Abstract
We describe the identification of two bacterial pathogens from a culture-negative brain abscess by the use of broad-spectrum 16S rRNA gene PCR. Simultaneous detection of Fusobacterium nucleatum and Porphyromonas endodontalis was possible due to a 24-bp length difference of their partially amplified 16S rRNA genes, which allowed separation by high-resolution polyacrylamide gel electrophoresis.
Broad-spectrum 16S rRNA gene PCR is a universal and highly versatile technology for the detection and identification of bacterial pathogens (11). Microbiological examination of life-threatening brain infections relies on microscopic examination and culture of abscess material. A recent study by Al Masalma et al. (1) explored an expanded technology applying multiple 16S rRNA fragment cloning and subsequent sequencing to analyze mixed floras in brain abscesses and found that the number of species associated with brain abscesses is much larger than previously noted. Based on a recent case and an in silico feasibility study, we present a modified technique for the molecular analysis of polymicrobial infections. After broad-spectrum 16S rRNA gene amplification, amplicons are separated electrophoretically and excised from a polyacrylamide gel for subsequent sequence identification. In comparison to the previously described labor-intensive cloning and sequencing strategies used to resolve polymicrobial infections by PCR (1, 4), the technique presented herein is less time-consuming and less expensive, further allowing its introduction in the routine diagnostic laboratory.
Case report.
A 29-year-old male was hospitalized after a first seizure, which developed in the context of frontal sinusitis. Three days before hospitalization, the patient was diagnosed with acute frontal sinusitis, which initially was treated symptomatically. Because of severe persistent frontal headache, amoxicillin-clavulanic acid (875 mg/125 mg) every 12 h was prescribed the day before hospital admission. A computed tomography (CT) scan of the skull performed at the hospital showed pansinusitis with a bone defect of the dorsal wall of the frontal sinus and an adjacent brain abscess. The antibiotics ceftriaxone (2-g dose, twice daily) and metronidazole (500-mg dose, three times daily) were given intravenously. On the fourth day of hospitalization, because of the progressive nature of the infection, surgical excision of the brain abscess was performed. Intraoperatively, three samples (designated I, II, and III) of abscess material were obtained and investigated by conventional and molecular bacteriological diagnostics (Table 1). Microscopic analysis showed the presence of leukocytes in samples I and III but no microorganisms in any of the samples. Aerobic media (Columbia blood agar, MacConkey agar, CNA blood agar, and Crowe agar) as well as anaerobic media (Brucella agar, kanamycin-vancomycin agar, phenylethyl alcohol agar, and thioglycolate medium) inoculated with the samples showed no bacterial growth after 3 days of incubation under aerobic (ambient atmosphere and 5% CO2 at 37°C) and anaerobic (atmosphere of 85% N2, 10% CO2, and 5% H2 at 37°C) conditions. A single PCR for broad-spectrum bacterial 16S rRNA genes using primers 5′-AGT TTG ATC MTG GCT CAG-3′ (BAK11w; Escherichia coli rrsA nucleotide [nt] positions 10 to 27) and 5′-GGA CTA CHA GGG TAT CTA AT-3′ (BAK2; E. coli rrsA nt positions 787 to 806) was performed as previously described (2). PCR resulted in two distinguishable fragments each for samples I and III, respectively, as visualized by CleanGel (GE Healthcare, Zurich, Switzerland) polyacrylamide gel electrophoresis (Fig. 1). The two PCR products of the two samples were purified after gel excision and reamplified in a seminested PCR using primers BAK11w and BAK553r (5′-TTA CCG CGG CTG CTG GCA C-3′, E. coli rrsA nt positions 515 to 533) and sequenced using the amplification primers. DNA sequence homology analyses were done using the SmartGene IDNS database and software (SmartGene, Zug, Switzerland). The sequences of the two PCR products reamplified using primers BAK11w and BAK553r showed highest homologies to the 16S rRNA gene of Fusobacterium nucleatum (1 mismatch in 499 bp, 1 ambiguous base indicating the presence of multiple 16S rRNA gene copies in the genome; GenBank accession number FJ638888.2) and Porphyromonas endodontalis (1 mismatch in 507 bp; GenBank accession number FJ638887.2), respectively.
TABLE 1.
Analysis of patient samples taken from the cerebral abscess
| Sample | Microscopy results |
Culture resultc | Identification by bacterial 16S rRNA gene PCR analysis | No. of mismatches/total no. | % Homology | |
|---|---|---|---|---|---|---|
| Leukocytesa | Microorganismsb | |||||
| I | >25 leukocytes | Negative | Negative | Fusobacterium nucleatum | 1/499 | 99.8 |
| Porphyromonas endodontalis | 1/507 | 99.8 | ||||
| II | Negative | Negative | Negative | NDd | ||
| III | Negative | Negative | Negative | Fusobacterium nucleatum | 1/499 | 99.8 |
| Porphyromonas endodontalis | 1/5071 | 99.8 | ||||
Average of 10 visual fields at a 100× magnification.
Average of 10 visual fields at a 1,000× magnification.
Samples were streaked on seven different agar media (Columbia blood agar, MacConkey agar, CNA blood agar, Crowe agar, brucella agar, kanamycin-vancomycin agar, and phenylethyl alcohol agar) and incubated under aerobic and anaerobic conditions as described in Materials and Methods.
ND, not done due to insufficient sample volume.
FIG. 1.
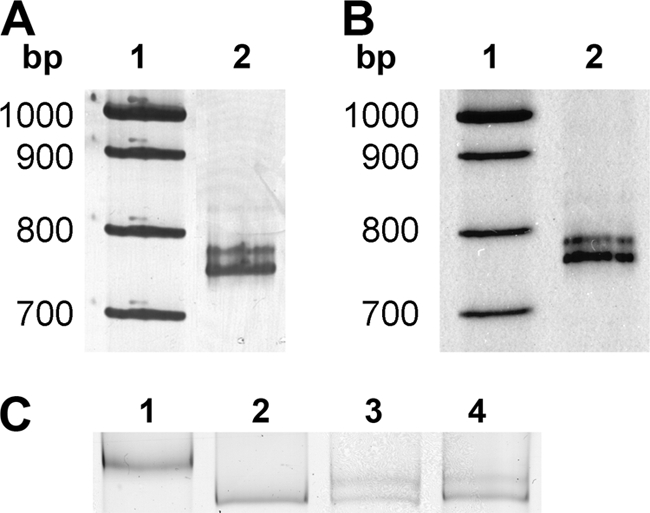
Polyacrylamide gel electrophoresis of 16S rRNA gene PCR products. (A) Lane 1, molecular mass ladder XIV (Roche Diagnostics); lane 2, 16S rRNA gene amplicons obtained with patient sample I. (B) The setup for sample III is identical to that described for panel A. (C) Lane 1, amplicon of Porphyromonas endodontalis ATCC 35406T culture; lane 2, amplicon of Fusobacterium nucleatum subsp. nucleatum culture; lane 3, amplicons obtained with patient sample I; lane 4, amplicons obtained with patient sample III.
In silico calculation of amplification fragment lengths using the BLASTN algorithm for F. nucleatum and P. endodontalis resulted in lengths of 774 and 798 bp, respectively. In polyacrylamide gel electrophoresis, the calculated lengths correspond to the amplicons of a direct PCR from a pure culture of either species (Fig. 1C, lanes 1 and 2). In previous studies, 16S rRNA gene amplifications resulting in two different fragments could not be resolved due to poor fragment separation (6). The progress in separation of the primary amplicons was achieved by using polyacrylamide instead of agarose gel electrophoresis for the improved resolution of amplicons. Our study shows that polymicrobial infections can be identified by broad-range bacterial 16S PCR when variable amplicon lengths allow electrophoretical separation. In silico calculation of the partial 16S rRNA gene amplicon of 156 human pathogenic bacterial species revealed that the fragments amplified by the universal primers BAK11w and BAK2 can range between 738 and 910 bp (22 species are listed in Table 2; see the supplemental material for a list of all 156 species), providing the opportunity for efficient separation in a significant number of polymicrobial infections.
TABLE 2.
16S rRNA gene amplicon length variability of 22 human pathogens
| Taxonomic name | Amplicon length (bp)a | NCBI accession no. |
|---|---|---|
| Agrobacterium radiobacter K84 | 738 | NC_011985.1 |
| Bacillus anthracis strain A0248 | 806 | NC_012659.1 |
| Bartonella henselae strain Houston-1 | 741 | BX897699.1 |
| Bordetella pertussis strain Tohama I | 791 | BX640418.1 |
| Borrelia burgdorferi ZS7 | 792 | CP001205.1 |
| Brucella melitensis ATCC 23457 | 738 | NC_012441.1 |
| Campylobacter jejuni RM1221 | 771 | CP000025.1 |
| Clostridium cellulolyticum H10 | 910 | NC_011898.1 |
| Clostridium difficile 630 | 758 | NC_009089.1 |
| Corynebacterium diphtheriae NCTC 13129 | 772 | NC_002935.2 |
| Enterococcus faecalis strain HN-S7 | 815 | FJ378704.1 |
| Escherichia coli BW2952 | 797 | NC_012759.1 |
| Francisella tularensis subsp. tularensis WY96-3418 | 785 | NC_009257.1 |
| Mycobacterium leprae Br4923 | 797 | NC_011896.1 |
| Mycoplasma hominis | 786 | M96660.1 |
| Mycoplasma pneumoniae M129 | 793 | U00089.2 |
| Nocardia asteroides strain ATCC 19247 | 749 | DQ659898.1 |
| Nocardia farcinica IFM 10152 | 769 | AP006618.1 |
| Pseudomonas aeruginosa LESB58 | 791 | FM209186.1 |
| Salmonella enterica subsp. enterica serovar Paratyphi C | 797 | NC_012125.1 |
| Staphylococcus aureus strain ATCC 14458 | 804 | DQ997837.1 |
| Streptococcus pneumoniae 70585 | 801 | NC_012468.1 |
Amplicon lengths were determined using universal primers BAK11w and BAK3 as start and end sequences, respectively.
Anaerobes play an important role in brain abscess formation and infections spreading from chronic sinusitis (3). In most mucous membranes, anaerobic Gram-negative bacilli (AGNB) outnumber aerobic and facultative bacteria in ratios ranging from 10:1 to 10,000:1 (5). F. nucleatum and P. endodontalis belong to the group of strictly anaerobic bacteria (8) presumably associated with periodontal disease (12). In our patient, brain abscess arose from acute sinusitis and is a well-known complication of this infection. Progression of the abscess under empirical antibiotic treatment is due to either inadequate antibiotic diffusion into the abscess or inappropriate antimicrobial spectrum of the prescribed antibiotics. Ceftriaxone and metronidazole show penetration in the brain, are active against the commonly described pathogens responsible for brain abscess, and are generally recommended as empirical therapy (9). In progressive disease despite correct antibiotic prescription, surgical drainage or excision, which also permits sampling for microbiological analyses, is mandatory.
Bacterial 16S rRNA gene PCR often outperforms anaerobic culture in the detection of fastidious and anaerobic pathogens in brain abscess material (7, 10, 11). This may be explained due to the difficult and time-critical sample treatment in plating for anaerobic cultures. Microorganisms were not found by microscopy. This is mainly due to the low sensitivity of the microscopic technique (cutoff, >104 bacteria per ml) and might also be attributable to bacterial lysis in the presence of granulocytic proteinases. Microscopy showed differing amounts of leukocytes, indicating different levels of inflammation in the material sampled (Table 1). Even though great care and rapid manipulations were employed during plating, cultures remained sterile in our case. The failure to grow the corresponding pathogens is more likely due to the previous antibiotic treatment of the patient (1). The causative agents in this case of a life-threatening brain abscess could be identified only by broad-spectrum PCR and a successful separation of the PCR amplicons. The patient was treated for 6 weeks with ceftriaxone and metronidazole and recovered fully with the exception of residual headache.
Supplementary Material
Acknowledgments
We thank the laboratory technicians for their expert help and Erik C. Böttger for continuous support and encouragement. We thank Rudolf Gmür for providing the ATCC type strain of Porphyromonas endodontalis.
This study was funded by the University of Zurich.
The authors declare no conflicts of interest.
P.M.K., S.K.R., and G.V.B. wrote the manuscript. P.M.K. and G.V.B. performed the 16S rRNA sequence analyses, and S.K.R. reviewed the clinical data and examined the patient history. All authors commented and agreed on the final manuscript.
Footnotes
Published ahead of print on 14 April 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Al Masalma, M., F. Armougom, W. M. Scheld, H. Dufour, P. H. Roche, M. Drancourt, and D. Raoult. 2009. The expansion of the microbiological spectrum of brain abscesses with use of multiple 16S ribosomal DNA sequencing. Clin. Infect. Dis. 48:1169-1178. [DOI] [PubMed] [Google Scholar]
- 2.Bosshard, P. P., A. Kronenberg, R. Zbinden, C. Ruef, E. C. Bottger, and M. Altwegg. 2003. Etiologic diagnosis of infective endocarditis by broad-range polymerase chain reaction: a 3-year experience. Clin. Infect. Dis. 37:167-172. [DOI] [PubMed] [Google Scholar]
- 3.Brook, I. 2006. The role of anaerobic bacteria in sinusitis. Anaerobe 12:5-12. [DOI] [PubMed] [Google Scholar]
- 4.Fenollar, F. V. Roux, A. Stein, M. Stein, M. Drancourt, and D. Raolt. 2006. Analysis of 525 samples to determine the usefulness of PCR amplification and sequencing of the 16S rRNA gene for diagnosis of bone and joint infections. J. Clin. Microbiol. 44:1018-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finegold, S. M. 1977. Anaerobic bacteria in human disease. Academic Press, New York, NY.
- 6.Goldenberger, D., A. Kunzli, P. Vogt, R. Zbinden, and M. Altwegg. 1997. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J. Clin. Microbiol. 35:2733-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kupila, L., K. Rantakokko-Jalava, J. Jalava, S. Nikkari, R. Peltonen, O. Meurman, R. J. Marttila, E. Kotilainen, and P. Kotilainen. 2003. Aetiological diagnosis of brain abscesses and spinal infections: application of broad range bacterial polymerase chain reaction analysis. J. Neurol. Neurosurg. Psychiatr. 74:728-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray, P. R., and E. J. Baron. 2007. Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 9.Sanford, J. P., D. N. Gilbert, R. C. Moellering, G. M. Eliopoulos, and M. A. Sande. 2008. The Sanford guide to antimicrobal therapy 2008, 38th ed. Antimicrobial Therapy, Sperryville, VA.
- 10.Tsai, J. C., L. J. Teng, and P. R. Hsueh. 2004. Direct detection of bacterial pathogens in brain abscesses by polymerase chain reaction amplification and sequencing of partial 16S ribosomal deoxyribonucleic acid fragments. Neurosurgery 55:1154-1162. [DOI] [PubMed] [Google Scholar]
- 11.Woo, P. C., S. K. Lau, J. L. Teng, H. Tse, and K. Y. Yuen. 2008. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 14:908-934. [DOI] [PubMed] [Google Scholar]
- 12.Ximenez-Fyvie, L. A., A. D. Haffajee, and S. S. Socransky. 2000. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J. Clin. Periodontol. 27:648-657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


