Abstract
In genetic analysis of bovine Staphylococcus aureus isolates that are recognized as an important pathogenic bacterium in bovine mastitis, multilocus sequence typing (MLST) showed strong correlation to the results of pulsed-field gel electrophoresis, coa PCR-restriction fragment length polymorphism (RFLP), spa typing, and the coagulase serotyping method. According to MLST results, strains derived from sequence type 97 (ST97) and ST705 were suggested as not only dominant bovine S. aureus lineages in Japan but also pandemic bovine S. aureus lineages. Although both lineages seem to be distantly related to each other by phylogenetic analysis, both had common characteristics, i.e., lukM/lukF′-PV and coagulase serotype VI. These characteristics were very rare among minor bovine strains and human strains and may contribute to the host specificity of these lineages. Four methicillin-resistant S. aureus (MRSA) isolates were first confirmed from bovine milk in Japan; these isolates showed geno- and serotypes that were identical or similar to those of human MRSA isolates in Japan (ST5, staphylococcal cassette chromosome mec type II [SCCmec II], Spa type t002 or t375, and coagulase serotype II, and ST89, SCCmec IIIa, Spa type t5266, and coagulase serotype I). ST5 and ST89 are uncommon among bovine isolates in the world, whereas these STs are common among human MRSA isolates in Japan.
Staphylococcus aureus is a major causal bacterium in contagious bovine mastitis. Although S. aureus has been isolated from heifer body sites and in the dairy farm environment, the lactating mammary gland is the primary reservoir of S. aureus involved in bovine mastitis (35, 36). Analyzing the genetic variation among S. aureus isolates from bovine milk is essential in bovine mastitis studies such as the identification of protective antigens for vaccine development and elucidation of the mechanism of pathogenesis. Furthermore, investigation of the relationship with isolates from environmental and other origins in dairy farms will lead to the identification of the sources of contagion on dairy farms and ways to prevent contagion from occurring.
There have been reports of the isolation of methicillin-resistant S. aureus (MRSA) from bovine milk for a long time (7). The emergence of MRSA infection on dairy farms is of great concern for animal and public hygiene. MRSA-contaminated livestock products, including bovine milk, may become causal agents for human MRSA infection (27). In addition, the spread of MRSA in herds will cause a delay in the treatment of not only mastitis but also other bovine diseases, because most MRSA strains in Japan exhibit multidrug resistance (37). Therefore, screening for MRSA from bovine milk and genetic analysis of isolates are indispensable, and these analyses will lead to elucidation of the mechanisms of MRSA emergence or invasion in dairy farms.
Many different typing methods have been developed for epidemiological studies or the analysis of genetic characteristics and relationships, but each method has drawbacks and advantages, so it is important that an optimal typing method be selected depending on the purpose. The correlation among different genotyping results, the relations with the phenotype, and the discriminatory power become reference points to select a genotyping method. Pulsed-field gel electrophoresis (PFGE), which shows high discriminatory power, is considered “the gold standard” for the typing of S. aureus isolates, but it is difficult to establish a precise database with PFGE (3). However, multilocus sequence typing (MLST) and X region of protein A gene (spa) typing are established and have websites with very useful resources (www.mlst.net and www.spaserver.ridom.de) for sharing and analyzing substantial databases of genotypes (9, 14). Other than these methods, multiple-locus variable-number tandem-repeat analysis (MLVA), which is based on the number of direct repeats of staphylococcal interspersed repeat units (SIRUs), has demonstrated high discriminatory power for human S. aureus isolates (19). Furthermore, staphylococcal cassette chromosome mec (SCCmec) typing is indispensable for the full characterization of MRSA (31).
This study analyzed the genetic variation among S. aureus isolates from bovine milk in Japan using different typing methods. Consequently, the genetic characteristics of bovine milk isolates were evaluated along with their relationship with foreign bovine milk isolates and human MRSA isolates. Furthermore, the discriminatory powers of the different typing methods for bovine S. aureus isolates were evaluated and compared.
MATERIALS AND METHODS
S. aureus isolates.
A total of 363 S. aureus isolates were collected from bovine milk from 260 Japanese dairy farms located in 21 prefectures, and 359 methicillin-susceptible S. aureus (MSSA) isolates and 4 MRSA isolates were obtained between May 1998 and May 2005. Furthermore, 9 previously reported human MRSA isolates (11) were added to evaluate the relationship between bovine isolates and human MRSA isolates. Species identification was demonstrated by detection of a species-specific 442-bp fragment using Voges-Proskauer test-positive and coagulase test-positive staphylococcal isolates (28). The sequence of the 16S rRNA gene was confirmed for isolates that could not be identified (8). MRSA isolates were confirmed in accordance with the Clinical and Laboratory Standards Institute method (6), and the possession of the penicillin-binding protein 2a gene (mecA) was confirmed by PCR with the primers described previously (21).
Genotyping and serotyping methods.
All of the isolates were analyzed by PFGE, which was performed as described by Ichiyama et al. (17). Chromosomal DNA included in the thinly sliced agarose gel was digested with 10 U of SmaI or XhoI for 18 h and then electrophoresed through a 1% pulsed-field certified agarose (Bio-Rad) gel in TBE buffer (44.5 mM Tris, 44.5 mM boric acid, 1 mM EDTA [pH 8.0]) at 10°C using a CHEF-DRII system (Bio-Rad). The conditions for electrophoresis for SmaI digestions were 5.7 V/cm for 25 h, with pulse times ranging from 5 to 40 s, and for XhoI digestions were 6.0 V/cm for 24 h, with pulse times ranging from 0.1 to 8 s. Low Range PFG marker (New England Biolabs) was used as the size standard. The PFGE pulsotypes were analyzed by visual inspection using BioNumerics software (version 5.10) according to the criteria described previously (30, 38). Briefly, a dendrogram was created using the Dice coefficient with optimization and a position tolerance setting of 1% based on the banding patterns of each pulsotype, and the pulsotypes that showed more than 80% similarity in this dendrogram were defined as the same cluster (30). MLST was performed with the primers described previously (9), and the products were purified using a QIAquick PCR purification kit (Qiagen) and sequenced on a 3130 genetic analyzer (Applied Biosystems) using a BigDye Terminator (version 3.1) ready reaction cycle sequencing kit (Applied Biosystems). Allele number and sequence type (ST) were determined using the MLST website (www.mlst.net). The founder and clonal complex (CC) of each ST were determined by using the enhanced version of Based Upon Related Sequence Types (eBURST) (10). A phylogenetic tree was constructed using the ClustalW website (http://clustalw.ddbj.nig.ac.jp/top-e.html), based on the 3,198 bp of the seven target loci sequences, which were concatenated in the order arcC, aroE, glpF, gmk, pta, tpi, and yqiL. spa typing was performed with the primers described previously (14), and numeric spa repeats and Spa type codes were determined using the Ridom SpaServer website (www.spaserver.ridom.de). MLVA typing was performed with the primers (13) that amplified the 6 SIRU loci (SIRU01, -05, -07, -13, -15, and -21). PCR-based restriction fragment length polymorphism (RFLP) analysis of the coagulase gene (coa) was performed with the primers described previously (16), and representative PCR products of each coa PCR-RFLP type were sequenced for determination of the precise restriction fragment lengths. The SCCmec types were determined by multiplex PCR with the primers described previously (29, 31, 41). The coagulase serotype was confirmed using a Seiken coagulase typing kit (Denka Seiken, Japan).
Detection of staphylococcal exotoxin genes.
The detection of gamma hemolysin, LukE/LukD, LukM/LukF′-PV, and the Panton-Valentine leukocidin genes (hlg, lukE/lukD, lukM/lukF′-PV, and pvl) was performed with the primers described previously (20). The following primers were used for the detection of the streptolysin-associated protein SagD-like protein gene (sagD) and the SagB-like protein gene (sagB): SagD-F (5′TTG AAT AAT CAA AAA AGT AAT3′), SagD-R (5′TTA AAG GTT ATC ATT TTC TAA3′), SagB-F (5′GTG GCT CAA AAG GAC ATA AAC3′), and SagB-R (5′CTA TTC CTT CCC ACA TAT A3′). The sizes of the PCR products were 564 bp and 309 bp, respectively.
Evaluation of the discriminatory power and correlation among different typing methods.
The discriminatory power of the typing methods and correlation among typing methods were calculated by using EpiCompare version 1.0 (Ridom GmbH, Wurzburg, Germany), as was the determination of Adjusted Rand's index and Wallace's coefficients (19).
RESULTS
Genetic and serological analysis of dominant S. aureus strains.
The DNA of the 372 S. aureus isolates after digestion with SmaI showed 144 pulsotypes in 23 clusters (A to W). Furthermore, the tested isolates were segmented into 191 pulsotypes by adding PFGE results following XhoI digestion. Eighty-four isolates (22.6%) from 16 prefectures showed 38 pulsotypes in cluster N, and 178 isolates (47.8%) from 21 prefectures showed 58 pulsotypes in cluster O (Table 1). A total of 191 representative S. aureus isolates from individual pulsotypes were analyzed by further typing.
TABLE 1.
Genetic and serological characteristics of Staphylococcus aureus isolates from bovine milk and humansa
| MLST |
Spa type | Coagulase serotype | coa PCR-RFLP genotype | PFGE pulsotype(s) | Toxin gene profile | No. of RUsb in SIRU locus: |
SCCmec type | No. of locations where foundc | Prefecture(s) where found | Source(s) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | ST | 1 | 5 | 7 | 13 | 15 | 21 | |||||||||
| 5 | 5 | t001 | II | 676H | B6 | hlg, lukE/lukD | 2 | 30< | 1 | 3 | 999 | 9 | II | 1 | Tochigi | Human wound |
| 5 | 5 | t002 | II | 676B | D | hlg, lukE/lukD | 2 | 3 | 1 | 3 | 1 | 9 | II | 1 | Unknown | Bovine milk |
| 5 | 5 | t179 | II | 676B | B1, B2, B3 | hlg, lukE/lukD | 2 | 3 | 1 | 3 | 1 | 9 | 3 | Fukuoka | Bovine milk | |
| 5 | 5 | t375 | II | 676B | E1, E2 | hlg, lukE/lukD | 2 | 2 | 1 | 3 | 999 | 9 | II | 1 | Unknown | Bovine milk |
| 5 | 5 | t375 | II | 676B | E3 | hlg, lukE/lukD | 2 | 2 | 1 | 3 | 999 | 9 | II | 1 | Ibaraki | Human pus |
| 5 | 5 | t5259 | II | 676B | B4, B5 | hlg, lukE/lukD | 2 | 3 | 1 | 3 | 1 | 6 | 1 | Fukuoka | Bovine milk | |
| 6 | 1362 | t701 | IV | 676D | Q11 | hlg, lukE/lukD | 2 | 7 | 999 | 4 | 1 | 9 | 1 | Ishikawa | Bovine milk | |
| 6 | 6 | t2360 | IV | 676D | Q10 | hlg, lukE/lukD | 2 | 7 | 999 | 4 | 1 | 10 | 1 | Tottori | Bovine milk | |
| 7 | 789 | t091 | III | 757B | H1, H2 | hlg, lukE/lukD, sagB, sagD | 999 | 999 | 3 | 2 | 3 | 9 | 2 | Iwate, Saga | Bovine milk | |
| 8 | 8 | t008 | III | 595B | G2 | hlg, lukE/lukD | 3 | 3 | 1 | 0 | 1 | 9 | 1 | Iwate | Bovine milk | |
| 8 | 8 | t008 | III | 595C | G6 | hlg, lukE/lukD | 3 | 3 | 2 | 0 | 1 | 9 | 1 | Hokkaido | Bovine milk | |
| 8 | 8 | t008 | III | 352A | G3 | hlg, lukE/lukD | 3 | 3 | 3 | 0 | 1 | 9 | 1 | Ishikawa | Bovine milk | |
| 8 | 8 | t4133 | III | 514A | G1 | hlg, lukE/lukD | 3 | 3 | 3 | 0 | 1 | 9 | 1 | Gunma | Bovine milk | |
| 8 | 72 | t4359 | V | 676F | M | hlg, lukE/lukD | 3 | 9 | 999 | 3 | 7 | 4 | 1 | Ishikawa | Bovine milk | |
| 8 | 630 | t377 | III | 595B | G4, G5 | hlg, lukE/lukD | 4 | 6 | 2 | 0 | 1 | 7 | 1 | Iwate | Bovine milk | |
| 12 | 12 | t160 | VII | 676D | R5 | hlg, lukE/lukD | 1 | 7 | 1 | 3 | 999 | 6 | 1 | Chiba | Bovine milk | |
| 12 | 12 | t160 | VII | 676D | R2 | hlg, lukE/lukD | 1 | 7 | 1 | 3 | 1 | 6 | 1 | Saga | Bovine milk | |
| 12 | 12 | t3418 | VII | 676D | R3 | hlg, lukE/lukD | 1 | 7 | 1 | 4 | 1 | 5 | 1 | Tochigi | Bovine milk | |
| 12 | 1369 | t160 | VII | 676D | R4 | hlg, lukE/lukD | 1 | 7 | 1 | 2 | 999 | 6 | 1 | Saitama | Bovine milk | |
| 12 | 1369 | t160 | VII | 676D | R1 | hlg, lukE/lukD | 1 | 7 | 1 | 2 | 1 | 6 | 1 | Saitama | Bovine milk | |
| 15 | 1 | t1775 | VII | 514B | Q7 | hlg, lukE/lukD, pvl | 1 | 999 | 3 | 1 | 2 | 9 | 1 | Tochigi | Bovine milk | |
| 15 | 81 | t127 | VII | 514B | Q3 | hlg, lukE/lukD | 1 | 999 | 2 | 0 | 2 | 6 | 1 | Ishikawa | Bovine milk | |
| 15 | 81 | t127 | VII | 514B | Q8 | hlg, lukE/lukD | 1 | 999 | 3 | 1 | 1 | 6 | 1 | Iwate | Bovine milk | |
| 15 | 81 | t127 | VII | 595F | Q6 | hlg, lukE/lukD, pvl | 1 | 999 | 2 | 0 | 1 | 6 | 1 | Hokkaido | Bovine milk | |
| 15 | 81 | t127 | VII | 595F | Q1, Q5 | hlg, lukE/lukD | 1 | 999 | 2 | 1 | 1 | 6 | 2 | Iwate, Gunma | Bovine milk | |
| 15 | 81 | t127 | VII | 595F | Q2 | hlg, lukE/lukD | 1 | 999 | 2 | 1 | 2 | 6 | 1 | Kochi | Bovine milk | |
| 15 | 81 | t127 | VII | 595F | Q4 | hlg, lukE/lukD | 1 | 999 | 3 | 1 | 1 | 6 | 1 | Fukuoka | Bovine milk | |
| 15 | 81 | t127 | VII | 595F | Q9 | hlg, lukE/lukD | 1 | 999 | 3 | 1 | 2 | 6 | 1 | Nara | Bovine milk | |
| 15 | 188 | t189 | V | 676A | K6 | hlg, lukE/lukD | 1 | 999 | 2 | 1 | 4 | 5 | 1 | Tochigi | Bovine milk | |
| 15 | 188 | t189 | V | 676E | K11 | hlg, lukE/lukD | 1 | 999 | 1 | 2 | 4 | 5 | 1 | Hokkaido | Bovine milk | |
| 15 | 188 | t189 | V | 676E | K1, K2, K4, K5, K7, K8, K9, K10, K12, K13 | hlg, lukE/lukD | 1 | 999 | 3 | 2 | 4 | 5 | 8 | Hokkaido, Iwate, Saitama, Shizuoka, Gifu, Fukuoka | Bovine milk | |
| 15 | 188 | t1858 | V | 676E | K3 | hlg, lukE/lukD | 1 | 999 | 3 | 1 | 4 | 6 | 1 | Tottori | Bovine milk | |
| 20 | 20 | t164 | VIII | 595E | U1, U4, U7 | hlg, lukE/lukD | 3 | 10 | 2 | 1 | 4 | 7 | 3 | Iwate | Bovine milk | |
| 20 | 20 | t164 | VIII | 595E | U11 | hlg | 3 | 10 | 2 | 1 | 4 | 7 | 1 | Hiroshima | Bovine milk | |
| 20 | 20 | t164 | VIII | 595E | U5 | hlg, lukE/lukD | 3 | 11 | 2 | 0 | 4 | 7 | 1 | Gunma | Bovine milk | |
| 20 | 20 | t164 | VIII | 595E | U10 | hlg | 3 | 11 | 2 | 1 | 4 | 7 | 1 | Tokyo | Bovine milk | |
| 20 | 20 | t693 | VIII | 595E | U9 | hlg, lukE/lukD | 3 | 10 | 2 | 1 | 4 | 0 | 1 | Tokyo | Bovine milk | |
| 20 | 20 | t2109 | VIII | 595E | U15 | hlg, lukE/lukD | 3 | 10 | 2 | 1 | 4 | 3 | 1 | Ishikawa | Bovine milk | |
| 20 | 20 | t3277 | VIII | 595E | U2, U18 | hlg, lukE/lukD | 3 | 10 | 2 | 1 | 4 | 6 | 2 | Hokkaido, Ishikawa | Bovine milk | |
| 20 | 20 | t3332 | VIII | 595E | U3 | hlg, lukE/lukD | 3 | 10 | 2 | 1 | 4 | 5 | 1 | Hokkaido | Bovine milk | |
| 20 | 20 | t3929 | VIII | 514B | U13, U14 | hlg, lukE/lukD | 3 | 8 | 2 | 1 | 4 | 3 | 1 | Hyogo | Bovine milk | |
| 20 | 20 | t4542 | VIII | 595E | U8 | hlg, lukE/lukD | 3 | 10 | 2 | 1 | 4 | 3 | 1 | Saitama | Bovine milk | |
| 20 | 20 | t5267 | VIII | 595E | U6 | hlg, lukE/lukD | 3 | 10 | 2 | 1 | 4 | 10 | 1 | Niigata | Bovine milk | |
| 20 | 20 | t5412 | VIII | 595E | U12 | hlg, lukE/lukD | 3 | 10 | 2 | 1 | 4 | 11 | 1 | Hokkaido | Bovine milk | |
| 20 | 1368 | t3277 | VIII | 595E | U17 | hlg, lukE/lukD | 3 | 10 | 2 | 1 | 4 | 6 | 1 | Hyogo | Bovine milk | |
| 20 | 1370 | t881 | VIII | 595E | U16 | hlg, lukE/lukD | 3 | 10 | 2 | 0 | 4 | 6 | 1 | Ishikawa | Bovine milk | |
| 25 | 25 | t078 | II | 595A | V5 | hlg, lukE/lukD | 3 | 5 | 3 | 2 | 4 | 8 | 1 | Hokkaido | Bovine milk | |
| 25 | 25 | t258 | II | 514B | V4 | hlg, lukE/lukD | 3 | 5 | 3 | 2 | 3 | 9 | 1 | Fukuoka | Bovine milk | |
| 25 | 26 | t287 | II | 595A | V1, V2 | hlg, lukE/lukD | 3 | 5 | 2 | 2 | 4 | 2 | 1 | Tokyo | Bovine milk | |
| 25 | 1372 | t258 | II | 514B | V3 | hlg, lukE/lukD | 3 | 3 | 3 | 2 | 3 | 9 | 1 | Gunma | Bovine milk | |
| 30 | 243 | t021 | IV | 676C | J | hlg, pvl | 2 | 3 | 2 | 2 | 2 | 8 | 1 | Hokkaido | Bovine milk | |
| 45 | 508 | t050 | VII | 676G | T1, T3 | hlg | 999 | 999 | 2 | 0 | 1 | 9 | 2 | Gunma, Fukuoka | Bovine milk | |
| 45 | 508 | t362 | VII | 676G | T4 | hlg | 999 | 999 | 2 | 0 | 1 | 1 | 1 | Kochi | Bovine milk | |
| 45 | 508 | t630 | VII | 676G | T2 | hlg | 999 | 999 | 2 | 0 | 1 | 8 | 1 | Fukuoka | Bovine milk | |
| 59 | 59 | t216 | VII | 757E | S | hlg | 2 | 999 | 4 | 1 | 4 | 7 | 1 | Iwate | Bovine milk | |
| 88 | 88 | t1028 | III | 757A | F3, F4 | hlg, lukE/lukD | 2 | 999 | 3 | 2 | 0 | 3 | 1 | Shizuoka | Bovine milk | |
| 88 | 88 | t1028 | III | 757A | F5 | hlg, lukE/lukD | 2 | 999 | 3 | 3 | 0 | 3 | 1 | Shizuoka | Bovine milk | |
| 88 | 88 | t5264 | III | 757A | F1 | hlg, lukE/lukD | 1 | 999 | 3 | 2 | 2 | 7 | 1 | Ishikawa | Bovine milk | |
| 88 | 1360 | t729 | III | 757A | F2 | hlg, lukE/lukD | 2 | 999 | 3 | 1 | 2 | 9 | 1 | Hokkaido | Bovine milk | |
| 97 | 97 | t224 | VI | 838A | O43 | hlg, lukE/lukD, lukM/lukF′-PV | 3 | 999 | 4 | 3 | 2 | 7 | 1 | Hyogo | Bovine milk | |
| 97 | 97 | t224 | VI | 838A | O56 | hlg, lukE/lukD | 3 | 999 | 4 | 3 | 2 | 7 | 1 | Hyogo | Bovine milk | |
| 97 | 97 | t267 | VI | 838A | O58 | hlg, lukE/lukD | 4 | 999 | 1 | 3 | 2 | 9 | 1 | Hyogo | Bovine milk | |
| 97 | 97 | t267 | VI | 838A | O38 | hlg, lukE/lukD | 4 | 999 | 2 | 2 | 2 | 9 | 1 | Okinawa | Bovine milk | |
| 97 | 97 | t359 | VI | 838A | O57 | hlg, lukE/lukD | 4 | 999 | 1 | 3 | 2 | 8 | 1 | Hyogo | Bovine milk | |
| 97 | 97 | t1109 | VI | 838A | O47, O48 | hlg, lukE/lukD | 4 | 999 | 3 | 3 | 2 | 2 | 1 | Saitama | Bovine milk | |
| 97 | 97 | t1234 | VI | 838A | O42 | hlg, lukE/lukD | 4 | 999 | 2 | 3 | 2 | 8 | 1 | Gunma | Bovine milk | |
| 97 | 124 | t224 | VI | 838A | O46 | hlg, lukE/lukD | 4 | 999 | 2 | 5 | 1 | 7 | 1 | Hyogo | Bovine milk | |
| 97 | 124 | t189 | VI | 838A | L | hlg, lukE/lukD | 4 | 999 | 3 | 5 | 999 | 5 | 1 | Tochigi | Bovine milk | |
| 97 | 124 | t224 | VI | 838A | O55 | hlg, lukE/lukD | 3 | 999 | 3 | 4 | 1 | 7 | 1 | Gunma | Bovine milk | |
| 97 | 124 | t224 | VI | 838A | O53 | hlg, lukE/lukD | 4 | 999 | 1 | 3 | 1 | 7 | 1 | Nara | Bovine milk | |
| 97 | 124 | t458 | VI | 838A | O50 | hlg, lukE/lukD | 4 | 999 | 3 | 4 | 999 | 1 | 1 | Nara | Bovine milk | |
| 97 | 124 | t458 | VI | 838A | O51, O52 | hlg, lukE/lukD | 4 | 999 | 3 | 4 | 1 | 1 | 1 | Niigata | Bovine milk | |
| 97 | 124 | t2453 | VI | 838A | P | hlg, lukE/lukD | 4 | 999 | 2 | 4 | 2 | 2 | 1 | Tochigi | Bovine milk | |
| 97 | 124 | t5265 | VI | 838B | O49 | hlg, lukE/lukD | 4 | 999 | 3 | 2 | 2 | 4 | 1 | Tochigi | Bovine milk | |
| 97 | 352 | t203 | VI | 595E | O23 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 4 | 3 | 2 | 7 | 1 | Yamagata | Bovine milk | |
| 97 | 352 | t267 | VI | 352B | O22 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 4 | 3 | 2 | 9 | 1 | Hiroshima | Bovine milk | |
| 97 | 352 | t267 | VI | 595E | O6 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 4 | 2 | 2 | 9 | 1 | Hokkaido | Bovine milk | |
| 97 | 352 | t267 | VI | 595E | O1, O4, O10, O11, O16, O17, O18, O32 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 4 | 3 | 2 | 9 | 6 | Hokkaido, Tochigi, Chiba, Kyoto, Hyogo | Bovine milk | |
| 97 | 352 | t267 | VI | 757C | O7 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 4 | 2 | 2 | 9 | 1 | Ishikawa | Bovine milk | |
| 97 | 352 | t267 | VI | 757C | O21 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 4 | 4 | 2 | 9 | 1 | Hyogo | Bovine milk | |
| 97 | 352 | t267 | VI | 838A | O45 | hlg, lukE/lukD, lukM/lukF′-PV | 999 | 999 | 4 | 2 | 2 | 9 | 1 | Tottori | Bovine milk | |
| 97 | 352 | t267 | VI | 838A | O41 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 2 | 2 | 2 | 9 | 1 | Okinawa | Bovine milk | |
| 97 | 352 | t267 | VI | 838A | O3 | hlg, lukE/lukD | 4 | 999 | 3 | 3 | 1 | 9 | 1 | Hyogo | Bovine milk | |
| 97 | 352 | t267 | VI | 838A | O36, O39, O40, O44 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 4 | 2 | 2 | 9 | 3 | Fukuoka, Okinawa | Bovine milk | |
| 97 | 352 | t267 | VI | 838A | O2 | hlg, lukE/lukD | 4 | 999 | 4 | 3 | 2 | 9 | 1 | Hokkaido | Bovine milk | |
| 97 | 352 | t267 | VI | 838A | O9, O12, O13, O31, O34, O35 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 4 | 3 | 2 | 9 | 5 | Tokyo, Hyogo, Hiroshima, Kochi | Bovine milk | |
| 97 | 352 | t359 | VI | 838A | O24 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 3 | 2 | 2 | 8 | 1 | Hokkaido | Bovine milk | |
| 97 | 352 | t359 | VI | 838A | O5, O20, O30 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 4 | 3 | 2 | 8 | 3 | Tochigi, Hyogo, Kochi | Bovine milk | |
| 97 | 352 | t521 | VI | 595E | O19 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 4 | 3 | 2 | 10 | 1 | Hyogo | Bovine milk | |
| 97 | 352 | t1201 | VI | 838A | O15 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 2 | 2 | 2 | 5 | 1 | Hyogo | Bovine milk | |
| 97 | 352 | t1201 | VI | 838A | O28, O29 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 3 | 2 | 2 | 5 | 2 | Tochigi, Hyogo | Bovine milk | |
| 97 | 352 | t1201 | VI | 676F | O26 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 3 | 2 | 2 | 5 | 1 | Niigata | Bovine milk | |
| 97 | 352 | t1201 | VI | 757D | O27 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 3 | 2 | 2 | 5 | 1 | Ishikawa | Bovine milk | |
| 97 | 352 | t2844 | VI | 838A | O25 | hlg, lukE/lukD, lukM/lukF′-PV | 3 | 999 | 3 | 2 | 2 | 4 | 1 | Tokyo | Bovine milk | |
| 97 | 352 | t2844 | VI | 838A | O8 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 3 | 2 | 2 | 4 | 1 | Chiba | Bovine milk | |
| 97 | 352 | t3782 | VI | 838A | O37 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 3 | 2 | 2 | 4 | 1 | Okinawa | Bovine milk | |
| 97 | 352 | t5352 | VI | 838A | O14 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 4 | 3 | 2 | 7 | 1 | Gunma | Bovine milk | |
| 97 | 1366 | t1109 | VI | 838A | O33 | hlg, lukE/lukD, lukM/lukF′-PV | 4 | 999 | 4 | 3 | 2 | 2 | 1 | Niigata | Bovine milk | |
| 97 | 1367 | t521 | VI | 838A | O54 | hlg, lukE/lukD | 4 | 999 | 3 | 2 | 2 | 10 | 1 | Tochigi | Bovine milk | |
| 509 | 89 | t375 | I | 676A | A3 | none | 2 | 999 | 1 | 1 | 1 | 7 | IIIa | 1 | Tokyo | Human wound |
| 509 | 89 | t375 | I | 676A | A4, A6 | hlg | 2 | 999 | 1 | 1 | 1 | 7 | IVE | 2 | Ibaraki | Human phlegm, human synovial fluid |
| 509 | 89 | t375 | I | 676A | A5, A7, A8 | hlg | 2 | 999 | 1 | 1 | 1 | 7 | Untypeable | 3 | Ibaraki, Gunma | Human eye mucus, human synovial fluid |
| 509 | 89 | t5266 | I | 676A | A1 | hlg, sagB | 2 | 999 | 1 | 0 | 1 | 6 | IIIa | 1 | Unknown | Bovine milk |
| 509 | 1373 | t5262 | I | 676A | A2 | hlg | 2 | 999 | 1 | 1 | 1 | 7 | Untypeable | 1 | Ibaraki | Human pus |
| 705 | 705 | t529 | VI | 595D | N33 | hlg, lukE/lukD, lukM/lukF′-PV, sagB, sagD | 1 | 999 | 999 | 999 | 999 | 1 | 1 | Chiba | Bovine milk | |
| 705 | 705 | t529 | VI | 595D | N5, N11, N12 | hlg, lukE/lukD, lukM/lukF′-PV, sagB, sagD | 1 | 999 | 999 | 999 | 0 | 1 | 3 | Gunma, Ishikawa, Okinawa | Bovine milk | |
| 705 | 705 | t529 | VI | 595D | N1, N2, N4, N7, N9, N10, N13, N14, N15, N16, N17, N18, N20, N22, N24, N27, N28, N29, N30, N31, N32, N34, N36 | hlg, lukE/lukD, lukM/lukF′-PV, sagB, sagD | 1 | 999 | 2 | 999 | 0 | 1 | 23 | Hokkaido, Tochigi, Saitama, Gunma, Ishikawa, Kochi, Hiroshima, Fukuoka, Okinawa | Bovine milk | |
| 705 | 705 | t529 | VI | 595D | N3, N8, N19, N37, N38 | hlg, lukE/lukD, lukM/lukF′-PV, sagB, sagD | 1 | 999 | 2 | 999 | 999 | 1 | 5 | Tokyo, Ishikawa, Gifu, Saga | Bovine milk | |
| 705 | 705 | t529 | VI | 595D | N25 | hlg, lukE/lukD, lukM/lukF′-PV, sagB, sagD | 1 | 999 | 3 | 2 | 0 | 1 | 1 | Hokkaido | Bovine milk | |
| 705 | 705 | t529 | VI | 595D | N6 | hlg, lukE/lukD, lukM/lukF′-PV, sagB, sagD | 1 | 999 | 4 | 999 | 0 | 1 | 1 | Ishikawa | Bovine milk | |
| 705 | 1363 | t529 | VI | 595D | N21, N23 | hlg, lukE/lukD, lukM/lukF′-PV, sagB, sagD | 1 | 999 | 999 | 999 | 0 | 1 | 2 | Tochigi, Okinawa | Bovine milk | |
| 705 | 1364 | t529 | VI | 595D | N26 | hlg, lukE/lukD, lukM/lukF′-PV, sagB, sagD | 1 | 999 | 999 | 999 | 0 | 1 | 1 | Tochigi | Bovine milk | |
| 705 | 1365 | t529 | VI | 595D | N35 | hlg, lukE/lukD, lukM/lukF′-PV, sagB, sagD | 1 | 999 | 999 | 999 | 999 | 1 | 1 | Hokkaido | Bovine milk | |
| 1359 | t5263 | II | 919A | C | hlg, sagB | 3 | 999 | 1 | 2 | 3 | 8 | 1 | Hyogo | Bovine milk | ||
| 1361 | t5260 | Untypeable | 919B | I | hlg, sagB, sagD | 999 | 999 | 3 | 999 | 3 | 7 | 1 | Iwate | Bovine milk | ||
| 1371 | t5261 | IV | 1000A | W | hlg | 3 | 999 | 3 | 999 | 3 | 8 | 1 | Hiroshima | Bovine milk | ||
MRSA isolates are highlighted in boldface. The major bovine lineages were CC97 and CC705.
“999” indicates that there was no amplification of SIRUs.
Locations were dairy farms or hospitals.
Representative isolates were grouped into 34 STs in 15 CCs and, independently, into 3 STs by MLST (Table 2). All of the representative isolates belonging to cluster O showed 5 STs (ST97, ST124, ST352, ST1366, and ST1367) that are members of CC97, the predicted ancestral ST of which is ST97 (Table 1). Two isolates belonging to clusters L and P also showed ST124 of CC97. All of the representative isolates belonging to cluster N showed 4 STs (ST705, ST1363, ST1364, and ST1365) that are members of CC705, the predicted ancestral ST of which is ST705 (Table 1).
TABLE 2.
Simpson's index of diversity and 95% confidence interval values of the methods used to characterize the 191 S. aureus isolates from bovine milk and humans
| Typing method | No. of different types found | Discriminatory index | 95% Confidence interval |
|---|---|---|---|
| MLVA (SIRU1, -5, -7, -13, -15, -21) | 91 | 0.971 | 0.96-0.982 |
| spa typing | 59 | 0.928 | 0.906-0.95 |
| MLST (STs) | 37 | 0.903 | 0.879-0.926 |
| coa PCR-RFLP | 28 | 0.88 | 0.855-0.905 |
| MLST (CCs) | 15 | 0.83 | 0.796-0.863 |
| Coagulase serotype | 8 | 0.699 | 0.636-0.763 |
According to spa typing, the representative isolates were grouped into 59 Spa types (Table 2). Representative isolates that are members of CC97 showed 15 Spa types (t189, t203, t224, t267, t359, t458, t521, t1109, t1234, t2101, t2453, t2844, t3782, t5265, and t5352); many of these Spa types consist of repeat units of shared or analogous sequences, and the order of repeat units is also similar (www.spaserver.ridom.de) (Table 1). The representative isolates that are members of CC705, however, showed an identical Spa type (t529) (Table 1).
Representative isolates produced coa PCR amplification products of 352, 514, 595, 676, 757, 838, 919, or 1,000 bp. Furthermore, these coa PCR products were segmented into 28 genotypes by AluI digestion (Table 2). The genotypes and their respective restriction fragments (by length [bp]) from coa PCR-RFLP were as follows: 352A, 214-138; 352B, 352; 514A, 214-162-138; 514B, 300-214; 595A, 381-214; 595B, 214-162-138-81; 595C, 376-138-81; 595D, 595; 595E, 300-295; 595F, 300-214-81; 676A, 457-219; 676B, 295-243-138; 676C, 381-214-81; 676D, 243-219-214; 676E, 300-214-162; 676F, 381-295; 676G, 676; 676H, 295-162-138-81; 757A, 457-219-81; 757B, 757; 757C, 381-376; 757D, 381-214-162; 757E, 324-295-138; 838A, 381-295-162; 838B, 381-295-81-81; 919A, 214-138-81-81-81-81-81-81-81; 919B, 214-162-162-138-81-81-81; and 1000A, 243-219-214-162-81-81. Representative isolates that are members of CC97 showed 7 genotypes (352B, 595E, 676F, 757C, 757D, 838A, and 838B), and representative isolates that are members of CC705 showed an identical genotype (595D) (Table 1).
Representative isolates showed 91 MLVA profiles (Table 2), and members of CC97 and CC705 showed 28 and 6 profiles, respectively (Table 1).
By coagulase serotype, representative isolates showed 8 serotypes (Table 2), and members of CC97 and CC705 showed coagulase serotype VI (Table 1). Furthermore, most of the CCs other than CC15 and CC8 showed a single coagulase serotype (Table 2).
As for the staphylococcal exotoxin gene profiles, 9 combinations were confirmed in representative isolates (Table 1). lukM/lukF′-PV was only detected in members of CC705 and CC97 (Table 1). sagB and sagD were detected in all of the representative isolates that are members of CC705, ST789, and ST1361 (Table 1). Additionally sagB was also detected in representative isolates of ST89 and ST1359 (Table 1). Of the bovine milk isolates, there were only 3 isolates, ST1, ST81, and ST243, detected by pvl (Table 1). On the other hand, hlg was detected in all representative isolates from bovine milk, and lukE/lukD was also detected in all representative isolates from bovine milk, except for the 12 isolates of ST20, ST59, ST89, ST243, ST508, ST1359, ST1361, and ST1371 (Table 1).
Comparison of different typing methods.
The discriminatory powers of the five typing methods (MLST, spa typing, coagulase serotype, coa PCR-RFLP, and MLVA) other than PFGE were determined by calculating the Simpson's index of diversity with 95% confidence intervals of the isolates typed by these methods (Table 2). MLVA showed the highest discriminatory power (0.971) of the five typing methods. In addition, spa typing and MLST also showed high discriminatory power, with a discriminatory index of 0.9 or higher (0.928 and 0.903, respectively). Adjusted Rand's and Wallace's coefficients were calculated to explore the correlation between typing methods (Tables 3 and 4). MLST (CCs) showed high adjusted Rand's coefficient (0.941) and Wallace's coefficients (0.911 and 0.993) with PFGE (clusters). Furthermore, MLST (CCs) showed comparatively high adjusted Rand's coefficients and Wallace's coefficients with coa PCR-RFLP (0.721, 0.658, and 0.903), spa typing (0.546, 0.425, and 0.977), and coagulase serotyping (0.597, 0.959, and 0.553). MLVA showed high Wallace's w1 coefficients for every typing method. spa typing also showed high Wallace's w1 coefficients for every typing method except for MLVA.
TABLE 3.
Adjusted Rand's coefficients for the methods used to characterize the 191 S. aureus isolates from bovine milk and humansa
| Typing method | MLVA | spa typing | MLST (STs) | coa PCR-RFLP | PFGE clusters | MLST (CCs) | Coagulase serotype | Staphylococcal exotoxin gene profile |
|---|---|---|---|---|---|---|---|---|
| MLVA (SIRU1, -5, -7, -13, -15, -21) | ||||||||
| spa typing | 0.540 | |||||||
| MLST (STs) | 0.413 | 0.663 | ||||||
| coa PCR-RFLP | 0.289 | 0.543 | 0.576 | |||||
| PFGE clusters | 0.286 | 0.589 | 0.733 | 0.719 | ||||
| MLST (CCs) | 0.262 | 0.546 | 0.705 | 0.721 | 0.941 | |||
| Coagulase serotype | 0.131 | 0.297 | 0.404 | 0.418 | 0.585 | 0.597 | ||
| Staphylococcal exotoxin gene profile (hlg, lukE/lukD, lukM/lukF′-PV, pvl, sagB, sagD) | 0.115 | 0.249 | 0.350 | 0.257 | 0.345 | 0.358 | 0.129 |
The methods are listed in order of discriminatory power, with the highest to lowest discriminatory powers from left to right and top to bottom.
TABLE 4.
Wallace's coefficients for the methods used to characterize the 191 S. aureus isolates from bovine milk and humansa
| Typing method | MLVA | spa typing | MLST (STs) | coa PCR-RFLP | PFGE clusters | MLST (CCs) | Coagulase serotype | Staphylococcal exotoxin gene profile |
|---|---|---|---|---|---|---|---|---|
| MLVA (SIRU1, -5, -7, -13, -15, -21) | 0.971 | 0.956 | 0.829 | 0.994 | 1 | 1 | 0.939 | |
| spa typing | 0.393 | 0.813 | 0.781 | 0.977 | 0.977 | 0.977 | 0.886 | |
| MLST (STs) | 0.285 | 0.6 | 0.694 | 0.976 | 1 | 1 | 0.926 | |
| coa PCR-RFLP | 0.2 | 0.467 | 0.562 | 0.854 | 0.903 | 0.903 | 0.685 | |
| PFGE clusters | 0.191 | 0.464 | 0.628 | 0.679 | 0.993 | 0.993 | 0.723 | |
| MLST (CCs) | 0.176 | 0.425 | 0.59 | 0.658 | 0.911 | 0.959 | 0.711 | |
| Coagulase serotype | 0.097 | 0.235 | 0.327 | 0.364 | 0.504 | 0.553 | 0.4 | |
| Staphylococcal exotoxin gene profile (hlg, lukE/lukD, lukM/lukF′PV, pvl, sagB, sagD) | 0.089 | 0.207 | 0.294 | 0.268 | 0.356 | 0.385 | 0.39 |
Each value in the body of the table is Wallace's w1 coefficient of the typing method given in the first column to the typing method given in the column head and is also Wallace's w2 coefficient of the typing method given in the column head to the typing method given in the first column.
Genetic characteristics of major lineages from bovine milk in Japan, and relationship with foreign bovine milk isolates.
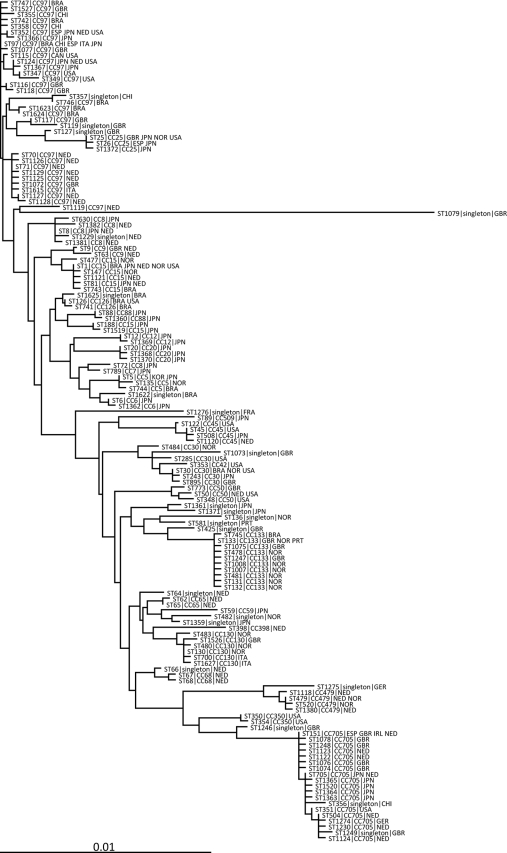
S. aureus strains of CC97 and CC705 are dominant lineages from bovine milk in Japan. Using many genotyping methods (MLVA, coa PCR-RFLP, and spa typing), the CC97 strains showed more genetic variation than the CC705 strains, as previously noted (Table 1). Newbould 305 is the strain that was isolated in 1958 from a clinical mastitis case in Canada (32), and this strain shows ST115, which is the ST of CC97 (Fig. 1). All isolates in CC705 showed an identical genetic background (t529 for Spa type, 595D for coa PCR-RFLP, and detection of hlg, lukE/lukD, lukM/lukF′-PV, sagB, and sagD) and coagulase serotype VI (Table 1). Moreover, the sequenced Irish bovine S. aureus strain RF122 (ET3-1) also shows ST151, which is a member of CC705 (Fig. 1) (15).
FIG. 1.
Phylogenetic tree based on the seven locus sequences (arcC, aroE, glpF, gmk, pta, tpi, and yqiL) that have been reported from bovine S. aureus isolates worldwide by the MLST website and previous reports (2, 15, 18, 22, 26, 33, 35, 36). The sequence type (ST), clonal complex (CC), and country or countries that each ST was detected from are shown. Country abbreviations: BRA, Brazil; CAN, Canada; CHI, Chile; ESP, Spain; FRA, France; GBR, United Kingdom; GER, Germany; IRL, Ireland; ITA, Italy; JPN, Japan; KOR, Republic of Korea; NED, Netherlands; NOR, Norway; PRT, Portugal; USA, United States. S. aureus strain Newbould 305 from clinical bovine mastitis in Canada shows ST115, and the sequenced Irish bovine strain RF122 shows ST151 (15). The scale bar indicates the rate of nucleotide substitution.
According to the MLST website and eBURST, 30 of 42 STs of CC97 (ST70, ST71, ST97, ST115, ST116, ST117, ST118, ST124, ST347, ST349, ST352, ST355, ST358, ST742, ST746, ST747, ST1072, ST1077, ST1119, ST1125, ST1126, ST1127, ST1128, ST1129, ST1366, ST1367, ST1527, ST1615, ST1623, and ST1624) have been confirmed in bovine isolates, while all 17 STs of CC705 (ST151, ST351, ST504, ST705, ST1074, ST1076, ST1078, ST1122, ST1123, ST1124, ST1230, ST1248, ST1274, ST1363, ST1364, ST1365, and ST1520) have only been confirmed in bovine isolates. CC97 strains have been isolated from bovine milk in Brazil, Canada, Chile, Italy, Norway, Netherlands, Spain, the United Kingdom, and the United States (Fig. 1). CC705 strains have been isolated from bovine milk in Chile, Germany, Ireland, Netherlands, Spain, the United Kingdom, and the United States (Fig. 1).
Genetic analysis of MRSA isolates from bovine milk.
Four bovine MRSA isolates were obtained from different bovine mastitis cases from 1999 to 2005 (Table 1). Of 4 bovine MRSA isolates, 3 isolates exhibited ST5-SCCmec II (Table 1). Two bovine MRSA isolates with pulsotypes E1 and E2 and a human MRSA isolate with pulsotype E3 showed identical genotype results (t375 for Spa type, 676B for coa PCR-RFLP, 2-2-1-3-999-9 in the MLVA profile, and the detection of hlg and lukE/lukD) and coagulase serotype II (Table 1). One bovine MRSA isolate with pulsotype D also showed similar genotype results (t002 for Spa type, 676B for coa PCR-RFLP, 2-3-1-3-1-9 in the MLVA profile, and the detection of hlg and lukE/lukD) and coagulase serotype II (Table 1). One remaining bovine MRSA isolate with pulsotype A1 exhibited ST89-SCCmec IIIa (Table 1). The ST89-SCCmec IIIa isolate had a different genotype from hospital-acquired MRSA (HA-MRSA) pandemic clones, such as the New York/Japan clone, pediatric clone (ST5-SCCmec IV), Berlin clone (ST45-SCCmec IV), Iberian clone (ST247-SCCmec Ia), Brazilian-Hungarian clone (ST239-SCCmec III), epidemic MRSA strain 15 (EMRSA-15) (ST22-SCCmec IV), and EMRSA-16 (ST36-SCCmec II). However, ST89-SCCmec IIIa is confirmed in a human MRSA isolate with pulsotype A3 (Table 1). MRSA isolates which are members of CC509 are members of PFGE cluster A and showed an identical coa PCR-RFLP genotype (676A) and coagulase serotype I. Moreover, they showed analogous Spa types (t375 and t5226) and MLVA profiles (2-999-1-1-1-7 and 2-999-1-0-1-6) (Table 1).
DISCUSSION
Weighing the results of various typing methods for bovine S. aureus isolates, three typing methods (MLVA, spa typing, and MLST) showed comparatively high discriminatory power. The discriminatory power of MLVA was the highest among these methods. Furthermore MLVA and spa typing showed high Wallace's w1 coefficients for various typing methods. These results show that both methods have high discriminatory power and that there is strong correlation between them. When there is a need to complement more elaborate genotyping results, both methods will be useful. On the other hand, MLST showed high adjusted Rand's coefficients and Wallace's coefficients with various typing methods for S. aureus, especially PFGE. This suggests that the genotyping results of MLST almost accord with the genotyping results of PFGE. As for correlation with other typing methods, isolates belonging to the same CC showed genotypes that were identical or analogous to each other. Moreover, there are established, useful websites for sharing and analyzing substantial databases of MLST genotypes (9), so MLST may become the main genotyping method for cluster analysis of bovine S. aureus isolates.
According to the results of various genotyping and coagulase serotyping methods, S. aureus strains as a causal bacterium of bovine mastitis in Japan mainly consist of specific lineages, CC97 and CC705. These results supported our previous speculation that two major S. aureus lineages spread as bovine mastitis causal strains (12). Both lineages have also been isolated from bovine milk worldwide. In particular, CC97 strains have become the dominant lineage in Chile (36), Brazil (2, 33), Japan, Netherlands (18), and the United States (36). CC705 strains have become the dominant lineage in Japan, Netherlands (18), and the United Kingdom (36). In addition to CC97 and CC705 strains, the CC126, CC133, and CC479 lineages are dominant in Brazil (33), Norway (22), and the Netherlands (18), respectively. Strains of these CCs have never been confirmed in Japan but have been widely isolated in South and North America and Europe and may spread to Asia in the future with the movement of cattle. According to the MLST website, CC97, CC126, CC133, CC479, and CC705 strains have been isolated mainly from ruminant samples, but strains of other CCs have not. As for CC705 and CC479 strains, these strains have only been isolated from bovine samples.
The CC97 strains showed multiple genotypes with the coa PCR-RFLP and spa typing, whereas the CC705 strains showed an identical genotype. Moreover, a phylogenetic analysis based on the seven locus sequences of MLST seems to suggest that the CC97 lineage emerged earlier than the CC705 lineage, and bovine strains worldwide seem to derive from CC97. Newbould 305, a member of the CC97 lineage, was isolated from bovine mastitic milk in 1958 (32). These facts led us to predict that CC97 is a bovine mastitis causal lineage that has existed for at least 50 years. However, CC705 seems to be the most newly established bovine lineage (Fig. 1), and the identical genotype results of CC705 strains seemed to support this speculation.
As this study elucidated, certain hypotheses have been proposed to explain why specific S. aureus lineages are frequently isolated from bovine samples. Herron-Olson et al. advocate the idea that host specialization of S. aureus depends on gene mutation, transfer, and decay. As noted previously, specific gene variations may play an important role in in vivo bacterial pathogenicity and colonization (15). According to phylogenetic analysis based on MLST data, CC97 and CC705, which are dominant lineages from bovine milk, are distantly related to each other (Fig. 1). However, lukM/lukF′-PV and coagulase serotype VI were nominated as common characteristics of both strains by our study. lukM/lukF′-PV is associated with horizontal gene transmission by temperate bacteriophages of bovine origin (40), so lukM/lukF′-PV will be widely found in these strains. LukM/LukF′-PV has highly active cytotoxicity on bovine neutrophils in comparison with that of other S. aureus leukocidins (4), and LukM/LukF′-PV may contribute to the neutralization of immunity by neutrophils in mammary organs. Moreover, LukM/LukF′-PV is a family of pore-forming toxins which may be involved with escape from the phagosome after phagocytosis and intracellular survival, which is an important capability for the maintenance of S. aureus intramammary infection (23). Intracellular survival is considered to be related to rebelliousness, which is a characteristic mark of bovine mastitis due to S. aureus, because most antibiotics cannot act in a cell, except for macrolides. Judging from the viewpoint of iron acquisition, obligate intracellular conditions will be favorable for the survival of S. aureus (15). Many of the biological activities of LukM/LukF′-PV have never been elucidated, including its participation in bovine mastitis, so further analysis of this toxin will be required in the future.
Coagulase, which is recognized as one of the virulence factors of S. aureus, causes coagulation of plasma. The antigenic diversity of coagulase is a major phenotypic determinant and is used as a characteristic marker, with 12 types having been discriminated so far (24). Coagulase serotype VI is dominant only in bovine isolates and seems to be extremely rare in human isolates (11, 24). The antigenic variation in coagulase may result from adaptation to prothrombin in different animal species in order to elude the host immune response (24). Thus, the antigenicity of coagulase may be an important factor for evading the host defense system, as well as for adapting to animals that are infected by S. aureus.
Staphylococcus protein A is a virulence factor participating in humoral immunity restraint. According to spa typing, CC705 strains showed an identical genotype (t529), and CC705 strains in the Netherlands also showed the same genotype (18). CC97 strains showed multiple genotypes, but many of these genotypes (t189, t203, t224, t267, t359, t521, t1109, t1234, t2453, and t3782) are closely related to each other (Table 1). Of these genotypes, t224 and t521 were confirmed in CC97 strains in Netherlands (18) and t267 and t359 were confirmed in CC97 strains in Brazil (2). spa of bovine strain RF122 contains stop codons and is, therefore, a pseudogene (15). The relationship between point mutations or mutations resulting in a pseudogene of spa and the pathogenicity of S. aureus strains is an interesting point for further examination.
Herron-Olson et al. suggested several genetic characteristics of bovine strain RF122. The streptolysin homolog genes (sagB and sagD), pyrogenic toxin super antigen TSST-1 gene (tst), and the bovine variant of the staphylococcal enterotoxin C gene (sec) are genetic characteristics of bovine strain RF122 (15). Previous studies also suggested that not only leukocidin but also superantigens are important during mastitis pathogenesis due to their immunomodulatory effects (34). However, this study and the previous studies suggested that these genes are rarely present in CC97 strains, so these exotoxins will not be fundamental in bovine mastitis (11, 12).
As for dominant foreign lineages from bovine milk, such as CC126, CC133, and CC479, the bacteriological characteristics of these CCs have not yet been sufficiently elucidated. Further analysis of these CCs will be indispensable for examining the universal characteristics of dominant bovine strains.
The New York/Japan MRSA clone has mainly spread as HA-MRSA in Japan and Korea, and ST5-SCCmec II is the specific genotype of this clone (25). Our study indicated that bovine MRSA isolates in Japan showed the specific genotype of the New York/Japan MRSA clone. Of them, 2 bovine MRSA isolates showed genotypes identical to the human MRSA isolate, and one bovine MRSA isolate also showed similar genotype results. One remaining bovine MRSA isolate, with ST89-SCCmec IIIa, is not identical to genotypes of the human MRSA isolate with ST89-SCCmec IIIa; however, both isolates showed analogous genotype results. Therefore, we cannot be sure how this bovine MRSA isolate arrived at the bovine mammary organ. However, the fact that this bovine MRSA isolate shows genotype results similar to those of a human MRSA isolate is interesting. Moreover ST5 and ST89 are uncommon among bovine isolates in the world, whereas both STs are common among human MRSA isolates in Japan. These analytical results and information lead us to guess that bovine MRSA emerged elsewhere than in the dairy environment. In Korea, Kwon et al. reported MRSA isolation from bovine milk and performed genetic analysis of these isolates (26). In this report, all 14 isolates showed ST5-SCCmec IVg. As to the origin of these isolates, pediatric clone ST5-MSSA and isolates with ST5-SCCmec II were the expected source. In any case, the dominant bovine MRSA isolates CC97 and CC705 observed in the present study have never been confirmed from bovine samples until now. In the first place, MRSA isolation from bovine samples is uncommon; this paper is the first report of MRSA isolation from bovine milk in Japan. However, eight STs of CC97 (ST97, ST458, ST953, ST1174, ST1179, ST1379, ST1419, and ST1476) have been confirmed in human or swine MRSA isolates, and it will therefore be necessary to take measures to prevent the appearance of MRSA strains that may be suitable for the in vivo environment of cattle. According to the comparative analysis of genomes and mobile genetic elements between bovine isolate RF122 and MRSA252, horizontal gene transfer (HGT) between the two isolates has been suggested, because RF122 and MRSA252 shared multiple genes in related genomic positions. Moreover, MRSA252 also shared enzyme genes with Listeria monocytogenes and Staphylococcus saprophyticus plasmid pSSP2 (5). Bovine MSSA strains may also acquire methicillin resistance by HGT from human MRSA or other bacteria. Because of their relatively small size, SCCmec types IV and V seem to have higher mobility than the other SCCmec types, i.e., I, II, and III (1). This feature may be responsible for these SCCmec types being found among community-acquired MRSA isolates worldwide (39). Moreover, SCCmec type IV was also detected among four STs of CC97 (ST97, ST953, ST1174, and ST1179). To prevent the coexistence and contact of bovine MSSA and MRSA strains, measures should be taken to prevent MRSA from spreading to dairy farms. Unauthorized persons and ambulatory patients who can be assumed to be sources of MRSA should be prohibited from entering dairy farms. Moreover, personnel are important sources or carriers of S. aureus on dairy farms, and thus hand washing and changing into clean clothing should be enforced as routine control measures (35, 36).
Acknowledgments
We thank Y. Tamada of the Hokkaido Shiribeshi Livestock Hygiene Service Center, T. Endô of the Yamagata Chuô Livestock Hygiene Service Center, H. Nakabayashi of the Niigata Chuô Livestock Hygiene Service Center, S. Koike of the Tochigi Kenpoku Livestock Hygiene Service Center, S. Nozue of the Gunma Livestock Health Laboratory, S. Kawaji of the Saitama Chuô Livestock Hygiene Service Center, Y. Kondô of the Chiba Prefectural Federation of Agricultural Mutual Aid Associations (Chiba NOSAI), T. Minamiura of the Tokyo Livestock Hygiene Service Center, H. Ide of the Ishikawa Nanbu Livestock Hygiene Service Center, K. Kawashima of the Shizuoka Chubu Livestock Hygiene Service Center, H. Kobayashi of the Gifu Livestock Hygiene Service Center, A. Ichihoshi of the Kyoto Chuô Livestock Hygiene Service Center, N. Fujii of the Nara Livestock Hygiene Service Center, T. Akiyama of the Wadayama Livestock Hygiene Service Center, K. Fujita of the Kurayoshi Livestock Hygiene Service Center, C. Kawamoto of the Higashihiroshima Livestock Hygiene Service Center, E. Mizuno of the Kochi Chuô Livestock Hygiene Service Center, T. Ogawa of the Fukuoka Chuô Livestock Hygiene Service Center, A. Miyamoto of the Saga Chubu Livestock Hygiene Service Center, N. Gushi of the Okinawa Chuô Livestock Hygiene Service Center, and M. Higashide, E. Ishikawa, and T. Sasaki of Kotobiken Medical Laboratories, Inc. for their help with the collection of isolates from bovine milk and humans. We would also like to thank A. Iijima for technical assistance.
This study was supported in part by grants from the National Agricultural Research Organization of Japan and Grants-in-Aid for the Regional R&D Proposal-Based Program from the Northern Advancement Center for Science & Technology of Hokkaido, Japan.
Footnotes
Published ahead of print on 14 April 2010.
REFERENCES
- 1.Aires de Sousa, M., and H. de Lencastre. 2003. Evolution of sporadic isolates of methicillin-resistant Staphylococcus aureus (MRSA) in hospitals and their similarities to isolates of community-acquired MRSA. J. Clin. Microbiol. 41:3806-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aires-de-Sousa, M., C. E. Parente, O. Vieira-da-Motta, I. C. Bonna, D. A. Silva, and H. de Lencastre. 2007. Characterization of Staphylococcus aureus isolates from buffalo, bovine, ovine, and caprine milk samples collected in Rio de Janeiro State, Brazil. Appl. Environ. Microbiol. 73:3845-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannerman, T. L., D. Hancock, F. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrio, M. B., P. Rainard, and G. Prévost. 2006. LukM/LukF′-PV is the most active Staphylococcus aureus leukotoxin on bovine neutrophils. Microbes Infect. 8:2068-2074. [DOI] [PubMed] [Google Scholar]
- 5.Brody, T., A. S. Yavatkar, Y. Lin, J. Ross, A. Kuzin, M. Kundu, Y. Fann, and W. F. Odenwald. 2008. Horizontal gene transfers link a human MRSA pathogen to contagious bovine mastitis bacteria. PLoS One 3:e3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2008. CLSI document M31-A3. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals, 3rd ed. Approved standard. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Devriese, L. A., and J. Hommez. 1975. Epidemiology of methicillin-resistant Staphylococcus aureus in dairy herds. Res. Vet. Sci. 19:23-27. [PubMed] [Google Scholar]
- 8.Dorsch, M., and E. Stackebrandt. 1992. Some modifications in the procedure of direct sequencing of PCR amplified 16S rRNA. J. Microbiol. Methods 16:271-279. [Google Scholar]
- 9.Enright, M. C., N. P. J. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hata, E., K. Katsuda, H. Kobayashi, K. Nishimori, I. Uchida, M. Higashide, E. Ishikawa, T. Sasaki, and M. Eguchi. 2008. Bacteriological characteristics of Staphylococcus aureus isolates from humans and bulk milk. J. Dairy Sci. 91:564-569. [DOI] [PubMed] [Google Scholar]
- 12.Hata, E., K. Katsuda, H. Kobayashi, T. Ogawa, T. Endô, and M. Eguchi. 2006. Characteristics and epidemiologic genotyping of Staphylococcus aureus isolates from bovine mastitic milk in Hokkaido, Japan. J. Vet. Med. Sci. 68:165-170. [DOI] [PubMed] [Google Scholar]
- 13.Hardy, K. J., B. A. Oppenheim, S. Gossain, F. Gao, and P. M. Hawkey. 2006. Use of variations in staphylococcal interspersed repeat units for molecular typing of methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 44:271-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harmsen, D., H. Claus, W. Witte, J. Rothgänger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herron-Olson, L., J. R. Fitzgerald, J. M. Musser, and V. Kapur. 2007. Molecular correlates of host specialization in Staphylococcus aureus. PLoS One 2:e1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hookey, J. V., J. F. Richardson, and B. D. Cookson. 1998. Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. J. Clin. Microbiol. 36:1083-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ichiyama, S., M. Ohta, K. Shimokata, N. Kato, and J. Takeuchi. 1991. Genomic DNA fingerprinting by pulsed-field gel electrophoresis as an epidemiological marker for study of nosocomial infections caused by methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 29:2690-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikawaty, R., E. C. Brouwer, M. D. Jansen, E. van Duijkeren, D. Mevius, J. Verhoef, and A. C. Fluit. 2009. Characterization of Dutch Staphylococcus aureus from bovine mastitis using a multiple locus variable number tandem repeat analysis. Vet. Microbiol. 136:277-284. [DOI] [PubMed] [Google Scholar]
- 19.Ikawaty, R., R. J. Willems, A. T. Box, J. Verhoef, and A. C. Fluit. 2008. Novel multiple-locus variable-number tandem-repeat analysis method for rapid molecular typing of human Staphylococcus aureus. J. Clin. Microbiol. 46:3147-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarraud, S., C. Mougel, J. Thioulouse, G. Lina, H. Meugnier, F. Forey, X. Nesme, J. Etienne, and F. Vandenesch. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonas, D., M. Speck, F. D. Daschner, and H. Grundmann. 2002. Rapid PCR-based identification of methicillin-resistant Staphylococcus aureus from screening swabs. J. Clin. Microbiol. 40:1821-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jørgensen, H. J., T. Mørk, D. A. Caugant, A. Kearns, and L. M. Rørvik. 2005. Genetic variation among Staphylococcus aureus strains from Norwegian bulk milk. Appl. Environ. Microbiol. 71:8352-8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerro Dego, O., J. E. van Dijk, and H. Nederbragt. 2002. Factors involved in the early pathogenesis of bovine Staphylococcus aureus mastitis with emphasis on bacterial adhesion and invasion. A review. Vet. Q. 24:181-198. [DOI] [PubMed] [Google Scholar]
- 24.Kinoshita, M., N. Kobayashi, S. Nagashima, M. Ishino, S. Otokozawa, K. Mise, A. Sumi, H. Tsutsumi, N. Uehara, N. Watanabe, and M. Endo. 2008. Diversity of staphylocoagulase and identification of novel variants of staphylocoagulase gene in Staphylococcus aureus. Microbiol. Immunol. 52:334-348. [DOI] [PubMed] [Google Scholar]
- 25.Ko, K. S., J. Y. Lee, J. Y. Suh, W. S. Oh, K. R. Peck, N. Y. Lee, and J. H. Song. 2005. Distribution of major genotypes among methicillin-resistant Staphylococcus aureus clones in Asian countries. J. Clin. Microbiol. 43:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon, N. H., K. T. Park, J. S. Moon, W. K. Jung, S. H. Kim, J. M. Kim, S. K. Hong, H. C. Koo, Y. S. Joo, and Y. H. Park. 2005. Staphylococcal cassette chromosome mec (SCCmec) characterization and molecular analysis for methicillin-resistant Staphylococcus aureus and novel SCCmec subtype IVg isolated from bovine milk in Korea. J. Antimicrob. Chemother. 56:624-632. [DOI] [PubMed] [Google Scholar]
- 27.Lee, J. H. 2003. Methicillin (oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl. Environ. Microbiol. 69:6489-6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martineau, F., F. J. Picard, P. H. Roy, M. Ouellette, and M. G. Bergeron. 1998. Species-specific and ubiquitous-DNA-based assays for rapid identification of Staphylococcus aureus. J. Clin. Microbiol. 36:618-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milheiriço, C., D. C. Oliveira, and H. de Lencastre. 2007. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: “SCCmec IV multiplex.” J. Antimicrob. Chemother. 60:42-48. [DOI] [PubMed] [Google Scholar]
- 30.Mørk, T., T. Tollersrud, B. Kvitle, H. J. Jørgensen, and S. Waage. 2005. Comparison of Staphylococcus aureus genotypes recovered from cases of bovine, ovine, and caprine mastitis. J. Clin. Microbiol. 43:3979-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prasad, L. B., and F. H. Newbould. 1968. Inoculation of the bovine teat duct with Staph. aureus: the relationship of teat duct length, milk yield and milking rate to development of intramammary infection. Can. Vet. J. 9:107-115. [PMC free article] [PubMed] [Google Scholar]
- 33.Rabello, R. F., B. M. Moreira, R. M. Lopes, L. M. Teixeira, L. W. Riley, and A. C. Castro. 2007. Multilocus sequence typing of Staphylococcus aureus isolates recovered from cows with mastitis in Brazilian dairy herds. J. Med. Microbiol. 56:1505-1511. [DOI] [PubMed] [Google Scholar]
- 34.Schuberth, H. J., C. Krueger, H. Zerbe, E. Bleckmann, and W. Leibold. 2001. Characterization of leukocytotoxic and superantigen-like factors produced by Staphylococcus aureus isolates from milk of cows with mastitis. Vet. Microbiol. 82:187-199. [DOI] [PubMed] [Google Scholar]
- 35.Smith, E. M., L. E. Green, G. F. Medley, H. E. Bird, and C. G. Dowson. 2005. Multilocus sequence typing of Staphylococcus aureus isolated from high-somatic-cell-count cows and the environment of an organic dairy farm in the United Kingdom. J. Clin. Microbiol. 43:4731-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, E. M., L. E. Green, G. F. Medley, H. E. Bird, L. K. Fox, Y. H. Schukken, J. V. Kruze, A. J. Bradley, R. N. Zadoks, and C. G. Dowson. 2005. Multilocus sequence typing of intercontinental bovine Staphylococcus aureus isolates. J. Clin. Microbiol. 43:4737-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soo Ko, K., K. R. Peck, W. Sup Oh, N. Y. Lee, K. Hiramatsu, and J. H. Song. 2005. Genetic differentiation of methicillin-resistant Staphylococcus aureus strains from Korea and Japan. Microb. Drug Resist. 11:279-286. [DOI] [PubMed] [Google Scholar]
- 38.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamada, T., N. Tochimaru, S. Nakasuji, E. Hata, H. Kobayashi, M. Eguchi, J. Kaneko, Y. Kamio, T. Kaidoh, and S. Takeuchi. 2005. Leukotoxin family genes in Staphylococcus aureus isolated from domestic animals and prevalence of lukM-lukF-PV genes by bacteriophages in bovine isolates. Vet. Microbiol. 110:97-103. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, K., J. A. McClure, S. Elsayed, T. Louie, and J. M. Conly. 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5026-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]