Abstract
We evaluated the SD Bioline Influenza Ag A/B/A(H1N1) Pandemic test kit and compared it with real-time reverse transcriptase PCR (RT-PCR) for its ability to detect H1N1 2009. The sensitivity and specificity of the test kit for H1N1 2009 were 77% and 100%, respectively.
In March and early April of 2009, the pandemic influenza A/H1N1 2009 virus (H1N1 2009) was detected in Mexico and the United States, followed by a rapid worldwide person-to-person spread (1, 6). The detection of influenza virus-specific RNA via reverse transcriptase PCR (RT-PCR) is the current method of choice for the detection of influenza virus (3, 5). Due to the requirement for specialized equipment and the long turnaround times, RT-PCR for the detection of H1N1 2009 is currently available only in limited clinical settings. Rapid antigen tests (RAT) have been conducted in a variety of clinical settings for the detection of H1N1 2009, due to the rapid results and ready availability of those tests. Recently, a new RAT [SD Bioline Influenza Ag A/B/A(H1N1) Pandemic; Standard Diagnostics, Inc., Yongin-si, South Korea] was developed for the specific detection of H1N1 2009. We evaluated the new RAT in comparison with the real-time RT-PCR assay in terms of its ability to detect H1N1 2009.
In an effort to compare the new RAT with real-time RT-PCR, nasopharyngeal swab specimens were collected using flocked swabs (Copan Diagnostics, Murrieta, CA) from suspected H1N1 2009 patients in November 2009. The ages of the patients ranged between 2 weeks and 83 years (average, 13.5 years). The clinical specimens on 561 paired swabs (2 per patient) were each placed into pairs of test tubes—one containing 300 μl of buffer solution for the antigen test and one containing 1 ml of viral transport medium (VTM) for real-time RT-PCR. One hundred ninety-eight unpaired swabs (1 per patient) were inserted into test tubes containing 1 ml of VTM for RAT and real-time RT-PCR. In an effort to evaluate the cross-reactivity of RAT, 16 different influenza virus suspensions (influenza A-15 subtypes and influenza B-1 subtype) and 117 virus-positive clinical specimens of nasopharyngeal aspirates (seasonal influenza A virus, adenovirus, coronavirus, human rhinovirus A, human metapneumovirus, and parainfluenza virus) were analyzed. Twenty seasonal influenza A virus-positive samples (9 for H1N1, 10 for H3N2, and 1 untypeable strain), which were obtained during the 2008-2009 influenza season, were included in the sample of clinical specimens.
The new RAT, which has 4 lines for the detection of H1N1 2009, influenza A virus, influenza B virus, and controls, was conducted using approximately 90 μl of samples. The viral RNA was extracted using a QIAamp viral RNA minikit (Qiagen, Hilden, Germany) from 140 μl of samples, in accordance with the manufacturer's instructions. The RNA was eluted from columns with 50 μl of nuclease-free water. Each reaction mixture for real-time RT-PCR was prepared in accordance with the manufacturer's instructions and included 5 μl of RNA extraction and PCR reagents (Influenza A/H1N1 Detection set; Roche Applied Science, Mannheim, Germany). Amplification and detection were conducted on a LightCycler 480 (Roche Applied Science, Mannheim, Germany).
Among the 561 paired specimens, 241 tested positive via real-time RT-PCR. Among the 241 PCR-confirmed cases, 186 (77%) were positive for H1N1 2009 and 152 (63%) were positive for influenza A virus by the new RAT. Among them, 37 cases were positive for H1N1 2009 but negative for influenza A virus on RAT and 3 cases were negative for H1N1 2009 but positive for influenza A virus. Among 320 PCR-negative cases, only one was positive for influenza A virus by RAT. Among the 198 unpaired specimens, 72 tested positive on PCR. Among the PCR-confirmed samples, 55 (76%) tested positive for H1N1 2009 by RAT and 42 (58%) tested positive for influenza A virus by RAT. Thirteen samples were positive for H1N1 2009 by RAT and PCR but negative for influenza A virus by RAT. Among the 126 PCR-negative samples, only one tested positive for H1N1 2009 by RAT. The overall sensitivities of the H1N1 2009 component and influenza A virus component in the new RAT were 77% and 62%, respectively (Table 1). The crossing-point (Cp) values of the 72 samples that tested positive on real-time RT-PCR but negative on H1N1 2009 RAT ranged between 20.4 and 37.7 (median value, 30.3), whereas the Cp values of samples testing positive on H1N1 2009 RAT and PCR ranged between 15.3 and 34.7 (median value, 22.5). The sensitivity of the H1N1 2009 RAT was 79% (55/70) in the 0- to 5-year-old group, 77% (156/204) in the 5- to 20-year-old group, and 77% (30/39) in the over-20-year-old group.
TABLE 1.
Performance of the new RATa compared to real-time RT-PCR for the detection of pandemic influenza A/H1N1 2009 virus
| Detection component of the new RAT | No. of cases |
% Sensitivity (95% CIf) | % Specificity (95% CI) | PPVb (%) | NPVc (%) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Total | PCR+ RAT+ | PCR+ RAT− | PCR− RAT+ | PCR− RAT− | |||||
| Detection component for 2009 H1N1 | |||||||||
| Paired samplesd | 561 | 186 | 55 | 0 | 320 | 77 (72-82) | 100 (99-100) | 100 | 85 |
| Unpaired samplee | 198 | 55 | 17 | 1 | 125 | 76 (65-84) | 99 (96-100) | 98 | 88 |
| Overall | 759 | 241 | 72 | 1 | 445 | 77 (72-81) | 100 (99-100) | 100 | 86 |
| Detection component for influenza A virus | |||||||||
| Paired samples | 561 | 152 | 89 | 1 | 319 | 63 (57-69) | 100 (98-100) | 99 | 78 |
| Unpaired sample | 198 | 42 | 30 | 0 | 126 | 58 (47-69) | 100 (97-100) | 100 | 81 |
| Overall | 759 | 194 | 119 | 1 | 445 | 62 (57-67) | 100 (99-100) | 100 | 79 |
As determined by SD Bioline Influenza Ag A/B/A(H1N1) Pandemic.
PPV, positive predictive value.
NPV, negative predictive value.
Two nasopharyngeal swabs (placed in 2 test tubes—one containing buffer solution and one containing viral transport medium) from the same patient at the same time were prepared for RAT and real-time RT-PCR.
One nasopharyngeal swab (placed in a tube containing viral transport medium) was used for RAT and real-time RT-PCR.
CI, confidence interval.
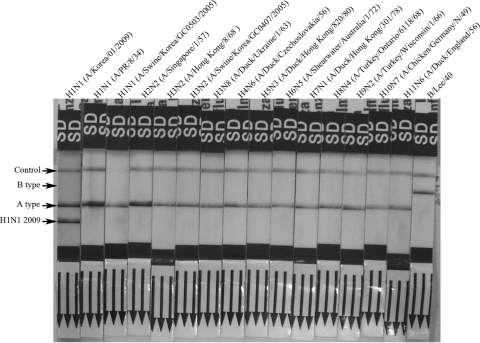
In the cross-reactivity test using the viral suspensions, A/Korea/01/2009 (H1N1) was strongly positive for H1N1 2009 by RAT and A/Swine/Korea/GC0503/2005 (H1N1) was weakly positive. The others were negative for H1N1 2009 (Fig. 1). Among the 117 clinical specimens, only one specimen (seasonal influenza A H1N1 virus) was weakly positive for H1N1 2009 by RAT, and the others were negative.
FIG. 1.
Cross-reactivity testing results for the new RAT against 16 different influenza viruses.
The RAT for the detection of H1N1 2009 revealed a broad range of sensitivity and specificity (9.6 to ∼75% and 80 to ∼100%, respectively) (4, 5, 7, 8). Because of its low sensitivity, the clinical utility of RAT remains a subject of debate (2, 8, 9). Our results show that the new RAT features relatively high sensitivity for the detection of H1N1 2009. We presume that the difference of antibody affinity and targeting sites (hemagglutinin and nucleoprotein) of detection components may affect different sensitivities. Additionally, in the majority of cases, the new RAT can distinguish between seasonal influenza virus and H1N1 2009, although our results are preliminary due to the very limited number of samples, and further specificity testing of the RAT with more seasonal influenza viruses may be required. Because more than 99% of the current seasonal H1 strains are resistant to oseltamivir, diagnostic tests to distinguish the pandemic strain from seasonal influenza virus may be important for clinical management (9). Also, RAT has the advantage of providing rapid results. In our laboratory, the average turnaround times of RAT and real-time RT-PCR were 0.9 and 14.9 h, respectively.
We suggest that SD Bioline Influenza Ag A/B/A(H1N1) Pandemic may be a useful diagnostic tool for the detection of H1N1 2009 in appropriate clinical settings, although a negative RAT may require confirmatory assays of greater sensitivity. Additionally, further studies may be necessary to validate the assay using other sample types and to determine the sensitivity of the assay for pandemic H1N1 2009, using viral stocks whose titers have been determined.
Acknowledgments
SD Bioline Influenza Ag A/B/A(HINI) Pandemic was developed by the research arm (2009-E00669-00) of the Korea Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 31 March 2010.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2009. Swine influenza A (H1N1) infection in two children—Southern California, March-April 2009. Morb. Mortal. Wkly. Rep. 58:400-402. [PubMed] [Google Scholar]
- 2.Drexler, J. F., A. Helmer, H. Kirberg, U. Reber, M. Panning, M. Müller, K. Höfling, B. Matz, C. Drosten, and A. M. Eis-Hübinger. 2009. Poor clinical sensitivity of rapid antigen test for influenza A pandemic (H1N1) 2009 virus. Emerg. Infect. Dis. 15:1662-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Druce, J., T. Tran, H. Kelly, M. Kaye, D. Chibo, R. Kostecki, A. Amiri, M. Catton, and C. Birch. 2005. Laboratory diagnosis and surveillance of human respiratory viruses by PCR in Victoria, Australia, 2002-2003. J. Med. Virol. 75:122-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ginocchio, C. C., F. Zhang, R. Manji, S. Arora, M. Bornfreund, L. Falk, M. Lotlikar, M. Kowerska, G. Becker, D. Korologos, M. de Geronimo, and J. M. Crawford. 2009. Evaluation of multiple test methods for the detection of the novel 2009 influenza A (H1N1) during the New York City outbreak. J. Clin. Virol. 45:191-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kok, J., C. C. Blyth, H. Foo, J. Patterson, J. Taylor, K. McPhie, V. M. Ratnamohan, J. R. Iredell, and D. E. Dwyer. 2010. Comparison of a rapid antigen test with nucleic acid testing during cocirculation of pandemic influenza A/H1N1 2009 and seasonal influenza A/H3N2. J. Clin. Microbiol. 48:290-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith, G. J., D. Vijaykrishna, J. Bahl, S. J. Lycett, M. Worobey, O. G. Pybus, S. K. Ma, C. L. Cheung, J. Raghwani, S. Bhatt, J. S. Peiris, Y. Guan, and A. Rambaut. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122-1125. [DOI] [PubMed] [Google Scholar]
- 7.Vasoo, S., J. Stevens, and K. Singh. 2009. Rapid antigen tests for diagnosis of pandemic (swine) influenza A/H1N1. Clin. Infect. Dis. 49:1090-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watcharananan, S., S. Kiertiburanakul, and W. Chantratita. 2010. Rapid influenza diagnostic test during the outbreak of the novel influenza A/H1N1 2009 in Thailand: an experience with better test performance in resource limited setting. J. Infect. 60:86-87. [DOI] [PubMed] [Google Scholar]
- 9.Welch, D. F., and C. C. Ginocchio. 2010. Role of rapid immunochromatographic antigen testing in diagnosis of influenza A virus 2009 H1N1 infection. J. Clin. Microbiol. 48:22-25. [DOI] [PMC free article] [PubMed] [Google Scholar]