Abstract
Knowledge of circulating Chlamydia trachomatis serovars can be beneficial for sexual network surveillance, monitoring treatment success, and associating specific clinical manifestations. Typically, C. trachomatis serovars are predicted by nucleotide sequencing of four variable domains within the ompA gene. However, sequencing procedures can be labor-intensive, are not readily available, and can lack the capacity to identify multiple serovars. This study describes the development and evaluation of a quantitative real-time PCR (qPCR) test algorithm for the rapid prediction of C. trachomatis serovars, including ocular (A to C) and anogenital (D to L3) strains. This test comprises a primary qPCR to confirm C. trachomatis positivity and the phylogenetic group(s) present and a secondary set of qPCRs to determine specific serovars. Cell culture isolates from 15 prototypic C. trachomatis serovars were correctly identified using this assay, with no cross-reactivity observed among serovars or with other common pathogenic microorganisms. Five hundred clinical specimens (previously diagnosed as being C. trachomatis positive) were evaluated by qPCR, with their results compared to results obtained by conventional sequencing. The qPCR identified 88.9% (423/476) complete matches (95% confidence interval [CI], 86 to 92%) of serovars compared to the results obtained using the sequence-based approach. Among the anogenital specimens, 2.4% (12/494) (95% CI, 1.3 to 4.2%) contained multiple serovars, categorized as single-serovar infections by conventional sequencing. Overall, this test exhibited high discriminatory success for predicting C. trachomatis serovars, particularly among anogenital infections. This is the first report of a qPCR typing assay offering differentiation of C. trachomatis serovars associated with both anogenital and ocular diseases.
Chlamydia trachomatis is the causative agent for one of the most common sexually transmissible infections worldwide. The majority of infections are asymptomatic and thereby left untreated, which can lead to a variety of human diseases across multiple organ systems. Urogenital infections are the most predominant, often associated with urethritis and epididymitis in men and cervicitis and pelvic inflammatory disease, with the potential outcome of ectopic pregnancy or tubal infertility, in women (3, 4, 5, 21, 30). C. trachomatis infections are also responsible for trachoma-associated blindness (16) and lymphogranuloma venereum (LGV), characterized by genital ulcers, inguinal lymphadenopathy, or proctocolitis (13).
C. trachomatis strains are divided into 15 distinct serovars based on their antigenic reactivity with specific monoclonal antibodies (17, 29). These different serovars are associated with characteristic clinical manifestations; notably, serovars A to C are associated with ocular disease (trachoma and conjunctivitis), serovars D to K with typical urogenital disease (6), and serovars L1 to L3 with systemic LGV (27, 29). Currently, C. trachomatis serovar prediction relies on nucleotide sequencing of the ompA gene, which encodes the major outer membrane protein (MOMP). The ompA gene contains four symmetrically spaced variable domains (VDs; VDI to VDIV), encoding major antigenic determinants flanked and interspaced by five conserved domains (1, 24, 32). Distinct nucleotide sequence variations within the ompA VDs of different serovars have been shown to facilitate C. trachomatis serovar prediction (2, 10, 12, 27). C. trachomatis serovars can be subdivided into three distinct phylogenetic clades based on the ompA gene: the B group (comprising B/Ba, D, E, L1, and L2), the C group (comprising A, C, H, I, J, K, and L3), and the intermediate (I) group (comprising F and G). Although there is no correlation of phylogenetic clades with characteristic disease, these clades/groups have value for the development of more rapid screening assays for predicting serovars (19, 20, 33).
Determination of circulating C. trachomatis serovars within a population can provide information on the epidemiology and pathogenesis of infection (7), including mapping sexual networks, can allow for monitoring treatment success, and may play a role in developing strategies for improved disease control, such as vaccine design. This study describes the development and evaluation of a quantitative real-time PCR (qPCR) approach for use in the rapid determination of infecting C. trachomatis serovars to alleviate the laborious demands associated with nucleotide sequencing and analysis.
MATERIALS AND METHODS
Bacterial strains.
Cell culture isolates representative of 15 prototypic C. trachomatis serovars were obtained from the ATCC Global Bioresource Center (Manassas, VA). These isolates were used to assess the analytical sensitivity and specificity of the C. trachomatis-specific primers and probes. The isolates included the following: A-Har-13 (VR-571B), B-HAR-36 (VR-573), Ba-Apache-2 (VR-347), C-TW-3 (VR-1477), D-UW-3/Cx (VR-885), E-Bour (VR-348B), F-IC-Cal-3 (VR-346), G-UW-57/Cx (VR-878), H-UW-43/Cx (VR-879), I-UW-12/Ur (VR-880), J-UW-36/Cx (VR-886), K-UW-31/Cx (VR-887), L1-440 (VR-901B), L2-434 (VR-902B), and L3-404 (VR-903). Cell cultures inoculated with these serovar isolates were propagated and used for the preparation of DNA extracts, using the MagNA Pure LC automated system (Roche Diagnostics, Indianapolis, IN). The C. trachomatis copy number was determined for each DNA extract by qPCR in conjunction with a quantified C. trachomatis standard (Advanced Biotechnologies Inc., Columbia, MD).
Clinical specimens.
DNA extracts obtained from C. trachomatis-positive anogenital specimens were used to assess the qPCR assays for clinical evaluation. These specimens were identified as C. trachomatis positive by the BD ProbeTec ET Chlamydia trachomatis assay (Becton Dickinson, Sparks, MD) or the COBAS Amplicor analyzer (Roche Diagnostics). Specimens (n = 494) were randomly selected from 612 consecutive positive samples obtained from men who had had sex with another man (MSM) in the previous 12 months who attended the Melbourne and Sydney Sexual Health Centres, Australia, during 2004 to 2008. These specimens comprised first-pass urine (n = 210) (42.3%) and anal swabs (n = 284) (57.7%). In addition, six DNA extracts obtained from eye swab specimens collected during the 2002 outbreak of trachoma in the Northern Territory (25) were selected from a collection of 31 specimens to evaluate detection of the A to C serovars. All clinical specimens (anal swabs, urine, and eye swabs) were stored at −80°C until processing. DNA extracts obtained from each specimen were kept at 4°C for immediate testing (<2 days) or stored at −30°C for a longer term. All specimens had been characterized by ompA sequencing as part of former independent studies. All DNA extracted from clinical specimens was deidentified (including specimen type and sequencing data), prior to testing by qPCR, in an effort to exclude bias in serovar prediction.
DNA extraction.
Total DNA was extracted from 200-μl volumes of the bacterial isolates and clinical specimens using the MagNA Pure LC system (Roche Diagnostics), in conjunction with the total DNA isolation kit (Roche Diagnostics). DNA was eluted into final volumes of 100 μl of the kit elution buffer and stored at −20°C until testing. In the event of primary/secondary PCR failure, extraction was repeated with the addition of 33.3 μg/ml poly(A) carrier RNA (Roche Diagnostics) to the lysis buffer, according to recommendations for improving extraction efficiencies of samples with low concentrations of DNA.
DNA sequencing.
DNA extracts obtained from the clinical specimens were sequenced across the four variable domains (VDs) of the ompA gene. Sequencing of the eye swab DNA extracts has been previously described (25). For the purpose of orientation, the following primer positions are based on the ompA gene of serovar A strains. DNA extracts obtained from anogenital specimens were amplified using the primer pair MS-1F (5′-GTGTGACGCTATCAGCATGC-3′; 162 to 181 bp) and MS-1R (5′-GTTCCTACTGCAATACCGCAAG-3′; 1,076 to 1,097 bp), spanning an approximately 936-bp region across the four VDs. The PCR comprised 10 μl of template DNA, 1× Green GoTaq Flexi buffer (Promega, Madison, WI), 2 mM MgCl2 (Promega), 0.2 mM each deoxynucleoside triphosphate (dNTP; Invitrogen, Carlsbad, CA), 1.25 units of GoTaq DNA polymerase (Promega), and 0.5 μM each primer in a 50-μl reaction volume. Amplification was performed using a GeneAmp PCR system 9700 (Applied Biosystems), with the following parameters: one cycle at 94°C for 10 min; 45 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s; and a final extension step at 72°C for 7 min. PCR products were visualized on a 2% agarose gel. In cases where amplification did not result in sufficient amplicons for sequencing, smaller overlapping PCR products were targeted using the following nested primer pairs: MS-1F/CT6-R (655 to 679 bp), 518-bp fragment spanning VDI to -II; CT6-F (655 to 679 bp)/MS-1R, 443-bp fragment spanning VDIII to -IV; and NL-F (394 to 413 bp) and NL-R (856 to 875 bp), 482-bp fragment spanning VDII to -III. The nucleotide sequences of these primers have been described previously (25). The PCR cycling conditions used were as described above with the exception of a lowered annealing temperature of 50°C and use of 5 μl of purified PCR template. PCR product purification was performed using the QIAquick PCR purification kit (Qiagen, Inc., Valencia, CA) on a small scale or AMPure (Agencourt Bioscience Corp., Beverly, MA) on a larger scale, as per the manufacturer's instructions. DNA concentrations were estimated using a Qubit fluorometer (Invitrogen).
Sanger nucleotide sequencing of purified PCR products was performed using a CEQ 8000 genetic analysis system (Beckman Coulter, Inc., Fullerton, CA). Twenty-microliter sequencing reactions consisted of 4 μl DTCS Quick Start mix, 2 μl of primer at a concentration of 1.6 pmol/μl, 10 μl of distilled water (dH2O), and 4 μl of purified DNA at a concentration of approximately 50 ng/reaction. The sequencing reaction was performed in GeneAmp PCR system 9700 (Applied Biosystems), with the following parameters: 30 cycles at 96°C for 20 s, 50°C for 20 s, and 60°C for 4 min. The dye-labeled PCR products were purified using CleanSEQ (Agencourt Bioscience) by following the manufacturer's recommendations. For high-throughput sequencing, PCR products were sequenced using the services of the Australian Genome Research Facility (Parkville, Victoria, Australia), which uses an ABI 3730xl DNA analyzer (Applied Biosystems). Both strands from each PCR product were sequenced to ensure sufficient sequence overlap and fidelity.
Data analysis.
Nucleotide sequence chromatograms were analyzed with DNASTAR following export of the raw sequence data. Sequences were aligned, and contiguous sequences were generated for each sample tested. DNA sequence data were submitted to the standard nucleotide-nucleotide BLAST search engine at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST) for comparative analyses with ompA sequences of known C. trachomatis strains to determine the consensus serovars.
qPCR assay.
All qPCR primers and 5′-nuclease hydrolysis (TaqMan) probes were designed by alignment of the ompA nucleotide sequences of the following C. trachomatis reference strains derived from GenBank (accession numbers are shown in parentheses): A/Sa1/OT (M58938), B/Alpha-95 (U80075), B-Jali-20 (M33636), Ba/Apache-2 (AF063194), C/TW3/OT (M17343), D/B120 (X62918), D/B185 (X62919), D/IC-Cal8 (X62920), E/Bour-1990 (X52557), F/IC-Cal3 (X52080), G/UW57/Cx (AF063199), H/Wash (X16007), I/UW-12 (AF063200), J/UW36/Cx (AF063202), K/UW31/Cx (AF063204), L1/440-Bu (M36533), L2/434-Bu (M14738), and L3/404-Bu (X55700). All qPCRs were performed using the LightCycler 480 real-time instrument (Roche Diagnostics), with each reaction comprised of a 20-μl volume, containing 1× LightCycler 480 probes master mix, 1.0 μM each primer, 200 nM each probe (100 nM B-group probe), and 5 μl of DNA template. DNA extracted from the bacterial isolates was used as the positive control, and nuclease-free water was used as the negative control. All PCR cycling conditions are listed in Table 1.
TABLE 1.
qPCR primer/probe sequences and amplification parameters
| PCR group type | Specificity | Sequences (5′-3′)a |
PCR conditions | |
|---|---|---|---|---|
| Primer | Probe | |||
| CT | CT | CATGARTGGCAAGCAAGTTTA | HEX-TGTTCACTCCYTACATTGGAGT-BHQ1 | 1 cycle at 95°C for 10 min |
| C group | GCAATACCGCAAGATTTTCTAG | FAM-TCCTTTACCAGCGATGGT-BHQ1 | 55 cycles at 95°C for 10 s, 55°C for 20 s, and 65°C for 40 s | |
| B group | LC610-TTTCACMTCGCCAGCTCC-BHQ2 | 1 cycle at 40°C for 10 s | ||
| I group | Cy5-CTACACTGCCGCATCCTG-BHQ3 | |||
| I | F | CATGARTGGCAAGCAAGTTTA | FAM-CTTCCGTGTTAGCTCCAGCTAC-BHQ1 | 1 cycle at 95°C for 10 min |
| G | GCAATACCGCAAGATTTTCTAG | Cy5-CTTCCGAGTTAGCTGCGACTAC-BHQ3 | 50 cycles at 95°C for 10 s, 58°C for 15 s, and 65°C for 20 s; 1 cycle at 40°C for 10 s | |
| B | B/Ba | CAAATGCCGCTTGCATGG | FAM-TTTCCGAATAACCCCACTAAATTGA-BHQ1 | 1 cycle at 95°C for 10 min |
| D | GTTGCACATCCACATTCCCACA | Cy5-TACAGACTCCGCTTTGACCGTT-BHQ3 | 50 cycles at 95°C for 10 s, 58°C | |
| E | GACATATGCAGGATGCTGAGATG | FAM-AACTTTACACAGATACTGCCTTCTC-BHQ1 | for 10 s, and 65°C for 20 s; 1 | |
| L1 | TATTGGAAAGAAGCSCCTAAAGT | HEX-TACAGCATCCTTTTTGACCGT-BHQ1 | cycle at 40°C for 10 s | |
| L2 | Cy5-ACAAGCTTACTATCTGAAACTGTAG-BHQ3 | |||
| C | H | CCACTTGGTGTGACGCTATCAGCAT | Cy3-TTGTAAGTCTGCTGCATCGTTGGTA-BHQ2 | 1 cycle at 95°C for 10 min |
| I | GCATCTTGCATGTGTTTGCCATAAGC | FAM-TCTAAGCCTGCTAYATCCTTGGTAG-BHQ1 | 55 cycles at 95°C for 10 s and 70°C for 45 s | |
| L3 | Cy5-TCGTTTGATAAGCCCGCTGTATCGC-BHQ3 | 1 cycle at 40°C for 10 s | ||
| A | ACTCCGCTTCCTTCAACTTAGTTGGA | FAM-ACAATATTCGCTGTATCAAAGCCAG-BHQ1 | 1 cycle at 95°C for 10 min | |
| K | CCTAACGTTGCACACCCACATTC | CY5-CCACAGCTCGATCCAAAG-BHQ3 | 55 cycles at 95°C for 10 s, 60°C for 10 s, 72°C for 20 s; 1 cycle at 40°C for 10 s | |
| J | AGCCTTATGATCGACGGAATTCT | FAM-AGGATCTCCACCGAAACC-BHQ1 | 1 cycle at 95°C for 10 min | |
| GTTGTTGGATCGTTTTGTAAGCC | 55 cycles at 95°C for 10 s, 65°C | |||
| C | CATGARTGGCAAGCAAGTTTA | Cy3-TTATCGGTTCCGGCAGAGACCACACTT-BHQ2 | for 15 s, and 72°C for 10 s; 1 | |
| GCAATACCGCAAGATTTTCTAG | cycle at 40°C for 10 s | |||
Nucleotide bases among the primers and probes that are underlined represent locked nucleic acid bases. Nucleotide sequences for consensus primers/probes have been described previously (26). FAM, 6-carboxyfluorescein; HEX, 5-hexachlorofluorescein; Cy3, cyanine 3; Cy5, cyanine 5; LC610, LightCycler Red 610; BHQ1 to -3, Black Hole Quencher 1 to 3.
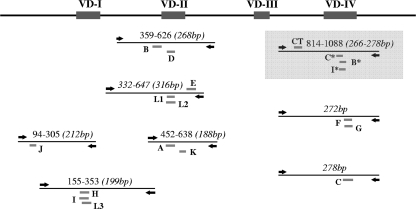
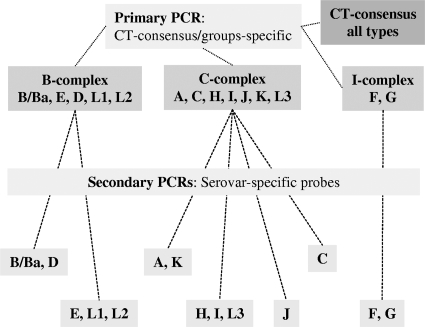
The primary C. trachomatis group-specific multiplex PCR used two primers and four probes, specific to all C. trachomatis types, including the B group (B, E, D, L1, and L2), C group (A, C, H, I, J, K, and L3) or intermediate group (F and G) serovars (Table 1). This PCR was directed to amplify a 266- to 278-bp region spanning VDIV; the consensus probe targeted a conserved region upstream of VDIV (Fig. 1). Analytical specificity of the primary PCR for C. trachomatis was assessed by testing a panel of DNA extracted from commonly detected pathogenic microorganisms, including Chlamydophila pecorum, Chlamydophila abortus, Chlamydophila pneumoniae, Neisseria gonorrhoeae, Neisseria meningitidis, Staphylococcus aureus (methicillin-sensitive and -resistant strains), Streptococcus pyogenes, Streptococcus pneumoniae, Escherichia coli, Enterobacter aerogenes, Enterococcus faecium, Enterococcus faecalis, and Trichomonas vaginalis. These strains were derived from clinical specimens. The primary group-specific PCR was used for determining which set of secondary serovar-specific PCRs to perform (Fig. 2). The serovar-specific PCRs comprise a various number of monoplex/multiplex reactions. The B-group serovars were distinguished by two multiplex PCRs (B- and D-group PCR and E-, L1-, and L2-group PCR), the C-group serovars by two multiplex PCRs (A- and K-group PCR and H-, I-, and L3-group PCR) and two monoplex PCRs (C-group PCR and J-group PCR), and I-group serovars by a single multiplex PCR. These PCRs were directed to amplify regions from 188 bp to 316 bp in length spanning the VDI, -II, or -IV, as detailed in Fig. 1. All clinical specimens were tested by qPCR, with the operator blinded from previous sequencing data and therefore those for C. trachomatis serovar, despite knowing the positivity of the specimens. All qPCR data were analyzed using LightCycler 480 software using the second derivative method. Appropriate color compensation data used for distinguishing the different fluorophore wavelengths and minimizing cross talk between channels were created, according to the manufacturer's recommendations. Color compensation was applied to all multiprobe qPCR tests.
FIG. 1.
Schematic representation of the ompA gene regions amplified by qPCRs for C. trachomatis serovar determination. Primers are shown by left/right arrows, and probes are indicated by serovar letters, with those above annealing to the sense strand and those below annealing to the antisense strand. *, C. trachomatis group-specific probes (primary PCR is shaded).
FIG. 2.
Schematic representation of the qPCR testing algorithm used for determining C. trachomatis serovars.
Mixed infections.
The capacity to detect multiple C. trachomatis infections was evaluated through mixing DNA extracts from the reference chlamydia strains. A DNA extract representative from each of the three groups was mixed with extracts from other group serovars of various copy numbers (102 and 104 copies per reaction) and tested in triplicate. There is currently no data on the difference in C. trachomatis bacterial load within a mixed infection; as such, the 100-fold difference in C. trachomatis copy numbers between serovars was arbitrarily chosen to emulate a mixed infection. Testing was performed with/without the presence of A549 human alveolar basal epithelial cell line DNA.
RESULTS
qPCR specificity/sensitivity.
The primary consensus/group-specific PCR was evaluated using nonquantified DNA (10−1 to 10−4 dilutions of MagNA Pure extracts) from the 15 C. trachomatis serovar reference strains. All serovars were correctly identified by the consensus probe and subsequently by the three group-specific probes. This primary PCR demonstrated specificity for only C. trachomatis, as there was no cross-reactivity observed between the primers/probes and other pathogenic microorganisms tested, including species of the closely related genus Chlamydophila. The secondary (serovar-specific) PCRs accurately identified the targeted serovars, with no cross-reactivity detected between serovars. The analytical sensitivity of the primary PCR was determined by testing in triplicate serial dilutions of quantified DNA extracted from the 15 reference strains. The limit of detection for C. trachomatis and their specific-groups ranged from 10 to 40 genome copies per reaction (Table 2).
TABLE 2.
Detection limits of the primary qPCR
| Serovar | Detection limitsa |
|||||
|---|---|---|---|---|---|---|
| Consensus C. trachomatis |
Group-specific C. trachomatis |
|||||
| Copies per reaction | Cq value | SD | Copies per reaction | Cq value | SD | |
| A | 10 | 41.0 | 0.1 | 10 | 40.3 | 0.1 |
| B | 10 | 40.4 | * | 10 | 37.7 | * |
| Ba | 10 | 41.0 | 1.5 | 10 | 39.8 | 1.5 |
| C | 10 | 42.2 | 1.6 | 10 | 41.6 | 1.6 |
| D | 20 | 39.3 | 0.4 | 20 | 38.2 | 0.4 |
| E | 10 | 40.2 | 1.0 | 10 | 37.5 | 1.1 |
| F | 10 | 41.2 | 1.7 | 10 | 40.1 | 1.7 |
| G | 20 | 40.4 | 1.0 | 20 | 39.3 | 1.0 |
| H | 20 | 41.8 | 1.0 | 20 | 41.1 | 1.1 |
| I | 10 | 42.2 | * | 10 | 38.5 | * |
| J | 10 | 42.3 | * | 10 | 41.6 | * |
| K | 20 | 40.8 | 0.8 | 20 | 39.7 | 1.0 |
| L1 | 40 | 39.2 | 0.4 | 40 | 37.9 | 0.5 |
| L2 | 20 | 40.8 | * | 20 | 37.1 | * |
| L3 | 20 | 40.6 | 0.6 | 20 | 40.1 | 0.7 |
Detection limits for the 15 reference C. trachomatis serovars, as determined by primary consensus/group-specific PCRs. Quantification cycle (Cq) values indicate the highest cycle numbers with which serovars were detected. *, samples were tested singly; thus, no standard deviation was calculated.
Mixed infections.
C. trachomatis serovars were mixed to mimic multiple infections and evaluated in triplicate using the primary consensus/group-specific PCR. All combinations of serovars were successfully detected, with accurate identification of serovars present at low levels (1 × 102 copies per reaction) in the presence of others found at higher levels (1 × 104 copies per reaction). Notably, serovars present in low copy numbers were detected approximately 13 cycles later than those at higher copy numbers, indicative of some level of PCR competition, given that a 2-log factor would typically be reflected by an approximate 6- to 7-cycle difference. Nonetheless, the standard deviation of the qPCR crossing point was typically under one cycle for serovars present in either high or low copy numbers (data not shown), providing assurance as to the reproducibility of detecting mixed infections, at least within the concentrations assessed.
Clinical specimen evaluation.
A total of 500 clinical specimens (494 anogenital and 6 eye swab specimens), previously identified as C. trachomatis positive, were evaluated by DNA sequencing (in an earlier study) and qPCR (in the current study). Of these specimens, C. trachomatis was identified by sequencing analysis among 487 (97.4%) specimens, and of these, 476 (95.2%) were able to have their serovar determined by sequence. Only sequence data obtained from fragments of sufficient length (from the 936-bp amplicon [VDI to -IV]) or from three smaller fragments, 518 bp (VDI to -II), 443 bp (VDIII to -IV), and 482 bp (VDIII to -IV) in length, were used for predicting C. trachomatis serovars by sequence analysis. In contrast, 455 specimens (91.0%) generated positive C. trachomatis amplification curves using the consensus/group-specific qPCR, and of these, 433 (86.6%) had a serovar predicted that matched the corresponding qPCR C. trachomatis group(s) (Table 3). Of only those specimens having a C. trachomatis serovar determined by sequencing, considered the “typing gold standard” for purposes of comparison (n = 476), a matching C. trachomatis group was detected in 439 (92.2%) specimens by qPCR, with 423 (88.9%) identified with a matching serovar, with a resultant sensitivity of 88.9% (95% confidence interval [CI], 85.7% to 91.5%). The 16 specimens with an identical CT-group result (qPCR versus sequencing) but no serovar data were categorized as “indeterminate,” likely due to low C. trachomatis copy numbers in these samples (data not shown). Since DNA sequencing identifies the most predominant serovar within a specimen, a match was inclusive as to whether a single serovar (by sequencing) corresponded with a serovar among a mixed infection by qPCR. There were five instances whereby a group-specific qPCR mixed infection did not match the qPCR serovar prediction; all of which were positive by B- and I-group probing, though only positive for a B-group serovar (three positive for serovar D and two positive for serovar E). Of the 11 specimens with insufficient sequence data necessary for accurate serovar prediction (Table 3), serovars were determined for 6 of these by qPCR (including a multiple infection with serovars E and J). Of 13 specimens unable to be sequenced, 4 had a serovar predicted by qPCR (3 positive for serovar G and 1 positive for serovar J).
TABLE 3.
Evaluation of the qPCR assay using 500 clinical specimensa
| Serovar | No. (%) of predicted matches |
||
|---|---|---|---|
| DNA sequencing | qPCR |
||
| CT group | CT serovar | ||
| A | 0 | 0 | 0 |
| B/Ba | 3 | 3 | 3 |
| C | 4 | 4 | 4 |
| D | 163 | 150 | 143 |
| E | 48 | 47 | 46 |
| F | 19 | 18 | 16 |
| G | 154 | 140 | 137 |
| H | 4 | 4 | 4 |
| I | 1 | 0 | 0 |
| J | 76 | 70 | 67 |
| K | 4 | 3 | 3 |
| L1 | 0 | 0 | 0 |
| L2 | 0 | 0 | 0 |
| L3 | 0 | 0 | 0 |
| Subtotal | 476 (95.2) | 439 (92.2) | 423 (88.9) |
| CT-POSb | 11 | 10 | 6 |
| CT-NEGc | 13 | 6 | 4 |
| Total | 500 | 455 (91.0) | 433 (86.6) |
Evaluation of the qPCR algorithm for C. trachomatis serovar prediction using clinical specimens. DNA sequencing was used as the gold standard for typing, to which the PCR results were compared. Anogenital specimens (n = 494) and ocular specimens (n = 6) were used for this comparison. Specimens were identified by qPCR as matching the CT group (B, C, or I group) and/or the CT serovar, as determined by sequencing.
Eleven specimens tested positive for C. trachomatis by sequencing (CT-POS) without serovar identification. Ten of these were positive by qPCR group-specific PCR.
Thirteen specimens tested negative for C. trachomatis by sequencing (CT-NEG). Six of these were positive by qPCR group-specific PCR.
Of the 494 anogenital specimens tested, mixed serovar infections were identified by qPCR among 12 (2.4%) individuals. However, only six specimens were characterized by serovars matching the group-specific result. Mixed infections were comprised of the following: two B/C-group serovars (serovars E/J and D/J), two C/I-group serovars (serovars G/J and G/J), and two C-group serovars (serovars H/K and J/K). The six specimens with nonmatching serovars were from the B/I group (three positive for serovar D, two positive for serovar E, and one undetermined serovar).
DISCUSSION
Traditional approaches for determining or predicting C. trachomatis serovars have involved either nucleotide sequencing of the ompA gene or restriction fragment length polymorphism (RFLP) analyses, which can be labor-intensive and time-consuming. Furthermore, these techniques do not allow for the identification of multiple serovar infections, at least without the additional requirement of laborious cloning procedures. Therefore, relatively simple, rapid, and on-hand approaches for predicting C. trachomatis serovars would prove invaluable.
In the current study, a qPCR assay has been designed and evaluated for the purpose of rapid prediction of a C. trachomatis serovar(s) from specimens diagnosed as C. trachomatis positive. This assay targets the ompA gene of C. trachomatis, since substantial genetic variability exists within the four VDs among serovars, which can be exploited for epidemiological analyses. All primers target sequences within the five conserved regions (apart from the serovar J-specific reverse primer), while the hydrolysis probes, for serovar identification, anneal to different VDs (with the exception of VDIII).
When comparing sequencing to the qPCR approach, it should be emphasized that the former utilized nested PCR for 49.7% (242/487) of C. trachomatis-positive confirmations, thereby biasing the positivity of sequencing rather than that of qPCR. Specimens whose serovar did not match (n = 2), or remained undetermined (n = 51), were most likely the result of the combination of low-copy-number C. trachomatis specimens and the nonnested nature of the qPCR assay. With this, a notable limitation of this qPCR assay was its marginally lower success rate in predicting C. trachomatis serovars compared to that of sequencing. Several of the published protocols for C. trachomatis serovar prediction incorporate an initial nested PCR followed by various methods of detection, including sequencing, qPCR, blotting, and microsphere suspension array, to improve assay sensitivity (7, 8, 9, 14, 31, 33). Therefore, as a step to potentially enhancing the current assay, it is possible that a nested PCR precedes the primary consensus/group-specific PCR. Nonetheless, it should be emphasized that this qPCR assay was not intended to be used as a primary diagnostic test but more as a relatively simple and rapid screening tool for predicting C. trachomatis serovars, particularly multiple serovars, while minimizing the requirements for DNA sequencing. In instances of ambiguous qPCR results, DNA sequencing is still recommended and has the additional benefit of detecting genotypic variants.
Evaluation of the qPCR with clinical specimens was limited to those serovars most prevalent in the populations studied. There was a limited quantity of DNA from the trachoma strains, including no A serovars and very few to nil H, I, K, and LGV serovars among the anogenital specimens. Therefore, further evaluation could be warranted for a more thorough and widespread validation of this assay with clinical samples, particularly with those serovars causing ocular trachoma and the more invasive LGV-associated disease. Outbreaks of LGV serovar L2, and in particular LGV biovar L2b, throughout Europe since 2003 have highlighted the importance of C. trachomatis serovar identification (28). The LGV outbreak has occurred predominantly among high-risk MSM populations, including HIV-positive individuals (23), whose presence has been noted in Australia (22). Given the significance of LGV serovars and the severity of their associated disease, close monitoring for early detection is considered highly relevant. Significantly, no LGV serovars were detected among specimens assessed in the current study. The current qPCR assay could prove beneficial for LGV identification, although it cannot distinguish further than L1, L2, and L3.
This assay has demonstrated the capacity to identify multiple serovars within a given specimen, which sequencing was unable to do, including correctly distinguishing serovars among synthetically mixed preparations. Importantly, these mixed preparations were arbitrarily prepared with a 100-fold difference in bacterial load, and it should be emphasized that a natural mixed infection could comprise a severalfold-higher difference between coinfecting serovars, which could ultimately affect the detection of a serovar at lower levels. The frequency of multiple serovars identified in this study (2.4%) is comparable to those in other C. trachomatis typing studies of between 1.5% and 5.4% (8, 15, 19, 20, 33). Interestingly, some studies have described multiple infections with frequencies as high as 8.7% and 13% (14, 31) which principally involved use of PCR followed by reverse line blot procedures.
C. trachomatis strains from the C group appeared to be the most common among the mixed infections, with serovar J detected in five of the six mixed infections (according to the serovar-specific qPCRs). Among six mixed infections (as per group-specific qPCR) with nonmatching serovar(s), all were comprised of B/I-group serovars, with the B-group serovar identified in five of these infections. This could possibly be explained by the presence of a low copy number for the I-group serovar. It is unlikely the result of a sensitivity concern, given that the serovar D PCR has shown a lower sensitivity than either of the PCRs for the F or G serovar (data not shown). Interestingly, these six mixed infections (with nonmatching serovar[s]) were all derived from urine samples with possibly lower DNA yields, while three of the six mixed infections that had serovars determined were derived from anal swab samples. Although not performed in the current study, this assay could also potentially be utilized for quantifying C. trachomatis DNA within clinical specimens in an effort to identify associations between bacterial load and clinical disease.
Aside from ompA sequence-based typing, which still predominates, there have been a limited number of alternative assays described for more rapid C. trachomatis serovar prediction. These have included multilocus sequence-based approaches (11, 18) and use of a microsphere suspension array (8). A real-time PCR system offering typing of urogenital strains (D to K) and (L1 to L3) has been reported previously (9). However, this assay does not allow the determination of the ocular serovars A, B/Ba, and C and tested a smaller-sized validation panel. In the current study, a much larger sample set of 500 specimens was evaluated, with serovars predicted for 86.4% of specimens, compared to those predicted for 80% of specimens using a published nonnested qPCR assay (9).
In conclusion, the qPCR assay described in the current study offers a comparatively simple and rapid approach (qPCR versus PCR/purification/sequencing/analysis and half of a day versus 1 to 2 days, respectively) for high-throughput prediction of C. trachomatis serovars, as opposed to traditional sequence-based methods. The ability to detect and quantify multiple serovars could allow for the study of associations with clinical disease and could facilitate the monitoring of serovar circulation and partner tracing within given populations.
Acknowledgments
This study was generously supported by an Australian National Health and Medical Research Council Project grant (454620).
Footnotes
Published ahead of print on 14 April 2010.
REFERENCES
- 1.Baehr, W., Y. X. Zhang, T. Joseph, H. Su, F. E. Nano, K. D. Everett, and H. D. Caldwell. 1988. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc. Natl. Acad. Sci. U. S. A. 85:4000-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandea, C. I., K. Kubota, T. M. Brown, P. H. Kilmarx, V. Bhullar, S. Yanpaisarn, P. Chaisilwattana, W. Siriwasin, and C. M. Black. 2001. Typing of Chlamydia trachomatis strains from urine samples by amplification and sequencing the major outer membrane protein gene (omp1). Sex. Transm. Infect. 77:419-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cates, W., Jr., and J. N. Wasserheit. 1991. Genital chlamydial infections: epidemiology and reproductive sequelae. Am. J. Obstet. Gynecol. 164:1771-1781. [DOI] [PubMed] [Google Scholar]
- 4.Cohen, C. R., and R. C. Brunham. 1999. Pathogenesis of Chlamydia induced pelvic inflammatory disease. Sex. Transm. Infect. 75:21-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garland, S. M., and B. Johnson. 1989. Chlamydia trachomatis infections—The Royal Women's Hospital experience. Med. J. Aust. 150:174-177. [DOI] [PubMed] [Google Scholar]
- 6.Garland, S. M., A. Malatt, S. Tabrizi, D. Grando, M. I. Lees, J. H. Andrew, and H. R. Taylor. 1995. Chlamydia trachomatis conjunctivitis. Prevalence and association with genital tract infection. Med. J. Aust. 162:363-366. [PubMed] [Google Scholar]
- 7.Geisler, W. M., C. M. Black, C. I. Bandea, and S. G. Morrison. 2008. Chlamydia trachomatis ompA genotyping as a tool for studying the natural history of genital chlamydial infection. Sex. Transm. Infect. 84:541-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang, C. T., W. W. Wong, L. H. Li, C. C. Chiang, B. D. Chen, and S. Y. Li. 2008. Genotyping of Chlamydia trachomatis by microsphere suspension array. J. Clin. Microbiol. 46:1126-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jalal, H., H. Stephen, S. Alexander, C. Carne, and C. Sonnex. 2007. Development of real-time PCR assays for genotyping of Chlamydia trachomatis. J. Clin. Microbiol. 45:2649-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jurstrand, M., L. Falk, H. Fredlund, M. Lindberg, P. Olcen, S. Andersson, K. Persson, J. Albert, and A. Backman. 2001. Characterization of Chlamydia trachomatis omp1 genotypes among sexually transmitted disease patients in Sweden. J. Clin. Microbiol. 39:3915-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klint, M., H. H. Fuxelius, R. R. Goldkuhl, H. Skarin, C. Rutemark, S. G. Andersson, K. Persson, and B. Herrmann. 2007. High-resolution genotyping of Chlamydia trachomatis strains by multilocus sequence analysis. J. Clin. Microbiol. 45:1410-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lister, N. A., S. N. Tabrizi, C. K. Fairley, A. Smith, P. H. Janssen, and S. Garland. 2004. Variability of the Chlamydia trachomatis omp1 gene detected in samples from men tested in male-only saunas in Melbourne, Australia. J. Clin. Microbiol. 42:2596-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mabey, D., and R. W. Peeling. 2002. Lymphogranuloma venereum. Sex. Transm. Infect. 78:90-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molano, M., C. J. Meijer, S. A. Morré, R. Pol, and A. J. van den Brule. 2004. Combination of PCR targeting the VD2 of omp1 and reverse line blot analysis for typing of urogenital Chlamydia trachomatis serovars in cervical scrape specimens. J. Clin. Microbiol. 42:2935-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mossman, D., K. W. Beagley, A. L. Landay, M. Loewenthal, C. Ooi, P. Timms, and M. Boyle. 2008. Genotyping of urogenital Chlamydia trachomatis in Regional New South Wales, Australia. Sex. Transm. Dis. 35:614-616. [DOI] [PubMed] [Google Scholar]
- 16.Muñoz, B., and S. West. 1997. Trachoma: the forgotten cause of blindness. Epidemiol. Rev. 19:205-217. [DOI] [PubMed] [Google Scholar]
- 17.Ossewaarde, J. M., M. Rieffe, A. de Vries, R. P. Derksen-Nawrocki, H. J. Hooft, G. J. van Doornum, and A. M. van Loon. 1994. Comparison of two panels of monoclonal antibodies for determination of Chlamydia trachomatis serovars. J. Clin. Microbiol. 32:2968-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedersen, L. N., L. Pødenphant, and J. K. Møller. 2008. Highly discriminative genotyping of Chlamydia trachomatis using omp1 and a set of variable number tandem repeats. Clin. Microbiol. Infect. 14:644-652. [DOI] [PubMed] [Google Scholar]
- 19.Quint, K., C. Porras, M. Safaeian, P. González, A. Hildesheim, W. Quint, L. J. van Doorn, S. Silva, W. Melchers, M. Schiffman, A. C. Rodríguez, S. Wacholder, E. Freer, B. Cortes, R. Herrero, and the Costa Rican Vaccine Trial Group. 2007. Evaluation of a novel PCR-based assay for detection and identification of Chlamydia trachomatis serovars in cervical specimens. J. Clin. Microbiol. 45:3986-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quint, K. D., L. J. van Doorn, B. Kleter, M. N. de Koning, H. A. van den Munckhof, S. A. Morre, B. ter Harmsel, E. Weiderpass, G. Harbers, W. J. Melchers, and W. G. Quint. 2007. A highly sensitive, multiplex broad-spectrum PCR-DNA-enzyme immunoassay and reverse hybridization assay for rapid detection and identification of Chlamydia trachomatis serovars. J. Mol. Diagn. 9:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stamm, W. E. 1999. Chlamydia trachomatis infections: progress and problems. J. Infect. Dis. 179:380-383. [DOI] [PubMed] [Google Scholar]
- 22.Stark, D., S. van Hal, R. Hillman, J. Harkness, and D. Marriott. 2007. Lymphogranuloma venereum in Australia: anorectal Chlamydia trachomatis serovar L2b in men who have sex with men. J. Clin. Microbiol. 45:1029-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stary, G., and A. Stary. 2008. Lymphogranuloma venereum outbreak in Europe. J. Dtsch. Dermatol. Ges. 6:935-940. [DOI] [PubMed] [Google Scholar]
- 24.Stephens, R. S., E. A. Wagar, and G. K. Schoolnik. 1988. High-resolution mapping of serovar-specific and common antigenic determinants of the major outer membrane protein of Chlamydia trachomatis. J. Exp. Med. 167:817-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevens, M. P., S. N. Tabrizi, R. Muller, V. Krause, and S. M. Garland. 2004. Characterization of Chlamydia trachomatis omp1 genotypes detected in eye swab samples from remote Australian communities. J. Clin. Microbiol. 42:2501-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stevens, M. P., S. E. Tan, L. Horvath, C. K. Fairley, S. M. Garland, and S. N. Tabrizi. 2008. Absence of a Chlamydia trachomatis variant, harbouring a deletion in the cryptic plasmid, in clients of a sexually transmissible infection clinic and antenatal patients in Melbourne. Commun. Dis. Intell. 32:77-81. [DOI] [PubMed] [Google Scholar]
- 27.Stothard, D. R., G. Boguslawski, and R. B. Jones. 1998. Phylogenetic analysis of the Chlamydia trachomatis major outer membrane protein and examination of potential pathogenic determinants. Infect. Immun. 66:3618-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van de Laar, M. J. 2006. The emergence of LGV in Western Europe: what do we know, what can we do? Euro Surveill. 11:146-148. [PubMed] [Google Scholar]
- 29.Wang, S. P., C. C. Kuo, R. C. Barnes, R. S. Stephens, and J. T. Grayston. 1985. Immunotyping of Chlamydia trachomatis with monoclonal antibodies. J. Infect. Dis. 152:791-800. [DOI] [PubMed] [Google Scholar]
- 30.Wein, P., M. Kloss, and S. M. Garland. 1990. Postabortal pelvic sepsis in association with Chlamydia trachomatis. Aust. N. Z. J. Obstet. Gynaecol. 30:347-350. [DOI] [PubMed] [Google Scholar]
- 31.Xiong, L., F. Kong, H. Zhou, and G. L. Gilbert. 2006. Use of PCR and reverse line blot hybridization assay for rapid simultaneous detection and serovar identification of Chlamydia trachomatis. J. Clin. Microbiol. 44:1413-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan, Y., Y. X. Zhang, N. G. Watkins, and H. D. Caldwell. 1989. Nucleotide and deduced amino acid sequences for the four variable domains of the major outer membrane proteins of the 15 Chlamydia trachomatis serovars. Infect. Immun. 57:1040-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng, H. P., L. F. Jiang, D. Y. Fang, Y. H. Xue, Y. A. Wu, J. M. Huang, and Z. Y. Ou. 2007. Application of an oligonucleotide array assay for rapid detecting and genotyping of Chlamydia trachomatis from urogenital specimens. Diagn. Microbiol. Infect. Dis. 57:1-6. [DOI] [PubMed] [Google Scholar]