Abstract
High-resolution melt analysis PCR (HRM PCR) for diagnosis of Old World Leishmania was developed using the 7SL RNA gene. Cutaneous leishmaniasis samples were analyzed. Sensitivity and specificity of HRM PCR were significantly better (P < 0.001) than those of internal transcribed spacer 1 PCR and similar to those of kinetoplast DNA PCR.
Leishmania causes cutaneous (CL), mucocutaneous, and visceral leishmaniasis (VL), and the diagnosis, treatment, and prognosis are dependent on accurate identification of the infecting parasite species. Microscopic and culture-based diagnostic methods are being replaced by rapid, highly sensitive and specific molecular techniques based on PCR that target coding and noncoding regions of the Leishmania genome (8). At minimum, these assays require post-PCR examination by agarose gel electrophoresis (1), and species identification requires additional processing by restriction fragment length polymorphism (RFLP) (8), hybridization (5), DNA sequencing (12), etc.
High-resolution melt (HRM) analysis is a relatively new technique that allows direct characterization of PCR amplicons in a closed system. It measures changes in the fluorescence intensity of a DNA intercalating dye during dissociation from double-stranded DNA to single-stranded DNA and can differentiate between single nucleotide polymorphisms (SNP). HRM has been used for analysis of genomic mutations (7), SNP genotyping (4), Mycobacterium tuberculosis rifampin susceptibility (3), Staphylococcus aureus typing (10), etc. HRM is simpler, less expensive, and faster than conventional PCR assays that require post-PCR processing.
A previous study comparing Leishmania 7SL RNA gene DNA sequences suggested that the gene might be a good diagnostic target (12). An HRM PCR assay for Old World Leishmania based on this gene (7SL-HRM) is described here that can be used to differentiate between L. tropica, L. major, and the L. donovani complex. The sensitivity and specificity of this assay, as well as species identification using clinical samples, were compared with those of assays frequently used for parasite diagnosis, internal transcribed spacer 1 (ITS1) and kinetoplast DNA (kDNA) PCR.
Cultured Leishmania strains (n = 77) were grown in M199 medium containing 10% fetal calf serum (FCS) and antibiotics. L. infantum (MHOM/PS/1999/LRC-L773), L. major (MHOM/IL/1980/Freidlin), L. tropica genotype I (MHOM/IL/2006/LRC-L1286), and L. tropica genotype IV (IARA/IL/2000/LRC-L810) were used as reference strains. Clinical samples on filter paper for routine diagnosis by ITS1 PCR were prepared as described previously (1), and DNA was extracted using the High Pure PCR template preparation kit (Roche, Germany). All samples were analyzed by ITS1 PCR, and if positive, species were determined by RFLP (8). Some samples were analyzed by kDNA PCR (1). The Helsinki Committee for Human Research of the Hadassah Hospital, Ein Kerem, Jerusalem, Israel, approved this study.
7SL RNA gene sequences of L. donovani complex (GenBank accession numbers AY722733, AY722734, AY722735, AY781795, and AY781792), L. tropica (AY722722, AY722723, and AY781789), and L. major (AY722713, AY722714, and AY722715) were compared by multialignment (http://bioinfo.genotoul.fr/multalin/multalin.html) (2). The PCR primers CJ7SLF (5′-ACG TGG ACC AGC GAG GGT-3′) and QRT7SLR (5′-CGG TTC CCT CGC TTC AAC-3′), in conserved regions flanking a 119-bp polymorphic internal region, were designed using the primer3 software program (http://frodo.wi.mit.edu/primer3/) (6).
7SL RNA PCR was carried out as follows: DNA samples or controls (2 μl) were added to 2× Thermo-Start PCR master mix (10 μl; Thermo Scientific), Syto 9 (1.5 μM; Invitrogen), primers (0.5 μM [each] final concentration), and ultra-pure PCR-grade water (Fisher); final volume, 20 μl/PCR. Amplification conditions were as follows: 15 min of denaturation at 95°C, followed by 40 cycles of denaturation for 5 s at 95°C; annealing for 10 s at 55°C; and a final extension for 2 min at 72°C. A 119-bp amplicon was observed with the Leishmania reference strains. HRM ramping was carried out at 0.1°C/s from 75 to 95°C. HRM PCR and analysis were performed using a Rotor-Gene 6000 real-time thermal analyzer (Corbett Life Science). Positive-control (reference strain DNA, 20 ng/reaction) and negative-control reactions were included in each experiment. PCR efficiency was evaluated using the threshold cycle (CT), and a normalized melt window, 88 to 92°C, was used in analyzing HRM curves. Statistical analysis was performed using the SPSS computer software program, v.15.0.
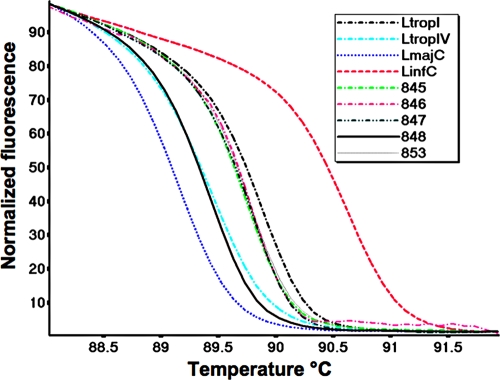
The sensitivity, specificity, and discrimination between species endemic to the Middle East (L. infantum, L. major, and L. tropica) were examined using reference strains (Fig. 1 and data not shown). Sensitivity was determined using 10-fold dilutions of each DNA. Plots of CT versus log DNA concentration were linear over 7 orders of magnitude (2 × 104 to 2 × 10−2 pg; r2 = 0.995), and detection limits were identical, 0.02 pg DNA/species. The average melting point (Tm) ± standard deviation (SD) for 30 7SL-HRM runs was as follows: L. major, 89.27 ± 0.06°C; L. tropica genotype IV, 89.49 ± 0.09°C; L. tropica genotype I, 89.78 ± 0.04°C; and L. infantum, 90.58 ± 0.02°C (indicating that each Tm is highly reproducible). Melting curves and Tm were unaffected by the quantity of DNA used and varied by ±0.06°C over the concentration range examined.
FIG. 1.
Comparison of normalized high-resolution melting (HRM) curves of the 7SL PCR amplicon obtained for Leishmania references species (Leishmania tropica genotype I [LtropI], MHOM/IL/2006/LRC-L1286; L. tropica genotype IV [LtropIV], IARA/IL/2000/LRC-L810; L. major [LmajC], MHOM/IL/1980/Freidlin; and L. infantum [LinfC], MHOM/PS/1999/LRC-L773) and five random patient samples (no. 845, 846, 847, 848, and 853).
Seventy-seven leishmanial strains (for L. tropica, n = 43; for L. major, n = 19; for L. infantum, n = 13; and for L. donovani, n = 2) from the Mediterranean Region (n = 45), Middle East (n = 10), Central Asia (n = 5), Africa (n = 13), and India (n = 4) were examined (see the supplemental material). HRM analysis correctly typed all the strains, regardless of parasite geographic or host origin. In addition, 7SL-HRM distinguished between L. tropica microsatellite genotypes I (n = 34) and IV (n = 9) found in Israel and the Palestinian Authority (9). DNA sequencing showed this was due to an SNP, C/T, at position 98 in the targeted gene sequence (data not shown). Nonleishmanial kinetoplastids, Sauroleishmania tarentolae (P10 strain), Trypanosoma brucei (BF427 strain), Crithidia fasiculata (ATCC 11745), Leptomonas seymouri (ATCC 30220), Phytomonas davidi (ATCC 50166), and Trypanosoma cruzi, were all negative (data not shown).
ITS1 PCR and 7SL-HRM were used to screen clinical samples (n = 192) (Fig. 1 and data not shown), and the results are summarized in Table 1. By ITS1 PCR, 49% (94/192) were positive, while the remaining 51% gave no PCR product, i.e., they were negative. Of the positive samples, 81/94 (86%) produced sufficient product for species determination by RFLP analysis: L. tropica (n = 37), L. major (n = 41), or L. donovani complex (n = 3). An additional 20 samples were positive when examined by 7SL-HRM (total number positive = 114/192 [59%]), all of which could be typed: L. tropica (n = 48), L. major (n = 61), or L. donovani complex (n = 4). Species identification of samples examined by both assays correlated 100%. The remaining samples (n = 78; 41%) were negative. Significant correlation (Fisher's exact test, two-tailed P value < 0.0001) and good agreement (κ = 0.78) were found between the assays. In addition, using HRM it was possible to distinguish between L. tropica genotypes I (n = 28) and IV (n = 20).
TABLE 1.
Comparison of human leishmaniasis diagnosis by ITS1 and 7SL-HRM PCR
| ITS1 result | No. of samples with 7SL-HRM result |
Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 93 | 1 | 94 |
| Negative | 20 | 78 | 98 |
| Total | 113 | 79 | 192 |
Samples (n = 20) positive by 7SL-HRM but missed by ITS1 PCR were examined by kDNA PCR. All were confirmed as positive by the latter method. Similarly, the HRM-negative samples (77/78) were confirmed as negative by kDNA PCR (data not shown).
7SL-HRM diagnosis of leishmaniasis is a rapid, simple technique that takes place without opening the tube, reducing the potential for operator error. It is more sensitive than ITS1 PCR, used in many laboratories, and doesn't require post-PCR processing in order to identify the parasite species. Turnaround time from sample receipt until species identification is only 4 h, 3 h less than needed for analysis by ITS1 PCR (1).
The choice of the gene used for HRM analysis is important in developing a successful assay, since it can take advantage of small differences in melting curves to distinguish between organisms with highly homologous sequences. However, it is generally better to choose a short DNA region with limited polymorphisms, since this gives better separation and simpler HRM curves (11). The 7SL-HRM can be used for diagnosis of human leishmaniasis, as well as epidemiological studies on potential reservoir hosts, sand fly vectors, and parasite genotypes. This assay may be further developed for diagnosis of New World Leishmania species, since the DNA sequences of the 7SL RNA gene from L. amazonensis, L. mexicana, and the L. viannia complex also show polymorphism (12). In conclusion, we have developed a new molecular assay for identifying Old World Leishmania species. It is rapid, sensitive, specific, and simple and can be used to directly diagnose parasites in the tissues of patients with leishmaniasis with a minimum of operator post-PCR manipulation and interpretation.
Supplementary Material
Acknowledgments
This research was supported by grant no. SCHO 448/8-1 from the Deutsche Forschungsgemeinschaft (DFG) as part of a German-Israeli-Palestinian Co-operative Project on Emergence of Cutaneous Leishmaniasis in the Middle East: an investigation of Leishmania tropica in the Palestinian Authority and Israel. C.L.J. holds the Michael and Penny Feiwel Chair in Dermatology.
We thank K. Azmi at Al-Quds University, Palestinian Authority, and Lionel Schnur at the Kuvin Centre for the Study of Infectious and Tropical Disease, IMRIC, Hebrew University Faculty of Medicine, Jerusalem, Israel, for providing several of the Leishmania strains.
Footnotes
Published ahead of print on 14 April 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Bensoussan, E., A. Nasereddin, F. Jonas, L. F. Schnur, and C. L. Jaffe. 2006. Comparison of PCR assays for diagnosis of cutaneous leishmaniasis. J. Clin. Microbiol. 44:1435-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoek, K. G., N. C. Gey van Pittius, H. Moolman-Smook, K. Carelse-Tofa, A. Jordaan, G. D. van der Spuy, E. Streicher, T. C. Victor, P. D. van Helden, and R. M. Warren. 2008. Fluorometric assay for testing rifampin susceptibility of Mycobacterium tuberculosis complex. J. Clin. Microbiol. 46:1369-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kristensen, L. S., and A. Dobrovic. 2008. Direct genotyping of single nucleotide polymorphisms in methyl metabolism genes using probe-free high-resolution melting analysis. Cancer Epidemiol. Biomarkers Prev. 17:1240-1247. [DOI] [PubMed] [Google Scholar]
- 5.Nasereddin, A., E. Bensoussan-Hermano, G. Schonian, G. Baneth, and C. L. Jaffe. 2008. Molecular diagnosis of Old World cutaneous leishmaniasis and species identification by use of a reverse line blot hybridization assay. J. Clin. Microbiol. 46:2848-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 7.Saitsu, H., M. Kato, T. Mizuguchi, K. Hamada, H. Osaka, J. Tohyama, K. Uruno, S. Kumada, K. Nishiyama, A. Nishimura, I. Okada, Y. Yoshimura, S. Hirai, T. Kumada, K. Hayasaka, A. Fukuda, K. Ogata, and N. Matsumoto. 2008. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat. Genet. 40:782-788. [DOI] [PubMed] [Google Scholar]
- 8.Schonian, G., A. Nasereddin, N. Dinse, C. Schweynoch, H. D. Schallig, W. Presber, and C. L. Jaffe. 2003. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn. Microbiol. Infect. Dis. 47:349-358. [DOI] [PubMed] [Google Scholar]
- 9.Schwenkenbecher, J. M., T. Wirth, L. F. Schnur, C. L. Jaffe, H. Schallig, A. Al-Jawabreh, O. Hamarsheh, K. Azmi, F. Pratlong, and G. Schonian. 2006. Microsatellite analysis reveals genetic structure of Leishmania tropica. Int. J. Parasitol. 36:237-246. [DOI] [PubMed] [Google Scholar]
- 10.Stephens, A. J., J. Inman-Bamber, P. M. Giffard, and F. Huygens. 2008. High-resolution melting analysis of the spa repeat region of Staphylococcus aureus. Clin. Chem. 54:432-436. [DOI] [PubMed] [Google Scholar]
- 11.Talmi-Frank, D., A. Nasereddin, L. F. Schnur, G. Schonian, S. O. Toz, C. L. Jaffe, and G. Baneth. 2010. Detection and identification of old world Leishmania by high resolution melt analysis. PLoS Negl. Trop. Dis. 4:e581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zelazny, A. M., D. P. Fedorko, L. Li, F. A. Neva, and S. H. Fischer. 2005. Evaluation of 7SL RNA gene sequences for the identification of Leishmania spp. Am. J. Trop. Med. Hyg. 72:415-420. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.