Abstract
We analyzed DNA eluted from FTA (Flinders Technology Associates) cards spotted with blood from human African trypanosomiasis (HAT) patients admitted at Lwala Hospital in eastern Uganda and Kaliua Health Centre in northwestern Tanzania. The aims were to evaluate loop-mediated isothermal amplification (LAMP) for detection of trypanosomal DNA in clinical samples and to characterize the infecting trypanosomes to the subspecies level. LAMP targeting the Trypanozoon conserved random inserted mobile element (RIME-LAMP) and that for the serum resistance-associated (SRA) gene (SRA-LAMP) were performed. For comparison, PCRs for the SRA gene specific for Trypanosoma brucei rhodesiense (SRA-PCR) and that to amplify the Trypanosoma brucei gambiense-specific surface glycoprotein (TgSGP-PCR) were done. Out of 128 samples analyzed, SRA-PCR was positive in 101 samples (78.9% sensitivity; 95% confidence interval [CI], 71.1 to 85.1%), SRA-LAMP was positive in 120 (93.8%; 95% CI, 88.2 to 96.8%), while RIME-LAMP revealed signals in 122 (95.3%; 95% CI, 90.2 to 97.8%). RIME-LAMP and SRA-LAMP were each significantly more sensitive than SRA-PCR (P values of 0.000 and 0.001, respectively; Fisher's exact test). There was poor agreement between RIME-LAMP and SRA-LAMP and the SRA-PCR, yielding kappa values of 0.31 and 0.40, respectively. Agreement between SRA-LAMP and RIME-LAMP was almost perfect (kappa value, 0.85; 95% CI, 0.64 to 1). All the 128 field samples were negative by TgSGP-PCR. Blood spots from three T. b. gambiense HAT cases from northwestern Uganda were positive by TgSGP-PCR and RIME-LAMP. PCR took five times longer to execute than LAMP. LAMP may be useful to monitor emerging HAT foci or to test travelers returning from countries where HAT is endemic. It should be evaluated in a case-control study to determine its utility as a HAT diagnostic.
Effective control of human African trypanosomiasis (HAT) is still hampered by unsatisfactory diagnostics and limited options for chemotherapeutic intervention. Diagnosis relies on demonstration of trypanosomes in blood, lymph, or cerebrospinal fluid by time-consuming, tedious, and complicated techniques of low sensitivity. Within health units, it is still not possible to distinguish between infections with Trypanosoma brucei rhodesiense (the acute form of eastern and southern Africa) and Trypanosoma brucei gambiense (the chronic form of central and western Africa). Thus, other parameters, mainly geographical locations of the patients, case histories, and clinical presentations, are used to determine the most-probable subspecies in question. This becomes problematic in areas close to borders between traditional ranges of the two diseases and also where new, nonhistorical foci emerge (5, 13). In Uganda, both T. b. rhodesiense HAT and T. b. gambiense HAT are reported in hitherto well-distinct foci in the eastern and northwestern parts, respectively, although the former continues to spread northwards (13). It is therefore increasingly important to monitor the epidemiology of trypanosomes in patients, the vector, and animal reservoirs, especially in new outbreak areas, in order to institute appropriate control measures.
Molecular tools have greatly facilitated unequivocal identification of human infective trypanosomes to the subspecies level. These are PCR based, targeting the serum resistance-associated (SRA) gene that is specific for T. b. rhodesiense (4, 6, 14) or the T. b. gambiense surface glycoprotein (TgSGP) that is specific for that subspecies (2, 15). The need for specialized equipment and a constant supply of electricity has hindered integration of those techniques into the diagnostic algorithm. Yet the type of infection directs the choice of treatment: for instance, late-stage T. b. rhodesiense infection cannot be treated with eflornithine due to innate resistance (7), but this drug is increasingly used against T. b. gambiense as a result of high numbers of melarsoprol treatment failures reported in some foci. When new HAT foci emerge, as is presently the case in Uganda, it is important to have highly sensitive, field-applicable tests to easily diagnose cases and to identify the infecting subspecies. Novel tests such as the PCR or nucleic acid sequence-based amplification assay (NASBA) coupled to oligochromatography (3, 10) have been devised and are ready for clinical evaluation. These tests amplify 18S ribosomal DNA or RNA, respectively, and as such cannot be used to distinguish between the two HAT types. Recently, loop-mediated isothermal amplification of DNA (LAMP) has been used for detection of members of the Trypanozoon subgenus as well as T. b. rhodesiense (11, 12) subsequent to earlier studies targeting various trypanosomal genes (8, 16). The reportedly high sensitivity, coupled with no need for specialized equipment, increases the likelihood of its adoption in resource-poor health centers where HAT is endemic. In this study, we aimed to evaluate LAMP for detection of trypanosomal DNA from Flinders Technology Associates (FTA) cards spotted with patient blood and to characterize the infecting trypanosomes to the subspecies level.
MATERIALS AND METHODS
Origin of patient blood spotted on FTA cards.
The samples were obtained from the Impamel III program (Improved Application of Melarsoprol) at Lwala Hospital, Kaberamaido District, in southeastern Uganda and from the Kaliua Health Centre, Urambo District, in northwestern Tanzania. Lwala Hospital is located in an area more than 100 km outside the historic T. b. rhodesiense foci (Busoga and Tororo). Kaliua Health Centre, a missionary hospital established in 1997, is located within the major area of northwestern Tanzania where sleeping sickness is endemic. A total of 128 samples (59 from Uganda and 69 from Tanzania) collected over a 2-year period, August 2006 to August 2008, were analyzed in this study.
Other samples included for reference.
Blood from three T. b. gambiense patients from Omugo, northwest Uganda, spotted on FTA cards (200 μl per spot) was also included. In addition, 200 μl (from suspensions of 1,000 trypanosomes/ml) of the following was spotted on FTA cards: cultured T. b. gambiense (Eliane), mouse blood spiked with T. b. brucei (GVR35), and human blood spiked with T. b. rhodesiense (UGA015).
Ethical considerations.
The Impamel III program received ethical clearance from the Ministry of Health, Uganda, the National Institute for Medical Research, Tanzania, and from Switzerland (Ethics Committees of both Cantons of Basel). These clearances refer to the entire Impamel III program, one of whose objectives was to collect blood samples for identification of infecting trypanosome subspecies that is here reported. The samples were collected after written informed consent was obtained from participants in the presence of independent witnesses.
Sample collection and DNA preparation.
Blood samples were collected on FTA cards (Whatman) by making 4 spots using the blood remaining after diagnostic procedures; only 1 such spot per patient was availed for this study. About 200 μl was applied on each spot, and the coded cards were allowed to air dry and then were enclosed in self-sealing bags containing silica and transported to the laboratory for storage at 4°C. They remained in storage for up to 1 year before DNA preparation for the tests. A 2.0-mm disc was punched from a dried blood spot using a Harris micropunch tool (Whatman) and processed with FTA purification reagent by following standard procedures. Briefly, each disc was washed three times (5 min each) with 200 μl of the reagent. It was then rinsed twice with Tris-EDTA (TE) buffer (5 min each) and allowed to air dry. From each card/patient, five such discs were prepared and finally pooled in a single tube per patient after being dried. The DNA was then eluted from the discs by incubating them in 100 μl of a 5% Chelex suspension at 90°C for 30 min (1). After a pulse spin at 13,000 × g, the eluted DNA was pipetted off and used immediately or stored at −20°C for use within 3 days of its preparation. In all subsequent amplifications, 4 μl of the DNA solution was added as the template.
PCR for the SRA and TgSGP genes.
A nested PCR for the SRA gene was carried out as described by Maina et al. (9) with the primers SRA-outer-s and SRA-outer-as and 4 μl of eluted DNA in a 25-μl reaction mixture using the PCR Master Mix (Abgene, United Kingdom). For the second PCR, 3 μl of the first product was included as the template in the reaction mixture with the primers SRA-inner-s and SRA-inner-as. Nested PCR for the T. b. gambiense-specific gene was also performed as described by Maina et al. (9) with the primers TgSGP-outer-s and TbsGP-outer-as as well as TbsGP-s and TgSGP-as. Initial denaturation was at 94°C for 5 min and was followed by 35 cycles of 94°C denaturation, 60°C annealing, and 72°C extension, each for 1 min. Final extension was at 72°C for 5 min. PCR products were loaded on 2% agarose gels stained with ethidium bromide.
LAMP of the SRA gene (SRA-LAMP) and the random insertion mobile element (RIME-LAMP).
For SRA-LAMP, we made use of primers recently described by Njiru et al. (11), namely, SRA-F3, SRA-B3, SRA-FIP, SRA-BIP, SRA-LF, and SRA-LB. The total reaction volume was 25 μl, into which the above-mentioned primers were added to final concentrations of 0.2 μM for F3 and B3, 2 μM for the forward inner primer (FIP) and the backward inner primer (BIP), and 0.8 μM for the forward loop (LF) and the backward loop (LB). In addition, 200 μM deoxynucleoside triphosphates (dNTPs), 0.8 M betaine, and 8 U of Bst polymerase (large fragment; New England Biolabs) were added to the mix containing 1× reaction buffer supplied with the enzyme. To this, 4 μl of DNA eluted from the FTA cards was added to achieve a 25-μl total reaction volume. The reaction mixture was incubated for 1 h at 62°C, followed by inactivation of the Bst polymerase at 80°C for 4 min. Two microliters of a 1/20 dilution of SYBR green in water was then added, and the tube was gently agitated while it was observed for color change.
Similar conditions and reagent concentrations were applied to RIME-LAMP using the primers described by Njiru et al. (12).
Data scoring and analysis.
For all tests performed in this study, each sample was analyzed twice. In the case of discrepancies, the test in question was repeated a third time, the results of which were considered final. Sensitivities for each test were determined as percentages with 95% confidence intervals (CI). Comparison of the sensitivities was by Fisher's exact test. Kappa values and concordance were also determined.
RESULTS
A total of 128 samples (59 from Lwala, Uganda, and 69 from Kaliua, Tanzania) were analyzed. Signals could be obtained from PCR only after nesting with products of the first-round amplification. Even then, TgSGP-PCR was positive only for the laboratory strain (Eliane) and the three confirmed T. b. gambiense patients from Omugo (Fig. 1.) but yielded no signal from any of the DNAs eluted from FTA cards from Lwala and Kaliua. There were three cases of discrepancy: samples 11012 and 12020 were positive by SRA-LAMP in the first run, were negative upon second testing, but turned out positive on the third attempt. On the other hand, sample 12025, which had been negative by SRA-PCR in the first run, was positive in the second and third runs.
FIG. 1.
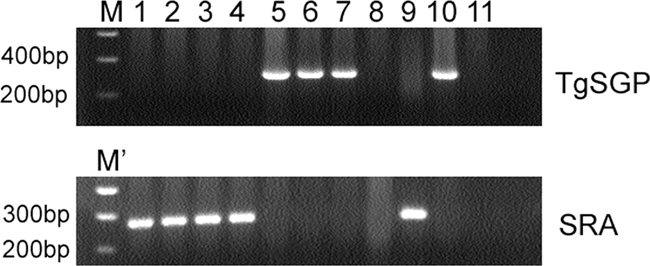
Representative gels to show signals obtained for SRA- and TgSGP-PCRs from corresponding templates. No TgSGP signal was obtained from any of the FTA cards spotted with patient blood from Lwala or Kaliua. Lanes: M, 0.2-kb to 10-kb Smart ladder; M', 100-bp ladder (NEB); 1 to 4, samples from patients at Kaliua (11001, 11006, 11012 and 1106); 5 to 7, samples from T. b. gambiense patients from Omugo (OM187, OM197 and OM203); 8 to 10, laboratory strains GVR35 (T. b. brucei), UGA015 (T. b. rhodesiense), and Eliane (T. b. gambiense), respectively; 11, negative control (where water was added in place of the template).
Overall, SRA-PCR was positive in 101/128 samples (78.9%; 95% CI, 71.1to 85.1%) but was less sensitive for samples from Tanzania (49/69; 71.0%, CI, 59.4 to 80.4%) than for Ugandan samples (52/59 [88.1%]; CI, 77.5 to 94.1%). The difference was significant (P = 0.029) and could not be attributed to the absence of parasites. For all cases, other than one from Uganda, trypanosomes had been previously demonstrated in stained smears during routine diagnosis (Table 1), indicating the presence of potentially detectable DNA within the samples. SRA-LAMP, on the other hand, was positive in 120/128 samples (93.8%; CI, 88.2 to 96.8%). SRA-LAMP was positive for 66/69 samples from Tanzania (95.7%; CI, 88.0 to 98.5%) and 54/59 of the Ugandan samples (91.5%; CI, 81.7 to 96.3%). RIME-LAMP detected the highest number of cases, 122/128, giving a sensitivity of 95.3% (95% CI, 90.2 to 97.8%), missing two samples from Tanzania and four from Uganda. In a comparison of the overall results of RIME-LAMP and SRA-PCR (Fisher's exact test), the former was significantly more sensitive (P = 0.000). The concordance was 83.6%, with a kappa value of 0.31 (95% CI, 0.02 to 0.51), indicating poor agreement (Table 2). Similarly, the difference in sensitivities of SRA-PCR (78.9%) and SRA-LAMP (93.8%) was highly significant (P = 0.001); the concordance was 85.2% and the kappa value was 0.40 (0.20 to 0.60), again indicating poor agreement. On the other hand, the difference between the sensitivities of the SRA-LAMP and RIME-LAMP was insignificant (P = 0.785); the concordance was 98.4%, and the kappa value was 0.85 (0.64 to 1), indicating almost perfect agreement. This means that either LAMP test can be reliably used to detect HAT due to T. b. rhodesiense.
TABLE 1.
Samples for which either the SRA-PCR, SRA-LAMP or RIME-LAMP were negativea
| Sample site or strain | No. of samples positive by indicated test/total no. of samples |
|||
|---|---|---|---|---|
| Stained smear | SRA-PCR | SRA-LAMP | RIME-LAMP | |
| Kaliua | 25/25 | 5/25 | 22/25 | 23/25 |
| Lwala | 9/10 | 3/10 | 5/10 | 6/10 |
| Omugo controls | ND* | 0/3 | 0/3 | 3/3 |
| T. b. rhodesiense lab strain | NA | 1/1 | 1/1 | 1/1 |
| T. b. brucei lab strain | NA | 0/1 | 0/1 | 1/1 |
| T. b. gambiense lab strain | NA | 0/1 | 0/1 | 1/1 |
Trypanosomes were demonstrated by stained smears in all samples except sample 12008 from Lwala, indicating the presence of DNAs that should have been detected by molecular methods. Only 93 samples had positive results for each of the three tests. All patient samples were negative by TgSGP-PCR, except the three from Omugo, northwestern Uganda. NA, not applicable; ND, not done. An asterisk indicates samples that were positive by the hematocrit centrifugation technique described by Woo et al. (17) routinely used at Omugo Health Center.
TABLE 2.
Comparison of the three tests for detection of trypanosomal DNA in patients from Lwala Hospital and Kaliua Health Centre
| Assays compared | Concordance (%) | Kappa value (95% CI) | Level of agreement | Difference in sensitivities (P value) |
|---|---|---|---|---|
| SRA-PCR and SRA-LAMP | 85.20 | 0.40 (0.20-0.60) | Poor | Highly Significant (0.001) |
| SRA-PCR and RIME-LAMP | 83.60 | 0.31 (0.02-0.51) | Poor | Highly Significant (0.000) |
| SRA-LAMP and RIME-LAMP | 98.40 | 0.85 (0.64-1) | Almost perfect | Insignificant (0.785) |
For all four methods used, no amplification was possible in three of the samples from Uganda (Lwala), although trypanosomes had been demonstrated in corresponding stained smears. Amplification results were consistent with those expected from the control/reference samples: SRA-PCR and both LAMPs gave positive results for the laboratory T. b. rhodesiense strain (UGA015), while RIME-LAMP but not SRA-PCR or SRA-LAMP gave positive results for the T. b. brucei strain (GVR35), Eliane (a T. b. gambiense laboratory strain), and the three T. b. gambiense cases from Omugo (Table 1; Fig. 2 shows representative LAMP reactions).
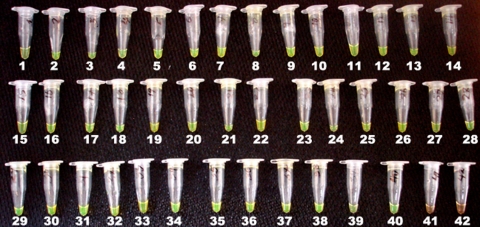
FIG. 2.
Representative RIME-LAMP reactions from selected patient samples. Templates in tubes 1 to 18 are samples from Tanzania, while tubes 19 to 38 contain samples from Uganda. Tube 39 contains the laboratory T. b. rhodesiense strain (UGA015), tube 40 contains the laboratory T. b. brucei strain (GVR35), and tubes 41 and 42 are negative controls in which 4 μl water was added as the template.
DISCUSSION
Differential diagnosis of infections with Trypanosoma brucei rhodesiense (the acute form in eastern and southern Africa) and T. b. gambiense (the chronic form in central and western Africa) has never been so important, since previously each was confined to geographically defined and distinct foci. The emergence of nonhistorical foci and the ever-reducing gap between the traditional ranges of the two forms of HAT renders subspecies identification an important step in control, since the two diseases have different diagnostic and treatment algorithms. Lwala Hospital caters to the new outbreak areas stretching as far north as Lira District, where a T. b. rhodesiense was isolated within 150 km of known T. b. gambiense foci (13). In this study, we have not encountered any T. b. gambiense organisms in samples collected at the two centers up to August 2008, when the last samples here tested were collected, as determined by subspecies-specific PCRs.
In this study, nested PCR was necessary to amplify the single-copy gene targets of the subspecific PCRs, rendering it more time-consuming (at least 5 h to perform both PCRs), compared to only 1 h required for the LAMP method. The overall sensitivity of SRA-PCR (78.9%) was unexpectedly low, given that trypanosomes had been demonstrated in most samples at diagnosis. This low sensitivity could have resulted from a decrease in DNA quality during storage, as up to 1 year elapsed between sampling and execution of the tests. The observed low sensitivity could also have resulted from the presence of inhibitory molecules of host origin in the DNA eluted from the FTA cards. Due to the need for sophisticated laboratories, it is unlikely that PCR-based methods will in the near future be integrated into routine point-of-care diagnosis, although they remain invaluable for epidemiological studies.
The higher sensitivity of LAMP observed in this study (compared to PCR-based methods) is worthy of note, especially given that it was obtained with samples whose collection does not require cold storage. Furthermore, the high level of concordance (98.4%) and agreement (kappa value, 0.85; 95% CI, 0.64 to 1) between SRA-LAMP and RIME-LAMP provide the possibility that either test could be used to reliably diagnose HAT due to T. b. rhodesiense. The major advantages of LAMP were the need for just a single reaction to obtain results within 1 h of initiation by adding SYBR green (11, 12). LAMP therefore has great potential application in resource-poor settings with basic facilities, since it can be done in a water bath, eliminating the need for a thermocycler. Given the ease of its execution, LAMP could be used to monitor for emerging HAT foci or to test blood from travelers returning from countries where the disease is endemic. What would be required is the reduction of the numerous manipulations involved in making reaction mixtures by the provision of kits in which all reactants (preferably lyophilized) are included. In this way, personnel at the diagnostic center would need only to add the reaction buffer and patient sample before starting the reaction, thereby minimizing the potential for contamination. Nevertheless, our results warrant larger case-control studies to generate in particular more data on sensitivity and specificity of LAMP in areas where the disease is endemic.
It must be borne in mind, however, that even in the presence of molecular diagnostics of undeniable sensitivity, the definition of a HAT case will continue to be based on the demonstration of trypanosomes in blood, lymph node aspirates, or cerebrospinal fluids. The molecular methods will nevertheless go a long way in detecting aparasitemic cases on whom extra effort should be made by health personnel to search for trypanosomes before treatment recommendations can be made.
Acknowledgments
We thank Emma P. Hhary, and Betty Akello as well as the entire teams at Lwala Hospital and Kaliua Health Centre for their active participation during sampling. We also thank Abbas Kakembo at the Uganda Ministry of Health, Allan Mpairwe, and Andrew Edielu of Lwala Hospital, as well as Lucas Matemba, for their untiring support throughout the project.
Funding was provided by the Swiss Agency for Development and Cooperation (SDC) grant number 7F-01977.02 (phase extension), the International Consortium for Parasitic Drug Discovery (CPDD) led by the University of North Carolina, Chapel Hill, NC, which is supported by the Bill & Melinda Gates Foundation, the Swiss Tropical and Public Health Institute, and the Makerere University Faculty of Veterinary Medicine.
Footnotes
Published ahead of print on 21 April 2010.
REFERENCES
- 1.Becker, S., J. R. Franco, P. P. Simarro, A. Stich, P. M. Abel, and D. Sterverding. 2004. Real-time PCR for detection of Trypanosoma brucei in human blood samples. Diagn. Microbiol. Infect. Dis. 50:193-199. [DOI] [PubMed] [Google Scholar]
- 2.Berberof, M., D. Perez-Morga, and E. Pays. 2001. A receptor-like flagellar pocket glycoprotein specific to Trypanosoma brucei gambiense. Mol. Biochem. Parasitol. 113:127-138. [DOI] [PubMed] [Google Scholar]
- 3.Deborggraeve, S., F. Claes, T. Laurent, P. Mertens, T. Leclipteux, J. C. Dujardin, P. Herdewijn, and P. Buscher. 2006. Molecular dipstick test for diagnosis of sleeping sickness. J. Clin. Microbiol. 44:2884-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Greef, C., E. Chimfwembe, J. Kihang'a Wabacha, E. Bajyana Songa, and R. Hamers. 1992. Only the serum resistant bloodstream forms of Trypanosoma brucei rhodesiense express the serum resistance associated (SRA) protein. Ann. Soc. Belg. Med. Trop. 72:13-21. [PubMed] [Google Scholar]
- 5.Fevre, E. M., P. G. Coleman, M. Odiit, J. Magona, S. C. Welburn, and M. E. J. Woolhouse. 2001. The origins of a new sleeping sickness outbreak (caused by Trypanosoma brucei infection) in eastern Uganda. Lancet 358:625-628. [DOI] [PubMed] [Google Scholar]
- 6.Gibson, W., T. Backhouse, and A. Griffiths. 2002. The human serum resistance associated gene is ubiquitous and conserved in Trypanosoma brucei rhodesiense throughout East Africa. Infect. Genet. Evol. 1:207-214. [DOI] [PubMed] [Google Scholar]
- 7.Iten, M., E. Matovu, R. Brun, and R. Kaminsky. 1995. Innate lack of susceptibility of Ugandan Trypanosoma brucei rhodesiense to dl-α-difluoromethylornithine (DFMO). Trop. Med. Parasitol. 46:190-194. [PubMed] [Google Scholar]
- 8.Kuboki, N., N. Inoue, T. Sakurai, F. Cello, D. J. Grab, H. Suzuki, C. Sugimoto, and I. Igarashi. 2003. Loop-mediated isothermal amplification for detection of African trypanosomes. J. Clin. Microbiol. 41:5517-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maina, N. W. N., M. Oberle, C. Otieno, C. Kunz, P. Mäser, J. M. Ndungu, and R. Brun. 2007. Isolation and propagation of Trypanosoma brucei gambiense from sleeping sickness patients in south Sudan. Trans. Roy. Soc. Trop. Med. Hyg. 101:540-546. [DOI] [PubMed] [Google Scholar]
- 10.Mugasa, C. M., T. Laurent, G. J. Schoone, P. A. Kager, G. W. Lubega, and H. D. F. H. Schallig. 2009. Nucleic acid sequence-based amplification with oligochromatography (NASBA-OC) for the detection of Trypanosoma brucei in clinical samples. J. Clin. Microbiol. 47:630-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Njiru, Z. K., A. S. J. Mikosza, T. Armstrong, J. C. Enyaru, J. M. Ndung'u, and A. R. C. Thompson. 2008. Loop-mediated isothermal amplification (LAMP) method for rapid detection of Trypanosoma brucei rhodesiense. PLoS Negl. Trop. Dis. 2:e147. doi: 10.1371/journal.pntd.0000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Njiru, Z. K., A. S. J. Mikosza, E. Matovu, J. C. K. Enyaru, J. O. Ouma, S. M. Kibona, R. C. A. Thompson, and J. M. Ndung'u. 2008. African trypanosomiasis: sensitive and rapid detection of the sub-genus Trypanozoon by loop-mediated isothermal amplification (LAMP) of parasite DNA. Int. J. Parasitol. 38:589-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Picozzi, K., E. M. Fevre, M. Odiit, M. Carrington, M. C. Eisler, I. Maudlin, and S. C. Welburn. 2005. Sleeping sickness in Uganda: a thin line between two fatal diseases. Br. Med. J. 331:1238-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radwanska, M., M. Chamekh, L. Vanhamme, F. Claes, S. Magez, E. Magnus, P. de Baetselier, P. Buscher, and E. Pays. 2002. The serum resistance-associated gene as a diagnostic tool for the detection of Trypanosoma brucei rhodesiense. Am. J. Trop. Med. Hyg. 67:684-690. [DOI] [PubMed] [Google Scholar]
- 15.Radwanska, M., F. Claes, S. Magez, E. Magnus, D. Perez-Morga, E. Pays, and P. Buscher. 2002. Novel primer sequences for polymerase chain reaction-based detection of Trypanosoma brucei gambiense. Am. J. Trop. Med. Hyg. 67:289-295. [DOI] [PubMed] [Google Scholar]
- 16.Thekisoe, O. M. M., N. Kuboki, A. Nambota, K. Fujisaki, C. Sugimoto, I. Igarashi, J. Yasuda, and N. Inoue. 2007. Species-specific loop-mediated isothermal amplification (LAMP) for diagnosis of trypanosomosis. Acta Trop. 102:182-189. [DOI] [PubMed] [Google Scholar]
- 17.Woo, P. T. K. 1970. The haematocrit centrifugation technique for the diagnosis of African trypanosomiasis. Acta Trop. 27:384-386. [PubMed] [Google Scholar]