Abstract
A loop-mediated isothermal amplification (LAMP) assay for the rapid detection of cytomegalovirus (CMV) was developed and evaluated. The LAMP assay specifically amplified only CMV DNA, and no cross-reactivity with the DNA of herpes simplex virus type 1, varicella-zoster virus, adenovirus, Aspergillus flavus, or Staphylococcus aureus was observed. The sequences of the LAMP assay-positive CMV products were perfectly (100%) matched with the CMV sequence deposited in the GenBank database. The sensitivity of the LAMP assay was found to be 10 copies/μl of CMV DNA. Vitreous samples from 40 patients with suspected retinitis were subjected to LAMP and real-time PCR for the detection of CMV. Of 40 patients with suspected viral retinitis, 10 tested positive for CMV by the real-time PCR and LAMP assays. A 100% concordance was observed between the results of the two methods. The LAMP assay is a rapid, highly specific, and sensitive method for the diagnosis of retinitis caused by CMV.
Viral retinitis is commonly caused by herpes simplex virus type 1 (HSV-1), HSV-2, varicella-zoster virus (VZV), cytomegalovirus (CMV), and occasionally, Epstein-Barr virus (EBV) (7). In patients with atypical features and in the early stages of ocular manifestations, clinical differentiation between cases of retinitis associated with CMV and other herpesvirus infections is often difficult (6). The differentiation of CMV retinitis from HSV and VZV retinitis is very important early in the course of the disease, as the therapeutic agent to be used for treatment differs from virus to virus (7). Conventional methods for the diagnosis of viral retinitis include the detection of viral antigen and virus isolation from intraocular specimens (2). These tests have been shown to have low sensitivities for the detection of viruses and are not currently recommended for use for the diagnosis of viral retinitis (2).
PCR has proved to be of great utility for the diagnosis of viral retinitis (3, 4, 5, 6, 8). However, owing to the expensive systems required, PCR is still not a very common diagnostic test. Notomi et al. have reported on a novel nucleic acid amplification assay termed the loop-mediated isothermal amplification (LAMP) assay (10). The assay amplifies the DNA under isothermal conditions (63 to 65°C) with high degrees of specificity, efficiency, and speed. The assay can be conducted in the laboratory in a water bath or heating block (10). Thus, the thermal cycling needs of a PCR are avoided. The assay can be used for the rapid detection of pathogens in peripheral health care settings in developing countries. The present study describes the development and evaluation of a simple and cost-effective LAMP assay for the rapid detection of CMV DNA in patients with viral retinitis.
MATERIALS AND METHODS
Clinical samples, positive controls, and DNA extraction.
The study was approved (reference no. LEC 08013) by the Institutional Review Board of the L. V. Prasad Eye Institute, Hyderabad, India. Vitreous samples from 40 patients with viral retinitis were included in the study. The viral DNA was extracted from clinical samples with a DNA minikit (Qiagen, Hilden, Germany). The viral DNA was finally eluted in 75 μl of elution buffer and stored at −20°C. The DNA of CMV strain AD169 was used as a positive control. All the patient samples were tested by the LAMP assay and by a real-time PCR assay. To address the reproducibility of the LAMP assay, the LAMP and real-time PCR assays were run in duplicate with all 40 patient samples.
LAMP reaction.
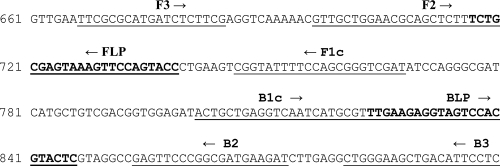
The LAMP reaction was conducted as described by Notomi et al. (10).The LAMP method requires a set of four primers that recognize a total of six distinct sequences in the target DNA and two loop primers that increase the amplification efficiency. The set of four primers include two outer primers (forward outer primer F3 and backward outer primer B3) and two inner primers (the forward inner primer [FIP] and the backward inner primer [BIP]). The loop primers include the forward loop primer (FLP) and the backward loop primer (BLP). FIP consists of the complementary sequence of the F1 region and the sense sequence of the F2 region. BIP consists of the complementary sequence of the B1 region and the sense sequence of the B2 region. The FLP and BLP primers were composed of the sequences that are complementary to the sequences between the F1 and F2 regions and the B1 and B2 regions, respectively. The primers used for CMV LAMP were designed against the sequence of the glycoprotein B (gB) gene by using Primer Explorer (version 4) software (Fujitsu, Tokyo, Japan). The location and the sequence of each primer and shown in Fig. 1 and Table 1.
FIG. 1.
Oligonucleotide primers used for LAMP assay of cytomegalovirus (GenBank accession no. U66425) within the gB gene. F1c and B1c, sequences complementary to the F1 and B1 regions, respectively; arrows, orientations of the various primers used in the CMV LAMP assay.
TABLE 1.
Primers used for cytomegalovirus LAMP assay
| Primer name | Sequence |
|---|---|
| F3 | TTCGCGCATGATCTCTTCG |
| B3 | GAGGAATGTCAGCTTCCCAG |
| FIP | ATCGACCCGCTGGAAAATACCGTTTTGTTGCTGGAACGCAGCTCTT |
| BIP | ACTGCTGAGGTCAATCATGCGTTTTTATCTTCATCGCCGGGAACTC |
| LPF | GGTACTGGAACTTTACTCGCAGA |
| LPB | TTGAAGAGGTAGTCCACGTACTC |
The LAMP assay was conducted with a 25-μl reaction mixture consisting of 1 μl of each of the outer primers (5 μM), 1 μl of each of the inner primers (40 μM), 1 μl of each of the loop primers (20 μM), 2.5 μl of 10× Bst DNA polymerase reaction buffer (New England Biolabs, MA), 1.5 μl of 50 mM MgSO4, 5.0 μl of 5 M betaine (Sigma Aldrich, St. Louis, MO), 2.5 μl deoxynucleoside triphosphates (1.4 mM), 1.0 μl of 8 U/μl Bst DNA polymerase (New England Biolabs), 5.0 μl of target DNA, and 1.5 μl of autoclaved Milli-Q water. The reaction mixture was incubated in a water bath at 64°C for 1 h and heated at 80°C for 2 min to terminate the reaction. Positive and negative controls were included in each run. The amplified product was detected by adding 1 μl of 1,000× SYBR green dye to each reaction tube. After a 15-min incubation in the dark at room temperature, a yellowish green color indicated a positive reaction, while a reddish orange (the color of the unbound dye) indicated a negative reaction. The color change in the reaction tubes was examined under natural light conditions. In addition, the LAMP products were detected by agarose gel (2%) electrophoresis with UV light transillumination.
Sensitivity and specificity of LAMP assay.
The specificity of the LAMP assay was evaluated by using CMV DNA (strain AD169), VZV DNA (Oka vaccine strain), HSV-1 DNA, adenovirus DNA, DNA extracted from Aspergillus flavus isolates from clinical samples, and Staphylococcus aureus (ATCC 25923). The AD169 strain of CMV used in this study was obtained from Christian Medical College, Vellore, India. The identity of the CMV isolate was further confirmed in our laboratory by bidirectional sequencing of the CMV DNA. The specificity of the LAMP assay was further tested by bidirectional sequencing of the CMV-positive LAMP product. The amplified LAMP products were purified with a PCR purification kit (Macherey Nagel, GmbH & Co., Germany) and were used as the templates for DNA sequencing. Sequencing was performed with a BigDye Terminator cycle sequencing kit (forward primer, 5′-TTCGCGCATGATCTCTTCG-3′; reverse primer, 5′-GAGGAATGTCAGCTTCCCAG-3′; the human cytomegalovirus gB gene region) on an ABI 3130 XI automated sequencer, according to the manufacturer's instructions (PE Applied Biosystems). The sequences were analyzed and identified by using the MEGABLAST search program of the GenBank database. The sensitivities of the LAMP assay and the commercial real-time PCR assay kit were tested by using 10 copies/μl to 10,000 copies/μl of CMV DNA. A further 10-fold dilution of the 10 copies/μl of CMV DNA was made, and the dilution was tested by the real-time PCR and LAMP assays.
Real-time PCR.
Real-time PCR was used to confirm the presence of CMV DNA in the clinical samples. The real-time PCR was performed with an Artus CMV TM PCR kit (Qiagen) on an ABI Prism 7900 apparatus (Applied Biosystems), according to the manufacturer's instructions.
RESULTS
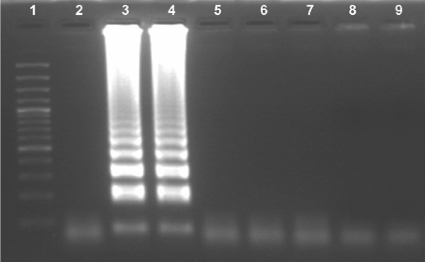
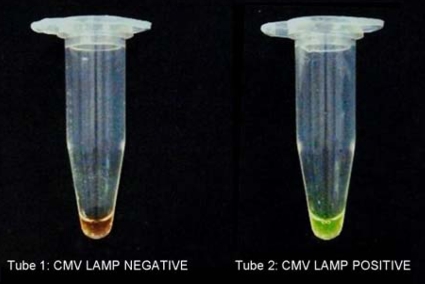
The LAMP primers amplified only CMV DNA. No LAMP products were detected with the DNAs of HSV-1, VZV, adenovirus, Aspergillus flavus, and Staphylococcus aureus (Fig. 2). The LAMP reaction tubes were examined with the naked eye to detect a positive reaction after addition of the SYBR green dye. Upon addition of the SYBR green dye to the LAMP reaction tubes, the color changed to yellowish green in a positive reaction (Fig. 3, tube 2) and remained reddish orange (the color of the unbound dye) in the negative reactions (Fig. 3, tube 1). All the samples that were positive by visual detection of a color change were also positive by gel electrophoresis (and vice versa). The sequences of the CMV LAMP products were perfectly (100%) matched with the CMV sequence deposited in the GenBank database. The CMV LAMP assay was positive in reactions with the CMV DNA at concentrations of 10 copies/μl, 100 copies/μl, 1,000 copies/μl, and 10,000 copies/μl (Fig. 4). No positivity was observed with the 10-fold dilution of the 10 copies/μl of CMV DNA by either the LAMP assay or the real-time PCR assay. The sensitivity of the LAMP assay was found to be 10 copies/μl of CMV DNA.
FIG. 2.
Specificity of LAMP assay. Lane 1, 100-bp DNA ladder; lane 2, negative control; lanes 3 and 4, CMV, for which LAMP assay products were obtained; lanes 5, 6, 7, 8, and 9, other organisms (HSV-1, VZV, adenovirus, Aspergillus flavus, and Staphylococcus aureus, respectively), for which no LAMP products were obtained. The products amplified by the LAMP assay exhibit a typical ladder-like pattern on electrophoresis, indicating the formation of stem-loop DNA with inverted repeats.
FIG. 3.
The tube with a positive reaction (tube 2) shows a color change to yellowish green, which can be distinguished from the reddish orange color of a negative reaction (tube 1).
FIG. 4.
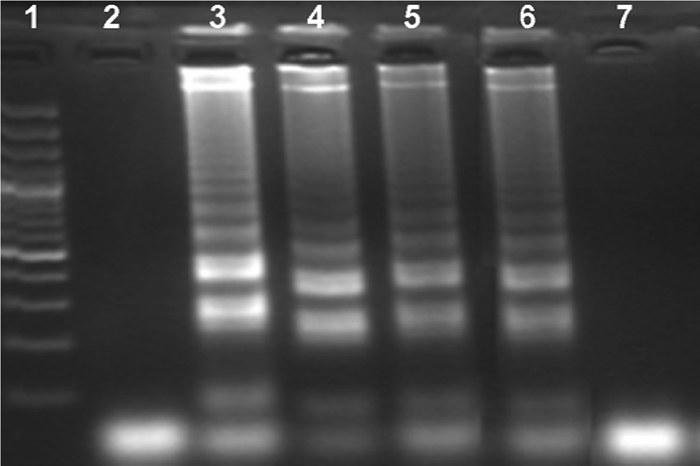
Sensitivity of LAMP assay. Lane 1, 100-bp DNA ladder; lane 2, negative control; lane 3, 10,000 copies/μl of CMV DNA; lane 4, 1,000 copies/μl of CMV DNA; lane 5, 100 copies/μl of CMV DNA; lane 6, 10 copies/μl of CMV DNA; lane 7, 10-fold dilution of 10 copies/μl of CMV DNA.
Vitreous specimens from 40 patients with clinically suspected viral retinitis were screened for CMV by the real-time PCR and LAMP assays. The LAMP assay found 10 samples to be positive and 30 samples to be negative. The real-time PCR also found 10 samples to be positive and 30 samples to be negative. Similar results were obtained by the repeat LAMP and real-time PCR assays, and the LAMP assay proved highly reproducible. A concordance between the two techniques of 100% was observed (Table 2).
TABLE 2.
Detection of CMV in vitreous samples by real-time PCR and LAMP assays
| Real-time PCR result | No. of vitreous samples with the following LAMP assay result: |
|
|---|---|---|
| Positive | Negative | |
| Positive | 10 | 0 |
| Negative | 0 | 30 |
| Total | 10 | 30 |
DISCUSSION
The PCR is a highly sensitive, specific, and rapid technique for the detection of small quantities of viral DNA in intraocular fluids (3, 4, 6). The reported sensitivity and specificity of the PCR for the diagnosis of CMV retinitis are 95% and 99%, respectively (8). Despite the clinical utility of PCR-based techniques, they have the inherent disadvantage of being time-consuming, as well as requiring considerable operator skill and expensive equipment.
In the study described here, we developed a loop-mediated isothermal amplification assay for the rapid detection of CMV in vitreous samples. The LAMP assay is a simple molecular biology diagnostic tool, in which the reaction takes place in a single tube that contains buffer as well as target DNA, Bst DNA polymerase, and primers. The tube containing this reaction mixture is incubated at 64°C in a regular laboratory water bath or heat block that maintains a constant temperature. There is no need for a thermal cycler because the entire amplification cycle can be achieved at a constant temperature. Moreover, there is no need for an electrophoresis apparatus and gel documentation system for the detection of the product amplified by the LAMP assay. The final product amplified by the LAMP assay is detected by unaided visual examination after addition of SYBR green dye to the reaction tube. A positive reaction is indicated if the tube turns yellowish green in the reaction tubes and remains reddish orange (the color of the unbound dye) in the negative reaction tubes. The time required for amplification and detection of the product is about 75 min by the LAMP assay, whereas that for PCR is 4 h.
The LAMP assay was developed and evaluated for the detection of severe acute respiratory syndrome (SARS), West Nile fever, dengue, Japanese encephalitis, and Chikungunya and BK virus infections in clinical samples; and the assay was found to be highly sensitive and specific (1, 11, 12, 13, 14). The LAMP assay is highly specific because it uses four primers (excluding the loop primers) for the detection of six distinct regions on the target DNA (10). In the present study, the assay specifically amplified only CMV; and no cross-reactivity with other viruses, bacteria, or fungi was observed. On sequencing, the sequence of the amplified product showed similarity to the CMV sequences deposited in the GenBank database. The LAMP assay is highly sensitive; we found that the lower detection limit of the LAMP assay is 10 copies of CMV DNA per microliter. We found a good correlation between the results of the real-time PCR and LAMP assays for the detection of CMV. The initial findings of this work are very promising and suggest that this test should be further evaluated in different laboratories and with a larger cohort.
In conclusion, the CMV LAMP assay is a highly sensitive, specific, rapid, cost-effective, and reliable diagnostic tool for the detection of CMV in patients with retinitis.
Acknowledgments
This work was supported by the Department of Science and Technology, Government of India, New Delhi, India (grant SR/FT/LS-008/2008).
Footnotes
Published ahead of print on 29 March 2010.
REFERENCES
- 1.Bista, B. R., C. Ishwad, R. M. Wadowsky, et al. 2007. Development of a loop-mediated isothermal amplification assay for rapid detection of BK virus. J. Clin. Microbiol. 45:1581-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cordero-Coma, M., F. Anzaar, T. Yilmaz, and C. S. Foster. 2007. Herpetic retinitis. Herpes 14:4-10. [PubMed] [Google Scholar]
- 3.Cunningham, E. T., G. A. Short, A. R. Irvine, J. S. Duker, and T. P. Margolis. 1996. Acquired immunodeficiency syndrome associated herpes simplex virus retinitis. Clinical description and use of a polymerase chain reaction based assay as a diagnostic tool. Arch. Ophthalmol. 114:834-840. [DOI] [PubMed] [Google Scholar]
- 4.de Boer, J. H., C. Verhagen, M. Bruinenberg, et al. 1996. Serologic and polymerase chain reaction analysis of intraocular fluids in the diagnosis of infectious uveitis. Am. J. Ophthalmol. 121:650-658. [DOI] [PubMed] [Google Scholar]
- 5.Fox, G. M., C. A. Crouse, E. Chuang, et al. 1996. Detection of herpes virus DNA in vitreous and aqueous specimens by the polymerase chain reaction. Arch. Ophthalmol. 121:650-656. [DOI] [PubMed] [Google Scholar]
- 6.Knox, C. M., D. Chandler, G. A. Short, and T. P. Margolis. 1998. Polymerase chain reaction-based assays of vitreous samples for the diagnosis of viral retinitis. Ophthalmology 105:37-45. [DOI] [PubMed] [Google Scholar]
- 7.Madhavan, H. N., K. Priya, and J. Biswas. 2004. Current perspectives of herpes viral retinitis and choroiditis. Indian J. Pathol. Microbiol. 47:453-468. [PubMed] [Google Scholar]
- 8.McCann, J. D., T. P. Margolis, M. G. Wong, et al. 1995. A sensitive and specific polymerase chain reaction based assay for the diagnosis of cytomegalovirus retinitis. Am. J. Ophthalmol. 120:219-226. [DOI] [PubMed] [Google Scholar]
- 9.Reference deleted.
- 10.Notomi, T., H. Okayama, H. Masubuchi, et al. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parida, M. M., P. Guillermio, S. Inoue, F. Hasebe, and K. Morita. 2004. Real-time reverse transcription loop-mediated isothermal amplification assay for rapid detection of West Nile virus. J. Clin. Microbiol. 42:257-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parida, M. M., K. Horioke, H. Ishida, et al. 2005. Rapid detection and differentiation of dengue virus serotypes by real-time reverse transcription-loop-mediated isothermal amplification assay. J. Clin. Microbiol. 43:2895-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parida, M. M., S. R. Santhosh, P. K. Dash, et al. 2007. Rapid and real-time detection of Chikungunya virus by reverse transcription loop-mediated isothermal amplification assay. J. Clin. Microbiol. 45:351-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poon, L. L. M., B. W. Y. Wong, K. H. Chan, et al. 2005. Evaluation of real-time reverse transcriptase PCR and real-time loop-mediated amplification assays for severe acute respiratory syndrome coronavirus detection. J. Clin. Microbiol. 43:3457-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]