Abstract
Conventional methods of yeast identification are often time-consuming and difficult; however, recent studies of sequence-based identification methods have shown promise. Additionally, little is known about the diversity of yeasts identified from various animal species in veterinary diagnostic laboratories. Therefore, in this study, we examined three methods of identification by using 109 yeast samples isolated during a 1-year period from veterinary clinical samples. Comparison of the three methods—traditional substrate assimilation, fatty acid profile analysis, and sequence-based analysis of the region spanning the D1 and D2 regions (D1/D2) of the large ribosomal subunit—showed that sequence analysis provided the highest percent identification among the three. Sequence analysis identified 87% of isolates to the species level, whereas substrate assimilation and fatty acid profile analysis identified only 54% and 47%, respectively. Less-stringent criteria for identification increased the percentage of isolates identified to 98% for sequence analysis, 62% for substrate assimilation, and 55% for fatty acid profile analysis. We also found that sequence analysis of the internal transcribed spacer 2 (ITS2) region provided further identification for 36% of yeast not identified to the species level by D1/D2 sequence analysis. Additionally, we identified a large variety of yeast from animal sources, with at least 30 different species among the isolates tested, and with the majority not belonging to the common Candida spp., such as C. albicans, C. glabrata, C. tropicalis, and the C. parapsilosis group. Thus, we determined that sequence analysis of the D1/D2 region was the best method for identification of the variety of yeasts found in a veterinary population.
In both veterinary and human diagnostic laboratories, correct identification of yeasts is important for the care of patients. Traditionally, yeast identification has been performed using biochemical analysis, substrate assimilation methods, morphological examination, or various combinations of the three. To increase the ease of identification, commercial tests that use these methods have been created, but despite the convenience provided by these methods, identification of yeasts by these conventional applications can still be time-consuming and difficult. In addition, considerable variability in the efficacy of these methods has been reported for identification of clinically important yeast (11; also reviewed in references 12, 32, 36, and 42), attributed primarily to the limitations of the databases used for the comparison of clinical isolates, as well as the subjectivity involved in the interpretation of results. Recent studies have also examined the effectiveness of various molecular identification methods for yeasts by the use of rRNA genes, with the internal transcribed spacer 1 (ITS1) and ITS2 regions and the region spanning the D1 and D2 regions (D1/D2) shown to be the most useful for species-level identification of yeasts, as a result of the variability within these regions (reviewed in references 14 and 32). In particular, sequence analysis of these regions has shown great promise in the practice of clinical mycology, with several large-scale studies showing these regions to differentiate clinical yeast isolates obtained from humans to the species level (3, 4, 6, 13, 24, 26, 33).
In human medicine, the species of yeasts cultured from patients is limited, with Candida albicans being the predominant species isolated (19, 30). In contrast, veterinary yeast isolates can be cultured from a wide variety of animal species, allowing for the possibility of more diversity among the isolates identified. Though much work has been performed examining phenotypic and genotypic methods of yeast identification from veterinary sources, the majority of these studies have concentrated on a single species of either animal or yeast (2, 15, 16, 28, 29). To our knowledge, there has been only one large-scale examination of multiple yeast and animal species to assess yeast identification (5). That study used conventional phenotypic tests for identification of these organisms; no studies examining sequence-based analyses have been performed.
Therefore, in this work, we examined the variety of yeasts seen in a veterinary diagnostic laboratory during a 1-year time period and determined the feasibility and effectiveness of identification by a traditional phenotypic method, a method analyzing fatty acid profiles, and a sequence-based molecular method. We found that sequence analysis could provide identification to the genus and species level for a much higher percentage of the isolates tested than could either of the phenotypic methods. We also found that there is great diversity in the yeasts identified from veterinary sources, with at least 30 different species identified among the 109 isolates tested and only 48% belonging to the more common Candida spp., such as C. albicans, the C. parapsilosis group, C. glabrata, and C. tropicalis. Additionally, seven of the isolates were identified as Arxula adeninivorans, and this more unusual isolate was found only in horses and dolphins.
MATERIALS AND METHODS
Selection of yeasts and growth conditions.
Yeasts used in this study were obtained from the culture of clinical veterinary samples during the year 2007 by the Animal Health Diagnostic Center of Cornell University. All isolates that could be retrieved as pure cultures from frozen archived stocks were used, constituting 91% of the total isolates from that year. Isolates were plated initially onto Sabouraud dextrose agar, from which an isolated colony was subcultured onto the same medium for 24 h at 28°C. Cultures were then harvested for use with the identification methods described below.
Identification by nutrient assimilation.
Yeasts were identified using API 20 C AUX test strips (bioMérieux, Durham, NC) according to the recommendations of the manufacturer. For this study, we considered an identification classified by the manufacturer as acceptable or better to be a reliable identification. As a positive control, all samples were tested in conjunction with Candida guilliermondii ATCC 6260. This test produced a numerical profile that was then compared to a database provided by the manufacturer (bioMérieux).
Fatty acid-based identification.
Yeast isolates were harvested from the second and third quadrants of agar medium to obtain organisms in the log phase of growth. Fatty acid methyl esters were produced and then extracted using the protocol recommended by the manufacturer (MIDI, Inc., Newark, DE). Fatty acid analysis was performed using an Agilent Technologies 6890N gas chromatograph system. The Sherlock MIS YEAST28/YSTCLN software version 6.0 was used for analysis. Samples were accompanied by the reference strain Candida albicans ATCC 14053 as the positive control and a reagent blank as the negative control. The system was calibrated using a calibration mix supplied by the manufacturer (MIS no. 1200-A calibration standards kit; MIDI Inc.). This method generated a similarity index (SI), with an SI of 1.000 indicating complete identity. For this method, based on the manufacturer's recommendations, an SI of ≥0.500 with 0.100 separation from the next match was considered a good identification, while an SI of ≥0.300 with 0.100 separation was considered acceptable.
Sequence-based identification.
A small portion of a single yeast colony was placed in 5 μl of Lyse-and-Go reagent (Pierce Protein Research Products, Rockford, IL) and placed in the thermocycler, using the settings recommended by the manufacturer for genomic DNA. Upon completion, 45 μl of a PCR master mix containing 300 nM concentration of each primer was added to the Lyse-and-Go reaction. For amplification of the D1/D2 region of the large ribosomal subunit, primers NL1 and NL4 were used (20). For amplification of the ITS region, primers ITS5 and ITS4 were used (44). Samples were then placed in the thermocycler, using the following settings: 94°C for 2 min, then 30 cycles of 94°C for 30 s, 52°C (NL1 and NL4) or 54°C (ITS5 and ITS4) for 45 s, and 72°C for 90 s, with a final elongation step of 72°C for 10 min. PCRs were then visualized using a 1% agarose gel. PCR products were prepared for sequencing by using ExoSap-It (USB, Cleveland, OH), a product which removes remaining primers and deoxynucleoside triphosphates (dNTPs) by using a combination of exonuclease I and shrimp alkaline phosphatase. Sequencing was then performed by the Cornell University Sequencing Center by using primer Seq-NL1, ATCAATAAGCGGAGGAAAAG, or ITS3 (44). This protocol was used for all yeasts in this study, except for one isolate eventually identified as Candida lusitaniae. As a result of previously reported polymorphisms in the D1/D2 region (22), Seq-NL1 did not provide a sequence long enough to be used for identification, so the NL4 reverse primer was used to sequence this isolate. Sequences were then uploaded to the Ribosomal Database Project (RDP) Pipeline (8) for sequence editing and quality analysis and were then downloaded and further edited manually if necessary. Sequences were then compared to the nonredundant NCBI database by using BLASTN, with the default settings used to find the most similar sequence, and were sorted by the E score. To be considered a good identification to the species level, the identification had to meet the following criteria: (i) sequence identity of ≥99.0%; (ii) ≥1.0% separation from the next closest species; (iii) ≥90.0% sequence coverage for the matching sequence; and (iv) matching sequence published in a peer-reviewed journal article or submitted by the ATCC. For identification to the genus level, all of the above criteria were used, except that a sequence identity of ≥97.0% was used and 1.0% separation from the next species was not required. Identification criteria were based upon those proposed by the Clinical and Laboratory Standards Institute (CLSI) for the identification of microorganisms by DNA sequence analysis (7). The separation criterion was increased to ≥1.0% from the ≥0.8% proposed by the CLSI, as comparisons are provided to the nearest whole percentage by BLAST. The coverage criterion of 90% was chosen as a conservative standard to ensure the quality of compared sequences. NCBI and the Doctor Fungus website (http://www.doctorfungus.org) were used to determine whether various yeast names were anamorphs, teleomorphs, or synonyms. Additionally, as different naming schemes exist for various yeast species, we used the following rules to remain consistent among the three identification methods: (i) isolates of C. albicans and Candida africana were always called C. albicans; (ii) isolates of C. parapsilosis, Candida orthopsilosis, and Candida metapsilosis were all called members of the C. parapsilosis group; and (iii) all variants of Cryptococcus neoformans were called C. neoformans.
Creation of the phylogenetic tree.
A representative sequence from each species or isolate, in the cases where species-level identification was not reached, was aligned using CLUSTAL W2 for multiple alignment with the default settings (23) (http://www.ebi.ac.uk/Tools/clustalw2). The multiple-alignment file was then used to create a neighbor-joining phylogram with CLUSTAL W2. The C. lusitaniae sequence was excluded from the tree, since the sequence was obtained using the reverse primer and therefore would not align properly with the other sequences.
RESULTS
Sequence analysis of the D1/D2 region provides better identification of veterinary yeast isolates than does either substrate assimilation or fatty acid profile analysis.
In this study, yeast isolates were obtained from a variety of animals and sources, with representatives from mammals, reptiles, amphibians, and avians (Table 1). To determine the most effective method of identification for yeasts in the veterinary clinical laboratory, we used three methods of identification. We compared two commercially available phenotypic methods—nutrient assimilation using the API 20 C AUX test strip and fatty acid profile analysis using the Sherlock microbial identification system—to sequence analysis of the D1/D2 region of the large-subunit rRNA gene. Interpretive criteria for each of these tests are described in Materials and Methods. Using these methods and interpretive criteria, we found that 54% of the isolates tested using substrate assimilation, 47% using fatty acid profile analysis, and 87% using D1/D2 sequence analysis could be identified to the species level (Table 2). These results show that sequence analysis provided the highest percent identification to the species level of the three methods.
TABLE 1.
Animals included in study and most common yeast species isolated from each animal group
| Animal (n) | No. of different yeast species isolated | Most common yeast species isolated (% of total) |
|---|---|---|
| Avians (10) | 7 | Candida albicans (40) |
| Amphibians (1) | 1 | Debaryomyces nepalensis (100) |
| Mammals (84) | ||
| Bovines (8) | 4 | Candida glabrata (50) |
| Canines (10) | 4 | Candida albicans (70) |
| Dolphins (8) | 3 | Arxula adeninivorans (50) |
| Equines (42) | 20 | Candida parapsilosis group (14) |
| Felines (7) | 3 | Candida parapsilosis group (57) |
| Other (9) | 6 | Candida albicans (44) |
| Reptiles (14) | 11 | Candida tropicalis (21) |
TABLE 2.
Percent identification of yeast isolates by all methods tested
| Method | % Identification to: |
|
|---|---|---|
| Species levela | Genus level or lower stringencyb | |
| Substrate assimilation | 54 | 62 |
| Fatty acid profile analysis | 47 | 55 |
| D1/D2 sequence analysis | 87 | 98 |
| ITS2 sequence analysis | 79 | 90 |
Identification was considered reliable using the following criteria: for substrate assimilation, identification classified by the manufacturer as acceptable or greater to species level; for fatty acid profile analysis, SI of ≥0.500 with 0.100 separation between species; and for sequence analysis, ≥99% sequence identity with ≥1% separation between species.
Identification was considered reliable using the following criteria: for substrate assimilation, identification classified by the manufacturer as acceptable or greater to species or genus level; for fatty acid profile analysis, SI of ≥0.300 with 0.100 separation between species; and for sequence analysis, ≥97% sequence identity without the requirement for 1% separation if the two most similar sequences were of the same genus.
We next examined whether the use of less-stringent identification criteria would increase the percent identification for any of the methods. For substrate assimilation, we considered identification defined by the manufacturer as acceptable to either genus or species level; for fatty acid analysis, we used an SI of ≥0.300 with 0.100 separation; and for sequence analysis, we expanded the criteria to include sequence identity of ≥97% without the requirement of ≥1% separation if both the most similar and the second-most similar sequence were of the same genus. Using these less-stringent criteria, we found an increase in the number of yeast isolates identified by both substrate assimilation and fatty acid analysis, to 62% and 55% identification, respectively (Table 2). However, for D1/D2 sequence analysis, we found that we could identify almost all isolates to the genus level by using this criteria, with only two isolates not identified (Table 2). These results thus show that D1/D2 sequence analysis more frequently provides an identification of veterinary yeast isolates to both the species and genus level.
Sequence analysis of the ITS2 region of yeast isolates.
As we found that the identity of 13% of the yeast isolates could not be determined to the species level by sequence analysis of the D1/D2 region, we next examined whether sequence analysis using the ITS2 region would be useful for further identification of these isolates. We used a subset of the isolates described in this study, with 90 yeast isolates being selected, including the 14 isolates not previously identified to the species level by sequencing of the D1/D2 region. The entire ITS region was amplified, and the ITS2 region was sequenced. Using the same criteria for identification as those previously described for D1/D2 analysis, we found that sequence analysis of the ITS2 region could identify 79% of the 90 yeast isolates analyzed to the species level and 90% to the genus level (Table 2). We also found that sequence analysis of the ITS2 region could further identify 5 of the 14 yeast isolates not previously identified by D1/D2 analysis to the species level. Upon comparison, we found that the two methods showed agreement on the identification of isolates 91% of the time. In addition, the 9% that did not agree consisted of only two species; Geotrichum silvicola and Debaryomyces nepalensis isolates identified by analysis of the D1/D2 region were identified, respectively, as Galactomyces geotrichum and Debaryomyces hansenii by analysis of the ITS2 region. For the Geotrichum isolates, one explanation for the disagreement may be that, at the time of analysis, there were no ITS2 sequences available for Geotrichum silvicola, suggesting that the D1/D2 identification may be the correct one. For the Debaryomyces isolates, the disagreement in identification may occur because these species are very closely related, and previous work has shown that it is difficult to differentiate between the two with either the D1/D2 or the ITS region (27).
Comparison of yeast identifications derived from each method tested.
Although each of the three test methods examined provided acceptable species identification to differing degrees, the species named in each case sometimes differed, and thus the validity of the methods remained uncertain. To further examine these identification methods, we next compared the identifications determined by each. We first examined the concordance among the tests. We found that 53 isolates (49%) were named acceptably to the species level by both the substrate assimilation method and sequencing of the D1/D2 region, and among these, 49 (85%) were identified as the same species. The concordance among other test combinations was not as great. Forty-five isolates (41%) were named by both fatty acid analysis and sequencing, with 27 (60%) named in common, and 31 (28%) were identified by both fatty acid analysis and substrate assimilation, with 22 (71%) in common. The concordance also varied considerably based upon the species. For example, the 53 isolates named identically using substrate assimilation and DNA sequencing included 14 C. albicans, 12 C. parapsilosis, and 10 C. glabrata isolates, together comprising 68% of the total. Conversely, the group of 42 isolates named using sequencing but not by substrate assimilation consisted of 22 different species, with Arxula adeninivorans being most highly represented, with eight isolates (19%). These results thus suggest that more-common species, such as those of the genus Candida, can be more readily identified by all methods, but that rarer species provide discordant identifications. To determine which among these discordant identifications was likely to be correct, we next used the sequence analysis of the ITS region as an independent means of identification. We first examined 38 isolates that had been named to the species level by sequencing of the D1/D2 region, but not by the substrate assimilation method. We found that 28 (74%) were identically named by the two sequencing methods. Among the remaining, three (8%) were identified as the same species but failed to reach the level of confidence required in this study, while four (11%) showed ambiguity between the closely related species Debaryomyces hansenii and Debaryomyces nepalensis. Conversely, we had found that seven isolates could be acceptably named using substrate assimilation, but not by D1/D2 region sequencing. The identification of none of these seven, however, was supported by sequencing of the ITS region. These results thus show that in instances of discordant identification, sequencing of the D1/D2 region provided a more reliable method to identify the species of yeasts encountered in this study.
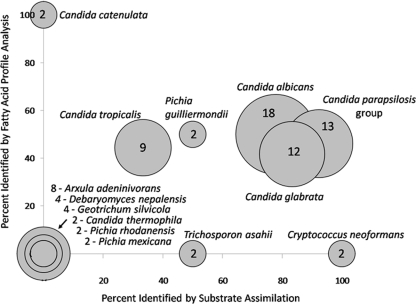
Based upon the performance of this test, we adopted sequence analysis as the reference method and compared the identifications determined by the other methods to the sequencing results by using isolates that had been identified to the species level by D1/D2 sequencing. For this analysis, we used yeasts for which we had identified at least two isolates for any given species, a total of 82 isolates. We found that substrate assimilation identified only 52% of the isolates to the species level correctly. Of the members of the genus Candida identified in this study, the most common of the genera, it identified correctly to the species level 14 of the 18 Candida albicans isolates, 12 of 13 Candida parapsilosis group isolates, 10 of 12 Candida glabrata isolates, 3 of 9 Candida tropicalis isolates, and 2 of 2 Cryptococcus neoformans isolates (Fig. 1). For Trichosporon asahii and Pichia guilliermondii, we found that nutrient assimilation could identify one of two isolates correctly for each species. All of the remaining 24 isolates identified by sequencing failed to be identified correctly by the nutrient assimilation method. These species ranged in prevalence from eight isolates (10%) for Arxula adeninivorans to two isolates each of Candida thermophila, Pichia mexicana, and Pichia rhodanensis species (Fig. 1). In comparison to DNA sequencing, fatty acid profile analysis identified 33% of the isolates correctly to the species level (Fig. 1). Only one species, Candida catenulata, was correctly identified in all cases, with both of the two isolates being identified (Fig. 1). However, for all other species, none was correctly identified more than 50% of the time. More specifically, correct identifications were obtained for 9 of 18 Candida albicans isolates, 6 of 13 Candida parapsilosis group isolates, 5 of 12 Candida glabrata isolates, 4 of 9 Candida tropicalis isolates, and 1 of 2 Pichia guilliermondii isolates (Fig. 1). The rest of the isolates, 26 of 82, were not identified correctly. These results thus suggest that although phenotypic methods may provide a species identification, such identifications may frequently be incorrect, as we found that 48% of those defined by nutrient assimilation and 67% by fatty acid analysis failed to agree with those determined by sequencing.
FIG. 1.
Comparison of identifications provided by substrate assimilation and fatty acid profile analysis to D1/D2 sequencing results. The x axis and y axis show the percentage of isolates for each species that substrate assimilation and fatty acid profile analysis identified concordantly with the D1/D2 sequencing results, respectively. The size of the bubbles is proportional to the number of yeast isolates for each species, and the numbers inside the bubbles or next to the bubbles give the exact number. Only isolates with a good identification to the species level by D1/D2 sequence analysis and where n is ≥2 were included in this comparison for a total of 82 isolates.
Diversity of yeasts in a veterinary population.
In this study, we also sought to examine the diversity of yeasts observed in a veterinary diagnostic laboratory setting. Using the results from both the D1/D2 and ITS2 sequence analyses, we identified 30 different species of yeasts among 109 isolates, which may underestimate the total, as it does not include the nine isolates that could not be identified to the species level. These results thus show that there is great variety in the yeasts identified from veterinary sources. In contrast to yeasts identified in human hospitals, only 17% of isolates were identified as Candida albicans. Additionally, less than 50% of the total belonged to the species Candida albicans, Candida tropicalis, and Candida glabrata and the Candida parapsilosis group. However, one point to note is that 39% of the yeasts were isolated from horses, and C. albicans was not isolated from any sample obtained from a horse in this study. In contrast, C. albicans was the predominant species identified in avians, canines, and other mammals (Table 1). Additionally, one yeast species that comprised 7% of the total yeast isolates was Arxula adeninivorans. Interestingly, it was isolated only from horses and dolphins, and for dolphins, it was the predominant species identified (Table 1).
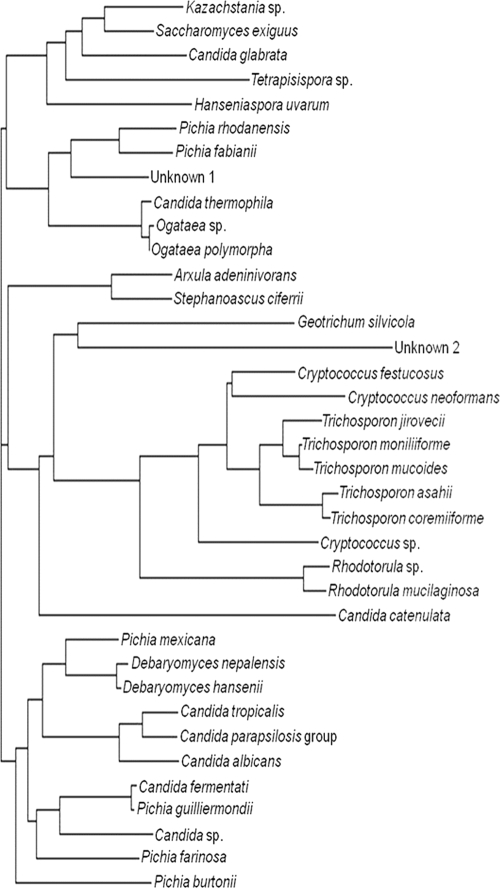
Even with use of sequence analysis of the D1/D2 region and the ITS2 region, there were still two isolates that could not be identified to the genus level by using the criteria implemented in this study. The first (Unknown 1) was isolated from a horse and had 96% sequence identity to Candida peoriaensis, and the second (Unknown 2) was isolated from a reptile and had 83% sequence identity to Candida ghanaensis. These percentages of identity were both based upon D1/D2 sequence analysis; in both cases, the percentages of identity for the ITS2 region were lower and to different organisms. These results suggest in both cases that these two isolates may represent uncharacterized species, showing that this method can be used to identify new and unusual yeast isolates. To better understand how these yeasts were related, we examined the phylogeny of representative species or isolates (in the cases where species identification was not made) identified in this study by using sequences derived from the D1/D2 region. As seen in Fig. 2, Unknown 1 falls into a group that contains various Ogataea spp., Pichia spp., and Candida spp.; interestingly all of the isolates in this particular grouping, including Unknown 1, came from equine sources. Unknown 2 falls in a group with Geotrichum silvicola, but the length of the branch indicates a distant relationship, consistent with the low percentage of identity to any known D1/D2 sequence (Fig. 2). These results therefore provide a phylogenetic framework that may be useful to the clinician when an exact identification is not possible.
FIG. 2.
Phylogenetic tree of yeast isolates. A neighbor-joining phylogram of representative D1/D2 sequences was created using the Clustal W2 multiple alignment tool.
DISCUSSION
In this study, we examined the usefulness of sequence analysis in comparison to more traditional phenotypic methods for identification of veterinary yeast isolates and found that sequence analysis provided more effective identification of the yeast isolates than did either of the phenotypic methods tested. Substrate assimilation identified only 54% of the isolates to the species level and 62% to the genus level. These results are not completely surprising since there are reports in the literature of 59.6% to 99.3% identification for the API 20 C (11, 36, 42). For fatty acid profile analysis, we could identify 47% of the isolates to the species level and 55% by using lower stringency for identification. These results are lower than previously reported; however, depending on the growth conditions, previous studies showed species-level identification of 49% to 71% (9, 17, 18). Thus, our results for the phenotypic methods were slightly lower than previously reported, but this may be a result of the diversity of veterinary yeast isolates. Significant to note is that the percentages of identification described above represent all of those considered to be acceptable to the species level by each method; they do not take into account whether the identification was correct. Upon comparison of these methods to the isolates that had good identifications to the species level by sequence analysis, we found that only 52% and 33% of the identifications made by substrate assimilation and fatty acid profile analysis, respectively, were consistent with the identification obtained using sequence analysis. These results thus suggest that veterinary isolate identifications by the use of these methods may frequently be incorrect.
Though sequence analysis of either the D1/D2 region or the ITS2 region provided higher percent identification than those provided by either of the phenotypic methods, there were still limitations with each of these methods. Sequence analysis of the D1/D2 and ITS2 regions provided identifications of only 87% and 79% of the isolates to the species level, respectively. The percentages of identification for both the D1/D2 region and ITS regions are lower than previous reports for human clinical isolates (6, 13, 24, 26, 33, 34). However, this may be attributed to the variety of yeasts isolated from animal sources, as well the number of uncommon isolates found in a veterinary population. For example, in this work we found that Candida albicans accounted for only 17% of the isolates and that other common Candida spp., such as Candida tropicalis, Candida glabrata, and the Candida parapsilosis group, accounted for only 31%. Thus, the majority of yeasts isolated from veterinary sources do not belong to the species most commonly isolated from human sources (30, 39). In fact, one yeast that comprised 7% of the total yeast isolates was Arxula adeninivorans, also known as Blastobotrys adeninivorans (21). It has previously been characterized as an anamorphic, nonpathogenic, halotolerant, and dimorphic yeast isolated from environmental sources such as soil, maize silage, and wood hydrolysates and has been studied to determine its usefulness in biotechnological applications (reviewed in reference 43). As mentioned above, it is not thought to be a pathogen, though recently a case of Blastobotrys proliferans was reported as the causative agent of peritonitis in a human (35). However, currently the clinical significance of these isolates is not known, but the ability to identify them may allow for future understanding of their relevance to veterinary medicine.
Of the isolates that could not be identified to the species level by using sequencing of the D1/D2 region, the lack of adequate separation between the two most similar species in the database was the most frequent cause. This may occur with closely related species, and previous reports suggest that the differentiation of closely related species may require sequence analysis of the ITS region (10, 38). However, in our study, we found that sequencing the ITS2 region provided identification of only 36% of the isolates not previously identified to the species level by D1/D2. Additionally, for the majority of isolates that could not be identified to the species level by sequencing of the ITS2 region, the inability to identify the organism could be attributed to low sequence identity to the sequences present in the NCBI database that met the criteria described in Materials and Methods. Therefore, in contrast to the D1/D2 region, where the difficulty in identification was a result of a lack of adequate separation between the two most similar species, the difficulty with identification using the ITS2 region was a result of a lack of available ITS2 sequences in the database for some of the species isolated in this study. Thus, based on the current database available, the best identification method for veterinary isolates is sequence analysis of the D1/D2 region of the large ribosomal subunit.
Another complication of sequence analysis is that the NCBI database is constantly growing and is not highly curated and, as such, includes sequences that may not be well supported. Additionally, naming conventions for yeasts are constantly changing. In addition to the confusion caused by certain yeasts having both anamorphic and teleomorphic names, there are also many yeasts that have synonymous names. Furthermore, with the advent of molecular phylogenetic analysis, there have been several new species suggested as subdivisions of traditional species. For example, a recent study has suggested that the species Candida africana may be a separate species from C. albicans (41), but there is still uncertainty about this distinction (1, 37). Similarly, recent work has shown that Candida parapsilosis groups II and III are actually separate species, Candida orthopsilosis and Candida metapsilosis, respectively (40), and that Cryptococcus neoformans has been described to have variants (25). These changes in naming conventions present significant obstacles to the correct identification of yeast isolates in the clinical setting. As all of the methods tested here rely upon comparisons to databases for proper identification, the maintenance of those databases to accurately reflect the currently accepted nomenclature is essential. Therefore, as the limitations involved in all methods, including sequencing, are a result of database maintenance, one way to address this issue may be to create databases of sequences derived from the NCBI collection within individual laboratories. By creating an in-house database, a laboratory might control the quality of the sequences and use the sequences of isolates most commonly seen in the particular laboratory setting. In this case, sequences from this study could be used to create a database specific for yeasts commonly isolated from veterinary sources.
One consideration for the implementation of sequence analysis in the clinical laboratory is cost-effectiveness. Examination of the three methods described in this work showed that consumables cost $12 for substrate assimilation using the API 20 C AUX kit, $3 for fatty acid profile analysis, and $5 to $6 for sequence analysis. However, other important cost considerations include a large capital investment in the gas chromatograph system for fatty acid profile analysis and, for sequence analysis, access to a reliable sequencing center that can provide the service at a reasonable cost. Additional considerations for all three methods include the turnaround time for each test. Using fatty acid analysis and sequence analysis, a turnaround time of 1 to 2 days is possible, whereas use of substrate assimilation requires 4 to 5 days. Therefore, individual laboratories must take into account the volume of testing, the need for expedience, and the availability of equipment or sequencing services for their individual labs to determine the best identification option. However, based on the results presented here and our analysis of costs, we found that sequence analysis of the D1/D2 region of the large ribosomal subunit was the most accurate and cost-efficient method of yeast identification for this veterinary diagnostic laboratory.
Acknowledgments
Funding for this study was provided by the Animal Health Diagnostic Center of Cornell University.
Footnotes
Published ahead of print on 14 April 2010.
REFERENCES
- 1.Alonso-Vargas, R., L. Elorduy, E. Eraso, F. J. Cano, J. Guarro, J. Ponton, and G. Quindos. 2008. Isolation of Candida africana, probable atypical strains of Candida albicans, from a patient with vaginitis. Med. Mycol. 46:167-170. [DOI] [PubMed] [Google Scholar]
- 2.Brito, E. H., R. S. Brilhante, R. A. Cordeiro, J. J. Sidrim, R. O. Fontenelle, L. M. Melo, E. S. Albuquerque, and M. F. Rocha. 2009. PCR-AGE, automated and manual methods to identify Candida strains from veterinary sources: a comparative approach. Vet. Microbiol. 139:318-322. [DOI] [PubMed] [Google Scholar]
- 3.Chen, Y. C., J. D. Eisner, M. M. Kattar, S. L. Rassoulian-Barrett, K. Lafe, U. Bui, A. P. Limaye, and B. T. Cookson. 2001. Polymorphic internal transcribed spacer region 1 DNA sequences identify medically important yeasts. J. Clin. Microbiol. 39:4042-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Y. C., J. D. Eisner, M. M. Kattar, S. L. Rassoulian-Barrett, K. LaFe, S. L. Yarfitz, A. P. Limaye, and B. T. Cookson. 2000. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes. J. Clin. Microbiol. 38:2302-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chengappa, M. M., R. L. Maddux, S. C. Greer, D. H. Pincus, and L. L. Geist. 1984. Isolation and identification of yeasts and yeastlike organisms from clinical veterinary sources. J. Clin. Microbiol. 19:427-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciardo, D. E., G. Schar, E. C. Bottger, M. Altwegg, and P. P. Bosshard. 2006. Internal transcribed spacer sequencing versus biochemical profiling for identification of medically important yeasts. J. Clin. Microbiol. 44:77-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CLSI. 2008. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; approved guideline. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crist, A. E., Jr., L. M. Johnson, and P. J. Burke. 1996. Evaluation of the Microbial Identification System for identification of clinically isolated yeasts. J. Clin. Microbiol. 34:2408-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fell, J. W., T. Boekhout, A. Fonseca, G. Scorzetti, and A. Statzell-Tallman. 2000. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 50(Pt. 3):1351-1371. [DOI] [PubMed] [Google Scholar]
- 11.Fenn, J. P., H. Segal, B. Barland, D. Denton, J. Whisenant, H. Chun, K. Christofferson, L. Hamilton, and K. Carroll. 1994. Comparison of updated Vitek Yeast Biochemical Card and API 20C yeast identification systems. J. Clin. Microbiol. 32:1184-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freydiere, A. M., R. Guinet, and P. Boiron. 2001. Yeast identification in the clinical microbiology laboratory: phenotypical methods. Med. Mycol. 39:9-33. [DOI] [PubMed] [Google Scholar]
- 13.Hall, L., S. Wohlfiel, and G. D. Roberts. 2003. Experience with the MicroSeq D2 large-subunit ribosomal DNA sequencing kit for identification of commonly encountered, clinically important yeast species. J. Clin. Microbiol. 41:5099-5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwen, P. C., S. H. Hinrichs, and M. E. Rupp. 2002. Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Med. Mycol. 40:87-109. [DOI] [PubMed] [Google Scholar]
- 15.Kano, R., Y. Hattori, K. Okuzumi, Y. Miyazaki, R. Yamauchi, H. Koie, T. Watari, and A. Hasegawa. 2002. Detection and identification of the Candida species by 25S ribosomal DNA analysis in the urine of candidal cystitis. J. Vet. Med. Sci. 64:115-117. [DOI] [PubMed] [Google Scholar]
- 16.Kano, R., Y. Sakamoto, A. Hanahachi, H. Kamata, Y. Fukuda, K. Fujiwara, and A. Hasegawa. 2001. Molecular identification of Candida parapsilosis from crop mucosa in a cockatiel. J. Vet. Diagn. Invest. 13:437-439. [DOI] [PubMed] [Google Scholar]
- 17.Kellogg, J. A., D. A. Bankert, and V. Chaturvedi. 1998. Limitations of the current microbial identification system for identification of clinical yeast isolates. J. Clin. Microbiol. 36:1197-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kellogg, J. A., D. A. Bankert, and V. Chaturvedi. 1999. Variation in Microbial Identification System accuracy for yeast identification depending on commercial source of Sabouraud dextrose agar. J. Clin. Microbiol. 37:2080-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kojic, E. M., and R. O. Darouiche. 2003. Comparison of adherence of Candida albicans and Candida parapsilosis to silicone catheters in vitro and in vivo. Clin. Microbiol. Infect. 9:684-690. [DOI] [PubMed] [Google Scholar]
- 20.Kurtzman, C. P., and C. J. Robnett. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurtzman, C. P., and C. J. Robnett. 2007. Multigene phylogenetic analysis of the Trichomonascus, Wickerhamiella and Zygoascus yeast clades, and the proposal of Sugiyamaella gen. nov. and 14 new species combinations. FEMS Yeast Res. 7:141-151. [DOI] [PubMed] [Google Scholar]
- 22.Lachance, M. A., H. M. Daniel, W. Meyer, G. S. Prasad, S. P. Gautam, and K. Boundy-Mills. 2003. The D1/D2 domain of the large-subunit rDNA of the yeast species Clavispora lusitaniae is unusually polymorphic. FEMS Yeast Res. 4:253-258. [DOI] [PubMed] [Google Scholar]
- 23.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 24.Leaw, S. N., H. C. Chang, H. F. Sun, R. Barton, J. P. Bouchara, and T. C. Chang. 2006. Identification of medically important yeast species by sequence analysis of the internal transcribed spacer regions. J. Clin. Microbiol. 44:693-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, X., and J. Heitman. 2006. The biology of the Cryptococcus neoformans species complex. Annu. Rev. Microbiol. 60:69-105. [DOI] [PubMed] [Google Scholar]
- 26.Linton, C. J., A. M. Borman, G. Cheung, A. D. Holmes, A. Szekely, M. D. Palmer, P. D. Bridge, C. K. Campbell, and E. M. Johnson. 2007. Molecular identification of unusual pathogenic yeast isolates by large ribosomal subunit gene sequencing: 2 years of experience at the United Kingdom Mycology Reference Laboratory. J. Clin. Microbiol. 45:1152-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martorell, P., M. T. Fernandez-Espinar, and A. Querol. 2005. Sequence-based identification of species belonging to the genus Debaryomyces. FEMS Yeast Res. 5:1157-1165. [DOI] [PubMed] [Google Scholar]
- 28.Milde, K., V. Kostka, E. F. Kaleta, H. Willems, and C. Jager. 2000. Multiplex-PCR-based differentiation and characterization of Candida-isolates derived from tortoises (Testudinidae). Vet. Microbiol. 76:395-402. [DOI] [PubMed] [Google Scholar]
- 29.Ozawa, H., K. Okabayashi, R. Kano, T. Watari, S. Watanabe, and A. Hasegawa. 2005. Rapid identification of Candida tropicalis from canine cystitis. Mycopathologia 160:159-162. [DOI] [PubMed] [Google Scholar]
- 30.Paulo, C., C. Mourao, P. M. Veiga, J. M. Marques, G. Rocha, A. F. Alves, A. Querol, A. A. Melico-Silvestre, I. Goncalves, O. Flores, C. Clemente, and T. Goncalves. 2009. Retrospective analysis of clinical yeast isolates in a hospital in the centre of Portugal: spectrum and revision of the identification procedures. Med. Mycol. 19:1-10. [DOI] [PubMed] [Google Scholar]
- 31.Reference deleted.
- 32.Pincus, D. H., S. Orenga, and S. Chatellier. 2007. Yeast identification—past, present, and future methods. Med. Mycol. 45:97-121. [DOI] [PubMed] [Google Scholar]
- 33.Pryce, T. M., S. Palladino, I. D. Kay, and G. W. Coombs. 2003. Rapid identification of fungi by sequencing the ITS1 and ITS2 regions using an automated capillary electrophoresis system. Med. Mycol. 41:369-381. [DOI] [PubMed] [Google Scholar]
- 34.Putignani, L., M. G. Paglia, E. Bordi, E. Nebuloso, L. P. Pucillo, and P. Visca. 2008. Identification of clinically relevant yeast species by DNA sequence analysis of the D2 variable region of the 25-28S rRNA gene. Mycoses 51:209-227. [DOI] [PubMed] [Google Scholar]
- 35.Quirin, N., M. Desnos-Ollivier, J. F. Cantin, J. C. Valery, Y. Doussy, R. Goursaud, F. Dromer, and J. M. Tivollier. 2007. Peritonitis due to Blastobotrys proliferans in a patient undergoing continuous ambulatory peritoneal dialysis. J. Clin. Microbiol. 45:3453-3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramani, R., S. Gromadzki, D. H. Pincus, I. F. Salkin, and V. Chaturvedi. 1998. Efficacy of API 20C and ID 32C systems for identification of common and rare clinical yeast isolates. J. Clin. Microbiol. 36:3396-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romeo, O., and G. Criseo. 2009. Morphological, biochemical and molecular characterisation of the first Italian Candida africana isolate. Mycoses 52:454-457. [DOI] [PubMed] [Google Scholar]
- 38.Scorzetti, G., J. W. Fell, A. Fonseca, and A. Statzell-Tallman. 2002. Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2:495-517. [DOI] [PubMed] [Google Scholar]
- 39.Swinne, D., N. Nolard, P. Van Rooij, and M. Detandt. 2009. Bloodstream yeast infections: a 15-month survey. Epidemiol. Infect. 137:1037-1040. [DOI] [PubMed] [Google Scholar]
- 40.Tavanti, A., A. D. Davidson, N. A. Gow, M. C. Maiden, and F. C. Odds. 2005. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J. Clin. Microbiol. 43:284-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tietz, H. J., M. Hopp, A. Schmalreck, W. Sterry, and V. Czaika. 2001. Candida africana sp. nov., a new human pathogen or a variant of Candida albicans? Mycoses 44:437-445. [DOI] [PubMed] [Google Scholar]
- 42.Verweij, P. E., I. M. Breuker, A. J. Rijs, and J. F. Meis. 1999. Comparative study of seven commercial yeast identification systems. J. Clin. Pathol. 52:271-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wartmann, T., and G. Kunze. 2000. Genetic transformation and biotechnological application of the yeast Arxula adeninivorans. Appl. Microbiol. Biotechnol. 54:619-624. [DOI] [PubMed] [Google Scholar]
- 44.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-320. In M. A. Innis, D. H. Gelfrand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, CA.