Abstract
Determination of hepatitis D virus (HDV) viremia represents the “gold standard” for the diagnosis of HDV infection. Hepatitis B virus (HBV)-HDV coinfection frequently leads to end-stage liver disease and hepatocellular carcinoma. No commercial assay for HDV RNA quantification that includes automated nucleic acid extraction is available, and in-house PCR tests are not well standardized. However, knowledge of HDV RNA levels may give important information for patient management and could be a useful tool for monitoring the response to antiviral therapies. One platform that is widely used for HBV DNA or HCV RNA quantification is the Cobas Ampliprep/TaqMan system. Using the utility channel of this platform, we established a novel protocol for TaqMan-based HDV RNA quantification after automatic extraction of RNA by the Ampliprep system. The assay was specific and showed linearity over a wide range from 3 × 102 to 107 copies/ml. Reproducibility was demonstrated by determination of the interrun and intrarun variabilities, which were similar to those achieved with the commercially available Cobas TaqMan assays for HCV RNA and HBV DNA. HDV RNA levels were stable in whole blood (n = 4), plasma (n = 3), and serum (n = 3) samples at room temperature for up to 6 days. Importantly, HDV RNA viremia showed only minor fluctuations, with the log10 coefficient of variation being between 1.3 and 11.2% for hepatitis delta patients studied every 2 weeks for up to 3 months (n = 6), while a rapid viral decline was observed early during treatment with pegylated alfa-2a interferon (n = 6). In conclusion, this novel automated HDV RNA assay is a useful tool for monitoring HDV-infected patients both before and during antiviral therapy.
Hepatitis delta is the most severe form of chronic viral hepatitis in humans. The hepatitis delta virus (HDV) is a defective RNA virus which requires the hepatitis B virus (HBV) surface antigen (HBsAg) for complete replication and transmission, although the full extent of the HBV helper function has not been unexplored (27, 34). The HDV genome is a small, 1,678-nucleotide single-stranded RNA with a circular configuration that can form a rod-like structure with at least 70% paired bases (16, 35). The HDV RNA encodes small and large hepatitis delta antigens as the sole proteins (34).
Hepatitis delta occurs only in HBsAg-positive individuals either as an acute coinfection or as a superinfection in patients with chronic hepatitis B (9). Several studies have shown that chronic HDV infection leads to more severe liver disease than chronic HBV monoinfection, with the course of progression of the fibrosis being accelerated, the risk of hepatocellular carcinoma being increased, and early decompensation occurring in the setting of established cirrhosis (9, 12, 13, 36). Simultaneous HBV and HDV infection has also been shown to be more severe than infection with HBV alone in chimpanzees (6).
The current treatment options for patients with delta hepatitis are very limited, as alfa interferon is able to clear HDV only in a minority of patients (23). High doses of alfa interferon have been associated with a beneficial long-term outcome in a small cohort of Italian hepatitis delta patients (10, 11). Pegylated alfa interferon has also been used in small trials to treat delta hepatitis, and the sustained virological response rates were about 20% (2, 7, 21). The nucleoside and nucleotide analogues used for the treatment of HBV infection are ineffective against HDV (22, 23, 39, 40, 42). No study has systematically investigated the effect of tenofovir or entecavir, two potent HBV polymerase inhibitors that have recently been approved for use for the treatment of hepatitis B, on HDV replication (5, 8).
HDV RNA determination enables the diagnosis of active viremia in case of the detection of anti-HDV in patients with chronic hepatitis B. Moreover, a possible correlation between HDV RNA levels and disease activity has been proposed (38); however, that correlation was not confirmed by others (43). In addition, HDV RNA quantification represents a useful tool for monitoring the response to treatment in patients with delta hepatitis receiving antiviral therapy. Several in-house quantitative HDV RNA PCR assays have been developed (2, 17, 38). However, these assays are not well standardized and quantitative values are difficult to compare.
HBV DNA or HCV RNA quantification is usually performed with commercial, fully automated PCR-based or branched DNA-based assays. These commercial systems also include automated nucleic acid extraction. One of those is the Cobas Ampliprep/TaqMan assay. Until recently, it was not possible to run in-house-based PCRs on that platform. By using AMPLILINK software (version 3.2) and utility channel applications (version 3.0), it is now possible to design in-house PCR protocols. The aim of the work described here was to (i) establish a protocol for TaqMan-based HDV RNA quantification with optimized primers after the automatic extraction of RNA by the Ampliprep system that can be used for routine diagnostics and (ii) evaluation of the protocol for longitudinal HDV RNA determinations in both untreated patients and patients receiving pegylated alfa-2a interferon.
MATERIALS AND METHODS
Patient samples.
In the present study we included plasma and serum samples from consecutive patients with delta hepatitis (n = 17) who presented at our liver outpatient clinic. All patients were anti-HIV negative. Anti-HCV-positive patients tested negative for HCV RNA.
To test for specificity, we used samples anonymously obtained from each of the following groups: apparently healthy employees of our hospital (Hannover Medical School) (n = 20), HBV-monoinfected patients (anti-HDV negative) (n = 20), HCV-monoinfected patients (anti-HBc negative) (n = 20), and HIV-monoinfected patients (n = 20).
Blood collection.
Blood (up to 50 ml) from patients with delta hepatitis was drawn in tubes containing EDTA potassium or serum (Sarstedt, Germany) and centrifuged, and the plasma/serum was stored at −20°C until it was further processed.
Nucleic acid extraction.
Nucleic acids were extracted from 200 μl plasma or serum by the Cobas AmpliPrep Instrument (Roche Diagnostics, Mannheim, Germany) for automated specimen processing by use of the total nucleic acid isolation kit (TINAI), according to the manufacturers' instructions (Roche Diagnostics). TINAI is part of a certified system that has been evaluated and that is also used for the standard routine detection and quantification of HBV DNA and HCV RNA. After the extraction by the TINAI kit, the total amount of extracted nucleic acids was solved in 80 μl.
HDV RNA quantification.
HDV RNA was quantified by a Cobas TaqMan-based in-house PCR (TaqMan 48; Roche Diagnostics). The forward primers (forward primer 1, 5′-TGGACGTKCGTCCTCCT-3′ [positions 837 to 853], where K is guanine or thymine; forward primer 2, 5′-TGGACGTCTGTCCTCCTT-3′ [positions 837 to 854]) were newly designed by one of the authors (T.B.) and bind to a conserved region of the genome that encodes the hepatitis delta antigen. The reverse primer (5′-TCTTCGGGTCGGCATGG [positions 891 to 907]) and the probe [5′-(5/6-carboxyfluorescein-ATG CCC AGG TCG GAC) (5/6-carboxytetramethylrhodamine)-3′ (positions 858 to 872)] were previously published elsewhere (17). The resulting PCR product was a 71-bp fragment (positions 837 to 907).
The following material was used for each quantification: 45.5 μl of H2O, 15 μl of 5× virus master mix (QuantiTect virus kit; Qiagen, Hilden, Germany), 0.75 μl of 100× virus reverse transcription (RT) mix (QuantiTect virus kit; Qiagen), and 3.75 μl of the 20× primer-probe mix to 65 μl. The primers and probe (high-pressure liquid chromatography purification grade; concentration, 0.1 nmol/μl) were purchased from IBA Gene TAGnologies (Göttingen, Germany).
The final concentrations of the primers and probe were 0.4 μM forward primer 1, 0.4 μM forward primer 2, 0.4 μM reverse primer, and 0.2 μM probe.
For quantification, 10 μl of the extracted RNA was added to the 65 μl of reagent. Hence, the total volume of the mixture and the extracted RNA was 75 μl.
The RNA was first transcribed to cDNA by reverse transcription at 50°C for 20 min, followed by denaturation at 95°C for 5 min. Afterwards, 50 cycles were performed, with each cycle consisting of denaturation at 95°C for 15 s and extension at 60°C for 45 s. Following each cycle, fluorescence was detected at a temperature of 60°C for 20 s.
We used a cDNA clone containing one copy of the complete HDV RNA sequence in a pSVL expression vector (20, 44) at concentrations of 1,000 fg/μl, 100 fg/μl, and 10 fg/μl (duplicates) as a standard. A negative control and a positive control were included. The raw data were processed with the Cobas TaqMan 48 utility channel data translator (Roche Diagnostics), and the processed data were analyzed by the LightCycler instrument data analysis software (Roche Diagnostics) by applying the fit points proportional mode.
For calculation of the number of plasmid copies per ml, the measured number of copies was multiplied by a factor of 13.33 (1,000 μl/75 μl).
Qualitative HDV RNA detection.
The lower limit of detection (LOD) for the quantitative HDV RNA assay was determined by using our in-house routine qualitative HDV RNA PCR, which has been described previously (15, 43). The following primers were used: primer nested D1 (forward, 5′-CGGATGCCCAGGTCGGACCGC-3′), primer nested D2 (reverse, 5′-CTCAGGGGAGGATTCACCGAC-3′), primer nested D3 (forward, 5′-AAACCTGTGAGTGGAAACCCGC-3′), and primer nested D4 (reverse, 5′-ATCACCGACGAAGGAAGGCCCTCG-3′). Primers D1 and D2 were used for the first PCR, and primers D3 and D4 were used for the second PCR.
Analytical performance. (i) LOD.
To test for the lower limit of detection, the stock of the plasmid (10,000 fg/μl = 1,100,000 copies/μl) was diluted with sterile water to 0.02 fg/μl, 0.01 fg/μl, 0.005 fg/μl, 0.001 fg/μl, and 0.0001 fg/μl. Four replicates of each dilution and two negative controls (blanks) were then tested by qualitative and quantitative HDV RNA PCRs.
(ii) Linearity.
To test the linearity of the quantitative HDV RNA PCR, the stock of the plasmid at a concentration of 10,000 fg/μl, which is equal to 1,100,000 copies, was diluted stepwise (1:10, 1:5, or 1:2), and four to six replicates were tested per dilution.
Furthermore, plasma samples from different patients were diluted stepwise (1:10, 1:5, or 1:2), and two replicates per dilution were tested.
(iii) Interrun variability.
To test the interrun variability, samples of HDV RNA from patients with low levels (<104 copies/ml) and high levels (≥105 copies/ml) of viremia as well as the plasmid containing HDV RNA (HDV plasmid) were aliquoted and stored at −20°C (HDV RNA) or −80°C (HDV plasmid) before use. For each PCR run, an aliquot was thawed and used as a positive control for the determination of interrun variability.
(iv) Intrarun variability.
Intrarun variability was determined by measuring the amount of HDV RNA in 14 samples with low, 14 with medium, and 14 with high levels of viremia within one PCR run.
Stability of HDV RNA.
To evaluate the stability of the HDV RNA after the blood draw, EDTA-anticoagulated whole blood, plasma, and serum were kept at room temperature for up to 6 days. On each day, a part of the plasma or serum sample was then stored at −20°C until the detection of HDV RNA.
The stability of the HDV RNA in plasma and serum and after the extraction of nucleic acids from plasma was analyzed after up to eight cycles of thawing and freezing. Thawing of the samples was performed at room temperature for 4 h, followed by freezing at −20°C for at least 20 h and then the next thaw-freeze cycle.
Course of HDV RNA levels in untreated and treated patients.
Six patients without antiviral treatment were evaluated biweekly for up to 12 weeks for their HDV RNA levels to analyze the fluctuation in HDV RNA levels over the natural course of delta hepatitis. The course of the HDV RNA levels during antiviral treatment in six patients receiving pegylated alfa-2a interferon was studied for 12 weeks.
Statistical methods.
Statistical analyses were performed with Prism software (version 5; GraphPad Software, Inc., La Jolla, CA).
RESULTS
Lower limit of detection.
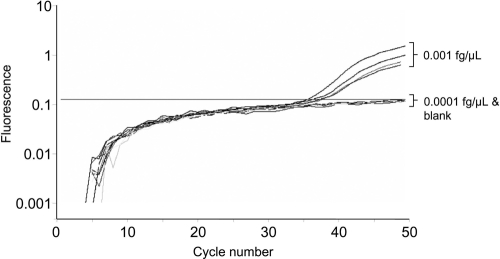
We first determined the LOD for the plasmid in a nested qualitative in-house HDV RNA PCR which has been described previously (15, 43). By use of this assay, 75 copies per ml (0.005 fg/μl) were detected in all four samples, while only one of four samples containing 15 copies/ml (0.001 fg/μl) tested positive. In contrast, the new Cobas TaqMan HDV PCR was more sensitive, as all samples containing 15 copies/ml (0.001 fg/μl) showed signals (Fig. 1). All samples containing 1.5 copies/ml tested HDV RNA negative by both PCRs (Table 1). Only samples with at least 300 copies per ml (0.02 fg/μl) revealed reproducible quantitative values (Fig. 2).
FIG. 1.
Determination of lower limit of detection of Cobas TaqMan quantitative HDV RNA PCR.
TABLE 1.
Lower limits of detection for nested qualitative routine PCR and the Cobas TaqMan quantitative HDV PCR
| No. of samples | Plasmid concn (fg/μl) | Calculated no. of copies per ml | No. of samples positive/no. tested by nested qualitative routine PCRa | No. of copies per ml (mean ± SD) measured by Cobas TaqMan quantitative HDV PCR |
|---|---|---|---|---|
| 4 | 0.02 | 300 | 4/4 | 240 ± 120 |
| 4 | 0.01 | 150 | 4/4 | 80 ± 67 |
| 4 | 0.005 | 75 | 4/4 | <15 (4/4 detectable) |
| 4 | 0.001 | 15 | 1/4 | <15 (4/4 detectable) |
| 4 | 0.0001 | 1.5 | 0/4 | Negative (0/4 detectable) |
| 2 | 0 | 0 | 0/2 | Negative (0/2 detectable) |
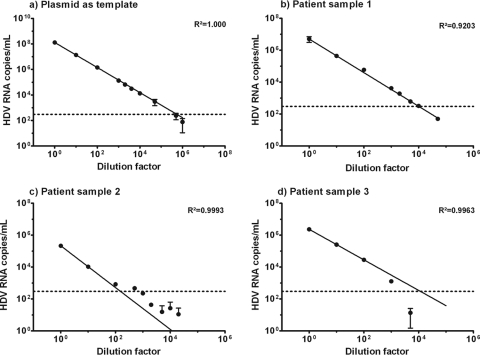
FIG. 2.
Linearity of TaqMan quantitative HDV PCR with the plasmid (a) or three different patient samples (b to d, respectively) as the template. Dotted lines, lower limit of linearity.
Linearity.
The quantitative HDV PCR was linear within a range of 3 × 102 to 1.5 × 108 copies per ml when the HDV plasmid was used as the template (Fig. 2a). Linearity was also confirmed for three patient plasma samples with high levels of viremia. After various dilutions, the nucleic acid was extracted and the HDV RNA was quantified. Linearity was observed from 3 × 102 up to 107 copies per ml in all three patient samples (Fig. 2b to d). The goodness of fit (R2) when the measured viral load was compared to the dilution factor was at least 0.9 for all patients.
Interrun variability.
To test the interrun variability, we used samples from two patients, one individual with a low level of viremia and another patient with high HDV RNA levels, and the HDV plasmid as a positive control. The samples were tested 17, 12, and 23 times, respectively. The results are shown in Table 2 and demonstrate reasonable interrun variability for under all conditions tested.
TABLE 2.
Interrun and intrarun variabilitiesa
| Parameter | Interrun variability |
Intrarun variability |
||||
|---|---|---|---|---|---|---|
| Low-viremia sample | High-viremia sample | HDV plasmid | Low-viremia sample | Medium-viremia sample | High-viremia sample | |
| No. of determinations | 17 | 12 | 23 | 14 | 14 | 14 |
| No. of copies per ml (mean ± SD) | 5.4 × 103 ± 1.2 × 103 | 2.0 × 105 ± 2.6 × 104 | 1.7 × 106 ± 4.9 × 105 | 5.1 × 102 ± 1.5 × 102 | 6.5 × 103 ± 2.6 × 103 | 2.6 × 105 ± 6.4 × 104 |
| % CV (SD/mean) | 22.4 | 13.3 | 28.7 | 29.8 | 40.5 | 24.7 |
| Log10 no. of copies per ml (mean ± SD) | 3.73 ± 0.09 | 5.29 ± 0.06 | 6.21 ± 0.16 | 2.71 ± 0.13 | 3.75 ± 0.28 | 5.40 ± 0.11 |
| Log10 % CV (SD/mean) | 2.5 | 1.1 | 2.6 | 5.0 | 7.5 | 2.1 |
Interrun variability was tested with two positive control samples (with low and high levels of viremia) and the HDV plasmid. Intrarun variability was tested with samples with low, medium, and high levels of viremia.
Intrarun variablity.
The intra-assay variability was determined by measuring the levels of HDV RNA in 14 aliquots of three different patient samples, including one patient with low HDV RNA levels, one with medium HDV RNA levels, and one with high HDV RNA levels, each in an individual PCR run. The results are presented in Table 2.
Specificity.
Specificity was tested by using 20 samples from each of the following groups: healthy controls, HBV-monoinfected patients (anti-HDV negative), HCV-monoinfected patients (anti-HBc negative), and HIV-monoinfected patients. All samples tested HDV RNA negative (100% specificity).
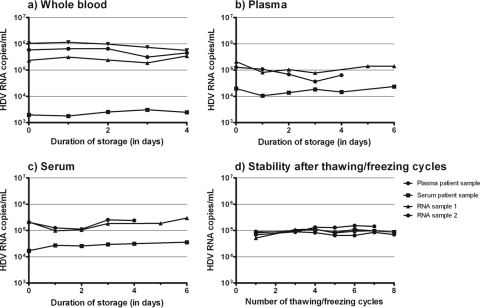
Stability of HDV RNA.
HDV RNA was stable at room temperature in whole blood (n = 4) for up to 4 days and in plasma (n = 3) and serum (n = 3) for up to 6 days (Fig. 3a to c). The HDV RNA levels in serum and plasma and the amount of extracted RNA did not decrease after up to eight cycles of thawing and freezing (Fig. 3d).
FIG. 3.
Stability of HDV RNA after storage at room temperature for up to 4 days in whole blood (a) and for up to 6 days in plasma (b) and serum (c). Each line in the three different graphs (a to c) represents a single patient sample. (d) Stability of HDV RNA in plasma and serum and of isolated RNA after up to eight thaw-freeze cycles.
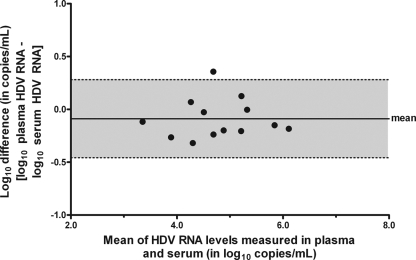
Comparison of HDV RNA levels in plasma and serum.
The HDV RNA levels were determined in the plasma as well as the serum of 13 patients. Figure 4 shows a Bland-Altman analysis demonstrating similar results for plasma and serum. The viral loads in the two sample types did not differ significantly (paired t test, P = 0.17).
FIG. 4.
Bland-Altman plot analysis of HDV RNA levels measured in plasma and serum of 13 patients by Cobas TaqMan HDV PCR. The difference between the HDV RNA levels in plasma and serum is represented as a function of the mean of the two values. The gray area corresponds to the 95% limit of agreement.
HDV RNA kinetics in untreated patients.
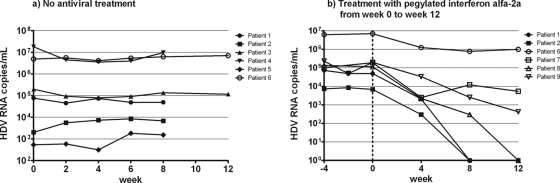
The mean HDV RNA levels in different patients ranged from 960 to 7.9 × 106 copies per ml. The HDV RNA levels were stable in five of six patients, with the log10 coefficient of variation (CV; standard deviation [SD]/mean) being between 1.25 and 6.66% for the individual patients; these values were in the range of the interrun and intrarun variabilities. The HDV RNA levels in the remaining patient (patient 5) were also relatively stable, but due to low viral loads of between 314 to 1,820 copies per ml, the log10 CV was 11.2% (Fig. 5a).
FIG. 5.
Kinetics of HDV RNA in untreated patients with chronic delta hepatitis for up to 12 weeks (a) and in patients receiving pegylated alfa-2a interferon for 12 weeks (b).
HDV RNA kinetics during antiviral treatment.
The HDV RNA levels 4 weeks prior to treatment (week −4) and at the baseline (week 0) were in the same range (Fig. 5b). During antiviral treatment, three of six patients with baseline viral loads of 6.86 × 103, 4.96 × 104, and 1.16 × 105 copies/ml cleared the HDV RNA after 8 weeks (patients 1 and 2) or 12 weeks (patient 8). Two of the remaining three patients (patients 7 and 9) demonstrated a decrease in the HDV RNA level of at least 1 log10 while they were receiving pegylated alfa-2a interferon, whereas patient 6 showed a decrease of only <1 log10 after 12 weeks (Fig. 5b).
DISCUSSION
We developed a novel quantitative HDV RNA assay by using the utility channel of the Cobas TaqMan platform. The Cobas TaqMan apparatus (Roche Diagnostics) is a real-time PCR platform which is widely used to quantify HBV DNA or HCV RNA (4, 24). The system allows the design of in-house real-time PCR assays. Another obvious advantage of using this platform for HDV quantification includes the automated extraction system, the use of which provides a minimal risk of contamination and a high degree of reproducibility (33). In addition, as the Cobas TaqMan system is well established, the steps and procedures used for the detection of RNA and DNA from other sources are similar for the detection of HDV RNA and would decrease the hands-on time and minimize potential sources of error (33).
The first problem to be overcome in the establishment of a quantitative HDV RNA assay was the lack of an international standard for calibration. We therefore used a cDNA clone containing one copy of the complete HDV RNA sequence in a pSVL expression vector as a standard (20, 44). The initial HDV RNA concentration in the plasmid (10 pg/μl) was diluted to generate a standard curve and to investigate the linearity and the lower limit of detection of the assay. Nevertheless, the availability of an international WHO standard for comparison of quantitative HDV RNA results would be highly desirable, as similar standards are well established for HBV (28) and HCV (29). Of note, in this assay a DNA clone is used to monitor the amplification of an RNA virus, and this has also been done by other groups (17). The quantification of the DNA clone does not include a reverse transcription step and might be a limitation. An alternative could be the use of synthetic RNA as the standard, as described previously (38); however, the limitation of synthetic RNA is that it may have secondary structures different from those of natural HDV RNA. Therefore, a recently published study using HDV RNA quantification generated a standard from a patient with a high HDV load after detection of viremia by the use of a cDNA standard (31). Indeed, we also recommend not only that the plasmid standard be run but also that defined patient-derived HDV RNA be used to validate the accuracies of the quantitative values obtained by this assay.
The quantitative HDV RNA assay developed in the present study showed good performance characteristics in terms of the limit of detection, linearity, reproducibility, and specificity. We found that the new assay had a lower limit of detection of 15 copies per ml, which was lower than the 75 copies/ml obtained by our previous in-house nested qualitative HDV RNA PCR (15, 43). The new assay also seems to be slightly more sensitive than other assays for HDV RNA quantification (2, 17, 38). The LOD could be further improved by increasing the volume of serum/plasma used for Ampliprep extraction, as is the case for commercially available assays; e.g., the manufacturer of the Cobas HBV DNA assay recommends the use of 1,050 μl (1).
Linearity was observed over a wide range, from 3 × 102 to 1.5 × 108 copies/ml for the plasmid and from 3 × 102 up to 107 copies per ml for the patient sample with the highest observed viral load. This linearity is well in line with the linearities of other assays performed on the Cobas TaqMan platform, including assays for HCV, HBV, cytomegalovirus, and HIV (1, 25, 32, 33). This linearity is sufficient, as in our experience about 90% of patients showed HDV RNA levels below 107 copies/ml, and the highest viral load that we ever detected was 8.4 × 107 copies/ml (43).
Reproducibility was assessed by analyzing interrun and intrarun variabilities. The log10 CVs were in the range of 1.1 to 7.5%, which, again, is well acceptable and in agreement with the reproducibilities of other commercially available Cobas TaqMan assays for HCV RNA (14, 30) and HBV DNA (1, 3) quantification. Finally, the mandatory requirement of 100% specificity was achieved, as none of the controls, including healthy donors and patients infected with HBV, HCV, or HIV, tested positive for HDV RNA. Nevertheless, the number of control samples needs to be increased to further confirm this finding.
The Cobas TaqMan HCV RNA assay revealed similar results for serum and plasma samples (33). In line with the experience for HCV, we found similar results when testing for HDV RNA in plasma and serum samples, confirming that our assay would also be applicable when only serum samples are available. Of note, we could also show that HDV RNA is equally stable over a period of several days in whole blood, plasma, and serum and as extracted RNA. HDV RNA was also stable after up to eight freeze-thaw cycles. Considering that HDV is an RNA virus, these findings might be somewhat surprising. However, these data on the stability of HDV are very well in line with those from our recent study in which we compared the stabilities of HCV RNA and HCV core antigen under different conditions (19). Also in that study, HCV RNA was rather stable over several days, even in whole-blood samples, unless the tubes were stored at 37°C. The finding that HDV RNA levels do not decline by more than 0.5 to 1 log10 copies/ml after 2 to 3 days has major practical implications for the logistics of the shipping of samples to the diagnostic laboratory. This would permit shipment from remote areas without the need for plasma-serum separation or special handling. Thus, there is no need to destroy blood samples if for some reason the shipment has been delayed. The information obtained can still be useful and can be communicated to the patient's physician.
Once the quantitative HDV RNA assay was established, we applied this tool to study the kinetics of HDV RNA over time in untreated patients with hepatitis delta and in individuals receiving antiviral treatment. While HCV RNA levels are rather stable over time and have very limited variability (26), HBV infection is known to be a highly dynamic disease (37). Thus, HBV DNA levels may show significant fluctuations within a short period of time. However, no information on whether HDV RNA levels in untreated patients may also fluctuate over a short period of time is so far available. We therefore applied the Cobas TaqMan quantitative HDV PCR to samples obtained from six patients with chronic delta hepatitis and studied the HDV RNA levels biweekly for up to 12 weeks. Five of six patients demonstrated stable HDV RNA levels over time, and the remaining patient demonstrated very low viral loads and had minor fluctuations in HDV RNA levels. Overall, these first data on the longitudinal measurement of the HDV RNA loads in untreated patients may suggest that during antiviral therapy, a decrease in the HDV RNA load of at least 1 log10 copy per ml could be considered treatment related. Indeed, significant declines in the viral load of up to 5 log10 copies/ml were observed when some of the patients were treated for 8 to 12 weeks with pegylated alfa-2a interferon. Thus, these data clearly show the usefulness of this assay for the management of patients with delta hepatitis. Quantitative HDV RNA determinations might become even more important, as treatment durations could be based on the kinetics of HDV RNA during treatment (7, 41), a clinical practice that is well established for hepatitis C virus infection (18).
In summary, we developed a reliable quantitative HDV RNA assay that can be run on the widely available Cobas TaqMan platform. We suggest that this assay can be used both for the long-term management of HDV-infected individuals and for monitoring the response to antiviral therapy in patients with chronic delta hepatitis.
Acknowledgments
This work was supported by grants from the German Federal Ministry of Education and Research (reference numbers 01EO0802 and 01KI078). The study was part of the activities of the VIRGIL European Network of Excellence on Antiviral Drug Resistance supported by a grant (LSHM-CT-2004-503359) from the Priority 1 Life Sciences, Genomics, and Biotechnology for Health program in the 6th Framework Programme of the EU.
The contents of this article are the sole responsibility of the authors.
Footnotes
Published ahead of print on 29 March 2010.
REFERENCES
- 1.Allice, T., F. Cerutti, F. Pittaluga, S. Varetto, S. Gabella, A. Marzano, A. Franchello, G. Colucci, and V. Ghisetti. 2007. COBAS AmpliPrep-COBAS TaqMan hepatitis B virus (HBV) test: a novel automated real-time PCR assay for quantification of HBV DNA in plasma. J. Clin. Microbiol. 45:828-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castelnau, C., F. Le Gal, M. P. Ripault, E. Gordien, M. Martinot-Peignoux, N. Boyer, B. N. Pham, S. Maylin, P. Bedossa, P. Deny, P. Marcellin, and E. Gault. 2006. Efficacy of peginterferon alpha-2b in chronic hepatitis delta: relevance of quantitative RT-PCR for follow-up. Hepatology 44:728-735. [DOI] [PubMed] [Google Scholar]
- 3.Chevaliez, S., M. Bouvier-Alias, S. Laperche, and J. M. Pawlotsky. 2008. Performance of the Cobas AmpliPrep/Cobas TaqMan real-time PCR assay for hepatitis B virus DNA quantification. J. Clin. Microbiol. 46:1716-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chevaliez, S., and J. M. Pawlotsky. 2009. How to use virological tools for optimal management of chronic hepatitis C. Liver Int. 29(Suppl. 1):9-14. [DOI] [PubMed] [Google Scholar]
- 5.Cornberg, M., U. Protzer, M. M. Dollinger, J. Petersen, H. Wedemeyer, T. Berg, W. Jilg, A. Erhardt, S. Wirth, P. Schirmacher, W. E. Fleig, and M. P. Manns. 2007. Prophylaxis, diagnosis and therapy of hepatitis-B-virus-(HBV-) infection: upgrade of the guideline, AWMF-Register 021/011. Z. Gastroenterol. 45:525-574. (In German.) [DOI] [PubMed] [Google Scholar]
- 6.Dienes, H. P., R. H. Purcell, H. Popper, and A. Ponzetto. 1990. The significance of infections with two types of viral hepatitis demonstrated by histologic features in chimpanzees. J. Hepatol. 10:77-84. [DOI] [PubMed] [Google Scholar]
- 7.Erhardt, A., W. Gerlich, C. Starke, U. Wend, A. Donner, A. Sagir, T. Heintges, and D. Haussinger. 2006. Treatment of chronic hepatitis delta with pegylated interferon-alpha2b. Liver Int. 26:805-810. [DOI] [PubMed] [Google Scholar]
- 8.European Association for the Study of the Liver. 2009. EASL clinical practice guidelines: management of chronic hepatitis B. J. Hepatol. 50:227-242. [DOI] [PubMed] [Google Scholar]
- 9.Farci, P. 2003. Delta hepatitis: an update. J. Hepatol. 39(Suppl. 1):S212-S219. [DOI] [PubMed] [Google Scholar]
- 10.Farci, P., A. Mandas, A. Coiana, M. E. Lai, V. Desmet, P. Van Eyken, Y. Gibo, L. Caruso, S. Scaccabarozzi, and D. Criscuolo. 1994. Treatment of chronic hepatitis D with interferon alfa-2a. N. Engl. J. Med. 330:88-94. [DOI] [PubMed] [Google Scholar]
- 11.Farci, P., T. Roskams, L. Chessa, G. Peddis, A. P. Mazzoleni, R. Scioscia, G. Serra, M. E. Lai, M. Loy, L. Caruso, V. Desmet, R. H. Purcell, and A. Balestrieri. 2004. Long-term benefit of interferon alpha therapy of chronic hepatitis D: regression of advanced hepatic fibrosis. Gastroenterology 126:1740-1749. [DOI] [PubMed] [Google Scholar]
- 12.Fattovich, G., S. Boscaro, F. Noventa, E. Pornaro, D. Stenico, A. Alberti, A. Ruol, and G. Realdi. 1987. Influence of hepatitis delta virus infection on progression to cirrhosis in chronic hepatitis type B. J. Infect. Dis. 155:931-935. [DOI] [PubMed] [Google Scholar]
- 13.Fattovich, G., G. Giustina, E. Christensen, M. Pantalena, I. Zagni, G. Realdi, and S. W. Schalm. 2000. Influence of hepatitis delta virus infection on morbidity and mortality in compensated cirrhosis type B. The European Concerted Action on Viral Hepatitis (Eurohep). Gut 46:420-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forman, M. S., and A. Valsamakis. 2008. Performance characteristics of a quantitative hepatitis C virus RNA assay using COBAS AmpliPrep total nucleic acid isolation and COBAS TaqMan hepatitis C virus analyte-specific reagent. J. Mol. Diagn. 10:147-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heidrich, B., K. Deterding, H. L. Tillmann, R. Raupach, M. P. Manns, and H. Wedemeyer. 2009. Virological and clinical characteristics of delta hepatitis in Central Europe. J. Viral Hepat. 16:883-894. [DOI] [PubMed] [Google Scholar]
- 16.Kuo, M. Y. P., J. Goldberg, L. Coates, W. Mason, J. Gerin, and J. Taylor. 1988. Molecular cloning of hepatitis delta virus RNA from an infected woodchuck liver: sequence, structure, and applications. J. Virol. 62:1855-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Gal, F., E. Gordien, D. Affolabi, T. Hanslik, C. Alloui, P. Deny, and E. Gault. 2005. Quantification of hepatitis delta virus RNA in serum by consensus real-time PCR indicates different patterns of virological response to interferon therapy in chronically infected patients. J. Clin. Microbiol. 43:2363-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manns, M. P., H. Wedemeyer, and M. Cornberg. 2006. Treating viral hepatitis C: efficacy, side effects, and complications. Gut 55:1350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mederacke, I., H. Wedemeyer, S. Ciesek, E. Steinmann, R. Raupach, K. Wursthorn, M. P. Manns, and H. L. Tillmann. 2009. Performance and clinical utility of a novel fully automated quantitative HCV-core antigen assay. J. Clin. Virol. 46:210-215. [DOI] [PubMed] [Google Scholar]
- 20.Netter, H. J., T. T. Wu, M. Bockol, A. Cywinski, W. S. Ryu, B. C. Tennant, and J. M. Taylor. 1995. Nucleotide sequence stability of the genome of hepatitis delta virus. J. Virol. 69:1687-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niro, G. A., A. Ciancio, G. B. Gaeta, A. Smedile, A. Marrone, A. Olivero, M. Stanzione, E. David, G. Brancaccio, R. Fontana, F. Perri, A. Andriulli, and M. Rizzetto. 2006. Pegylated interferon alpha-2b as monotherapy or in combination with ribavirin in chronic hepatitis delta. Hepatology 44:713-720. [DOI] [PubMed] [Google Scholar]
- 22.Niro, G. A., A. Ciancio, H. L. Tillman, M. Lagget, A. Olivero, F. Perri, R. Fontana, N. Little, F. Campbell, A. Smedile, M. P. Manns, A. Andriulli, and M. Rizzetto. 2005. Lamivudine therapy in chronic delta hepatitis: a multicentre randomized-controlled pilot study. Aliment. Pharmacol. Ther. 22:227-232. [DOI] [PubMed] [Google Scholar]
- 23.Niro, G. A., F. Rosina, and M. Rizzetto. 2005. Treatment of hepatitis D. J. Viral Hepat. 12:2-9. [DOI] [PubMed] [Google Scholar]
- 24.Pawlotsky, J. M. 2003. Hepatitis B virus (HBV) DNA assays (methods and practical use) and viral kinetics. J. Hepatol. 39(Suppl. 1):S31-S35. [DOI] [PubMed] [Google Scholar]
- 25.Piiparinen, H., K. Hockerstedt, C. Gronhagen-Riska, and I. Lautenschlager. 2004. Comparison of two quantitative CMV PCR tests, Cobas Amplicor CMV Monitor and TaqMan assay, and pp65-antigenemia assay in the determination of viral loads from peripheral blood of organ transplant patients. J. Clin. Virol. 30:258-266. [DOI] [PubMed] [Google Scholar]
- 26.Pontisso, P., G. Bellati, M. Brunetto, L. Chemello, G. Colloredo, R. Di Stefano, M. Nicoletti, M. G. Rumi, M. G. Ruvoletto, R. Soffredini, L. M. Valenza, and G. Colucci. 1999. Hepatitis C virus RNA profiles in chronically infected individuals: do they relate to disease activity? Hepatology 29:585-589. [DOI] [PubMed] [Google Scholar]
- 27.Rizzetto, M. 1983. The delta agent. Hepatology 3:729-737. [DOI] [PubMed] [Google Scholar]
- 28.Saldanha, J., W. Gerlich, N. Lelie, P. Dawson, K. Heermann, and A. Heath. 2001. An international collaborative study to establish a World Health Organization international standard for hepatitis B virus DNA nucleic acid amplification techniques. Vox Sang. 80:63-71. [DOI] [PubMed] [Google Scholar]
- 29.Saldanha, J., N. Lelie, and A. Heath. 1999. Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. WHO Collaborative Study Group. Vox Sang. 76:149-158. [DOI] [PubMed] [Google Scholar]
- 30.Sarrazin, C., A. Dragan, B. C. Gartner, M. S. Forman, S. Traver, S. Zeuzem, and A. Valsamakis. 2008. Evaluation of an automated, highly sensitive, real-time PCR-based assay (COBAS Ampliprep/COBAS TaqMan) for quantification of HCV RNA. J. Clin. Virol. 43:162-168. [DOI] [PubMed] [Google Scholar]
- 31.Schaper, M., F. Rodriguez-Frias, R. Jardi, D. Tabernero, M. Homs, G. Ruiz, J. Quer, R. Esteban, and M. Buti. 5 March 2010. Quantitative longitudinal evaluations of hepatitis delta virus RNA and hepatitis B virus DNA shows a dynamic, complex replicative profile in chronic hepatitis B and D. J. Hepatol. [Epub ahead of print.] [DOI] [PubMed]
- 32.Schumacher, W., E. Frick, M. Kauselmann, V. Maier-Hoyle, R. van der Vleit, and R. Babiel. 2007. Fully automated quantification of human immunodeficiency virus (HIV) type 1 RNA in human plasma by the COBAS AmpliPrep/COBAS TaqMan system. J. Clin. Virol. 38:304-312. [DOI] [PubMed] [Google Scholar]
- 33.Sizmann, D., C. Boeck, J. Boelter, D. Fischer, M. Miethke, S. Nicolaus, M. Zadak, and R. Babiel. 2007. Fully automated quantification of hepatitis C virus (HCV) RNA in human plasma and human serum by the COBAS AmpliPrep/COBAS TaqMan system. J. Clin. Virol. 38:326-333. [DOI] [PubMed] [Google Scholar]
- 34.Taylor, J. M. 2006. Hepatitis delta virus. Virology 344:71-76. [DOI] [PubMed] [Google Scholar]
- 35.Wang, K. S., Q. L. Choo, A. J. Weiner, J. H. Ou, R. C. Najarian, R. M. Thayer, G. T. Mullenbach, K. J. Denniston, J. L. Gerin, and M. Houghton. 1986. Structure, sequence and expression of the hepatitis delta (delta) viral genome. Nature 323:508-514. [DOI] [PubMed] [Google Scholar]
- 36.Wedemeyer, H., and M. P. Manns. 2010. Epidemiology, pathogenesis and management of hepatitis D: update and challenges ahead. Nat. Rev. Gastroenterol. Hepatol. 7:31-40. [DOI] [PubMed] [Google Scholar]
- 37.Wursthorn, K., M. P. Manns, and H. Wedemeyer. 2008. Natural history: the importance of viral load, liver damage and HCC. Best Pract. Res. Clin. Gastroenterol. 22:1063-1079. [DOI] [PubMed] [Google Scholar]
- 38.Yamashiro, T., K. Nagayama, N. Enomoto, H. Watanabe, T. Miyagi, H. Nakasone, H. Sakugawa, and M. Watanabe. 2004. Quantitation of the level of hepatitis delta virus RNA in serum, by real-time polymerase chain reaction—and its possible correlation with the clinical stage of liver disease. J. Infect. Dis. 189:1151-1157. [DOI] [PubMed] [Google Scholar]
- 39.Yurdaydin, C., H. Bozkaya, H. Cetinkaya, T. Sahin, D. Karaoguz, M. Toruner, O. Erkan, A. O. Heper, E. Erden, A. M. Bozdayi, and O. Uzunalimoglu. 2005. Lamivudine vs lamivudine and interferon combination treatment of HBeAg(−) chronic hepatitis B. J. Viral Hepat. 12:262-268. [DOI] [PubMed] [Google Scholar]
- 40.Yurdaydin, C., H. Bozkaya, S. Gurel, H. L. Tillmann, N. Aslan, A. Okcu-Heper, E. Erden, K. Yalcin, N. Iliman, O. Uzunalimoglu, M. P. Manns, and A. M. Bozdayi. 2002. Famciclovir treatment of chronic delta hepatitis. J. Hepatol. 37:266-271. [DOI] [PubMed] [Google Scholar]
- 41.Yurdaydin, C., H. Bozkaya, F. O. Onder, H. Senturk, H. Karaaslan, M. Akdogan, H. Cetinkaya, E. Erden, O. Erkan-Esin, K. Yalcin, A. M. Bozdayi, R. F. Schinazi, J. L. Gerin, O. Uzunalimoglu, and A. Ozden. 2008. Treatment of chronic delta hepatitis with lamivudine vs lamivudine + interferon vs interferon. J. Viral Hepat. 15:314-321. [DOI] [PubMed] [Google Scholar]
- 42.Yurdaydin, C., H. Wedemeyer, G. Dalekos, A. Erhardt, Y. Cakaloglu, H. Degertekin, S. Gurel, S. Zeuzem, K. Zachou, H. Bozkaya, H. P. Dienes, T. Bock, and M. P. Manns. 2006. A multicenter randomised study comparing the efficacy of pegylated interferon-alfa-2a plus adevofir dipivoxil vs. pegylated interferon-alfa-2a plus placebo vs. adevofir dipivoxil for the treatment of chronic delta hepatitis: the Hep-Net/International Delta Hepatitis Intervention Trial (HID-IT). Hepatology 44:230A. [Google Scholar]
- 43.Zachou, K., C. Yurdaydin, U. Drebber, G. N. Dalekos, A. Erhardt, Y. Cakaloglu, H. Degertekin, S. Gurel, S. Zeuzem, H. Bozkaya, V. Schlaphoff, H. P. Dienes, T. C. Bock, M. P. Manns, and H. Wedemeyer. 15 October 2009. Quantitative HBsAg and HDV-RNA levels in chronic delta hepatitis. Liver Int. [Epub ahead of print.] [DOI] [PubMed]
- 44.Zheng, H., T. B. Fu, D. Lazinski, and J. Taylor. 1992. Editing on the genomic RNA of human hepatitis delta virus. J. Virol. 66:4693-4697. [DOI] [PMC free article] [PubMed] [Google Scholar]