5494-5516 | 10 |
| SLV5317 | Reverse | CTCGCCACCTACRAWGCBTGGTT | 5083-5105 | 10 |
Accession number of the reference strain used to indicate primer and probe positions, AY 237422.3 SaV Mc114.
Fluorescence measured at 610 nm. BHQ, black hole quencher.
Sequence analysis.
The BioNumerics software (Applied Maths BVBA, Sint-Martens-Latem, Belgium) was used for alignment of sequence data and clustering for genotype assignment. The different isolates used in this study were assigned to specific genogroups and subgenogroups according to nomenclature described in Fields Virology (6). Nine reference strains (GenBank accession numbers U65427, U73124, AF194182, AF435814, AY289803, AY289804, AF435812, U95645, AF182760) were included in the analysis to assign the genotypes of the clinical samples used in this study. Phylogenetic trees were generated using the neighbor-joining method in the TREECON software, version 1.3b (32). Statistical confidence for the trees was obtained by bootstrap analysis using 1,000 pseudoreplicates.
As this initial analysis suggested the emergence of SaVs in the Netherlands, laboratories in the Food-borne Viruses in Europe (FBVE) network were contacted to investigate GE outbreaks in their respective countries (n = 13). Sweden, Slovenia, and Hungary (23) reported SaV outbreaks and sequences from outbreak strains, and single or sporadic cases in those countries were included in the analyses, along with all available SaV sequences from GenBank (Table 2). For most outbreaks, two sequences were available. The representative strains from the different outbreaks, reference strains from GenBank, and strains from this study used in the phylogenetic analysis are presented in Table 2.
TABLE 2.
Summary of sequences from the Netherlands and Sweden used for comparison analysis of capsid sequences in Fig. 4
| Sequence source | Capsid sequences used for Fig. 4 |
|---|---|
| The Netherlands | Old strains: SaV-1999-0601, SaV-1999-1795, SaV-1999-1841, SaV-1999-2360, SaV-1999-2686, SaV-1999-2672, SaV-1999-2786, SaV-1999-3466, SaV-1999-3632, SaV-2005-045 (sequences 228-235), SaV-2005-045-227, SaV-2005-009-035, SaV-2006-087-591 |
| New strains from outbreaks: SaV-2008-259-2025, SaV-2008-142-1441, SaV-2007-401-2478, SaV-2008-303-2314, SaV-2008-20 (consensus of two sequences, 93 and 94), SaV-2008-72 (consensus of two sequences, 723 and 724), SaV-2008-117 (consensus of two sequences, 1222 and 1224), SaV-2008-126 (consensus of two sequences, 1318 and 1319), SaV-2008-152 (consensus of four sequences, 1503, 1504, 1505, and 1506), SaV-2008-157 (consensus of five sequences, 1524, 1525, 1541, 1549, and 1550), SaV-2008-163 (consensus of two sequences, 1583 and 1584), SaV-2008-185 (consensus of two sequences, 1678 and 1679), SaV-2008-186 (consensus of two sequences, 1672 and 1673), SaV-2008-191 (consensus of two sequences, 1706 and 1707), SaV-2007-373 (consensus of three sequences, 2238, 2343, and 2344), SaV-2007-387 (consensus of three sequences, 2350, 2351, and 2474) | |
| Other detected strains: SaV-2008-000-439, SaV-2008-000-1406, SaV-2008-000-1438, SaV-2008-000-2015, SaV-2008-57-0507, SaV-2008-146-1471, SaV-2008-148-1543, SaV-2008-168-1624, SaV-2008-278-2129, SaV-2008-323-2388, SaV-2009-39-0344, SaV-2009-39-0590, SaV-2009-045-0385, SaV-2009-GEOPS | |
| Sweden | Sweden1_08 (consensus of sequences 937_08, 939_08, 328_08, 329_08, 5896_08, and 9175_08 from Sweden), Sweden2_08 (consensus of sequences 11881_08, 11640_08, 12986_08, 1529_08, 12031_08, 1530_08, and 1529_08 from Sweden), Sweden3_08 (consensus of sequences 21414_08, 16216_08, 20468_08, 6499_08, and 6673_08 from Sweden), Sweden4_08 (consensus of sequences 8211_08 and 11535_08 from Sweden), Sweden5_08 (consensus of sequences 21413_08, 21717_08, 20656_08, 21091_08, and 21476_08 from Sweden), Sweden6_08 (consensus of sequences 7580_08 and 4878_08 from Sweden), Sweden19405_08, Sweden1645_08, Sweden12911_08, Sweden_1999-126, Sweden_1999-1422, Sweden_1999-1514 |
| GenBank (reference strain accession no.) | U65427, U73124, AF194182, AF435814, AY289803, AY289804, AF435812, U95645, AF182760 |
Analysis of evolutionary dynamics of new sapovirus strains.
The evolutionary dynamics of SaV strains were analyzed using capsid nucleotide sequences and the times of sampling of these strains in the BEAST program package (2). Evolutionary dynamics were estimated using a Bayesian Markov chain Monte Carlo (MCMC) approach implemented in the BEAST program (BEAST version 1.4.7). BEAST MCMC analysis estimates posterior distributions of the timed evolutionary history distribution, based on the incorporation of sampling times in a molecular-clock model, the substitution process, and demographic history. The general time-reversible (GTR) model of substitution, which allows different rates for all possible nucleotide changes and for different base frequencies, was used for the analysis. Additionally, we allowed for rate variation among all sites in the alignment and for invariant sites to exist in the alignment. The uncorrelated lognormal relaxed clock model was used, meaning that different branches can evolve at different rates. The resulting samples of trees were summarized using Tree-Annotator (distributed in the BEAST program package) in maximum-clade-credibility trees, which are scaled to a timescale shown on the x axis. The tips are aligned with the detection dates of the samples, whereas the node heights represent genetic distances.
RESULTS
Etiology of outbreaks from 1 November 2007 to 1 January 2009.
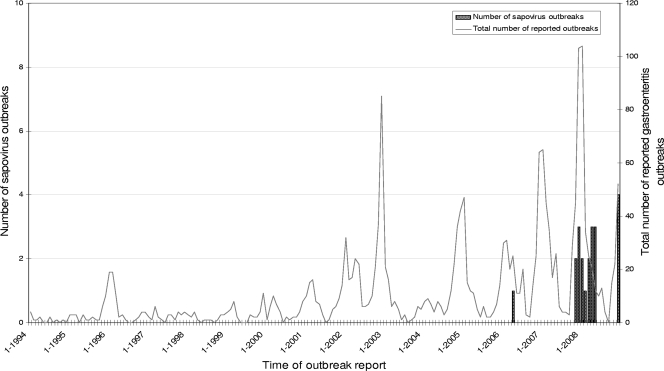
Of the 478 outbreaks sampled between November 2007 and January 2006, 74.6% were caused by noroviruses and 4.4% by rotaviruses, consistent with findings for previous seasons (Fig. 1) (29). No adenovirus or astrovirus outbreaks were identified in outbreaks during this period. Surprisingly, however, SaVs were detected as the cause of 19 outbreaks, which corresponds to 4.0% of all GE outbreaks reported in this study period (Fig. 2). Additionally, 14 SaV-positive samples were detected in individual patients from other outbreaks, including outbreaks attributed to other viruses. In total, 61 SaV-positive samples were obtained in the Netherlands (Fig. 2).
FIG. 1.
History of all reported gastroenteritis outbreaks and sapovirus outbreaks in the Netherlands.
FIG. 2.
Numbers of sapovirus cases and corresponding outbreaks (OB numbers). Sapovirus genotypes are indicated above the bars. The bars from OB2007323 to OB2008168 indicate single sapovirus infections in outbreaks, while bar OB2008000 indicates the sporadic cases. NT, not typed. In the key to bar patterns, “1” represents the number of positive samples and “0” represents the number of negative samples.
Epidemiology of outbreaks of sapoviruses in the Netherlands.
The most common setting of the 19 sapovirus-associated outbreaks was nursing homes (n = 12 [63%]), followed by hospitals (n = 5 [26%]) and child day care centers (n = 2 [11%]), which is not significantly different from the distribution of settings for all GE outbreaks of suspected viral etiology during this period.
The ages of patients infected with SaVs ranged from under 1 year of age up to 100 years old (Fig. 3). Most patients were older than 60 years (69%; 42 of 61 cases) but this probably reflects the bias in reporting and sampling, since 69% of all cases reported in this period were adults older than 60 years (Fig. 3). Furthermore, reporting of GE illness in nursing homes and health care settings is mandatory.
FIG. 3.
Age distribution (in years) of sapovirus patients involved in outbreaks and sporadic cases in the Netherlands, as detected through passive surveillance between 2007 and 2009. NK, not known.
Geographically, most outbreaks were reported from the middle and south of the Netherlands (n = 16 [84%]). Sapovirus-associated outbreaks occurred in 7 of 12 provinces (17 different cities), with 5 (26.3%) of 19 outbreaks occurring in the province Noord-Brabant, followed by Utrecht, with 4 outbreaks (21.1%). Sapovirus-associated outbreaks occurred in different cities (and provinces) around the same time.
SaV cases were identified between November 2007 and May 2008 (Fig. 1) and between November 2008 and April 2009.
Genetic characterization of sapovirus strains from outbreaks and sporadic cases.
All positive samples were characterized by sequence analysis. Genotype I.2 SaVs were detected in 11 (57.9%) of 19 outbreaks (one was in a hospital, two in child day care centers, and eight in nursing homes). Five outbreaks (26.3%) caused by SaV genotype IV.1 strains involved adults only, two outbreaks occurred in hospitals, and three outbreaks occurred in nursing homes. The strains from three outbreaks could not be typed. Sporadic SaV infections were caused by four different genotypes, I.1, I.2, II.1, and IV.1; of these, genotype I.2 was responsible for 36% of sporadic cases.
Genetic analysis.
Comparison of partial capsid sequences, performed for genotype I.2 and IV.1 sequences, showed that the genotype I.2 strains were highly similar (>99% nucleotide similarity over 312 nucleotides), whereas some diversity was observed for genotype IV.1 sequences (Fig. 4 and Table 2). A full comparison of capsid sequences was performed using all SaV sequences available from GenBank and sequences obtained from other countries that also noticed SaV outbreaks, including Sweden, Slovenia, and Hungary. This analysis showed high homology between genotype I.2 strains detected in different geographical regions (Sweden, Slovenia, Taiwan, Japan, and Russia) since 2007. Older strains detected in the Netherlands and Sweden were distinct, with the exception of one strain identified in 2004 in Thailand. For genotype IV.1, recent strain sequences from the Netherlands and Sweden, Japan, and Hong Kong clustered together, but more divergence was observed (97% nucleotide similarity over 312 nucleotides) (Fig. 4 and Table 2). Sequence analysis of polymerase gene segments of genotype I.2 strains from Hungary, Slovenia, China, and the Netherlands confirmed the tight clustering.
FIG. 4.
Comparison analysis of SaV partial capsid nucleotide sequences using neighbor-joining tree analysis, showing different genogroups and clusters. The numbers at the branches indicate bootstrap values for the genotypes and isolates. Names of new sapovirus strains isolated in the Netherlands are red, sapovirus outbreaks from the Netherlands are marked with an asterisk, new strains from Slovenia are in blue, and new strains from Sweden are in green. GenBank accession numbers (in black) for the reference strains are as follows: U65427 (Hu/SaV/GI.1/Sapporo/1982/JP), U73124 (Hu/SaV/GI.2/Parkville/1994/US), AF194182 (Hu/SaV/GI.3/Stockholm318/1997/SE), AF435814 (Hu/SaV/GIV.1/Hou7-1181/1990/US), AY289803 (Hu/SaV/GV.1/Argentina39/AR), AY289804 (Hu/SaV/GII.3/cruise ship/2000/US), AF435812 (Hu/SaV/GII.2/Mex340/1990/MX), U95645 (Hu/SaV/GII.1/London/1992/UK), AF182760 (Sw/SaV/GIII.1/PEC-Cowden/1980/US), where JP stands for Japan, Hu for human, US for United States, SE for Sweden, AR for Argentina, MX for Mexico, UK for United Kingdom, Sw for swine, and PEC for porcine enteric calcivirus.
Analysis of evolutionary dynamics of new sapovirus strains.
The evolutionary dynamics of partial capsid sequences of these two SaV genotypes were estimated and showed distinct phylogenies. The genotype IV.1 viruses diverged from a common node dating back to 1998 (Fig. 5, bottom), whereas for genotype I.2, the pattern suggested evolution of the dominant strain from an ancestor identified in 2002 (Fig. 5, top).
FIG. 5.
Evolutionary dynamics of sapovirus, based on partial capsid nucleotide sequences of GI.2 (top) and GIV.1 (bottom). The time scale is shown on the x axis, the tips are aligned with the dates of detection of samples, and node heights represent distances between the isolates.
DISCUSSION
Since the start of acute-viral-GE surveillance in the Netherlands in 1994 (29) through 2006, no GE outbreaks have been assigned to SaVs. However, SaV was detected in outbreaks after November 2007. As this observed increase in SaV detection coincided with the introduction of a revised molecular detection method, we initially suspected that it reflected the increased sensitivity of the PCR compared to those of previously used protocols. Therefore, samples from unexplained outbreaks from 2006 through 2007 were retested using the revised new method. The retrospective testing confirmed the absence of SaV in outbreaks that happened prior to November 2007, and thus, the emergence of SaV infections in the Netherlands is not an artifact of changes in the protocols used. A possible alternative explanation has to do with changes in the referral bias for outbreak samples. Specifically, as norovirus testing is becoming more and more routine in the Netherlands, the selection of patient samples that are received and used for surveillance has been changing. Changes in patient demography would affect our conclusions if the increased incidence of SaV outbreaks were observed in children. However, the data indicated a higher incidence of SaV outbreaks in adults and nursing homes, which have always been included in our surveillance (1, 10, 16). SaVs have been reported as causes of occasional outbreaks in adults, but their prevalence seems to be increasing. Similar findings were reported recently from Sweden (15), Canada (22), Japan (8, 10-13, 24), and Hungary (23). In addition, food-borne outbreaks and several novel recombinant SaV strains have been identified (8, 12, 17, 21, 31).
The increased reporting of SaV outbreaks was concomitant with the increased prevalence of SaV genotype I.2 (58% of SaV-associated outbreaks in the Netherlands, 27% in Sweden, 40% in Slovenia, and 100% in Hungary). SaV strains belonging to genotype I.2 have been causing infections worldwide recently (5, 11, 18, 34). Again, this could reflect changes in the sensitivities of the methods used; nevertheless, previously used PCR methods are based on relatively conserved targets within the polymerase region. Therefore, the observed increase of genotype I.2 SaV infections is most likely not a consequence of new detection assays. Furthermore, recent genotype I.2 sequences from all over the world show high similarity, suggesting that there is a common source for the virus, such as contaminated food, as it has been found in other SaV outbreaks (21, 31). The near-clonal nature of the outbreak strains belonging to genotype I.2 suggests that the specific lineage of this genotype has expanded more recently from a common ancestor. This was confirmed using a phylodynamic analysis, suggesting that recent strains emerged as drift variants, similar to what has been observed for norovirus genotype II.4 viruses, and expansion of a successful variant has been observed for some norovirus genotypes (27). In addition, calicivirus infections are increasing globally (27), and sapovirus genotype I.2 may have the same mechanisms of persistence in humans as norovirus genotype II.4, such as low infectious dose, high infectivity, high level of shedding, high environmental persistence, multiple transmission routes, and prolonged shedding after clinical recovery (20). Combined with data from Sweden, Hungary, and Slovenia, we conclude that SaV infections are increasing in Europe and, based on recent reports, are also emerging worldwide (5, 10, 11, 15, 18, 22, 34). Therefore, we recommend the inclusion of a sapovirus diagnostic assay in the analysis of outbreaks of suspected viral GE in other laboratories.
Acknowledgments
This work was supported financially by the European Commission, DG Research Quality of Life Program, 6th Framework (EVENT, grant SP22-CT-2004-502571).
We declare that we have no conflicting interests with respect to this work.
We thank Gábor Reuter (Hungary), Mateja Poljsak-Prijatelj (Slovenia), and Janet Zimsek Mijovski (Slovenia) for providing sapovirus sequences found in their countries. We also thank Karyna Rosario for critical reading of the manuscript.
Footnotes
Published ahead of print on 14 April 2010.
REFERENCES
- 1.Chiba, S., Y. Sakuma, R. Kogasaka, M. Akihara, K. Horino, T. Nakao, and S. Fukui. 1979. An outbreak of gastroenteritis associated with calicivirus in an infant home. J. Med. Virol. 4:249-254. [DOI] [PubMed] [Google Scholar]
- 2.Drummond, A. J., and A. Rambaut. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duizer, E., A. Pielaat, H. Vennema, A. Kroneman, and M. Koopmans. 2007. Probabilities in norovirus outbreak diagnosis. J. Clin. Virol. 40:38-42. [DOI] [PubMed] [Google Scholar]
- 4.Farkas, T., W. M. Zhong, Y. Jing, P. W. Huang, S. M. Espinosa, N. Martinez, A. L. Morrow, G. M. Ruiz-Palacios, L. K. Pickering, and X. Jiang. 2004. Genetic diversity among sapoviruses. Arch. Virol. 149:1309-1323. [DOI] [PubMed] [Google Scholar]
- 5.Gallimore, C. I., M. Iturriza-Gomara, D. Lewis, D. Cubitt, H. Cotterill, and J. J. Gray. 2006. Characterization of sapoviruses collected in the United Kingdom from 1989 to 2004. J. Med. Virol. 78:673-682. [DOI] [PubMed] [Google Scholar]
- 6.Green, K. Y. 2007. Caliciviridae: the Noroviruses, p. 949-980. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 7.Green, K. Y., T. Ando, M. S. Balayan, T. Berke, I. N. Clarke, M. K. Estes, D. O. Matson, S. Nakata, J. D. Neill, M. J. Studdert, and H. J. Thiel. 2000. Taxonomy of the caliciviruses. J. Infect. Dis. 181(Suppl. 2):S322-S330. [DOI] [PubMed] [Google Scholar]
- 8.Hansman, G. S., S. Ishida, S. Yoshizumi, M. Miyoshi, T. Ikeda, T. Oka, and N. Takeda. 2007. Recombinant sapovirus gastroenteritis, Japan. Emerg. Infect. Dis. 13:786-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansman, G. S., T. Oka, K. Katayama, and N. Takeda. 2007. Human sapoviruses: genetic diversity, recombination, and classification. Rev. Med. Virol. 17:133-141. [DOI] [PubMed] [Google Scholar]
- 10.Hansman, G. S., H. Saito, C. Shibata, S. Ishizuka, M. Oseto, T. Oka, and N. Takeda. 2007. Outbreak of gastroenteritis due to sapovirus. J. Clin. Microbiol. 45:1347-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansman, G. S., N. Takeda, K. Katayama, E. T. Tu, C. J. McIver, W. D. Rawlinson, and P. A. White. 2006. Genetic diversity of sapovirus in children, Australia. Emerg. Infect. Dis. 12:141-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansman, G. S., N. Takeda, T. Oka, M. Oseto, K. O. Hedlund, and K. Katayama. 2005. Intergenogroup recombination in sapoviruses. Emerg. Infect. Dis. 11:1916-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ike, A. C., K. Hartelt, R. M. Oehme, and S. O. Brockmann. 2008. Detection and characterization of sapoviruses in outbreaks of gastroenteritis in southwest Germany. J. Clin. Virol. 43:37-41. [DOI] [PubMed] [Google Scholar]
- 14.Ishida, S., S. Yoshizumi, M. Miyoshi, T. Ikeda, T. Okui, K. Katayama, N. Takeda, and T. Oka. 2008. Characterization of sapoviruses detected in Hokkaido, Japan. Jpn. J. Infect. Dis. 61:504-506. [PubMed] [Google Scholar]
- 15.Johansson, P. J., K. Bergentoft, P. A. Larsson, G. Magnusson, A. Widell, M. Thorhagen, and K. O. Hedlund. 2005. A nosocomial sapovirus-associated outbreak of gastroenteritis in adults. Scand. J. Infect. Dis. 37:200-204. [DOI] [PubMed] [Google Scholar]
- 16.Johnsen, C. K., S. Midgley, and B. Bottiger. 2009. Genetic diversity of sapovirus infections in Danish children 2005-2007. J. Clin. Virol. 46:265-269. [DOI] [PubMed] [Google Scholar]
- 17.Katayama, K., T. Miyoshi, K. Uchino, T. Oka, T. Tanaka, N. Takeda, and G. S. Hansman. 2004. Novel recombinant sapovirus. Emerg. Infect. Dis. 10:1874-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khamrin, P., N. Maneekarn, S. Peerakome, S. Tonusin, R. Malasao, M. Mizuguchi, S. Okitsu, and H. Ushijima. 2007. Genetic diversity of noroviruses and sapoviruses in children hospitalized with acute gastroenteritis in Chiang Mai, Thailand. J. Med. Virol. 79:1921-1926. [DOI] [PubMed] [Google Scholar]
- 19.Koopmans, M., J. Vinje, E. Duizer, M. de Wit, and Y. van Duijnhoven. 2001. Molecular epidemiology of human enteric caliciviruses in the Netherlands. Novartis Found. Symp. 238:197-218. [DOI] [PubMed] [Google Scholar]
- 20.Lindesmith, L. C., E. F. Donaldson, A. D. Lobue, J. L. Cannon, D. P. Zheng, J. Vinje, and R. S. Baric. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakagawa-Okamoto, R., T. Arita-Nishida, S. Toda, H. Kato, H. Iwata, M. Akiyama, O. Nishio, H. Kimura, M. Noda, N. Takeda, and T. Oka. 2009. Detection of multiple sapovirus genotypes and genogroups in oyster-associated outbreaks. Jpn. J. Infect. Dis. 62:63-66. [PubMed] [Google Scholar]
- 22.Pang, X. L., B. E. Lee, G. J. Tyrrell, and J. K. Preiksaitis. 2008. Epidemiology and genotype analysis of sapovirus associated with gastroenteritis outbreaks in Alberta, Canada: 2004-2007. J. Infect. Dis. 199:547-551. [DOI] [PubMed] [Google Scholar]
- 23.Pankovics, P., Z. Kugler, A. Katai, and G. Reuter. 2009. First gastroenteritis outbreak caused by sapovirus (GI2) in Hungary—part of an international epidemic? Orv. Hetil. 150:1223-1229. (In Hungarian.) [DOI] [PubMed] [Google Scholar]
- 24.Phan, T. G., Q. D. Trinh, F. Yagyu, K. Sugita, S. Okitsu, W. E. Muller, and H. Ushijima. 2006. Outbreak of sapovirus infection among infants and children with acute gastroenteritis in Osaka City, Japan during 2004-2005. J. Med. Virol. 78:839-846. [DOI] [PubMed] [Google Scholar]
- 25.Robinson, S., I. N. Clarke, I. B. Vipond, E. O. Caul, and P. R. Lambden. 2002. Epidemiology of human Sapporo-like caliciviruses in the South West of England: molecular characterisation of a genetically distinct isolate. J. Med. Virol. 67:282-288. [DOI] [PubMed] [Google Scholar]
- 26.Schuffenecker, I., T. Ando, D. Thouvenot, B. Lina, and M. Aymard. 2001. Genetic classification of “Sapporo-like viruses.” Arch. Virol. 146:2115-2132. [DOI] [PubMed] [Google Scholar]
- 27.Siebenga, J. J., H. Vennema, D. P. Zheng, J. Vinje, B. E. Lee, X. L. Pang, E. C. Ho, W. Lim, A. Choudekar, S. Broor, T. Halperin, N. B. Rasool, J. Hewitt, G. E. Greening, M. Jin, Z. J. Duan, Y. Lucero, M. O'Ryan, M. Hoehne, E. Schreier, R. M. Ratcliff, P. A. White, N. Iritani, G. Reuter, and M. Koopmans. 2009. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001-2007. J. Infect. Dis. 200:802-812. [DOI] [PubMed] [Google Scholar]
- 28.Svraka, S., E. Duizer, H. Egberink, J. Dekkers, H. Vennema, and M. Koopmans. 2009. A new generic real-time reverse transcription polymerase chain reaction assay for vesiviruses; vesiviruses were not detected in human samples. J. Virol. Methods 157:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svraka, S., E. Duizer, H. Vennema, E. de Bruin, B. van der Veer, B. Dorresteijn, and M. Koopmans. 2007. Etiological role of viruses in outbreaks of acute gastroenteritis in The Netherlands from 1994 through 2005. J. Clin. Microbiol. 45:1389-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svraka, S., B. van der Veer, E. Duizer, J. Dekkers, M. Koopmans, and H. Vennema. 2009. Novel approach for detection of enteric viruses to enable syndrome surveillance of acute viral gastroenteritis. J. Clin. Microbiol. 47:1674-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Usuku, S., M. Kumazaki, K. Kitamura, O. Tochikubo, and Y. Noguchi. 2008. An outbreak of food-borne gastroenteritis due to sapovirus among junior high school students. Jpn. J. Infect. Dis. 61:438-441. [PubMed] [Google Scholar]
- 32.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 33.Vinje, J., H. Deijl, R. van der Heide, D. Lewis, K. O. Hedlund, L. Svensson, and M. P. Koopmans. 2000. Molecular detection and epidemiology of Sapporo-like viruses. J. Clin. Microbiol. 38:530-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, F. T., T. Oka, N. Takeda, K. Katayama, G. S. Hansman, C. H. Muo, S. Y. Liang, C. H. Hung, D. Dah-Shyong Jiang, J. Hsin Chang, J. Y. Yang, H. S. Wu, and C. F. Yang. 2008. Acute gastroenteritis caused by GI/2 sapovirus, Taiwan, 2007. Emerg. Infect. Dis. 14:1169-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan, H., F. Yagyu, S. Okitsu, O. Nishio, and H. Ushijima. 2003. Detection of norovirus (GI, GII), sapovirus and astrovirus in fecal samples using reverse transcription single-round multiplex PCR. J. Virol. Methods 114:37-44. [DOI] [PubMed] [Google Scholar]