Abstract
Elevated HIV-1 viral load (VL) observed in specimens frozen in situ in plasma preparation tubes (PPTs) compared to EDTA plasma specimens may affect therapeutic monitoring of HIV-infected patients. The increase in viral load is cell associated and minimized when plasma from the PPT is aspirated or recentrifuged prior to freezing. This study investigates the contribution of integrated HIV-1 proviral DNA to elevated VL in the quantification of HIV-1 RNA in plasma. Fifty paired specimens collected in EDTA tubes and PPTs frozen in situ were used for analysis. HIV-1 VL was measured using the COBAS Amplicor Monitor ultrasensitive test version 1.5. Contaminating proviral DNA was detected using a nested PCR targeting the Alu repeat in human genomic DNA and HIV pol gene simultaneously. Treatment of the specimen with DNase resulted in significantly lower quantifiable HIV-1 RNA in specimens from PPTs compared to the corresponding EDTA tubes (P = 0.004). After the RNA was destroyed by heat treatment, the mean HIV-1 RNA VL decreased by 79% in the EDTA tube compared to 65% in the PPT. The nested PCR amplified integrated proviral DNA in nucleic acid extracted from plasma in PPT and EDTA specimens with high viral load values. Likewise, a semiquantitative densitometric analysis revealed that the total amount of genomic DNA in the PPT was higher than that in the EDTA tube. Our investigation clearly shows that both proviral DNA and intracellular RNA are amplified simultaneously in the COBAS Amplicor HIV-1 Monitor assay and that proviral DNA contributes to the elevated VL in plasma frozen in PPTs.
Discrepancies in viral load (VL) measurements obtained in different plasma collection tubes have underscored the importance of specimen collection and handling in the determination of accurate results in HIV viral load assays. Plasma preparation tubes (PPTs) are Vacutainer tubes that contain dehydrated K2EDTA and a gel separator which, upon centrifugation, forms a barrier that separates blood cells from plasma, allowing for collection, storage, freezing, and shipment of the plasma within the same tube. Use of the PPT also reduces the time required to process specimens, eliminates potential error in specimen labeling, and reduces the risk of HIV exposure associated with the transfer of plasma to secondary tubes for shipping.
Elevated HIV-1 viral loads in plasma specimens collected and frozen in PPTs and quantified in the standard and ultrasensitive Roche COBAS Amplicor HIV-1 Monitor assays (2, 4, 13) have led investigators to believe that it may have an impact on therapeutic management of HIV-infected patients (1, 8). Some reports attribute the observed VL blips to random biological and statistical variations, with limited clinical significance (6), while others have implied that the elevated HIV-1 VL may interfere in the assessment of virological suppression. Subsequent studies show that aspiration and transfer of plasma from a PPT to a storage tube after centrifugation or recentrifugation of the PPT after transportation reduces or eliminates the artificial elevation of VL observed in the PPT (9, 10, 11). Salimnia et al. (11) imply that the elevation may be due to nonparticulate genetic material released from cells associated with the gel in the PPT after the plasma was thawed. Additional investigations report that samples collected in PPTs can be centrifuged up to 6 h after phlebotomy and stored in situ at 4°C for up to 72 h without any effect on the measured HIV-1 VL (9).
In a recent prospective study, Kran et al. (5) observed a direct correlation between the number of cells present in the plasma, measured in the Advia 60 cell counter, and the amount of HIV-1 that was quantified using the Roche COBAS AmpliPrep-COBAS TaqMan (CAP/TAQ) HIV-1 test. Using paired specimens that were collected and centrifuged in PPTs, they found lower cell counts and reduced HIV-1 VL in the PPTs that were recentrifuged prior to HIV-1 quantification versus the PPTs that were centrifuged once, after phlebotomy. Using the Roche proviral DNA assay, the authors were able to detect HIV-1 proviral DNA in the specimens with higher cell counts and corresponding elevated VLs.
The aim of our investigation is to further examine the source of nucleic acid responsible for the elevated VL in the PPT. Our protocol specifically explores integrated HIV-1 proviral DNA as the major contributor to the elevation in viral load observed in PPT. In addition, we show that the Roche Amplicor HIV-1 RNA assay coamplifies both RNA and proviral DNA simultaneously in plasma that is frozen in PPT.
MATERIALS AND METHODS
Specimen processing.
A total of 50 consenting HIV-1-positive patients attending the adult infectious diseases clinic at the University of Medicine and Dentistry of New Jersey (UMDNJ) were used in the study. Fourteen of the 50 patients had <50 copies/ml of HIV-1 RNA in the EDTA plasma specimens. Twenty-eight patients had quantifiable VL values between 50 and 1,000 copies/ml, and five patients had VL values between 1,000 and 2,500 copies/ml quantified by the Roche COBAS Amplicor HIV-1 Monitor version 1.5 ultrasensitive test. Three subjects with HIV-1 VLs between 3,000 and 5,000 copies/ml were selected for analysis of HIV-1 proviral DNA. The study was approved by the UMDNJ Institutional Review Board (IRB 0120040082). Blood samples from each subject were collected in both EDTA tubes and plasma preparation tubes (PPTs) by standard venipuncture and maintained at room temperature until centrifugation within 2 h of collection. The EDTA tubes were centrifuged at 1,600 × g for 20 min, and the plasma samples were aspirated, transferred to cryogenic vials, and stored at −70°C until the day of testing. The PPTs were centrifuged at the suggested 1,100 × g for 10 min, and plasma samples were frozen at −70°C in situ until the day of testing.
HIV-1 RNA quantification.
All HIV-1 RNA quantification assays were performed using the Roche COBAS Amplicor HIV-1 Monitor version 1.5 ultrasensitive test (Roche Diagnostics, Branchburg, NJ) per the manufacturer's recommendations. Specimens from the same subject were run in triplicate, and the VL result was calculated from an average of the three values obtained. The nucleic acid extraction procedure was modified for quantification experiments, when the RNA or DNA was subject to variable experimental conditions as described below.
Amplification of HIV-1 proviral DNA.
Nested PCR was used for amplification and identification of integrated proviral HIV-1 DNA sequences in the specimens that were extracted for VL quantification (12). For the first round of amplification, a forward primer was selected from the conserved Alu repetitive element of the human genome (Alu primer, 5′-CTCACGCCTGTAATCCCAGCA), and the reverse primer (LA5 primer) represented the conserved gag-pol region of the HIV genome (LA5 primer, 5′-1907TGTATCATCTGCTCCTGTATC). Selection of the primer in the Alu element was based on the premise that HIV generally integrates randomly into the cellular DNA, and the probability of having Alu elements close enough to the integrated HIV genome was reasonably high. The first amplification reaction was carried out using AmpliTaq Gold polymerase (Applied Biosystems) in a GeneAmp PCR system 9700 (Applied Biosystems) in a total volume of 25 μl. After 35 cycles of enrichment, 5 μl of the product of the first round of PCR was used for amplification of HIV using a new forward primer, LA2, from the gag region of the HIV genome (LA2 primer, 5′-CCCTCTCAGAAGCAGGAGCCGA) and the same reverse primer (LA5) to yield a 143-bp fragment. This inner amplification of the nested PCR that targeted the HIV genome was carried out in the LightCycler real-time instrument (Roche Diagnostics) with Sybr green as the fluorescent dye. Amplicons were subsequently loaded on a 2% agarose gel for confirmation of the product based on the size of the amplicon and subsequently confirmed by sequencing. The β-globin primer and probe from the LightCycler FastStart DNA master hybridization probes control kit DNA (Roche Applied Science) was used as the internal control for amplification of genomic DNA.
HIV RNA quantification in modified specimens.
Specimens with HIV-1 VLs of >750 copies/ml were selected for this part of the analysis. HIV-1 quantification was performed using the Roche COBAS Amplicor HIV-1 Monitor version 1.5 ultrasensitive test (Monitor assay) using the modifications described below.
Extraction of RNA.
The data presented in Fig. 2 and Table 1 were obtained from EDTA and PPT in situ plasma samples. RNA was extracted using the QIAamp viral RNA kit (Qiagen, Inc., CA) as follows. The plasma sample (0.5 ml) was mixed with 4 volumes (2.0 ml) of QIAamp lysis buffer (AVL) supplemented with carrier RNA. After incubation and processing on QIAamp minicolumns per the manufacturer's recommendation, the specimen was eluted with 25 μl of preheated solution of 0.01 M Tris-HCl. Subsequently, 25 μl of diluent from the Roche COBAS Amplicor HIV-1 Monitor kit, reconstituted with twice the amount of the suggested quantitation standard (QS) from the HIV-1 Monitor assay kit, was added for a final volume of 50 μl. This was done to retain the suggested input QS copy number suggested by the manufacturer for HIV-1 quantification in a total volume of 50 μl. Extracted nucleic acid served as a control and was stored at −70°C until tested.
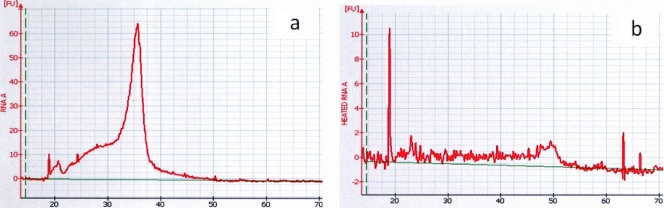
FIG. 2.
(a) Electropherogram of a high-quality eukaryotic total RNA sample showing a spike. The x axis represents the time (in seconds) at which the RNA is detected, and the y axis represents the fluorescence which reflects the quantity of RNA. (b) Electropherograms of the same RNA sample after heat inactivation showing total destruction of RNA. FU, fluorescence units.
TABLE 1.
Comparison of HIV-1 viral load in EDTA and PPT plasma specimens before and after removal of RNA by heat inactivation
| Tube and treatment | HIV-1 viral load (mean ± SD) (no. of copies/ml) | % RNA destroyeda | P valueb |
|---|---|---|---|
| EDTA tubes (n = 8) | |||
| Untreated | 1,431 ± 773 | 79.31 | 0.002 |
| Heat inactivated | 296 ± 193 | ||
| PPTs (n = 8) | |||
| Untreated | 1,792 ± 985 | 64.56 | 0.003 |
| Heat inactivated | 635 ± 422 |
Percentage of RNA that was destroyed after heat inactivation.
The P value shows the statistical significance of the different quantifiable VL values in untreated and heat-inactivated specimens for a given type of tube.
Heat inactivation of HIV-1 RNA.
For quantification experiments where the RNA was destroyed by heat treatment, specimens were extracted and eluted in 0.01 M Tris-HCl. Following extraction, the 25-μl aliquot was heated in boiling water for 30 min to destroy the RNA. Twenty-five microliters of diluent from the Roche COBAS Amplicor HIV-1 Monitor version 1.5 ultrasensitive test, reconstituted with twice the amount of the suggested QS (see above), was added to 25 μl of the heated aliquot of RNA to make a final volume of 50 μl.
DNase I treatment for removal of HIV-1 proviral DNA.
Extracted RNA, as specified above, was treated with DNase I (Invitrogen, Life Technologies, CA) for removal of HIV-1 proviral DNA. The RNA sample (20 μl) was added to 1.6 μl DNase I in 2.4 μl of 10× buffer according to the manufacturer's instructions. The mixture was incubated at room temperature for 15 min. One microliter of a 25 mM EDTA solution was then added to the mixture and incubated at 65°C for 10 min to stop the reaction. The 25-μl mixture was added to 25 μl of the diluent reconstituted with the appropriate QS (see above) to make a final volume of 50 μl.
For a control, 20 μl of untreated RNA was diluted with 30 μl of the QS-reconstituted diluent to make a final volume of 50 μl. The HIV-1 viral loads in the inactivated (heat-treated) RNA and DNase-treated specimens were compared to the respective control after quantification in the Roche COBAS Amplicor HIV-1 Monitor assay.
Agilent bioanalyzer.
The quality of RNA and DNA in treated and untreated specimens was evaluated using 1 μl of the respective specimen in the Agilent 2100 bioanalyzer (Agilent Technologies) with the RNA 6000 pico chip according to the manufacturer's instruction.
RESULTS
Comparison of HIV-1 VL in plasma samples collected in the EDTA tube versus PPT.
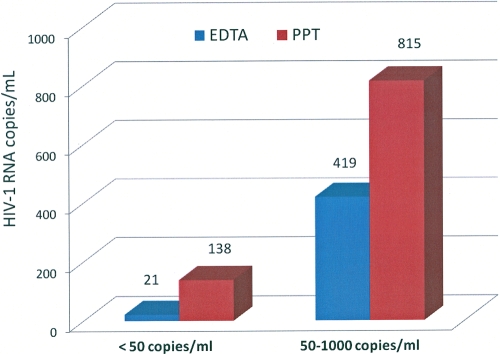
Fourteen of the 50 specimens had detectable VLs that were below the limit of quantification in the EDTA tube (<50 copies/ml). All 14 specimens collected in the PPTs had quantifiable HIV-1 RNA (mean, 138 copies/ml) as measured by the Roche COBAS Amplicor HIV-1 Monitor version 1.5 ultrasensitive assay. The mean VLs of the 28 specimens with VLs between 50 and 1,000 copies/ml were 419 copies/ml for the EDTA plasma specimens and 815 copies/ml in the corresponding PPT plasma specimens (Fig. 1). All specimens were run in triplicate at the time of testing. The difference in HIV-1 VLs of the two tubes was statistically significant (P = 0.002). Increases in VL, ranging from 5% and 439% were observed in 24 of the 28 specimens, and marginal decreases in VLs were apparent in the four remaining specimens.
FIG. 1.
HIV-1 viral load measured in EDTA tubes and the corresponding plasma preparation tubes (PPTs) in two different VL categories. The PPTs (n = 14) had an average viral load (VL) of 138 copies/ml compared to the EDTA tubes (n = 14) with VL values of <50 copies/ml. In specimens (n = 28) with higher VLs (50 to 1,000 copies/ml), the EDTA tubes and corresponding PPT tubes showed averages of 419 and 815 HIV-1 copies/ml, respectively.
HIV-1 VL in plasma from PPT in situ after removal of RNA.
To further investigate the reason for the increased HIV-1 VLs in PPTs, we obtained eight paired plasma specimens with VLs ranging from 750 and 3,200 copies/ml in PPTs and EDTA tubes and subjected the extracted RNA to heat inactivation as described in Materials and Methods. The effect of heat on the destruction of RNA is seen in Fig. 2, where the RNA was analyzed and its presence was visualized on the Agilent bioanalyzer. The sharp peak present in intact extracted HIV-1 RNA seen in Fig. 2a is absent after heat treatment as seen in Fig. 2b. Both untreated and heat-treated RNA specimens were quantified in triplicate for HIV VL, using the Roche COBAS Amplicor HIV-1 Monitor version 1.5 ultrasensitive assay. The results seen in Table 1 show that despite the apparent absence of RNA, a quantifiable VL was present in both EDTA and PPT plasma specimens. The average amount of detectable HIV RNA, as indicated by the amount of RNA destroyed, was 79.31% in EDTA plasma specimens and 64.56% in plasma specimens collected in PPTs. This difference was not statistically significant. The effect of heat treatment of the extracted RNA on the viral load within each tube type is also seen in Table 1. There is a statistical difference between the HIV-1 VLs obtained before and after the destruction of RNA within the EDTA (P = 0.002) and PPT (P = 0.003) plasma specimens, respectively. The amount of quantifiable HIV-1 after apparent destruction of RNA was higher in the extracted specimen from the PPT compared to the specimen from the EDTA tube.
Estimation of HIV viral load after treatment with DNase I.
Treatment of the extracted nucleic acid with DNase I significantly reduced the HIV-1 VL measured in plasma from the PPT compared to the EDTA tube (Table 2). Each viral load value in Table 2 represents an average of three specimens collected in EDTA tubes and PPTs, assayed in triplicate. Specimens with higher HIV-1 VLs that showed discrepant values in EDTA tubes and PPTs were selected for analysis. In addition to treatment with DNase I, a duplicate of the extracted specimen was subjected to heat inactivation to destroy the RNA. Removal of DNA using DNase I resulted in a 6.3% loss of quantified HIV-1 RNA in the EDTA plasma specimen and a corresponding 40.1% loss in HIV-1 VL in the PPT plasma specimen. Similarly, removal of RNA by heat treatment resulted in a significant loss of quantifiable HIV-1, 77% and 58% in EDTA and PPT tubes, respectively.
TABLE 2.
Effect of DNase I treatment on HIV-1 viral load in specimens collected in EDTA tubes and PPTs
| Tube and treatment | Viral load (copies/ml)a | % loss of nucleic acid |
|---|---|---|
| EDTA tubes | ||
| None (control) | 3,190 | 0.0 |
| DNase I | 2,990 | 6.3b |
| Heat treatment | 740 | 76.8 |
| PPTs | ||
| None (control) | 3,960 | 0.0 |
| DNase I | 2,370 | 40.1b |
| Heat treatment | 1,680 | 57.5 |
Average values of three specimens in three replicate experiments.
The values for DNase-treated EDTA tubes and PPTs were statistically significantly different (P = 0.004).
Detection of genomic DNA in plasma specimens.
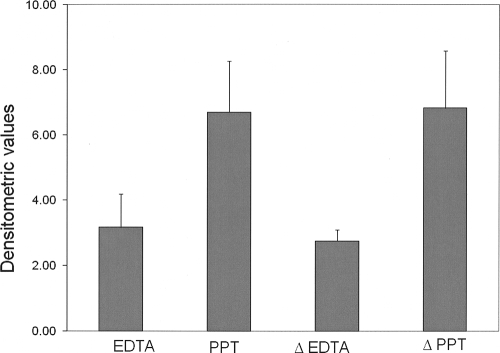
The presence of genomic DNA from both EDTA and PPT plasma specimens as seen in Fig. 3 confirms the presence of cell-associated nucleic acid in the extracted RNA specimens. The values represent an average of three experiments where β-globin, a eukaryotic housekeeping gene, was amplified from extracted RNA obtained from EDTA and PPT plasma specimens. The relative amount of β-globin present before and after heat treatment in both EDTA and PPT plasma specimens was estimated by densitometric analysis of the amplicons that were run on a 2% agarose gel. The values on the x axis represent arbitrary densitometric values obtained on analysis. The data clearly show that a significantly greater amount of cell-associated DNA as represented by β-globin is present in PPT plasma specimens compared to specimens extracted from EDTA plasma specimens. Furthermore, heat treatment of the extracted specimens for removal of RNA did not significantly alter the amount of cell-associated DNA present in both plasma specimens.
FIG. 3.
Densitometric analysis of amplified β-globin DNA in nucleic acid extracted from EDTA and PPT plasma specimens. ΔEDTA and ΔPPT represent extracted specimens that were heat inactivated for removal of RNA.
Amplification of HIV-1 proviral DNA in cell-associated nucleic acid from plasma specimens.
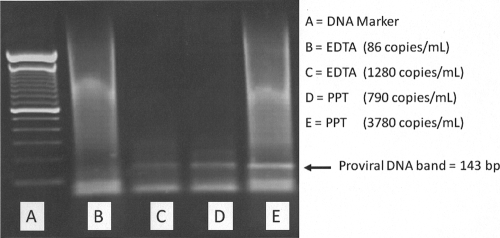
Proviral DNA was amplified in specimens extracted from EDTA and PPT tubes using the nested PCR described above. Figure 4 is a representative gel of two EDTA and two PPT specimens with different VL values. Proviral DNA bands (143-bp amplicons) were detected in both PPT specimens with VLs of 790 and 3,680 copies/ml. In contrast, no amplifiable HIV-1 proviral DNA was detected in the EDTA specimen with a VL of 86 copies/ml. The EDTA specimen with a higher VL of 1,280 copies/ml, however, did show the presence of proviral DNA.
FIG. 4.
Agarose gel electrophoresis of HIV-1 proviral DNA amplified from specimens in EDTA tubes and PPTs. Lane B does not show the specific band. Lanes C, D, and E had the 143-bp amplicon representing specific amplification of HIV-1 proviral DNA.
DISCUSSION
The accurate quantification of HIV-1 RNA particularly at the lower limit of quantification (LOQ) of the VL assay is of paramount significance in the assessment of virological clearance in response to highly active antiretroviral therapy (HAART). Artificially elevated HIV-1 RNA levels in PPTs are suggestive of HIV-1 viremia and consequently associated with clinical failure to HAART (1, 3, 4, 13). Recent reports have determined that elevated VLs in PPTs, linked to transient viremia or blips (14), are due to technical aberrations caused by cell-associated HIV nucleic acids (5, 6, 8). Our data clearly suggest that this artificial elevation in VL is due to the presence of HIV-1 proviral DNA from residual cells present in the plasma specimen.
Studies from our laboratory, confirmed by others, have reported that freezing plasma in PPTs in situ results in elevated VLs and this phenomenon is more noticeable in specimens with lower VL values (4, 9, 11). Transfer of the plasma soon after centrifugation (10, 12) or recentrifugation of unfrozen specimens after transportation (6, 11) was able to eliminate the artificial increase in VL, suggesting that cellular material is the possible reason for the elevation in HIV-1 VL. Kran et al. (5) further investigated the cellular subgroups in PPT and EDTA plasma specimens and show a strong correlation between the number of lymphocytes, in particular, CD4+ T cells and increased VL. Our own data (H. Fernandes and L. Rainen, unpublished data) show that cell counts in centrifuged PPT plasma were higher in specimens that were placed on a rocking platform to simulate transportation than in the corresponding stationary specimens, signifying the possible release of cells trapped on the surface of the gel in the PPT.
HIV quantification is dependent on optimal sample handling and storage conditions. In our experiments, the time and speed of centrifugation for the separation of plasma were 1,600 × g for 20 min for the EDTA tubes and 1,100 × g for 10 min for the PPT tubes. However, earlier studies conducted in our laboratory using similar conditions of 1,100 × g for 10 min for both PPT and EDTA tubes did not show significant differences in the VLs when the plasma was aspirated after centrifugation (9; L. Rainen, H. Fernandes, K. Abu-Lawi, P. Riska, and M. Brown, presented at the 21st Annual Clinical Virology Symposium, Clearwater Beach, FL, 8 to 11 May 2005) indicating that the time and speed of centrifugation did not contribute significantly to the increase in VL.
Removal of RNA by heat treatment, as seen in our results, results in a significant, but not total, loss of quantifiable HIV-1, in both the EDTA and PPT tubes when measured in the Amplicor assay. The loss of RNA resulting in decreased VL was greater in extracted nucleic acids from EDTA (79%) than in PPT (65%) frozen plasma specimens (Table 2). This implies that specimens from both EDTA tubes and PPTs have nucleic acid other than RNA that contributes to the quantifiable VL in the specimen, with the PPT having more of the heat-resistant material. The issue that needs to be addressed relates to the contribution of residual proviral DNA to HIV-1 quantification in the assay. As stated by Salimnia et al. (11), “A recent study by the Viral Quality Assurance Laboratory (NO1-AI-85354) determined that a single copy of HIV-1-specific proviral DNA could increase the detected viral load by 100 to 200 RNA copies (Cheryl Jennings [Rush Medical Center], personal communication).” Keeping this in mind, it is reasonable to assume that a small amount of contaminating proviral DNA present in the specimen could contribute significantly to HIV-1 VL. The VL quantified from proviral DNA therefore is not an accurate quantification and does not reflect the amount of proviral DNA present in the specimen.
Treating the extracted specimens with DNase I to eliminate any contaminating proviral DNA clearly shows that DNA is a major contributor to the elevated VLs seen in PPTs. Our results further demonstrate the increased levels of cellular DNA, as evident by amplification of the β-globin housekeeping gene, in plasma specimens from PPT compared to EDTA tubes. Confirmation of the presence of integrated proviral DNA in the extracted specimen is evident in the results of the nested PCR. All of the above factors clearly implicate HIV-1 proviral DNA as the reason for elevated HIV-1 VL values in PPTs.
The Roche COBAS Amplicor HIV-1 Monitor version 1.5 assay that is used for quantification of HIV-1 amplifies a 143-bp segment in the conserved gag (group antigen) region of the HIV-1 genome. Integrated proviral DNA of the HIV-1 viral genome contains the gag gene and is therefore amplified when present in the extracted RNA specimen. In fact, the Amplicor assay for the detection of HIV-1 proviral DNA uses the very same primers to target the same 143-bp region of the gag gene of the HIV-1 genome and can potentially amplify any virus-specific template, either DNA or RNA, that may be present (11). Yet another factor to consider in this analysis is the method of extraction of nucleic acid. The Amplicor assay uses a “total nucleic acid” method for the extraction of RNA whereby both RNA and DNA are extracted in the same specimen. The quantification assay is therefore amenable to amplification of RNA and contaminating DNA within the specimen, thus yielding an elevated VL. The effect of increased assay sensitivity, using such extraction methodology, is also evident in the recent report by Lima et al. (7), who show that the more sensitive HIV-1 TaqMan assay yields falsely elevated results compared to the Amplicor assay. Taken together, the amplification strategy together with the extraction methodology in the Amplicor HIV-1 assays have the potential to yield elevated HIV-1 VLs in samples quantified from PPTs in situ.
The data presented in this study have determined that HIV-1 proviral DNA present in cells is the reason for elevated HIV-1 RNA values in specimens collected and frozen in PPTs in situ quantified in the Roche COBAS Amplicor HIV-1 Monitor version 1.5 ultrasensitive assay. Specimen collection, extraction methodologies, targeted regions in the genome, and amplification assays utilized should be evaluated for accuracy in the precise quantification of HIV-1 RNA, especially for therapeutic management where viral clearance is a decisive determinant.
Footnotes
Published ahead of print on 24 February 2010.
REFERENCES
- 1.Alvarez-Arnao, D., A. Li, B. Canedo, and C. Baleeiro. 2005. HIV RNA measurement using EDTA or PPT tubes results in different degrees of undetectability: impact on clinical practice, abstr. MoPe15.2C06. Abstr. 3rd Int. AIDS Soc. Conf. HIV Pathog. Treatment. Rio de Janeiro, Brazil, 24 to 27 July 2005. International AIDS Society, Geneva, Switzerland. http://www.aegis.com/conferences/iashivpt/2005/MoPe15-2C06.html.
- 2.Elbeik, T., P. Nassos, P. Kipnis, B. Haller, and V. L. Ng. 2005. Evaluation of the VACUTAINER PPT plasma preparation tube for use with the Bayer VERSANT assay for quantification of human immunodeficiency virus type 1 RNA. J. Clin. Microbiol. 43:3769-3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.García-Bujalancea, S., C. Ladrón de Guevaraa, J. González-Garcíab, J. R. Arribasb, F. Zamorab, and A. Gutiérreza. 2007. Elevation of viral load by PCR and use of plasma preparation tubes for quantification of human immunodeficiency virus type 1. J. Microbiol. Methods 69:384-386. [DOI] [PubMed] [Google Scholar]
- 4.Griffith, B. P., and D. R. Mayo. 2006. Increased levels of HIV RNA detected in samples with viral loads close to the detection limit collected in plasma preparation tubes. J. Clin. Virol. 35:197-200. [DOI] [PubMed] [Google Scholar]
- 5.Kran, A.-M. B., T. O. Jonassen, M. Sannes, K. Jakobsen, A. Lind, A. Maeland, and M. Holberg-Petersen. 2009. Overestimation of human immunodeficiency virus type 1 load caused by the presence of cells in plasma from plasma preparation tubes. J. Clin. Microbiol. 47:2170-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee, P. K., T. L. Kieffer, R. F. Siliciano, and R. E. Nettles. 2006. HIV-1 viral load blips are of limited clinical significance. J. Antimicrob. Chemother. 57:803-805. [DOI] [PubMed] [Google Scholar]
- 7.Lima, V., R. Harrigan, and J. S. G. Montaner. 2009. Increased reporting of detectable plasma HIV-1 RNA levels at the critical threshold of 50 copies per milliliter with the Taqman assay in comparison to the Amplicor assay. J. Acquir. Immune Defic. Syndr. 51:3-6. [DOI] [PubMed] [Google Scholar]
- 8.Nettles, R. E., and T. L. Kieffer. 2006. Update on HIV-1 viral load blips. Curr. Opin. HIV AIDS 1:157-161. [DOI] [PubMed] [Google Scholar]
- 9.Rainen, L., H. Salimnia, M. Fairfax, H. Fernandes, and M. Brown. 2005. Sample handling parameters affecting performance of BD Vacutainer PPT and Plus K2 EDTA tubes with the Roche Amplicor and Cobas Amplicor HIV-1 Monitor test, version 1.5 ultrasensitive and standard specimen processing procedures. BD white paper. BD Diagnostics, Franklin Lakes, NJ.
- 10.Rebeiro, P. F., A. Kheshti, S. S. Bebawy, S. E. Stinnette, H. Erdem, Y. W. Tang, T. R. Sterling, S. P. Raffanti, and R. T. D'Aquila. 2008. Increased detectability of plasma HIV-1 RNA after introduction of a new assay and altered specimen-processing procedures. Clin. Infect. Dis. 47:1354-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salimnia, H., E. C. Moore, L. R. Crane, R. D. MacArthur, and M. R. Fairfax. 2005. Discordance between viral loads determined by Roche COBAS AMPLICOR human immunodeficiency virus type 1 Monitor (version 1.5) standard and ultrasensitive assays caused by freezing patient plasma in centrifuged Becton-Dickinson Vacutainer brand plasma preparation tubes. J. Clin. Microbiol. 43:4635-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharkey, M. K. T., D. R. Kuritzkes, and M. Stevenson. 2005. In vivo evidence for instability of episomal human immunodeficiency virus type 1 cDNA. J. Virol. 79:5203-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Squires, K., A. Lazzarin, J. M. Gatell, W. G. Powderly, V. Pokrovskiy, J.-F. Delfraissy, J. Jemsek, A. Rivero, W. Rozenbaum, S. Schrader, M. Sension, A. Vibhagool, A. Thiry, and M. Giordano. 2004. Comparison of once-daily atazanavir with efavirenz, each in combination with fixed-dose zidovudine and lamivudine, as initial therapy for patients infected with HIV. J. Acquir. Immune Defic. Syndr. 36:1011-1019. [DOI] [PubMed] [Google Scholar]
- 14.Stosor, V., F. J. Palella, Jr., B. Berzins, M. Till, A. Leake, J. S. Chmiel, and R. L. Murphy. 2005. Transient viremia in HIV-infected patients and use of plasma preparation tubes. Clin. Infect. Dis. 41:1671-1674. [DOI] [PubMed] [Google Scholar]