Abstract
The zygomycete genus Lichtheimia (syn. Absidia pro parte, Mycocladus) consists of saprotrophic fungi inhabiting soil or dead plant material. Lichtheimia corymbifera (syn. Absidia corymbifera, Mycocladus corymbifer) and Lichtheimia ramosa (syn. Absidia ramosa, Mycocladus ramosus) may cause fulminant infections in patients with impaired immunity. The present study investigated the species boundaries in Lichtheimia using genealogical concordance phylogenetic species recognition (by comparison of the genealogies of the internal transcribed spacer [ITS] sequence, the D1/D2 region of the large subunit [LSU], and actin), biological species recognition by mating tests, as well as morphological and physiological characteristics. The three molecular markers used were selected by evaluating the polymorphisms and paralogies of several loci, including those for β-tubulin, translation elongation factor 1α, the two largest subunits of the RNA polymerase II (RPB1 and RPB2), the mitochondrial cytochrome c oxidase subunit I (COI), and the mitochondrial small-subunit (mtSSU) rDNA, among four strains belonging to different putative species. Comparing the genealogies of the ITS, LSU, and actin genes, we recognized seven phylogenetic species. However, mating tests did not show intrinsic reproductive barriers for two pairs of the phylogenetic species. Therefore, we regard five species in Lichtheima to be confirmed: Lichtheimia corymbifera, L. ornata comb. nov., L. ramosa, L. hyalospora, and L. sphaerocystis sp. nov. Only the first three species seem to have clinical relevance. Lichtheimia blakesleeana is reduced to a synonym of Lichtheimia hyalospora. We provide a detailed description of Lichtheimia sphaerocystis sp. nov. and a key for the identification of all accepted species identified in the present study on the basis of their morphological traits and growth at different temperatures.
Mucormycoses, i.e., infections caused by members of the Mucoromycotina subphylum, are uncommon but often dramatic, requiring immediate action on the basis of an accurate diagnosis. The recently observed increase in case reports on mucormycosis (32) can be ascribed to the growing number of patients with risk factors, such as diabetes, neutropenia, bone marrow transplantation, and the long-term use of steroids. Although this trend was already observed prior to the availability of voriconazole in medical applications (17, 21), several studies related the increasing incidence of mucormycoses to voriconazole prophylaxis and treatment of aspergillosis infection in immunocompromised patients (18, 37, 43). According to Roden et al. (32), approximately 5% of cases of mucormycoses are caused by species of Lichtheimia (syn. Absidia pro parte [p.p.], Mycocladus). These authors reviewed 25 well-documented cases of Lichtheimia (as Absidia) infections that have occurred since 1940. However, the actual incidence of the species may have been underestimated, given the fact that no less than 53 clinical Lichtheimia strains were sent to the CBS Fungal Biodiversity Centre (Utrecht, Netherlands) and the Instituto de Salud Carlos III National Centre of Microbiology (CNM-CM; Madrid, Spain) in the past 10 years alone.
Members of the genus Absidia were originally characterized by the formation of pyriform sporangia with a distinct apophysis and branched sporangiophores. Subsequent phylogenetic and physiological studies showed that Absidia-like fungi represent three separate lineages (15): (i) Absidia sensu stricto, which shows decreased growth rates above 30°C and no growth above 40°C (mesophilic) and which has zygospores that are protected by long appendages of the suspensors; (ii) Lentamyces, which does not grow at temperatures above 30°C and which consists of potential parasites of other fungi (13); and (iii) Lichtheimia, which consists of thermotolerant species that show good growth at human body temperature and which produce zygospores with equatorial rings and suspensors without appendages. Only the last group has clinical relevance. It was first named Mycocladus, typified by Mycocladus verticillatus (15). However, the type strain of that species turned out to represent a mixed culture of Absidia s. str. and, possibly, a Lentamyces species; thus, it was not congeneric with any of the thermotolerant species. Therefore, this group had to be renamed with the oldest available genus name, Lichtheimia (14), typified by Lichtheimia corymbifera.
The genus Lichtheimia basically consists of saprotrophic species inhabiting soil and decaying plant material. According to Hoffmann et al. (14), the genus Lichtheimia contained four species: L. corymbifera (syn. Absidia corymbifera, Mycocladus corymbifer), L. ramosa (syn. A. ramosa, M. ramosus), L. blakesleeana (syn. A. blakesleeana, M. blakesleeanus), and L. hyalospora (syn. A. hyalospora, M. hyalosporus). Of these, only L. corymbifera and L. ramosa have been reported from human infections. Whether L. ramosa and L. corymbifera are separate species has been a controversial issue in the past, when studies addressing this question applied phenetic criteria. For example, Ellis and Hesseltine (4) treated L. corymbifera and L. ramosa as distinct species, whereas Nottebrock et al. (28) and Schipper (36) reduced L. ramosa to synonymy with L. corymbifera. Garcia-Hermoso et al. (8) recently reestablished L. ramosa as a separate species, on the basis of sequence analyses of the internal transcribed spacer (ITS) sequence, the D1/D2 region of the large subunit (LSU), and a partial sequence of the translation elongation factor 1α (EF-1α) gene, supplemented with phenotypic characteristics. However, evidence that the clade besides the L. corymbifera clade belongs to L. ramosa remained pending because their study did not include the neotype strain of L. ramosa. They assigned the clade to L. ramosa because two isolates included (isolates CBS 269.65 and CBS 270.65) were originally identified as such. The authors found considerable sequence divergence, especially in L. ramosa. One of the strains that was morphologically similar to L. corymbifera could not be assigned to a species by the use of sequence data. Our sequence analysis of strains from all species known in the genus Lichtheimia also revealed well-supported subgroups, suggesting the existence of additional taxa.
The present study aims to explore the species boundaries in the genus Lichtheimia, to evaluate the clinical importance of each species, and to provide discriminating characteristics of clinical as well as environmental species in order to allow a reliable identification. Genealogical concordance phylogenetic species recognition (GCPSR) (40), based on the genealogies of the genes for the ITS region, the D1/D2 region of LSU, and the partial actin gene, was used to define phylogenetic species. Mating tests were performed to recognize biological species (biological species recognition), and morphology and growth characteristics were used to develop taxonomic concepts and practical features for the identification of genus and species. After evaluation of the combined data, we propose the acceptance of five species in Lichtheimia, namely, L. corymbifera, L. ramosa, L. ornata comb. nov., L. hyalospora, and L. sphaerocystis sp. nov., of which only the first three are clinically relevant.
MATERIALS AND METHODS
Strains.
A total of 53 isolates of Lichtheimia comprising 19 environmental strains, 23 clinical strains, and 11 strains of unknown sources and including all former type strains of Lichtheimia species available at the reference collection of the CBS were studied (Table 1). All strains used in this study are deposited either at the CBS, at the CNM-CM, or at the Fungal Reference Centre (PRZ; Jena, Germany).
TABLE 1.
Strains used in this studya
| Strain | Species | Country | Source | GenBank accession no(s). |
|||
|---|---|---|---|---|---|---|---|
| ITS | LSU | Actin |
|||||
| Outparalog I | Outparalog II | ||||||
| CBS 100.17 | L. corymbifera | NA | NA | GQ342885 | GQ342942 | GQ342820 | GQ342715 |
| CBS 100.31 | L. corymbifera | NA | Aborted cow | GQ342879 | GQ342914 | GQ342825, GQ342822 | GQ342722, GQ342714 |
| CBS 100.51 | L. corymbifera | NA | NA | GQ342886 | GQ342939 | GQ342826, GQ342824 | GQ342719 |
| CBS 102.48 | L. corymbifera | India | Moldy shoe | GQ342888 | GQ342910 | GQ342821 | GQ342718 |
| CBS 429.75 | L. corymbifera (NT) | Afghanistan | Soil | GQ342878 | GQ342903 | GQ342831 | GQ342712 |
| CBS 519.71 | L. corymbifera (T of A. griseola) | Japan | Kurone | GQ342889 | GQ342904 | GQ342827, GQ342832 | |
| CBS 101040 | L. corymbifera | France | Human, keratomycosis | GQ342882 | GQ342918 | GQ342830, GQ342819 | GQ342723, GQ342721 |
| CBS 109940 | L. corymbifera | Norway | Human, finger tissue | GQ342881 | GQ342917 | GQ342817 | |
| CBS 115811 | L. corymbifera | Germany | Indoor air | GQ342887 | GQ342932 | GQ342833, GQ342829 | GQ342720 |
| CBS 120580 | L. corymbifera | France | Human, lung | GQ342884 | GQ342919 | GQ342828 | GQ342713 |
| CBS 120581 | L. corymbifera | France | Human, bronchus | GQ342883 | GQ342948 | GQ342823 | GQ342716 |
| CBS 120805 | L. corymbifera | France | Human, bone | GQ342880 | GQ342915 | GQ342818 | GQ342717 |
| CBS 100.28 | L. hyalospora (T of A. blakesleeana) | USA | Bertholletia excelsa, nut | GQ342896 | GQ342902 | GQ342748, GQ342749 | |
| CBS 100.36 | L. hyalospora | NA | NA | GQ342898, GQ342897 | GQ342943 | GQ342750, GQ342751 | |
| CBS 102.36 | L. hyalospora (T of A. cristata) | Ghana | Manihot esculenta, stem | GQ342895 | GQ342907 | GQ342752, GQ342753 | |
| CBS 173.67 | L. hyalospora (NT of Tieghemella hyalospora) | Philippines | Fermented food, taosi | GQ342893 | GQ342905 | GQ342755 | |
| CBS 518.71 | L. hyalospora (T of A. blakesleeana var. atrospora) | Japan | Kurone developed during the manufacture of soy sauce (koji) | GQ342894 | GQ342944 | GQ342754, GQ342756 | |
| CBS 291.66 | L. ornata (T of A. ornata) | India | Dung of bird | GQ342891 | GQ342946 | GQ342836, GQ342837 | GQ342724, GQ342725 |
| CBS 958.68 | L. ornata | NA | NA | GQ342890 | GQ342936 | GQ342834 | GQ342726 |
| CNM-CM4978 | L. ornata | Spain | Human, wound | GQ342892 | GQ342835 | GQ342727 | |
| AS 3.4808 | L. ramosa (T of A. idahoensis var. thermophila) | China | Soil | GQ342867 | GQ342955 | GQ342770, GQ342774 | |
| CBS 100.24 | L. ramosa | NA | NA | GQ342876 | GQ342941 | GQ342814, GQ342804 | |
| CBS 100.49 | L. ramosa | Indonesia | Cow dung | GQ342858 | GQ342940 | GQ342812, GQ342791 | |
| CBS 100.55 | L. ramosa | NA | NA | GQ342851 | GQ342938 | GQ342766, GQ342771 | |
| CBS 101.51 | L. ramosa | Netherlands | Guinea pig, lung | GQ342859 | GQ342945 | GQ342796, GQ342789 | GQ342747 |
| CBS 101.55 | L. ramosa | Switzerland | Human, cornea | GQ342865 | GQ342947 | GQ342788 | GQ342731 |
| CBS 103.35 | L. ramosa (T of A. gracilis) | NA | Musa sapientum, fruit | GQ342847 | GQ342908 | GQ342763 | |
| CBS 223.78 | L. ramosa | NA | Cocoa soil | GQ342877 | GQ342934 | GQ342807, GQ342811 | GQ342734, GQ342739 |
| CBS 269.65 | L. ramosa | NA | Hay | GQ342857 | GQ342949 | GQ342801, GQ342802 | GQ342738 |
| CBS 271.65 | L. ramosa | NA | NA | GQ342875 | GQ342937 | GQ342805, GQ342816 | GQ342746, GQ342740 |
| CBS 582.65 | L. ramosa (NT) | Ghana | Theobroma cacao, seed | GQ342874 | GQ342909 | GQ342809, GQ342815 | GQ342745 |
| CBS 649.78 | L. ramosa | India | Cultivated field soil | GQ342849 | GQ342912 | GQ342779, GQ342781 | GQ342728 |
| CBS 713.74 | L. ramosa | NA | NA | GQ342856 | GQ342935 | GQ342797 | GQ342737 |
| CBS 112528 | L. ramosa | Germany | Human, wound; double infection with Candida albicans | GQ342850 | GQ342913 | GQ342764, GQ342813 | |
| CBS 124197 | L. ramosa | Greece | Human | GQ342870 | GQ342951 | GQ342842, GQ342843, GQ342844, GQ342845, GQ342846 | GQ342839 |
| CBS 124198 | L. ramosa | Netherlands | Culture contaminant | GQ342848 | GQ342906 | GQ342840, GQ342841 | GQ342838 |
| CNM-CM1638 | L. ramosa | Spain | Human, gastric juice | GQ342866 | GQ342954 | GQ342800 | GQ342730 |
| CNM-CM2166 | L. ramosa | Spain | Human, sputum | GQ342863 | GQ342926 | GQ342798, GQ342792 | |
| CNM-CM3148 | L. ramosa | Spain | Human, corneal exudate | GQ342872 | GQ342925 | GQ342768, GQ342775, GQ342782 | |
| CNM-CM3590 | L. ramosa | Spain | Human | GQ342869 | GQ342924 | GQ342785, GQ342786, GQ342810 | GQ342741, GQ342744 |
| CNM-CM4119 | L. ramosa | Spain | Human, skin | GQ342862 | GQ342923 | GQ342793, GQ342803 | GQ342742 |
| CNM-CM4228 | L. ramosa | Spain | Human, skin | GQ342861 | GQ342922 | GQ342787, GQ342794 | GQ342729 |
| CNM-CM4253 | L. ramosa | Spain | Human, skin | GQ342860 | GQ342921 | GQ342780, GQ342795 | GQ342733 |
| CNM-CM4261 | L. ramosa | Spain | Human, lung | GQ342854 | GQ342953 | GQ342767, GQ342776 | |
| CNM-CM4337 | L. ramosa | Spain | Human, skin | GQ342852 | GQ342920 | GQ342765 | |
| CNM-CM4427 | L. ramosa | Spain | Human, bronchoaspirate | GQ342853 | GQ342931 | GQ342773 | |
| CNM-CM4537 | L. ramosa | Spain | Human, skin | GQ342873 | GQ342930 | GQ342772, GQ342777 | |
| CNM-CM4849 | L. ramosa | Spain | Human, skin | GQ342855, GQ342868 | GQ342929 | GQ342769, GQ342778 | |
| CNM-CM5111 | L. ramosa | Spain | Human, sputum | GQ342871 | GQ342928 | GQ342783, GQ342784, GQ342806, GQ342808 | GQ342735, GQ342736, GQ342743 |
| CNM-CM5171 | L. ramosa | Belgium | Human | GQ342864 | GQ342927 | GQ342790, GQ342799 | GQ342732 |
| CBS 420.70 | L. sphaerocystis | India | NA | GQ342900 | GQ342933 | GQ342760, GQ342761 | |
| CBS 647.78 | L. sphaerocystis | India | Dung of mouse | GQ342899 | GQ342911 | GQ342757, GQ342759 | |
| CBS 648.78 | L. sphaerocystis | India | Soil | GQ342901 | GQ342916 | GQ342758, GQ342762 | |
NA, not available; NT, neotype strain; T, type strain.
Molecular studies. (i) Extraction of genomic DNA.
Cultures were grown for 2 days on malt extract agar (MEA; 5%, malt extract agar produced by Oxoid, Badhoevedorp, Netherlands) at 24°C. Genomic DNA was extracted according to the procedure described by Möller et al. (25) but with several modifications. Briefly, fungal material was transferred to a tube containing two glass beads and 500 ml TES buffer (124 mM Tris, 12.8 mM sodium EDTA, 87 mM sodium dodecyl sulfate [SDS], pH 8). The samples were homogenized for 3 min at 30 Hz with a TissueLyser homogenizer (Qiagen, Venlo, Netherlands) and spun for 2 min at 14,000 rpm (20,400 relative centrifugal force [RCF]). Afterwards, 5.1 U of proteinase K (in 10 μl; Sigma-Aldrich Chemie BV, Zwijndrecht, Netherlands) was added, followed by 30 min of incubation in a water bath at 55°C. A total of 120 μl of 5 M sodium chloride and 1/10 volume of 10% cetyltrimethylammonium bromide were added to the material, followed by 60 min of incubation at 65°C. The samples were again homogenized for 3 min (30 Hz) by use of the TissueLyser homogenizer. One volume of SEVAG (chloroform and isoamyl alcohol [24:1, vol/vol]) was added, and the samples were spun for 5 min at 4°C at 14,000 rpm (20,400 RCF). The upper (aqueous) phase was transferred to a new tube, and a 0.55× volume of isopropanol was added to precipitate the DNA. After incubation at −20°C for at least 30 min, the DNA was pelleted at 14,000 rpm (20,400 RCF) for 10 min at 4°C. The supernatant was decanted, and the DNA pellet was washed twice with 700 ml 70% ethanol, dried, and resuspended in 50 μl of TE buffer (12.4 mM Tris, 1.34 mM sodium EDTA, pH 8.0). The genomic DNA was stored at −20°C.
(ii) Marker selection.
We searched for genomic regions with polymorphisms comparable to the polymorphism of the ITS region in order to apply GCPSR (40). Using the DNA extracts of four strains belonging to different putative species (Lichtheimia blakesleeana [later reduced to a synonym of L. hyalospora, strain CBS 100.28], L. corymbifera CBS 100.51, L. ornata CBS 958.68, L. ramosa CBS 582.65), we amplified and sequenced parts of the genes coding for the following products: actin, β-tubulin, EF-1α, ITS, the D1/D2 region of the nuclear ribosomal large subunit (nucLSU), the largest subunit of the RNA polymerase II (RPB1), the second largest subunit of the RNA polymerase II (RPB2), mitochondrial cytochrome c oxidase subunit I (COI), and the mitochondrial ribosomal small subunit (mtSSU). The primers used are given in Table 2. Direct sequencing was possible only for the PCR products comprising the COI, LSU, mtSSU, and ITS regions for all strains except strains CBS 100.36 and CNM-CM 4849. The PCR products of the remaining loci had to be cloned before they were sequenced in the JM109 competent cell line of Escherichia coli by use of the pGEM-T Easy vector (Promega, Leiden, Netherlands), according to the instructions of the manufacturer. Similarity values, based on the uncorrected distances for the six possible pairings between the four strains, were calculated for all genomic regions tested with BioNumerics (version 0.4.61) software (Applied Maths NV, Sint-Martens-Latem, Belgium).
TABLE 2.
Primers used for amplification and sequencing of nine different genomic regions in order to select marker for GCPSR
| Region | Name of primer, primer sequence (reference), used for: |
|
|---|---|---|
| PCR | Sequencing | |
| Actin | Act-1, 5′-TGGGACGATATGGAIAAIATCTGGCA-3′ (46) | Act-1, 5′-TGGGACGATATGGAIAAIATCTGGCA-3′ (46) |
| Act-4ra, 5′-TCITCGTATTCTTGCTTIGAIATCCACAT-3′ (46) | Act-4ra, 5′-TCITCGTATTCTTGCTTIGAIATCCACAT-3′ (46) | |
| β-Tubulin | B36f modified, 5′-CACCCACTCMCTYGGTGGTG-3′ (41) | B36f modified, 5′-CACCCACTCMCTYGGTGGTG-3′ (41) |
| B12r modified, 5′-CATGAAGAARTGRAGACGVGGGAA-3′ (41) | B12r modified, 5′-CATGAAGAARTGRAGACGVGGGAA-3′ (41) | |
| COI | M-cox1-fa, 5′-GATATGGCATTTCCTCGAT-3′a | M-cox1-fa, 5′-GATATGGCATTTCCTCGAT-3′a |
| M-cox1-rb, 5′-GGWACTGCAATAATCATTGTAGC-3′a | M-cox1-rb, 5′-GGWACTGCAATAATCATTGTAGC-3′a | |
| EF-1α | MEF-1, 5′-ATGGGTAAAGARAAGACTCACG-3′ (30) | MEF-1, 5′-ATGGGTAAAGARAAGACTCACG-3′(30) |
| MEF-4, 5′-ATGACACCRACAGCGACGGTTTG-3′ (30) | MEF-4, 5′-ATGACACCRACAGCGACGGTTTG-3′ (30) | |
| MEF-11, 5′-AAGAAGATTGGTTTCAACCC-3′ (30) | ||
| MEF-21, 5′-GGGTTGAAACCAATCTTCTT-3′ (30) | ||
| ITS | V9G, 5′-TTACGTCCCTGCCCTTTGTA-3′ (2) | V9G, 5′-TTACGTCCCTGCCCTTTGTA-3′ (2) |
| LR3, 5′-GGTCCGTGTTTCAAGAC-3′ (45) | ITS1, 5′-TCCGTAGGTGAACCTGCGG-3′ (47) | |
| ITS4, 5′- TCCTCCGCTTATTGATATGC-3′ (47) | ||
| LS266, 5′-GCATTCCCAAACAACTCGACTC-3′ (9) | ||
| mtSSU | mrSSU1, 5′AGCAGTGAGGAATATTGGTC-3′ (48) | mrSSU1, 5′AGCAGTGAGGAATATTGGTC-3′ (48) |
| mrSSU3r, 5′-ATGTGGCACGTCTATAGCCC-3′ (48) | mrSSU3r, 5′-ATGTGGCACGTCTATAGCCC-3′ (48) | |
| nucLSU | V9G, 5′-TTACGTCCCTGCCCTTTGTA-3′ (2) | NL1, 5′-GCATATCAATAAGCGGAGGAAAAG-3′ (29) |
| LR3, 5′-GGTCCGTGTTTCAAGAC-3′ (45) | LR3, 5′-GGTCCGTGTTTCAAGAC-3′ (45) | |
| RPB1 | RPB1-Df modified, 5′-TAYAACGCNGATTTCGATGG-3′ (38) | RPB1-Df modified, 5′-TAYAACGCNGATTTCGATGG-3′ (38) |
| RPB1-Fr modified, 5′-CCTTCACGACCACCCATAGC-3′ (38) | RPB1-Fr modified, 5′-CCTTCACGACCACCCATAGC-3′ (38) | |
| RPB2 | RPB2-6f modified, 5′-CCYGCWGAAACKCCMGAAGG-3′b | RPB2-6f modified, 5′-CCYGCWGAAACKCCMGAAGG-3′b |
| bRPB2-7r1, 5′-CCCATRGCYTGYTTMCCCATDGC-3′b | bRPB2-7r1, 5′-CCCATRGCYTGYTTMCCCATDGC-3′b | |
Designed for this study.
B. Matheny, 2006, PCR primers for amplification and sequencing of rpb2 (RNA polymerase II second largest subunit) in the phylum Basidiomycota (Fungi) (http://www.clarku.edu/faculty/dhibbett/rpb2%20primers.htm).
For several loci, different sequences for the same strains were obtained. In order to ascertain the origin of this diversity, single-spore cultures of three strains of L. ramosa (strains CBS 223.78, CBS 124197, and CNM-CM 5111) were achieved by suspending sporangiospores in sterile distilled water and plating several dilutions of these suspensions on MEA in petri dishes. The outgrowing mycelia were isolated. The DNA was extracted and used for PCR amplification, cloning, and sequencing, as described above.
(iii) Amplification, cloning, and sequencing of the selected marker.
DNA segments comprising either the complete ITS region and the D1/D2 region of the LSU or a large part of the actin gene were amplified with the primer sets listed in Table 2. For both PCR types, the reaction mixture (25 μl) contained 0.4 μM each primer, 0.185 mM each deoxynucleoside triphosphate (GC Biotech, Alphen a/d Rijn, Netherlands), 10× NH4 BioTaq reaction buffer (GC Biotech), a final concentration of 1.5 mM MgCl2, 0.8 U BioTaq DNA polymerase (GC Biotech), and about 20 ng of DNA. The cycling conditions for the ribosomal fragment included one initial cycle at 94°C for 5 min, followed by 35 cycles of 1 min at 94°C, 1 min at 53°C, and 2 min at 72°C and then one final cycle of 7 min at 72°C. For the actin gene fragment, the following PCR conditions were applied: predenaturation for 5 min at 95°C, followed by 35 cycles of 1 min at 95°C, 1 min at 58°C, and 2 min at 72°C and then one final cycle of 7 min at 72°C. The PCRs were performed on a thermal cycler (model 2720; Applied Biosystems, Nieuwerkerk a/d IJssel, Netherlands). The reactions products were analyzed in 1% agarose gels.
PCR products comprising the actin fragment or the ITS region (in the case of strains CBS 100.36 and CNM-CM 4849) were ligated into the pGEM-T Easy vector (Promega) and cloned in E. coli JM109 competent cells (Promega), according to the manufacturer's instructions. Colony PCRs were performed with the primer pair M13f (5′-GTAAAACGACGGCCAGT-3′) and M13r (5′-GGAAACAGCTATGACCATG-3′). In the first step, four clones of each strain were sequenced. In case these clones did not include the required outparalog (see Results), we sequenced additional clones until all strains were represented by at least one sequence of outparalog I in the alignment. Both strands of the products of the primary PCR (ITS, LSU) or the colony PCR (actin) were directly cycle sequenced by use of a sequencing kit (BigDye Terminator cycle sequencing ready reaction kit; Applied Biosystems) and the primer sets given in Table 2. The cycle sequencing products were analyzed on an ABI 3730XL automatic sequencer (Applied Biosystems).
(iv) Sequence analysis.
Consensus sequences were constructed by means of the SeqMan (version 7.2.2) program (Lasergene; DNAStar) (Table 1). The sequences were aligned by use of the server version of the MAFFT program (www.ebi.ac.uk/Tools/mafft) and manually corrected in the Se-Al (version 2.0a11) program (A. Rambaut, 2002; http://tree.bio.ed.ac.uk/software/seal/). In cases in which actin gene sequences belonging to the same outparalog of a single strain were different (inparalogs), the actin gene sequences of each strain that deviated the most were included in the alignment. Phylogenetic relationships were estimated for all alignments by maximum parsimony analysis done in PAUP (phylogenetic analysis using parsimony; version 4.0b10; D. L. Swofford, 2002, Sinauer Associates, Sunderland, MA) and by use of a Bayesian approach with Monte Carlo Markov chains performed with the MrBayes (version 3.1.2) computer program (33). In maximum parsimony analyses, a heuristic search was performed with 1,000 replicates and tree-bisection-reconnection (TBR) as the branch-swapping algorithm. Gaps were treated as missing data. The robustness of the trees was estimated by bootstrap analysis with 1,000 replicates. The Jukes-Cantor-69 model of DNA substitution was selected by use of the MrAIC (version 1.4.3) program (J. A. A. Nylander, 2004, Evolutionary Biology Centre, Uppsala University) for all three alignments and used in the Bayesian analyses. Four simultaneous Monte Carlo Markov chains were run over 2 million generations by using random starting trees and the default starting parameters of the DNA substitution model. The trees were sampled every 100 generations. After the first 8,000 trees sampled were discarded (burn-in), a posteriori probabilities were estimated by computing majority rule consensus trees. Dichotomocladium elegans was used as the outgroup in analyses of the LSU because it was shown to be closely related to Lichtheimia (as Absidia p.p.) by O'Donnell (29). The analyses of actin and ITS were performed without an outgroup because the inclusion of Dichotomocladium decreased the quality of the alignment due to large sequence differences.
Morphological study. (i) Study of culture characteristic and sporangiophore morphology.
Strains were cultivated on MEA at 24°C and on synthetic Mucor agar (SMA; dextrose [Merck, Amsterdam, Netherlands], asparagine [Sigma-Aldrich Chemie BV], thiamine chloride [Sigma-Aldrich Chemie BV], agar [Oxoid]), described by Chen and Zheng (1), at 28°C in the dark. After 7 days (MEA) and 3 weeks (SMA), the texture and the color of the colonies were described, and in the case of the newly described species, the colonies were rated by using the charts of Rayner (31). Mounts for microscopic examination of the morphology of the sporangiophores were made from 7-day-old MEA cultures in two different ways for each species: (i) by pressing a piece of adhesive tape on the colony and (ii) by squashing a small portion of the colony, including the submersed mycelium. The fungal material was mounted in lactic acid with cotton blue (2 mg cotton blue ml−1 lactic acid) and in lactic acid only and examined with a microscope (eclipse 80i; Nikon, Amstelveen, Netherlands). Measurements were performed with NIS-Elements D (version 3.0) software (Nikon).
(ii) Study of giant cells.
In the search for additional distinguishing characteristics, we tested the development and structures of giant cells on three media at different temperatures: on MEA at 24°C; on potato dextrose agar (PDA; dextrose [Merck], agar [Oxoid]) at 24°C, 33°C, and 37°C; and on yeast extract agar (YEA; yeast extract [Difco, Alphen a/d Rijn, Netherlands], malt extract [Difco], dextrose [Merck], agar [Oxoid]) at 33°C and 37°C, as described by Haynes et al. (10). In the first screening, 5 strains of Lichtheimia corymbifera (strains CBS 101040, CBS 120580, CBS 120581, CBS 429.75, and CBS 519.71), 5 strains of L. ramosa (strains CBS 100.55, CBS 103.35, CBS 223.78, CBS 123197, and CBS 124198), and all 11 strains of the remaining species (Table 1) were included. After the first results became available, we extended the study on YEA at 37°C to all strains of L. corymbifera. Every week the cultures were screened for giant cells, after removal of the aerial mycelium with a sterile needle under a stereomicroscope (SMZ 1500; Nikon). Giant cells were taken from the culture and mounted in lactic acid for microscopic examination.
(iii) Mating experiments.
Sporangiospore suspensions were prepared from 5-day-old cultures grown on MEA in petri dishes at 24°C by adding roughly 2 ml of sterile distilled water and by sucking the water several times into a pipette. One or 2 drops of the suspension were placed at a distances of approximately 1 to 2 cm from the drop(s) of the second strain on YEA, as described by Haynes et al. (10), and incubated at 31°C in the dark for 7 weeks. A total number of 168 contrasts were tested, including 73 intraspecific and 95 interspecific mating tests.
(iv) Growth kinetics.
MEA plates were inoculated with small blocks taken from the edge of 3-day-old colonies. The plates were incubated at the following temperatures: 24°C, 33°C, 37°C, 40°C, 43°C, 46°C, 49°C, and 52°C. The diameter was measured twice a day for 3 days. The diameter of the colony taken 8 h after inoculation was subtracted from all following measurements. The growth rate, measured in millimeters per hour, was calculated for each strain and temperature. Descriptive and comparative statistical analyses were performed. Comparisons were done by analysis of variance (ANOVA), including the Bonferroni post hoc test. A P value of ≤0.05 was considered statistically significant. Statistical analysis was done with the help of the SPSS (version 16.0) software package (SPSS S.L., Madrid, Spain). The growth rates of each species at the different temperatures were plotted on a graph prepared with the SigmaPlot (version 11.0) program (SPSS S.L.).
Nucleotide sequence accession numbers.
The consensus sequences have been deposited in the GenBank database under accession numbers GQ342712 to GQ342955.
Mycobank accession numbers.
Two nomenclatural novelties, namely, Lichtheimia ornata and L. sphaerocystis, have been deposited in MycoBank under accession numbers MB516506 and MB516505, respectively.
RESULTS
Molecular markers for species recognition in Lichtheimia.
Table 3 lists the similarity values for the genomic regions of the six pairs of strains analyzed. In cases in which more than a single sequence of a certain locus (paralogs) was found for each strain, the highest possible similarity value for each pair of strains is given in Table 3.
TABLE 3.
Maximal similarity values between six pairs of strains for nine different genomic regionsa
| Region | Similarity values (%) between the following pairs of strains: |
Paralogous conditions | |||||
|---|---|---|---|---|---|---|---|
| L. blakesleeana CBS 100.28 and L. corymbifera CBS 100.51 | L. blakesleeana CBS 100.28 and L. ornata CBS 958.68 | L. blakesleeana CBS 100.28 and L. ramosa CBS 582.65 | L. corymbifera CBS 100.51 and L. ramosa CBS 582.65 | L. corymbifera CBS 100.51 and L. ornata CBS 958.68 | L. ramosa CBS 582.65 and L. ornata CBS 958.68 | ||
| ITS | 86.0 | 82.0 | 79.2 | 81.3 | 92.4 | 81.1 | Polymorphism among copies (paralogs) detected in two strains only, remaining strains directly sequenced |
| ß-Tubulin | 91.2 | 90.0 | 91.2 | 88.4 | 92.4 | 88.1 | Several paralogs with large sequences differences, selection of coorthologous paralogs not possible |
| RPB2 | NAb | NA | NA | 90.0 | 97.5 | 89.7 | No outparalogs detected, several inparalogs with small sequence differences |
| Actin | 91.3 | 91.0 | 94.2 | 91.6 | 96.2 | 92.1 | Two outparalogs in three species, several inparalogs with small sequence differences |
| RPB1 | 92.4 | 92.6 | 91.1 | 91.8 | 96.7 | 92.1 | Two outparalogs, several inparalogs with small sequence differences |
| LSU | 93.9 | 93.7 | 93.4 | 95.5 | 98.1 | 95.3 | Probably no polymorphism among copies (paralogs) (directly sequenced) |
| EF-1α | 94.4 | 94.1 | 94.9 | 94.0 | 97.7 | 93.0 | No outparalogs detected, numerous inparalogs with large sequence differences forming subclades (at least two duplications after species determination) |
| COI | 97.8 | 98.2 | 98.4 | 97.9 | 98.2 | 98.1 | No paralogs detected |
| mtSSU | 100 | 100 | 100 | 100 | 100 | 100 | Probably no paralogs (directly sequenced) |
The nine different genomic regions are ITS, partial β-tubulin, partial RPB2, partial actin, partial RPB1, D1/D2 region of the LSU, partial EF-1α, partial COI, and partial mtSSU.
NA, not available.
ITS was by far the most variable among all nine regions tested. The second highest degree of polymorphism was found in the nucleotide sequences of β-tubulin, followed by RPB2, actin, RPB1, LSU, EF-1α, and COI. The mitochondrial SSU was strictly monomorphic. For some of the genes studied, we detected paralogs originating from different duplication events. In order to avoid confusion between these different paralog types, we introduced the terms “outparalogs,” which evolved via an ancient duplication process preceding speciation, and “inparalogs,” which, judging from the tree topology, evolved subsequent to the speciation event (19). On the basis of the gene genealogies and species concepts maintained in this paper, gene sequences were considered outparalogous if they formed two or more clades and each clade contained the sequences of more than a single species (see the small scale in Fig. 1b). If a clade consisted purely of paralogous sequences of a single species, an inparalogous relationship was indicated. Inparalogs of a certain species are considered coorthologous to inparalogs of other species if they form a monophyletic group. This evaluation is required to ascertain the use of the correct inparalogs for phylogenetic analyses.
FIG. 1.
Maximum parsimony phylograms of three different markers: ITS (a), actin (b) (the small-scale tree includes outparalog I [blue frame] and outparalog II [red frame]; the large-scale tree represents only outparalog I [blue frame in the small-scale tree]), and the D1/D2 region of the LSU (c). Branches with bootstrap values in the maximum parsimony analysis of 85 or higher are printed in boldface. Branch support values are indicated by the numbers near the branches (maximum parsimony bootstrap proportions/Bayesian posterior probabilities) but are given only for decisive branches. Strains forming a well-supported group in the ITS phylogram are marked in a certain color. The same color is also used for each strain in the LSU and the actin phylograms. In the ITS phylogram, clinical strains are underlined in red; names are given only for type strains. For each strain of L. ramosa, L. corymbifera, and L. ornata, the predominant spore shape is mapped on the ITS phylogram: white/spherical, subspherical to broadly ellipsoidal, subspherical predominant; light gray/broadly ellipsoidal, subspherical to broadly ellipsoidal; medium gray/broadly ellipsoidal, subspherical to broadly ellipsoidal, with broadly ellipsoidal being predominant; dark gray/ellipsoidal, broadly ellipsoidal to ellipsoidal; black/ellipsoidal, ellipsoidal to broadly cylindrical. The respective numbers of clones (c) are given after the strain numbers. Actin sequences belonging to the single-spore cultures obtained in this study are indicated by ss. T, type strain, NT, neotype strain.
Our main criteria for the selection of molecular markers for species recognition were variability and, in terms of paralog identities, absent or easy-to-discriminate outparalogs enabling the unambiguous assignment of coorthologous paralogs, and small sequence differences among inparalogs (Table 3). On the basis of these criteria, we chose ITS, LSU, and actin. The absence of outparalogs as well as small sequence differences among inparalogs would also have made the RPB2 gene a promising candidate for species recognition. However, we could not obtain editable trace files for strain CBS 100.28.
The LSU could be sequenced directly for all strains. Direct sequencing of the ITS region was hampered by polymorphisms among ITS copies in two strains. Most species of Lichtheimia possess two outparalogs of actin (outparalogs I and II) which differed in their intronic and exonic sequences and which were easily distinguished. Strains listed as Lichtheimia blakesleeana and L. hyalospora and their sibling species, L. sphaerocystis, a novel species that is described below, deviated in having only outparalog I. For nearly all strains studied, we found more than one inparalog originating from the duplication of outparalog I. In all species studied with the exception of L. ramosa, inparalogs of the same strain differed by 1 to 6 bp. In L. ramosa, inparalogs of the same strain varied by from 1 up to 21 bp. Each of the three single-spore isolates of L. ramosa strains CBS 223.78, CBS 124197, and CNM-CM 5111 possessed three different inparalogs originating from outparalog I, suggesting the existence of a gene family. Some of the inparalogs (e.g., clone 2 of CNM-CM4978 and clone 1 of CBS 112528) contained single-base deletions in their exonic sequences (pseudogenes), which indicates a loss of function. The inparalogs of all Lichtheimia species belonging to outparalog I of the actin gene formed a well-supported clade in the phylogenetic tree that matched the coorthologous relation of the inparalogs (Fig. 1b).
Molecular phylogenetic analyses.
Maximum parsimony analyses based on the ITS sequence alignment (a total of 899 characteristics, of which 383 were parsimony informative) resulted in 906 most parsimonious trees (tree length [TL] = 726 steps), the LSU sequence alignment (a total of 657 characteristics, of which 78 were parsimony informative) resulted in 185 most parsimonious trees (TL = 230 steps), and the actin sequence alignment (a total of 885 characteristics, of which 280 were parsimony informative) resulted in 228 most parsimonious trees (TL = 653). The tree topologies obtained by maximum parsimony and Bayesian Markov chain Monte Carlo analyses were largely similar for all three markers and did not contain conflicting well-supported groups. Therefore, the Bayesian trees are not shown, but Bayesian posterior probabilities are included in the maximum parsimony trees in Fig. 1.
In Fig. 1, the strain designations are attributed the same color if the strains belong to a single, well-supported group in the ITS genealogy. Because the same color coding is used in the LSU and actin trees, conflicts in gene genealogies of different loci are visualized by the intermixing of colors in the supported groups of the LSU and actin trees.
In Fig. 1b, the actin tree, including outparalogs I (blue frame) and II (red frame), is pictured on a small scale. The large-scale tree represents only the outparalog I part of the tree (blue frame). We detected only outparalog I in L. blakesleeana, L. hyalospora, and L. sphaerocystis, described below, although we sequenced at least eight clones of each strain. The similarity of the inparalogous sequences resulted in a lack of distinct subclades in nearly all groups, rendering a decision on the coorthologs that were valid unnecessary. In the L. ramosa clade, strains CBS 271.65 and CBS 582.65 possessed different inparalogous sequences that were included in two distinct and well-supported subclades, one single-colored clade (pink) (strains CBS 271.65, CBS 582.65, and CBS 100.24) and another multicolored clade (pink, black, lilac) in Fig. 1b. In this case, we selected the inparalogous sequences of the multicolored clade to be valid coorthologs because of their higher degrees of similarity to the inparalogs of the remaining strains of this species. This selection did not influence the boundaries of the phylogenetic species.
The tree topologies of the ITS, LSU, and actin trees were concordant except for groups of L. ramosa strains. Three groups of L. ramosa strains supported in the ITS genealogy (indicated as lilac, vinaceous, and light blue in Fig. 1a) were not detected in the genealogies of the LSU and actin genes. The group of L. ramosa marked in pink in the ITS tree was also supported in the LSU tree but was not detected in the actin tree because the inparalogous sequences constituting the well-supported pink group in the genealogy of the actin gene were treated as invalid coorthologs, on the basis of the criteria outlined above.
Considering that phylogenetic species do not necessarily have to be monophyletic at all loci due to different rates of lineage sorting (16), the following criteria (3) were applied to recognize clades that represent independent evolutionary lineages: (i) genealogical concordance, i.e., the presence of a clade in all single-locus genealogies, regardless of the level of support, and (ii) genealogical nondiscordance, i.e., a high degree of support for the clade in at least one single-locus genealogy without a contradiction supported in another single-locus genealogy. Only relatively distinct lineages were accepted as phylogenetic species in order to avoid the possibility that minor-tip clades would have to be recognized as phylogenetic species. The boundaries of phylogenetic species were determined in such a way that no strain was left to be unclassified (3). By applying these criteria, seven phylogenetic species in Lichtheimia were recognized (Fig. 1a), as follows: (i) L. ramosa subgroup I, which contains the former type strains of Absidia gracilis (strain CBS 103.35) and Absidia idahoensis var. thermophila (strain AS3.4808); (ii) L. ramosa subgroup II, which includes the neotype strain of L. ramosa (strain CBS 582.65); (iii) L. corymbifera, which includes the neotype strain of L. corymbifera (strain CBS 429.75) and the former type strain of Absidia griseola (strain CBS 519.71); (iv) L. ornata with the former type strain of Absidia ornata (strain CBS 291.66); (v) L. blakesleeana subgroup I, which contains the former type strains of L. hyalospora (strain CBS 173.67), A. blakesleeana var. atrospora (strain CBS 518.71), and A. cristata (strain CBS 102.36); (vi) L. blakesleeana subgroup II, which contains the former type strain of L. blakesleeana (strain CBS 100.36); and (vii) L. sphaerocystis, which contains three strains with a sporangiophore morphology similar to that for L. blakesleeana.
Strain CBS 269.65, which was used as a reference strain by Garcia-Hermoso et al. (8), belongs to subgroup I of L. ramosa. Strain CNRMA/F05-100, which could not be assigned to a species by Garcia-Hermoso et al. (8), was identified as L. ornata on the basis of its ITS sequence (data not shown).
Biological species recognition by successful mating.
Zygospores were found in 17 of 168 mating experiments, including 73 intraspecific matings (Fig. 2). Azygospores were observed in a single case (L. ramosa CBS 582.65 × L. ramosa CNM-CM3590). Twelve of the 17 mating pairs producing zygospores were intraspecific matings and 5 were interspecific matings, according to the taxonomic concepts of Lichtheimia maintained in this paper. Mature zygospores developing after intraspecific matings (Fig. 3a to c) were usually dark red-brown and 58 (rarely 46) to 77 (rarely 91) μm in length by 48 (rarely 38) to 67 (rarely 82) μm in width and possessed one to five equatorial rings. The suspensors were rough and frequently unequal in size. High numbers of zygospores were usually achieved in intraspecific matings. Zygospores resulting from interspecific matings (Fig. 3d to f) were often smaller and less intensively colored (orange-brown), and they often contained large oil droplets; the equatorial rings were frequently less pronounced or absent. Only a small number (<10) of zygospores was found in three of five interspecific matings. Consequently, we consider high numbers of large, darkly pigmented zygospores with distinct equatorial rings to be an indication of successful mating within the same species. Such zygospores were present in high numbers in five matings between the two Lichtheimia ramosa subgroups and in a single mating between the two L. blakesleeana subgroups (Fig. 2). Therefore, we consider these subgroups to belong to the same biological species. In contrast, we consider the phylogenetic species L. ornata and L. sphaerocystis to be separate species because we did not find conflicting mating results and because both species possess morphological traits that distinguish them from their phylogenetic sibling species, L. corymbifera and L. blakesleeana, respectively (see below).
FIG. 2.
Results of 73 intraspecific and 95 interspecific matings in Lichtheimia. X, no mating test was performed; white field, no zygospore formation; large gray field, formation of large dark red-brown zygospores, with less than 10 zygospores being formed; small gray field, formation of smaller orange brown zygospores, with less than 10 zygospores being formed; large black field, formation of large dark red-brown zygospores, with more than 10 zygospores being formed; small black field, formation of smaller orange-brown zygospores, with more than 10 zygospores being formed. NT, neotype strain; T, type strain.
FIG. 3.
Zygospores of Lichtheimia. (a to c) Intraspecific matings: L. blakesleeana CBS 100.28 × CBS 102.36 (a), L. ramosa CBS 124198 × CBS 100.24 (b), and L. ramosa CBS 271.65 × CBS 223.78 (c). (d to f) Interspecific matings: L. blakesleeana CBS 100.28 × L. ornata CBS 958.68 (d), L. ramosa CBS 124198 × L. ornata CBS 958.68 (e), and L. blakesleeana CBS 100.36 × L. ramosa CBS 100.49 (f). Scale bar, 50 μm.
Culture characteristic and sporangiospore morphology.
Pigmentation of the colony reverse on SMA was used by Hesseltine et al. (11) and Chen and Zheng (1) to distinguish two varieties of Absidia idahoensis. Because the former type strain of A. idahoensis var. thermophila was reidentified as L. ramosa, we tested the suitability of this character for species identification. However, intraspecific variability in the pigmentation of the mycelia and the colony reverse did not exceed that between species (data not shown). Therefore, this trait was regarded to be unsuitable for use for recognition of Lichtheimia species.
The morphologies of the sporangiophores of Lichtheimia corymbifera, L. ornata, and L. ramosa were very similar. The differences in spore shape between L. corymbifera and L. ramosa were not consistent. Strains with intermediate spore shapes existed in both species (Fig. 4c and d), although the majority of L. corymbifera strains formed subglobose to broadly ellipsoidal sporangiospores (Fig. 4a) and many of the L. ramosa strains produced ellipsoidal to broadly cylindrical spores (Fig. 4b). In Fig. 1a, the preponderant spore shape in each strain is mapped on the ITS tree, demonstrating the inapplicability of this character for species identification.
FIG. 4.
Spore shape in Lichtheimia corymbifera (a and c) and L. ramosa (b and d): subglobse to broadly ellipsoidal shape of L. corymbifera (strain CBS 429.75, neotype strain) (a), ellipsoidal to cylindrical shape of L. ramosa (strain CBS 582.65, neotype strain) (b), predominantly broadly ellipsoidal shape in L. corymbifera (strain CBS 100.31) (c), and predominantly broadly ellipsoidal shape in L. ramosa (strain CBS 112528) (d).
All strains of L. blakesleeana predominantly formed subglobose or, more rarely, broadly ellipsoidal sporangiospores that differed in size and color, depending on the strain. The two strains constituting subgroup II (strains CBS 100.28 and CBS 100.36) formed small (diameter, 3.8 to 5.2 μm), hyaline to brownish, smooth to rough sporangiospores. Subgroup I of L. blakesleeana includes the former type strain of A. cristata (strain CBS 102.36), which produces spores of the same size as subgroup II, and two strains with distinctly larger sporangiospores, namely, strain CBS 173.67, the former type strain of L. hyalospora, which forms large (diameter, 4.6 to 8.3 μm), hyaline to subhyaline, smooth to slightly rough sporangiospores, and strain CBS 518.71, the former type strain of A. blakesleeana var. atrospora, which produces large (diameter, 6.0 to 11 μm), hyaline to brownish, smooth to rough sporangiospores. Both strains with large spores originated from fermented food and have probably been cultivated and subcultured for a long time. This fact and their grouping with a strain with small spores suggest that L. hyalospora and A. blakesleeana var. atrospora are morphological mutants rather than separate lineages.
Giant cells.
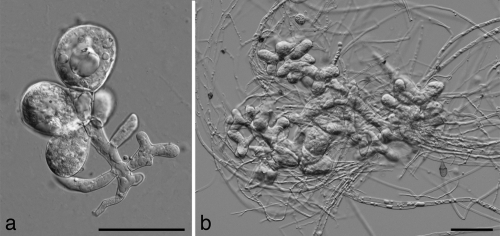
Giant cells are strongly swollen, branched or unbranched, often droplet-filled hyphae with thick, refractive walls. They are often septate, especially when they are branched, and the giant cells of some strains possess projections. Giant cells are common in all species of Lichtheimia, but their size and complexity depend on the medium and the growth temperature. On MEA at 24°C, numerous strains of all species developed giant cells. They were also common on PDA, on which the highest degrees of differentiation were noted at 24°C and with less differentiation noted at 33°C and 37°C. Cultivation on YEA at 37°C was especially appropriate for the discrimination between L. corymbifera and L. ornata. In L. blakesleeana, L. corymbifera, and L. ramosa, the size and shape of giant cells varied strongly according to the strain, the temperature, and the medium. In most strains they were slightly or distinctly branched. Two strains of L. ramosa (strains CBS 223.78 and CBS 124198) had globose, thick-walled giant cells with projections. This type of giant cell is characteristic for L. sphaerocystis (see below). L. ornata differed from its sister species, L. corymbifera, by forming large (380 to 760 [rarely 900] μm by 320 to 660 [rarely 770] μm), compact, densely branched giant cells (see Fig. 9) in 2-week-old YEA cultures. They were predominantly formed in the mycelial mat attached to the substrate and in the submerged mycelium, but they also occurred in the aerial mycelium, although they were smaller and less developed. The agar surface of the L. ornata isolates appeared to be characteristically granular on YEA, due to an abundance of large submerged giant cells. In contrast, we could not find any L. corymbifera giant cell in YEA cultures at 37°C.
FIG. 9.
Giant cells of Lichtheimia ornata formed in 2-week-old YEA cultures: younger giant cell formed by CBS 291.66 (a); mature giant cell formed by CNM-CM4978 (b). Scale bar, 100 μm.
Growth kinetics.
Figure 5 shows the mean growth rate (mm/h) and standard deviations for each species at each temperature analyzed. Significant P values (P ≤ 0.05) were found in all ANOVAs performed for each temperature. The largest distinctions between species were found at 40°C and 43°C. At 43°C, L. ramosa could clearly be distinguished from L. corymbifera and L. ornata by its growth rate, while L. hyalospora (including L. blakesleeana, which is reduced to synonymy below) and L. sphaerocystis did not grow at this temperature. L. ramosa was the species with the highest growth velocity at all temperatures. Twenty-six of 30 (86.7%) strains of L. ramosa grew at 49°C, but only 2 of 12 (16.7%) strains of L. corymbifera grew at 49°C. According to our experimental design, the maximum growth temperatures were 46°C for L. ornata, 40°C for L. blakesleeana, and 37°C for all L. sphaerocystis strains except a single strain that grew at 40°C.
FIG. 5.
Mean growth rate (mm/h) and standard deviations of Lichtheimia species at eight different temperatures.
Synopsis of the results and taxonomic conclusions.
On the basis of five successful matings suggesting the absence of an intrinsic reproductive barrier and a lack of diagnostic morphological and physiological characteristics, we considered the two subgroups of Lichtheimia ramosa to represent a single species. We also favored retention of the two phylogenetic entities of L. blakesleeana, subgroup I and subgroup II, within the same species, because of the formation of a high number of large, dark-colored zygospores found in a mating between the subgroups and because of the absence of phenotypically distinctive traits. The basionym of L. hyalospora is older than that of L. blakesleeana, and hence, the correct name for the resulting species is L. hyalospora. The strains within this species with large spores are considered mutants because they form a subgroup with environmental strain CBS 102.36, which has small spores, in all three genealogies.
The two phylogenetic entities L. corymbifera and L. ornata were distinct and well-differentiated in the genealogies of all three markers. In addition, the groups formed different types of giant cells. Therefore, we suggest that L. ornata be maintained as a separate species. L. sphaerocystis was also distinct and well differentiated in the genealogies of all three genes and differed morphologically from its sibling species, L. hyalospora, by the formation of consistently globose giant cells. For these reasons, we describe it as a new species below.
The following taxonomy is proposed (for more synonyms, see references 4 and 12).
Lichtheimia corymbifera (Cohn) Vuill., Bull. Soc. Mycol. Fr. 19:126 (1903). MycoBank accession number MB416447. Basionym: Mucor corymbifer Cohn, in Lichtheim, Z. Klin. Med. 7:149 (1884). Synonyms: Mycocladus corymbifera (Cohn) J. H. Mirza, in Mirza et al., Mucor. Pakistan (Faisalabad): 95 (1979), comb. inval., art. 36.1; Mycocladus corymbifer (Cohn) Vánová, Ceská Mykol. 45:26 (1991); Absidia griseola H. Naganishi & Hirahara, in Naganishi & Hirahara, Bull. Hiroshima Jogakuin Agric. Coll. 20:13 (1970).
Lichtheimia ramosa (Zopf) Vuill., Bull. Soc. Mycol. Fr. 19:126 (1903). Mycobank accession number MB416448. Basionym: Rhizopus ramosus Zopf, in Schenk, Handb. Botanik 4:587 (1890). Replaced synonym: Mucor ramosus Lindt, Arch. Exp. Pathol. Pharmacol. 21:269 (1886); nom. illegit., art. 53.1. Synonyms: Mycocladus ramosus (Zopf) J. H. Mirza, in Mirza et al., Mucor. Pakistan (Faisalabad): 95 (1979), comb. inval., art. 36.1; Mycocladus ramosus (Zopf) Váňová, Česká Mykol. 45:26 (1991); Absidia gracilis Linnem., Flora (Regensburg) 130:203 (1936), nom. inval., art. 36.1; Absidia idahoensis Hesseltine, M. K. Mahoney and S. W. Peterson var. thermophila G.-Q. Chen and R.-Y. Zheng, Mycotaxon 69:174 (1998).
Lichtheimia hyalospora (Saito) K. Hoffm., G. Walther and K. Voigt, Mycol. Res. 113:278 (2009). MycoBank accession number MB512830. Basionym: Tieghemella hyalospora Saito, Zentralbl. Bakteriol. Parasitenkd, Infektionskrankh. Hyg., Abt. 2, 17:103 (1906). Synonyms: Absidia hyalospora (Saito) Lendn., Mat. Fl. Cryptog. Suisse 3(1):142 (1908). Mycocladus hyalospora (Saito) J. H. Mirza, in Mirza et al., Mucor. Pakistan (Faisalabad): 97 (1979), comb. inval. Absidia blakesleeana Lendn., Bull. Soc. Bot. Genève, Sér. 2, 15:149 (1924). Mycocladus blakesleeanus (Lendn.) J. H. Mirza, in Mirza et al., Mucor. Pakistan (Faisalabad): 94 (1979). Lichtheimia blakesleeana (Lendn.) K. Hoffm., G. Walther, and K. Voigt, Mycol. Res. 113:278 (2009). Absidia blakesleeana Lendn. var. atrospora Schipper, Persoonia 14:141 (1990).
Lichtheimia ornata (A. K. Sarbhoy) A. Alastruey-Izquierdo & G. Walther, comb. nov. MycoBank accession number MB516506. Basionym: Absidia ornata A. K. Sarbhoy, Can. J. Bot. 43:999 (1965). Synonym: Absidia hesseltinei B. S. Mehrotra (as “hesseltinii”), Final Technical Report: 48 (1967), nom. inval., art. 32.1(c). Material studied: CBS 291.66 (IMI 115180, NRRL 10293), ex-type strain, India, Vindhyachal near Allahabad, from dung of bird, isolated by Botany Department, University Allahabad, no. M21, 1961, deposited by A. K. Sarbhoy, CBS H-6618; CBS 958.68 (ATCC 24263, NRRL A-11841, VKM F-1524), deposited by C. W. Hesseltine in December 1968, CNM-CM4978, Spain, Aragón, Hospital Miguel Servet, 2007, isolated from a wound of a 50-year-old male.
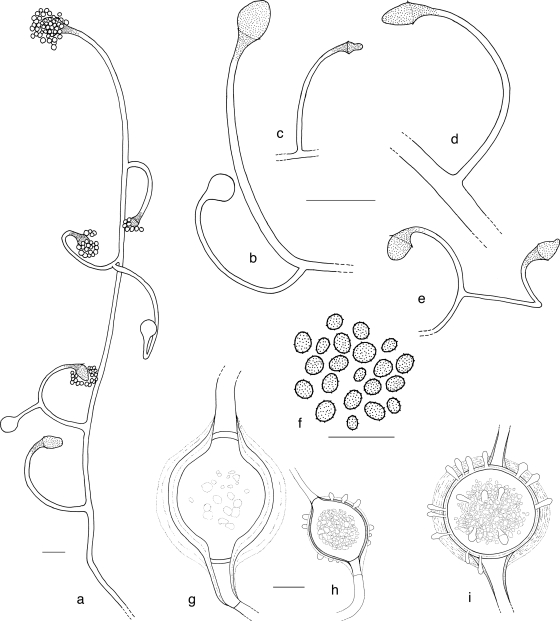
Lichtheimia sphaerocystis A. Alastruey-Izquierdo & G. Walther, sp. nov. MycoBank accession number MB516505 (Fig. 6a to i, Fig. 7a to w). Etymology: refers to the globose shape of the giant cells.
FIG. 6.
Lichtheimia sphaerocystis CBS 420.70T. (a) Aerial hypha-bearing sporangiophores (scale bar, 25 μm); (b to e) sporangiophores with columella (scale bar, 40 μm); (f) sporangiospores (scale bar, 20 μm); (g) mature giant cell; (h) young giant cell, strain CBS 648.78; (i) mature giant cell. (g to i) Scale bar, 40 μm.
FIG. 7.
Lichtheimia sphaerocystis. (a) Colony surface of strain CBS 420.70; (b) colony reverse of CBS 420.70; (c) colony surface of CBS 648.78 (predominant giant cell formation); (d) colony surface of CBS 647.78 (predominant sporangiospore formation); (e to g) CBS 647.78 sporangiophores (scale bar, 50 μm); (h to j) CBS 647.78 sporangiophores; (k to m) CBS 420.70, part of the sporangiophore with mature sporangium; (n to q) CBS 420.70 columella; (r) CBS 420.70 sporangiospores; (s to v) CBS 420.70, young (s and t) and mature (u and v) giant cells; (w) CBS 648.78 mature giant cell. (h to w) Scale bars, 20 μm.
Latin diagnosis.
Coloniae extensae, cotoneae vel coactae, albae vel cellulis giganteis copiosis cremeae vel sporangiophoris griseae, reversum ochraceum. Temperatura crescentiae optima 33°C, maxima 37°C vel 40°C. Sporangiophora simplicia vel ramosa, singula vel bina orientia, recta vel incurva vel circinata; septum subsporangiale plerumque absens. Sporangia globosa vel pyriformia, multispora, deliquescentia, atrobrunnea vel atra; sporangia maxima terminalia apophysi conica conspicua praedita, 16 vel 43 μm diameter; columellae elllipsoideae vel sursum angustatae vel raro subglobosae, rarissime una vel duabus projectionibus praeditae, 8.5 vel 33 per 6.8 vel 29 mm, collari praesente vel absente. Sporangiosporae leves vel asperulatae, hyalinae vel dilute brunneae, acervatae fuscae, subglobosae vel late ellipsoideae vel modice irregulares, 3.5 vel 7 μm diameter vel 4.2 vel 6.8 per 3.3 vel 5.5 μm. Cellulae giganteae intercalares, globosae, 60 vel 150 μm diameter (strato mucido excluso), saepe guttulatae, crassitunicatae, saepe projectionibus simplicibus vel ramosis praeditae. Stolones et rhizoidea praesentes. Species heterothallica. Zygosporae ignotae.
Colonies expanding on MEA at 24°C, cottony to felty, at first white, later, depending on the proportion of sporangia and giant cells: gray to dark gray (lavender gray to leaden gray, according to Rayner [31]) in colonies with predominant sporangiophores, consistently white to cream-colored in colonies with predominant giant cells, reverse ochreous. On MEA, optimal growth temperature 33°C, maximal growth temperature 37°C to 40°C, no growth at 43°C. Sporangiophores simple or branched, arising solitarily or in pairs but not in whorls, either directly from the substrate or from aerial hyphae, hyaline, often with light brown apophysis and columella, smooth or slightly rough, straight, bent or circinate. Subsporangial septa mostly absent. Sporangia spherical to pyriform (including apophysis), multispored, deliquescent, blackish brown to black, largest sporangia terminal, with conspicuous conical apophysis, 16 to 43 μm in diameter. Columella ellipsoidal, ellipsoidal-tapering or more rarely subglobose, occasionally with one or more projections, 8.5 to 33 by 6.8 to 29 μm, with or without collar. Sporangiospores smooth to rough walled, hyaline to light brown, dark brown in mass, subglobose to broadly ellipsoidal or slightly irregular, 3.6 to 7.0 μm in diameter or 4.2 to 6.8 by 3.3 to 5.5 μm. Large intercalary hyphal swellings (giant cells) in aerial hyphae and in the mycelium attached to the medium, but not in the mycelium permeating the medium (substrate mycelium), spherical, 60 to 150 μm in diameter (excluding the mucous layers), septate, often droplet filled, thick walled, often with simple or branched projections. Cell wall of the giant cells consisting of two refractive layers with a total thickness of 3 to 14 μm, enclosed by two to several mucous layers up to 31 μm thick. Projections 9 to 24 (rarely 40) μm long. Strain CBS 648.78 ceased to form giant cells after several transfers. Stolons and rhizoids present. Heterothallic. Zygospores not observed.
Holotype.
Strain CBS 420.70, India, March 1970, deposited by M. C. Srinivasan. Dried culture in herb. CBS, herbarium number CBS H-20402.
Other material studied: strain CBS 647.78, India, Uttar Pradesh, Gorakhpur, dung of mouse, March 1976, isolated and deposited by P. C. Misra, P.C.M. 596; strain CBS 648.78, India, Uttar Pradesh, Gorakhpur, from soil in Shorea robusta forest, March 1977, isolated and deposited by P. C. Misra, P.C.M. 623.
Key to the species.
On the basis of our morphological and physiological results, we developed the following key for the phenotypic identification of all accepted Lichtheimia species. Some characteristics for the discrimination of Absidia s. str. and Lentamyces were adopted from Hesseltine and Ellis (12) and Hoffmann and colleagues (13, 15):
1a. Subsporangial septa present; growth of aerial hyphae indeterminate; most of the sporangiophores in whorls; not thermotolerant; no or reduced growth at 37°C; zygospores with appendaged suspensors……………………...Absidia s. str.
1b. Subsporangial septa present; aerial hyphae generally ending in a sporangium; whorls of sporangiophores absent; not thermotolerant; no growth above 30°C; homothallic; zygospores warty, without appendaged suspensors……..Lentamyces
1c. Subsporangial septa absent or rare; aerial hyphae generally ending in a sporangium; whorls of sporangiophores present in some species but not obvious; thermotolerant; typically good growth at 37°C; heterothallic; zygospores with equatorial rings, without appendaged suspensors…………… ……………………………………………………..Lichtheimia, 2
2a. Sporangia dark brown or dark gray to black; no continued growth at 43°C (initial growth that may occur in rare cases stops after less than a day); mature sporangiospores rough and/or >6.5 μm in their longest extension……………………………3
2b. Sporangia light brownish gray; continued growth at 43°C; mature sporangiospores smooth and <6.5 μm in their longest extension…………….…………………………………..5
3a. Giant cells consistently globose, 60 to 150 μm in diameter (Fig. 6g to i, Fig. 7s to w) ……..Lichtheimia sphaerocystis
3b. Giant cells (if present) more hypha-like, irregularly swollen, simple to strongly branched, never consistently globose (Fig. 8) ………………………...Lichtheimia hyalospora (4)
FIG. 8.
Giant cells of Lichtheimia hyalospora strains CBS 102.36 (a) and CBS 100.36 (b) formed in PDA cultures. Scale bars, 100 μm.
4a. Mature sporangiospores small (<5.5 μm), rough, and brownish….small-spored variants of Lichtheimia hyalospora
4b. Mature sporangiospores larger (on the majority, >5.5 μm), smooth or rough, hyaline or brownish……..……………… ……………….large-spored variants of Lichtheimia hyalospora
5a. Colony diameter at 43°C after 72 h >40 mm (average growth rate, 1.3 mm/h; growth rate range, 0.5 to 3.2 mm/h); spores ellipsoidal to cylindrical or subglobose to broadly ellipsoidal….…………………………………….....Lichtheimia ramosa
5b. Colony diameter at 43°C after 72 h <27 mm (average growth rate, 0.4 mm/h; growth rate range, 0.1 to 1.0 mm/h); spores never consistently ellipsoidal to cylindrical…………...6
6a. Giant cells densely branched (Fig. 9), 380 to 760 (rarely 900) μm by 320 to 660 (rarely 770) μm, present in 2-week-old YEA cultures………………………………...Lichtheimia ornata
6b. Giant cells absent from 2-week-old YEA cultures…… …………………………………………….Lichtheimia corymbifera
Clinical relevance and distribution.
Clinical stains are underlined in red in the ITS tree (Fig. 1a). Judging from these data, only species with more pronounced thermotolerance, namely, L. corymbifera, L. ornata, and L. ramosa, are clinically relevant. Lichtheimia corymbifera and L. ramosa seem to be relatively common etiological agents, while L. ornata has been isolated from clinical sources only twice: once from the skin of a 51-year-old female undergoing an amputation in France (8) and a second time from a wound of a 50-year-old male in Spain. The data suggest similar distribution areas for all clinically relevant species, although only strains originating from Asia and Europe were available for study and the number of strains was too low to infer a geographic distribution.
DISCUSSION
Phylogenetic species recognition and molecular markers.
As far as we are aware, this is the first study that has applied GCPSR to the analysis of an entire genus of the Mucorales. We found multilocus analyses of Lichtheimia strains to be hampered by the common presence and the high number of paralogs, necessitating labor- and cost-intensive cloning steps. Another problem with the application of GCPSR is the limited variability of protein-coding genes compared to the variability of the ITS region. Hence, by the use of three molecular markers representing only two loci, the number of markers used in our study is relatively low for GCPSR. Nevertheless, we consider the resulting species concepts to be reliable because the molecular results are not in conflict with one another and, furthermore, are not in conflict with the recognized biological species that are supported by morphological and physiological findings.
The calculated error rate for the polymerase used in this study ranges from 0.02 to 0.11 errors per 1,000 bp, according to the literature cited by the manufacturer (http://www.bioline.com/faq/h_biotaqfaq.asp). However, in our study the level of polymorphism between the inparalogs was significantly higher than 1.0 per 1,000 in most cases. The low error rate of the polymerase used is further supported by the fact that all the ITS and COI sequences that were generated directly and via cloning were identical. For these reasons, we are convinced that the sequence differences detected are true polymorphisms. The abundance of paralogous sequences is also reported for Rhizopus arrhizus (as R. oryzae), another member of the Mucorales (20). The analysis of the genome sequence of R. arrhizus revealed an ancient whole-genome duplication event, together with recent gene duplications, that led to expanded gene families with a high number of paralogs.
While most eukaryotes, such as animals (26), plants (23), and slime molds (44), are known to have a small actin gene family, fungi were supposed to possess only a single copy of an actin-encoding gene, on the basis of results obtained with yeasts such as Saccharomyces cerevisiae (6) and Schizosaccharomyces pombe (24), and filamentous fungi, such as Aspergillus nidulans (5), Trichoderma reesei (22), and Neurospora crassa (42). Recently, however, the zygomycetous species Abidia glauca and the basidiomycetous fungi Schizophyllum commune and Suillus bovinus have been reported to carry two actin-encoding genes (39). Our data suggest the existence of a gene family for actin in the studied species of Lichtheimia. The two outparalogs of actin show a high degree of nucleotide polymorphisms, including exonic sequences, which suggests a duplication event before the process of diversification of the Mucorales. This fact should be taken into account when actin is used for phylogenetic analysis, especially at lower taxonomic levels, because the choice of the outparalog may influence the position of the species in the tree topology.
Sequence differences among actin inparalogs originating from the same outparalog were small, and several actin inparalogs were found in all species studied. Cases in which gene copies (repeat copies, or “inparalogs”) in a genome are more similar to each other than to their respective orthologous counterparts (repeats, or “coorthologous inparalogs”) in closely related species could be caused by (i) duplications following speciation in all species; (ii) duplications preceding speciation and the presence of mechanisms, such as concerted evolution retaining similarity among ancient duplicates; or (iii) both duplications before and after speciation, as in birth-and-death evolution combined with the permanent loss of gene copies (7, 27). We can only speculate on the origin of the paralogs (inparalogs) in the actin gene family. The most likely process is birth-and-death evolution, because it is assumed for many protein-encoding gene families (27), and our finding of pseudogenes supports this assumption. In this case, the term “inparalog” used here would be misapplied to some extent, but we kept this terminology for better understanding.
Although the presence of several inparalogs is an obvious disadvantage for use in phylogenetic studies, we think that the use of the actin gene was legitimate because of the comparatively low levels of sequence variations between actin inparalogs and the lack of an alternative locus.
Biological species recognition by mating test.
The relatively low number of successful intraspecific matings (16.4%) corresponds with the results of Ellis and Hesseltine (4), who found only 4.7% successful matings in L. corymbifera (as Absidia corymbifera). Successful interspecific matings were previously described for L. corymbifera and L. ramosa (5) and for L. ornata (strain CBS 958.68, as Absidia hesseltinei) and L. ramosa (strain CBS 270.65, as L. corymbifera) (36) and also for other genera, such as Mucor (35). As in the present study, Schipper (35) found the zygospores resulting from interspecific matings in the Mucor racemosus group to be less colored and reduced in number and size. The existence of interspecific zygospores reduces the significance of successful mating for species recognition, even though there is doubt about their occurrence under natural conditions, and necessitates a detailed analysis of the zygospores. However, mating test results are useful for the detection of the lack of an intrinsic reproductive barrier when the number, appearance, and size of the zygospores are taken into account. Obviously, mating barriers between species in the Mucorales are not as developed as they are in other fungal groups, allowing the formation of zygospores between species that share only 79.8% similarity in their ITS sequences, as in the case of the L. hyalospora CBS 100.36 × L. ramosa CBS 100.49 mating.
Diagnostic characteristics.
Our results show that sporangiophore morphology and spore shape are insufficient for the differentiation of L. corymbifera, L. ornata, and L. ramosa and that additional characteristics are needed. In contrast, Garcia-Hermoso et al. (8) found significant differences in the lengths, widths, and length/width ratios of the sporangiospores of L. corymbifera and L. ramosa. They described further discriminative characteristics, such as a different texture of the growth and slightly colored sporangiospores in L. ramosa, that we did not detect in our study. However, their morphological findings were based on the analysis of only five strains of each species, which possibly did not cover the variability of these species.
The presence of intermediate spore shapes in L. corymbifera and L. ramosa found in this study may explain why earlier mycologists tended to treat them as synonyms (28, 36). These intermediate spore shapes might also explain why studies that used the spore shape for discriminating both species encountered numerous intraspecific matings (28). The growth rates at certain temperatures as well as the appearance of giant cells provided additional valuable characteristics for species recognition.
The ITS region is the marker of choice for molecular identification because of its high degree of variability and the possibility of direct sequencing in most cases.
Taxonomy.
This study shows that L. corymbifera and L. ramosa are distinct species on the basis of molecular data, as already supposed by Ellis and Hesseltine (4), who used morphology and mating tests, and by Garcia-Hermoso (8), who used sequence analyses. Lichtheimia ornata was distinguished by the formation of “peculiar thick-walled globose to elongated bodies with very prominent finger-shaped appendages” (34, p. 999, as Absidia ornata). We consider these structures to be early stages of the giant cells that are characteristic for this species, although their size, described as 44 to 70 μm, is relatively small.
Schipper (36) already recognized L. sphaerocystis and L. hyalospora (as L. blakesleeana) as distinct taxa on the basis of morphological features, referring to strains of L. sphaerocystis as “Absidia aff. blakesleeana.” The degradation of L. blakesleeana to a synonym of L. hyalospora is supported by the presence of zygospores between strain NRRL 2983, formerly named A. hyalospora, and strain NRRL 1306, formerly named A. blakesleeana (12, p. 779). However, with respect to A. blakesleeana, the authors stated on page 782 of the same publication (12) that “the only closely related species is A. hyalospora which has much larger spores; it does not seem to fruit on Czapek's solution agar, and it does not mate with A. blakesleeana tester strains.” All three isolates of L. hyalospora studied by Hesseltine and Ellis (12) were isolated from fermented food (taosi, or soybeans). Continued subcultivation of single strains for fermentation might have enhanced the occurrence of mutants whose phenotypes can be characterized by an increase in spore size.
In conclusion, by comparing the gene genealogies on the basis of three molecular markers (ITS, actin, and LSU) and by considering the mating results and the differences in morphology and growth rates at different temperatures, we recognize five species in the genus Lichtheimia: L. corymbifera, L. ornata, L. ramosa, L. hyalospora, and L. sphaerocystis. Only the first three species seem to have clinical significance.
Acknowledgments
We are grateful to Walter Gams for providing the Latin diagnosis, for important advice in nomenclatural approaches and experimental design, and for critical reading of the manuscript. Grit Walther thanks Ursula Eberhardt for helpful discussions and advice. Kerstin Voigt thanks Ru-yong Zheng (Key Laboratory of Systematic Mycology and Lichenology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China) for providing the former type strain of Absidia idahoensis var. thermophila (strain AS 3.4808).
Footnotes
Published ahead of print on 31 March 2010.
REFERENCES
- 1.Chen, G.-Q., and R.-Y. Zheng. 1998. A new thermophilic variety of Absidia idahoensis from China. Mycotaxon 69:173-179. [Google Scholar]
- 2.de Hoog, G. S., and A. H. G. Gerrits van den Ende. 1998. Molecular diagnostics of clinical strains of filamentous basidiomycetes. Mycoses 41:183-189. [DOI] [PubMed] [Google Scholar]
- 3.Dettman, J. R., D. J. Jacobson, and J. W. Taylor. 2003. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57:2703-2720. [DOI] [PubMed] [Google Scholar]
- 4.Ellis, J. J., and G. W. Hesseltine. 1966. Species of Absidia with ovoid sporangiospores. II. Sabouraudia 5:59-77. [PubMed] [Google Scholar]
- 5.Fidel, S., J. H. Doonan, and N. R. Morris. 1988. Aspergillus nidulans contains a single actin gene which has unique intron locations and encodes a γ-actin. Gene 70:283-293. [DOI] [PubMed] [Google Scholar]
- 6.Gallwitz, D., and I. Sures. 1980. Structure of a split yeast gene: complete nucleotide sequence of the actin gene in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 77:2546-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganley, A. R. D., and T. Kobayashi. 2007. Highly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res. 17:184-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Hermoso, D., D. Hoinard, J.-C. Gantier, F. Grenouillet, F. Dromer, and E. Dannaoui. Molecular and phenotypic evaluation of Lichtheimia corymbifera (formerly Absidia corymbifera) complex isolates associated with human mucormycosis: rehabilitation of L. ramosa. J. Clin. Microbiol. 47:3862-3870. [DOI] [PMC free article] [PubMed]
- 9.Gerrits van den Ende, A. H. G., and G. S. de Hoog. 1999. Variability and molecular diagnostics of the neurotropic species Cladophialophora bantiana. Stud. Mycol. 43:151-162. [Google Scholar]
- 10.Haynes, W. C., L. J. Wickerham, and C. W. Hesseltine. 1955. Maintenance of cultures of industrially important microorganism. Appl. Microbiol. 3:361-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hesseltine, C. W., M. K. Mahoney, and S. W. Peterson. 1990. A new species of Absidia from an alkali bee brood chamber. Mycologia 82:523-526. [Google Scholar]
- 12.Hesseltine, C. W., and J. J. Ellis. 1966. Species of Absidia with ovoid sporangiospores. I. Mycologia 58:761-785. [PubMed] [Google Scholar]
- 13.Hoffmann, K., and K. Voigt. 2009. Absidia parricida plays a dominant role in biotrophic fusion parasitism among mucoralean fungi (Zygomycetes): Lentamyces, a new genus for A. parricida and A. zychae. Plant Biol. 11:537-554. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann, K., G. Walther, and K. Voigt. 2009. Mycocladus vs. Lichtheimia, a correction (Lichtheimiaceae fam. nov., Mucorales, Mucoromycotina). Mycol. Res. 113:277-278. [Google Scholar]
- 15.Hoffmann, K., S. Discher, and K. Voigt. 2007. Revision of the genus Absidia (Mucorales, Zygomycetes) based on physiological, phylogenetic, and morphological characters; thermotolerant Absidia spp. form a coherent group, Mycocladiaceae fam. nov. Mycol. Res. 111:1169-1183. [DOI] [PubMed] [Google Scholar]
- 16.Hudson, R. R., and J. A. Coyne. 2002. Mathematical consequences of the genealogical species concept. Evolution 56:1557-1565. [DOI] [PubMed] [Google Scholar]
- 17.Kaufman, C. 2004. Zygomycosis: re-emergence of an old pathogen. Clin. Infect. Dis. 39:588-590. [DOI] [PubMed] [Google Scholar]
- 18.Kontoyiannis, D. P., M. S. Lionakis, R. E. Lewis, G. Chamilos, M. Healy, C. Perego, A. Safdar, H. Kantarjian, R. Champlin, and T. J. Walsh. 2005. Zygomycosis in a tertiary-care cancer center in the era of Aspergillus-active antifungal therapy: a case-control observational study of 27 recent cases. J. Infect. Dis. 191:1350-1360. [DOI] [PubMed] [Google Scholar]
- 19.Koonin, E. V. 2005. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 39:309-338. [DOI] [PubMed] [Google Scholar]
- 20.Ma, L.-J., A. S. Ibrahim, C. Skory, M. G. Grabherr, G. Burger, M. Butler, M. Elias, A. Idnurm, B. F. Lang, T. Sone, A. Abe, S. E. Calvo, L. M. Corrochano, R. Engels, J. Fu, W. Hansberg, J.-M. Kim, C. D. Kodira, M. J. Koehrsen, B. Liu, D. Miranda-Saavedra, S. O'Leary, L. Ortiz-Castellanos, R. Poulter, J. Rodriguez-Romero, J. Ruiz-Herrera, Y.-Q. Shen, Q. Zeng, J. Galagan, B. W. Birren, C. A. Cuomo, and B. L. Wickes. 2009. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet. 5:e1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marr, K. A., R. A. Carter, F. Crippa, A. Wald, and L. Corey. 2002. Epidemiology and outcome of mould infections in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 34:909-917. [DOI] [PubMed] [Google Scholar]
- 22.Matheucci, E., Jr., F. Henrique-Silva, S. El-Gogary, C. H. B. Rossini, A. Leite, J. Escobar Vera, J. C. Carle Urioste, O. Crivellaro, and H. El-Dorry. 1995. Structure, organization and promoter expression of the actin-encoding gene in Trichoderma reesei. Gene 161:103-106. [DOI] [PubMed] [Google Scholar]
- 23.Meagher, R. B., E. C. McKinneya, and M. K. Kandasamy. 1999. Isovariant dynamics expand and buffer the responses of complex systems: the diverse plant actin gene family. Plant Cell 11:995-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mertins, P., and D. Gallwitz. 1987. A single intronless actin gene in the fission yeast Schizosaccharomyces pombe: nucleotide sequence and transcripts formed in homologous and heterologous yeast. Nucleic Acids Res. 15:7369-7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Möller, E. M., G. Bahnweg, H. Sandermann, and H. H. Geiger. 1992. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 22:6115-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mounier, N., and J. C. Sparrow. 1997. Structural comparisons of muscle and nonmuscle actins give insights into the evolution of their functional differences. J. Mol. Evol. 44:89-97. [DOI] [PubMed] [Google Scholar]
- 27.Nei, M., and A. P. Rooney. 2005. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 39:121-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nottebrock, H., H. J. Scholer, and M. Wall. 1974. Taxonomy and identification of mucormycosis-causing fungi. I. Synonymity of Absidia ramosa with A. corymbifera. Sabouraudia 12:64-74. [PubMed] [Google Scholar]
- 29.O'Donnell, K. 1993. Fusarium and its near relatives, p. 225-233. In D. R. Reynolds and J. W. Taylor (ed.), The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. CAB International, Wallingford, United Kingdom.
- 30.O'Donnell, K., F. M. Lutzoni, T. J. Ward, and G. L. Benny. 2001. Evolutionary relationships among mucoralean fungi (Zygomycota): evidence for family polyphyly on a large scale. Mycologia 93:286-296. [Google Scholar]
- 31.Rayner, R. W. 1970. A mycological colour chart. CMI and British Mycological Society, Kew, Surrey, England.
- 32.Roden, M. M., T. E. Zaoutis, W. L. Buchanan, T. A. Knudsen, T. A. Sarkisova, R. L. Schaufele, M. Sein, T. Sein, C. C. Chiou, J. H. Chu, D. P. Kontoyiannis, and T. J. Walsh. 2005. Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clin. Infect. Dis. 41:634-653. [DOI] [PubMed] [Google Scholar]
- 33.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 34.Sarbhoy, A. K. 1965. A new species of Absidia from India. Can. J. Bot. 43:999-1001. [Google Scholar]
- 35.Schipper, M. A. A. 1976. On Mucor circinelloides, Mucor racemosus and related species. Stud. Mycol. 12:1-40. [Google Scholar]
- 36.Schipper, M. A. A. 1990. Notes on Mucorales. 1. Observations on Absidia. Persoonia 14:133-148. [Google Scholar]
- 37.Siwek, G. T., K. J. Dodgson, M. de Magalhaes-Silverman, L. A. Bartelt, S. B. Kilborn, P. L. Hoth, D. J. Diekema, and M. A. Pfaller. 2004. Invasive zygomycosis in hematopoietic stem cell transplant recipients receiving voriconazole prophylaxis. Clin. Infect. Dis. 39:584-587. [DOI] [PubMed] [Google Scholar]
- 38.Stiller, J. W., and B. D. Hall. 1997. The origin of red algae: implications for plastid evolution. Proc. Natl. Acad. Sci. U. S. A. 94:4520-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarkka, M. T., R. Vasarab, M. Gorfera, and M. Raudaskoskia. 2000. Molecular characterization of actin genes from homobasidiomycetes: two different actin genes from Schizophyllum commune and Suillus bovinus. Gene 251:27-35. [DOI] [PubMed] [Google Scholar]
- 40.Taylor, J. W., D. J. Jacobson, S. Kroken, T. Kasuga, D. M. Geiser, D. S. Hibbett, and M. C. Fisher. 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 31:21-32. [DOI] [PubMed] [Google Scholar]
- 41.Thon, M. R., and D. J. Royse. 1999. Partial β-tubulin gene sequences for evolutionary studies in Basidiomycotina. Mycologia 91:468-474. [Google Scholar]
- 42.Tinsley, J. H., I. H. Lee, P. F. Minke, and M. Plamann. 1998. Analysis of actin and actin-related protein 3 (ARP3) gene expression following induction of hyphal tip formation and apolar growth in Neurospora. Mol. Gen. Genet. 259:601-609. [DOI] [PubMed] [Google Scholar]
- 43.Trifilio, S., S. Singhal, S. Williams, O. Frankfurt, L. Gordon, A. Evens, J. Winter, M. Tallman, J. Pi, and J. Mehta. 2007. Breakthrough fungal infections after allogeneic hematopoietic stem cell transplantation in patients on prophylactic voriconazole. Bone Marrow Transplant. 40:451-456. [DOI] [PubMed] [Google Scholar]
- 44.Vandekerckhove, J., and K. Weber. 1980. Vegetative Dictyostelium cells containing 17 actin genes express a single major actin. Nature 284:475-477. [DOI] [PubMed] [Google Scholar]
- 45.Vilgalys, R., and M. Hester. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172:4238-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voigt, K., and J. Wöstemeyer. 2000. Reliable amplification of actin genes facilitates deep-level phylogeny. Microbiol. Res. 155:179-195. [DOI] [PubMed] [Google Scholar]
- 47.White, T. J., T. Bruns, S. Lee, and J. W. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications, Academic Press, Inc., New York, NY.
- 48.Zoller, S., C. Scheidegger, and C. Sperisen. 1999. PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of Lichen-forming Ascomycetes. Lichenologist 31:511-516. [Google Scholar]