Abstract
Cryptosporidium spp., a common cause of diarrhea in children, were investigated in the first multisite study in India. Diarrheal stools from hospitalized children aged <5 years from Delhi, Trichy, and Vellore were analyzed by microscopy, PCR-restriction fragment length polymorphism (RFLP), and/or sequencing at the small-subunit (SSU) rRNA and Cpgp40/15 loci for species determination and subgenotyping, respectively. Seventy of 2,579 (2.7%) children, 75% of whom were <2 years old, had cryptosporidial diarrhea as determined by microscopy. Genotyping and subgenotyping showed that Cryptosporidium hominis was the most commonly identified species (59/67 children), and subgenotypes Ie, Ia, Ib, and Id were common in all centers. A novel C. parvum subgenotype, IIn, was identified in Vellore. Meteorological analysis revealed a higher rate of cryptosporidial positivity during hotter and drier weather in Delhi.
Cryptosporidium spp. are an important cause of endemic parasitic diarrhea in children in developing countries. In addition to causing symptoms associated with watery diarrhea, vomiting, and weight loss, early childhood cryptosporidiosis has been shown by studies to be associated with subsequent faltering of growth (reviewed in reference 11). Cryptosporidium hominis and C. parvum cause the majority of infections in children in developing countries, with C. hominis predominating and occasional reports of infection with zoonotic species such as C. felis, C. canis, C. meleagridis, and C. muris (30). C. hominis infection has been found to be associated with greater levels of oocyst shedding (4) and longer durations of oocyst shedding (31) and diarrhea (15) than C. parvum infection. In a recent community-based study in Vellore, we found increased levels of severity of diarrhea in C. hominis-infected children compared to the levels observed in children infected with other species (1).
Cryptosporidium spp. have been classified into several distinct subgenotypes based on extensive polymorphisms in the Cpgp40/15 (also referred to as GP60) locus by use of PCR-restriction fragment length polymorphism (RFLP) or sequencing of PCR products (reviewed in reference 30).
A number of studies from India have reported Cryptosporidium spp. in diarrheal stool samples from children, with positivity rates of up to nearly 20% (17) and asymptomatic infection rates of up to 10% (19), using stool microscopy for detection. However, only three studies have used molecular techniques for identification of cryptosporidiosis in children in India (9, 13, 22), suggesting that the actual infection rates may be significantly higher. In a previous hospital-based study in Vellore, we that found that PCR (15.2%) identified more than 3 times the number of cases of cryptosporidial diarrhea than microscopy (4.4%) (2). The aim of the present study was to identify the Cryptosporidium species and Cpgp40/15 subgenotypes associated with cryptosporidial diarrhea in hospitalized children from 3 centers in the country, since no studies have examined cryptosporidiosis using the same methods in more than one location.
MATERIALS AND METHODS
Study population and sample collection.
This study was performed on stool samples originally collected for a multicenter rotavirus surveillance program called the Indian Rotavirus Strain Surveillance Network from December 2005 to December 2008, where samples were tested for rotavirus but underwent no further testing under the surveillance protocol. Samples from 3 centers representing both southern and northern India, namely, Christian Medical College, Vellore, India (southern), St. Stephen's Hospital, Delhi, India (northern), and Child Jesus Hospital, Trichy, India (southern), were available for this study. Children less than 5 years of age presenting to one of the 3 study hospitals with acute gastroenteritis and requiring hospitalization for rehydration for at least 6 h were enrolled. Children not requiring supervised oral or intravenous rehydration were excluded. A detailed clinical evaluation of the episode of diarrhea, including duration, severity of diarrhea (maximum number of stools in a 24-h period), vomiting, fever, and degree of dehydration, was performed. Informed consent was obtained from the parent/guardian, and the study was approved by the Institutional Review Board of Christian Medical College, Vellore, India.
Laboratory procedures.
A stool specimen was collected from each child and tested for Cryptosporidium spp. by acid-fast staining and microscopy. Aliquots of positive samples were stored at −70°C for further characterization, and all further laboratory work was carried out at the Vellore center.
DNA extraction, PCR-RFLP, and sequencing.
DNA was extracted from the microscopy-positive stool samples using a QIAamp DNA stool kit (Qiagen, Inc., Valencia, CA) and then analyzed by PCR-RFLP at the small-subunit (SSU) rRNA (32) and Cpgp40/15 (8) loci, using previously described protocols. For samples where ambiguous results were obtained by PCR-RFLP at the Cpgp40/15 locus, purified PCR products were sequenced by the BigDye Terminator method at Christian Medical College or the Tufts University Core facility. Multiple sequence alignment of sequences from this study as well as nucleotide sequences representative of the known subgenotypes obtained from GenBank was carried out using MUSCLE, followed by phylogenetic analysis using the maximum likelihood method with PhyML (3, 14) and tree construction using TreeDyn (7) with the default settings in the Phylogeny.fr server (version 2) (10). Sequences were also analyzed for the number of tandem repeats of the serine-coding trinucleotides TCA, TCG, and TCT at the 5′ end of the gp40 gene sequence to further characterize subtypes within each family, as described by Xiao (30).
Meteorological data collection and analysis.
In order to assess the possible association between meteorological parameters (temperature, rainfall, and humidity) and cryptosporidial diarrhea, monthly data on the mean maximum and minimum temperatures, relative humidity levels at 8:30 a.m. and 5:30 p.m., and total rainfall were obtained separately for each of the 3 locations. Data for the Delhi region were obtained from the Regional Meteorological Center, New Delhi, India, and those for the Trichy and Vellore regions were obtained from the Regional Meteorological Center, Chennai, India. In order to adjust for the potential bias due to the variable number of stool samples screened during a particular month, the proportion of stool samples positive for Cryptosporidium spp. for that month was calculated by dividing the total number of samples positive for Cryptosporidium against the total number of diarrheal samples screened during that month.
Statistical analysis.
Data were analyzed using STATA 10.1 for Windows (StataCorp, College Station, TX). Differences in age and cryptosporidial positivity rates between the three centers were compared using the Mann-Whitney U test and Fisher's exact test, respectively. Comparison of clinical features among patients infected with different species and subgenotypes was performed using the Mann-Whitney U test for duration of diarrhea and Fisher's exact test for severity of diarrhea. Correlation between the cryptosporidial positivity rate and the meteorological parameter values were assessed using Spearman's rank order correlation coefficient test. A P value of <0.05 was considered statistically significant.
Nucleotide sequence accession numbers.
Sequences from this study were deposited in GenBank (accession numbers FJ897784-88 and GQ384437-44).
RESULTS
Prevalence and seasonality of cryptosporidial diarrhea.
Seventy of the 2,579 children enrolled in the study (2.7%) were found to have Cryptosporidium spp. in their stool samples by microscopy. Children from Delhi showed a higher prevalence (34/970 [3.5%]) (P = 0.055) than children from the 2 southern Indian centers in Vellore (20/1,018 [2.0%]) and Trichy (16/591 [2.7%]). Most children with cryptosporidiosis were less than 2 years of age (75.4%), with a median (interquartile range [IQR]) age of 13 (9 to 22) months. The median ages of children in Delhi (11 [IQR, 8 to 18] months) and Trichy (13 [IQR, 8.5 to 21] months) were lower than that of children in Vellore (17.5 [IQR, 12 to 25] months) (P = 0.024). Most children were male (67.1%). The median (IQR) duration of diarrhea was 3 (2 to 5) days, with 10 children having diarrhea for over a week, 3 of whom had diarrhea for more than 2 weeks. When assessed for severity of diarrhea based on the number of stools in a 24-h period, the median (IQR) number of stools was found to be 8 (5 to 15).
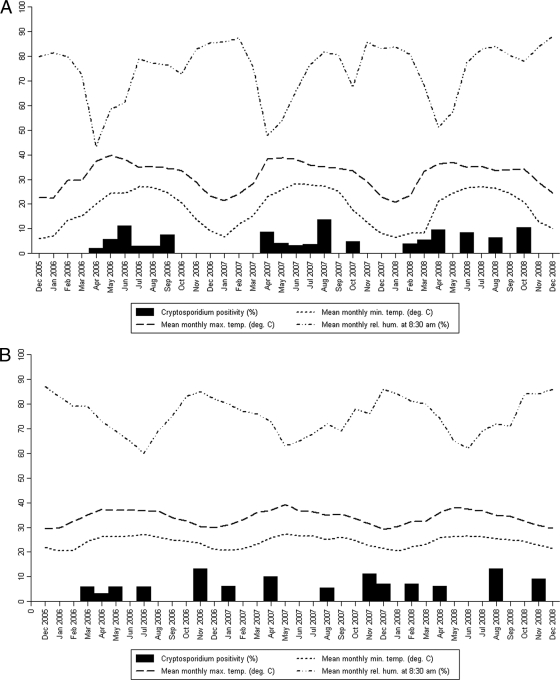
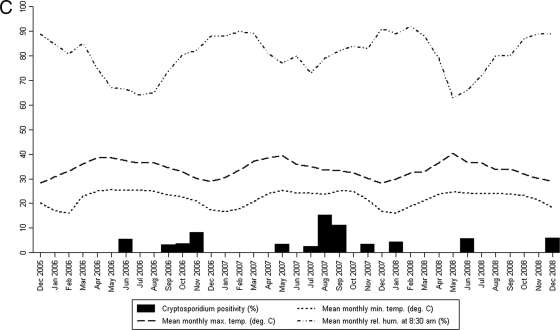
The meteorological data were analyzed to determine if there was any seasonal variation in the prevalences of cryptosporidiosis among children in each of the three centers over the 3 years of the study (Fig. 1). For the Delhi region, there was a statistically significant positive correlation between temperature, both minimum and maximum, and cryptosporidial positivity rates (P = 0.003 and P < 0.001, respectively). On the other hand, there was a significant negative correlation between relative humidity and cryptosporidial positivity (P = 0.009). No such correlation was observed for the Vellore and Trichy regions, which have less seasonal variation than Delhi (Table 1).
FIG. 1.
Monthly distribution of cryptosporidial positivity rates and the meteorological parameters for the Delhi (A), Trichy (B), and Vellore (C) centers.
TABLE 1.
Correlation between cryptosporidial positivity rates and values for the various meteorological parameters in the three centers
| Characteristic | Delhi |
Trichy |
Vellore |
|||
|---|---|---|---|---|---|---|
| Rhoa | P | Rho | P | Rho | P | |
| Mean monthly maximum temp | 0.608 | <0.001 | −0.15 | 0.384 | −0.094 | 0.58 |
| Mean monthly minimum temp | 0.475 | 0.003 | −0.093 | 0.59 | 0.097 | 0.569 |
| Total monthly rainfall | 0.218 | 0.201 | 0.195 | 0.253 | 0.233 | 0.165 |
| Mean monthly relative humidity at 8:30 a.m. | −0.487 | 0.003 | 0.193 | 0.26 | −0.149 | 0.378 |
| Mean monthly relative humidity at 5:30 p.m. | −0.091 | 0.596 | 0.131 | 0.446 | 0.255 | 0.128 |
Spearman's rank order correlation coefficient.
Species and subgenotypes.
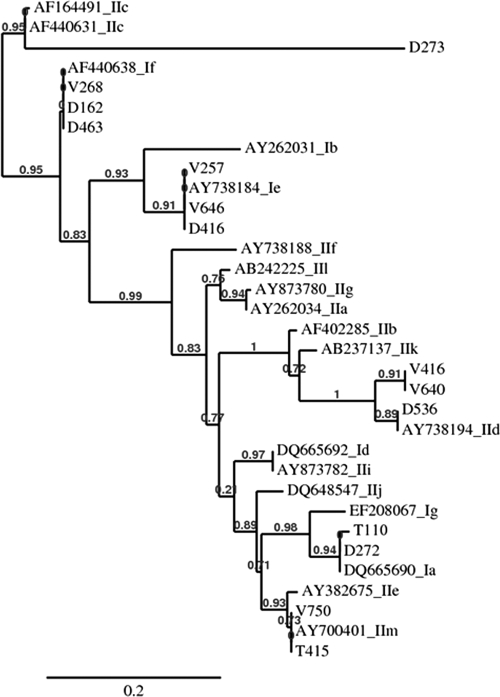
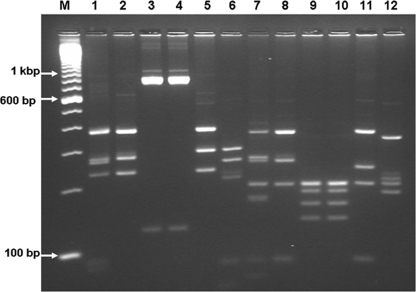
PCR products for the SSU rRNA locus could be amplified from stool DNA from 67 of 70 children. C. hominis was the most commonly identified species in all 3 centers and was detected in 88.1% (59/67) of children, followed by C. parvum, which was detected in 10.5% (7/67) of children. C. meleagridis was identified in only one child (from Trichy) (Table 2). PCR products for the Cpgp40/15 locus could be amplified from stool DNA from 56 of 59 children with C. hominis infection and 6 of 7 children with C. parvum infection. Subgenotypes Ia and Ie (16/56 of each) were the most commonly identified subgenotypes among the C. hominis-infected children, followed by subgenotypes Ib (11/56 children) and Id (7/56 children). The If subgenotype (5/56 children), however, was seen mainly in Delhi, with only one instance seen in Vellore. An RFLP pattern suggestive of mixed infection with Ia and If was also seen in one child from the Delhi center. Among the 7 children with C. parvum infection, one each with subgenotypes IIc and IId was identified in the Delhi center. Sequencing and phylogenetic analysis showed the presence of a newly identified subgenotype, IIm (previously identified in children from Bangladesh) (H. D. Ward, unpublished data), in the southern Indian centers in Vellore and Trichy (numbers T415 and V740) and another previously unreported subgenotype, named IIn in accordance with current nomenclature conventions (30), in the Vellore center (numbers V416 and V640) (Fig. 2). The unique PCR-RFLP patterns of these C. parvum subgenotypes are shown in Fig. 3. When the sequences were analyzed for tandem repeats of serine-coding nucleotides at the 5′ end of the gp40 gene sequence, both sequences from the C. hominis Ia family were found to be of the IaA18 and IaA19 subtypes, all 3 sequences from the C. hominis Ie family were found to be of the A11G3T3 subtype, and the sequences from the C. hominis If family were all found to be of the A13G1 subtype. Among the C. parvum sequences, that from the anthroponotic IIc family was of the A5G3 subtype and that from the zoonotic IId family was of the A15G1 subtype. Both sequences from the C. parvum IIm family were of the A7G1 subtype, which was identical to what was observed for the sequences identified in Bangladesh (Hira et al., submitted), and both sequences from the novel IIn family were of the A8 subtype.
TABLE 2.
Cryptosporidium species and subgenotypes identified in three centers in India
| Species or subgenotype | No. (%) for: |
|||
|---|---|---|---|---|
| Delhi | Vellore | Trichy | All | |
| Species (n = 67) | ||||
| C. hominis | 28 (90.3) | 17 (85) | 14 (87.5) | 59 (88.1) |
| C. parvum | 3 (9.6) | 3 (15) | 1 (6.3) | 7 (10.5) |
| C. meleagridis | 0 (0) | 0 | 1 (6.3) | 1 (1.5) |
| Subgenotype(s) (n = 62) | ||||
| Ia | 9 (31) | 2 (10.5) | 5 (35.7) | 16 (25.8) |
| Ib | 4 (13.7) | 5 (26.3) | 2 (14.2) | 11 (17.7) |
| Id | 3 (10.3) | 3 (15.8) | 1 (7.1) | 7 (11.3) |
| Ie | 6 (20.7) | 5 (26.3) | 5 (35.7) | 16 (25.8) |
| If | 4 (13.8) | 1 (5.2) | 0 (0) | 5 (8.1) |
| IIc | 1 (3.4) | 0 (0) | 0 (0) | 1 (1.6) |
| IId | 1 (3.4) | 0 (0) | 0 (0) | 1 (1.6) |
| IIm | 0 (0) | 1 (5.2) | 1 (7.1) | 2 (3.2) |
| IIn | 0 (0) | 2 (10.5) | 0 (0) | 2 (3.2) |
| Mixed | 1 (3.4) | 0 (0) | 0 (0) | 1 (1.6) |
FIG. 2.
Phylogenetic tree of sequences from a multisite study of cryptosporidial diarrhea in children in India compared to what was observed for known representative Cpgp40/15 subgenotypes. (The maximum likelihood method with PhyML and tree construction with TreeDyn were used with default settings in the Phylogeny.fr server [version 2]; the sequences for samples V416 and V640 were different from those for all other known subgenotypes and were named IIn.)
FIG. 3.
Cpgp40/15 subgenotypes of C. parvum identified in a multisite site on cryptosporidial diarrhea in India (M, 100-bp molecular weight marker; lanes 1 to 12, RFLP patterns observed following digestion with AluI [lanes 1 to 6] and with RsaI [lanes 7 to 12]; lanes 1 and 7, IIc; lanes 2 and 8, IId; lanes 3, 9, 4, and 10, IIm; lanes 5, 11, 6 and 12, IIn).
There were no significant differences in clinical features, including severity (P = 0.800), duration of diarrhea (P = 1.000), or presence of vomiting (P = 0.695), among children with C. hominis and C. parvum infections.
In addition, there was no significant difference in severity of diarrhea among children infected with C. hominis subgenotypes Ia, Ib, Id, and Ie. However, there was a trend toward association of shorter duration of diarrhea with subgenotype Id than with the other subgenotypes (P = 0.06) (Table 3). Similarly, there was a trend toward association of older age with subgenotype Ia (median [IQR] age, 18 [12 to 30] months) than with the other subgenotypes (median [IQR] age, 12 [9 to 18] months) (P = 0.063).
TABLE 3.
Association of clinical features with subgenotype
| C. hominis subtype | Duration of diarrhea (days)a for: |
P | Severity of diarrhea (no. of stools in 24 h)a for: |
P | Presence of vomiting (no. of subjects) for: |
P | |||
|---|---|---|---|---|---|---|---|---|---|
| Indicated subtype (n = 58) | Other subtypes | Indicated subtype (n = 56) | Other subtypes | Indicated subtype (n = 57) | Other subtypes | ||||
| Ia | 3.5 (2-7) | 3 (2-5) | 0.489 | 10 (6-15) | 8 (5-12) | 0.477 | 10 (62.5) | 21 (56.8) | 0.768 |
| Ib | 4 (2-6) | 3 (2-5) | 0.616 | 9 (6-10) | 8 (5-15) | 0.879 | 6 (54.6) | 25 (59.5) | 1.000 |
| Id | 2 (2-3) | 3 (2-7) | 0.060 | 8 (5-15) | 8 (6-12) | 0.946 | 5 (83.3) | 26 (55.3) | 0.382 |
| Ie | 3 (2-7) | 3 (2-5) | 0.876 | 7 (5-15) | 8 (6-12) | 0.800 | 9 (60.0) | 22 (57.9) | 1.000 |
| If | 3 (3-5) | 3 (2-5) | 0.880 | 7 (6-10) | 8 (5-15) | 0.674 | 1 (20.0) | 30 (62.5) | 0.147 |
Median (IQR).
DISCUSSION
In this study, we identified similarities and differences in infecting species, subgenotype, and seasonality of cryptosporidial diarrhea among children from 3 different centers in India. For all centers, C. hominis was the most commonly identified species among hospitalized children. This is in keeping with previous studies on cryptosporidial diarrhea among children from India, including our community-based birth cohort study (1) and two other hospital-based studies from Kolkata and Secunderabad (9, 22). In the current study, zoonotic species (C. parvum and C. meleagridis) were identified in only a few (12%) hospitalized children, which is also in keeping with the results of our community-based (19%) study (1) and those of the hospital-based studies in Kolkata and Secunderabad (9, 22). In contrast, we identified several adults (34%) infected with zoonotic species, including C. parvum, C. meleagridis, and C. felis, in our hospital-based study of HIV-infected patients with cryptosporidiosis in Vellore (21).
The most common subgenotypes were Ie, Ia, Ib, and Id, which were identified in all 3 centers. All the Ie alleles were found to be of the A11G3T3 subtype, which was the most commonly identified subtype in previous studies from developing countries (30). Among the C. parvum Cpgp40/15 sequences, single zoonotic IId and anthroponotic IIc subgenotypes were identified. The sequence from the IIc family was subtyped as A5G3, as expected, as there is no variation in this subtype family (30). A more recently described subgenotype, IIm, which was of the A7G1 subtype (Hira et al., submitted), was found exclusively in the southern Indian centers, and a novel subgenotype, called “IIn,” which was related to the IId subgenotype, was identified at the Vellore center. Interestingly, on phylogenetic analysis, the IIm subgenotype (Hira et al., submitted), which has been identified only in humans thus far, was found to be similar to the IIe subgenotype, also thought to be an anthroponotic subtype (27). There was also an increased level of diversity of C. hominis subgenotypes among hospitalized children, as seen in other studies from developing countries, unlike for the previous community-based study in Vellore, where there was a predominance of a single subgenotype, Ia (1). In addition, greater levels of diversity of C. parvum subgenotypes were identified in hospitalized children in this study and in a previous study of HIV-infected adults (21) in Vellore than in the community-based study, where only the anthroponotic IIc subgenotype was identified (1).
Analysis of the meteorological data showed an association of increased prevalence of cryptosporidiosis with increased temperature and low humidity for the Delhi center but not for the 2 southern Indian centers in Vellore and Trichy. This is similar to a peak in incidence of cryptosporidiosis which has been found to occur about a month after a peak in ambient temperatures in the United States (23). Interestingly, there was no correlation with rainfall in any of the centers, although a study from Kolkata reported that the highest incidence of cryptosporidiosis occurred in the rainy season from June to October (9). Endemic cryptosporidiosis has also been associated with the onset of the rainy season in Guinea Bissau (26), Uganda (29), Malawi (25), and Brazil (24). A recent study from Kenya also documented increased oocyst contamination of surface waters at the end of the rainy season, which was consistent with the timing of human infections in the region (20). However, cryptosporidiosis has been reported to occur in the spring season in South Korea (6), in the summer and autumn months in Israel (12), and in the cooler months of November to April in Kuwait (28). Differences in seasonality from different geographical areas can be explained by a recent meta-analysis of seasonality of cryptosporidiosis that found that increases in temperature and rainfall were predictors of increased cryptosporidiosis. However, there was some variation depending on the climate category, with rainfall being more important in the tropics and temperature more important in more-temperate climates (16). It is also possible that modes of transmission in tropical countries, such as India, may be different from those in temperate countries, resulting in different seasonal patterns.
The average ages of children with cryptosporidial diarrhea among hospitalized children in this study (17.5 months) and those in our previous community-based study (16.4 months) were similar (16). There were more male children in the current study than in the community-based study, in which there were no gender differences (1). The median durations of diarrhea in both studies (hospital-based study, 3 [2 to 5] days; community-based study, 3 [2 to 4] days) were also similar. In this study, we found no significant association between infecting species and clinical features. However, in our birth cohort study in Vellore (1) as well as in a previous birth cohort study from Peru (5) and a study of HIV-infected patients in Tanzania (15), C. hominis infection was found to be associated with greater duration and severity of diarrhea than infection with other species. In the current study, there was a trend toward an association between subgenotype Id and shorter duration of diarrhea as well as between subgenotype Ia and older age. Cama et al. reported significant associations between C. hominis subgenotypes Ia, Ib, Id, and Ie with diarrhea but only Ib with nausea, vomiting, and general malaise in a birth cohort study of cryptosporidiosis from Peru (5). In a study of cryptosporidiosis in HIV-infected children in South Africa, those infected with subgenotype IIc were significantly older than those infected with other subgenotypes (18).
In conclusion, this study documented the distribution of cryptosporidial species and subgenotypes in different regions of the country. A more detailed analysis of a greater number of subjects and continued monitoring of the incidence of cryptosporidiosis with temperature and rainfall are required to determine climatic associations with cryptosporidiosis in the Indian setting.
Acknowledgments
This study was supported by Fogarty International Research Cooperative agreement R03TW2711, International Research in Infectious Disease grant AI075452, and Global Infectious Disease Research Training grant D43 TW007392.
Footnotes
Published ahead of print on 14 April 2010.
REFERENCES
- 1.Ajjampur, S. S., B. P. Gladstone, D. Selvapandian, J. P. Muliyil, H. Ward, and G. Kang. 2007. Molecular and spatial epidemiology of cryptosporidiosis in children in a semiurban community in South India. J. Clin. Microbiol. 45:915-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ajjampur, S. S. R., P. Rajendran, I. Banerjee, S. Ramani, B. Monica, P. Sankaran, R. Vivek, R. Sarkar, H. Ward, and G. Kang. 2008. Closing the diagnostic gap in diarrhoea in Indian children by the application of molecular techniques. J. Med. Microbiol. 57:1364-1368. [DOI] [PubMed] [Google Scholar]
- 3.Anisimova, M., and O. Gascuel. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55:539-552. [DOI] [PubMed] [Google Scholar]
- 4.Bushen, O. Y., A. Kohli, R. C. Pinkerton, K. Dupnik, R. D. Newman, C. L. Sears, R. Fayer, A. A. Lima, and R. L. Guerrant. 2007. Heavy cryptosporidial infections in children in northeast Brazil: comparison of Cryptosporidium hominis and Cryptosporidium parvum. Trans. R. Soc. Trop. Med. Hyg. 101:378-384. [DOI] [PubMed] [Google Scholar]
- 5.Cama, V. A., C. Bern, J. Roberts, L. Cabrera, C. R. Sterling, Y. Ortega, R. H. Gilman, and L. Xiao. 2008. Cryptosporidium species and subtypes and clinical manifestations in children, Peru. Emerg. Infect. Dis. 14:1567-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chai, J. Y., N. Y. Kim, S. M. Guk, Y. K. Park, M. Seo, E. T. Han, and S. H. Lee. 2001. High prevalence and seasonality of cryptosporidiosis in a small rural village occupied predominantly by aged people in the Republic of Korea. Am. J. Trop. Med. Hyg. 65:518-522. [DOI] [PubMed] [Google Scholar]
- 7.Chevenet, F., C. Brun, A. L. Banuls, B. Jacq, and R. Christen. 2006. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen, S., F. Dalle, A. Gallay, M. Di Palma, A. Bonnin, and H. D. Ward. 2006. Identification of Cpgp40/15 Type Ib as the predominant allele in isolates of Cryptosporidium spp. from a waterborne outbreak of gastroenteritis in South Burgundy, France. J. Clin. Microbiol. 44:589-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das, P., S. S. Roy, K. Mitradhar, P. Dutta, M. K. Bhattacharya, A. Sen, S. Ganguly, S. K. Bhattacharya, A. A. Lal, and L. Xiao. 2006. Molecular characterization of Cryptosporidium spp. in children in Kolkata, India. J. Clin. Microbiol. 44:4246-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dereeper, A., V. Guignon, G. Blanc, S. Audic, S. Buffet, F. Chevenet, J. F. Dufayard, S. Guindon, V. Lefort, M. Lescot, J. M. Claverie, and O. Gascuel. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465-W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dillingham, R. A., A. A. Lima, and R. L. Guerrant. 2002. Cryptosporidiosis: epidemiology and impact. Microbes Infect. 4:1059-1066. [DOI] [PubMed] [Google Scholar]
- 12.Fraser, D., R. Dagan, L. Naggan, V. Greene, J. El-On, Y. Abu-Rbiah, and R. J. Deckelbaum. 1997. Natural history of Giardia lamblia and Cryptosporidium infections in a cohort of Israeli Bedouin infants: a study of a population in transition. Am. J. Trop. Med. Hyg. 57:544-549. [DOI] [PubMed] [Google Scholar]
- 13.Gatei, W., P. Das, P. Dutta, A. Sen, V. Cama, A. A. Lal, and L. Xiao. 2007. Multilocus sequence typing and genetic structure of Cryptosporidium hominis from children in Kolkata, India. Infect. Genet. Evol. 7:197-205. [DOI] [PubMed] [Google Scholar]
- 14.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 15.Houpt, E. R., O. Y. Bushen, N. E. Sam, A. Kohli, A. Asgharpour, C. T. Ng, D. P. Calfee, R. L. Guerrant, V. Maro, S. Ole-Nguyaine, and J. F. Shao. 2005. Short report: asymptomatic Cryptosporidium hominis infection among human immunodeficiency virus-infected patients in Tanzania. Am. J. Trop. Med. Hyg. 73:520-522. [PubMed] [Google Scholar]
- 16.Jagai, J. S., D. A. Castronovo, J. Monchak, and E. N. Naumova. 2009. Seasonality of cryptosporidiosis: a meta-analysis approach. Environ. Res. 109:465-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaur, R., D. Rawat, M. Kakkar, B. Uppal, and V. K. Sharma. 2002. Intestinal parasites in children with diarrhea in Delhi, India. Southeast Asian J. Trop. Med. Public Health 33:725-729. [PubMed] [Google Scholar]
- 18.Leav, B. A., M. R. Mackay, A. Anyanwu, R. M. O'Connor, A. M. Cevallos, G. Kindra, N. C. Rollins, M. L. Bennish, R. G. Nelson, and H. D. Ward. 2002. Analysis of sequence diversity at the highly polymorphic Cpgp40/15 locus among Cryptosporidium isolates from human immunodeficiency virus-infected children in South Africa. Infect. Immun. 70:3881-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathan, M. M., S. Venkatesan, R. George, M. Mathew, and V. I. Mathan. 1985. Cryptosporidium and diarrhoea in southern Indian children. Lancet ii(8465):1172-1175. [DOI] [PubMed]
- 20.Muchiri, J. M., L. Ascolillo, M. Mugambi, T. Mutwiri, H. D. Ward, E. N. Naumova, A. I. Egorov, S. Cohen, J. G. Else, and J. K. Griffiths. 2009. Seasonality of Cryptosporidium oocyst detection in surface waters of Meru, Kenya as determined by two isolation methods followed by PCR. J. Water Health 7:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muthusamy, D., S. S. Rao, S. Ramani, B. Monica, I. Banerjee, O. C. Abraham, D. C. Mathai, B. Primrose, J. Muliyil, C. A. Wanke, H. D. Ward, and G. Kang. 2006. Multilocus genotyping of Cryptosporidium sp. isolates from human immunodeficiency virus-infected individuals in South India. J. Clin. Microbiol. 44:632-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagamani, K., P. R. Pavuluri, M. Gyaneshwari, K. Prasanthi, M. I. Rao, and N. K. Saxena. 2007. Molecular characterisation of Cryptosporidium: an emerging parasite. Indian J. Med. Microbiol. 25:133-136. [DOI] [PubMed] [Google Scholar]
- 23.Naumova, E. N., J. S. Jagai, B. Matyas, A. DeMaria, Jr., I. B. MacNeill, and J. K. Griffiths. 2007. Seasonality in six enterically transmitted diseases and ambient temperature. Epidemiol. Infect. 135:281-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman, R. D., C. L. Sears, S. R. Moore, J. P. Nataro, T. Wuhib, D. A. Agnew, R. L. Guerrant, and A. A. Lima. 1999. Longitudinal study of Cryptosporidium infection in children in northeastern Brazil. J. Infect. Dis. 180:167-175. [DOI] [PubMed] [Google Scholar]
- 25.Peng, M. M., S. R. Meshnick, N. A. Cunliffe, B. D. Thindwa, C. A. Hart, R. L. Broadhead, and L. Xiao. 2003. Molecular epidemiology of cryptosporidiosis in children in Malawi. J. Eukaryot Microbiol. 50(Suppl):557-559. [DOI] [PubMed] [Google Scholar]
- 26.Perch, M., M. Sodemann, M. S. Jakobsen, P. Valentiner-Branth, H. Steinsland, T. K. Fischer, D. D. Lopes, P. Aaby, and K. Molbak. 2001. Seven years' experience with Cryptosporidium parvum in Guinea-Bissau, West Africa. Ann. Trop. Paediatr. 21:313-318. [DOI] [PubMed] [Google Scholar]
- 27.Soba, B., and J. Logar. 2008. Genetic classification of Cryptosporidium isolates from humans and calves in Slovenia. Parasitology 135:1263-1270. [DOI] [PubMed] [Google Scholar]
- 28.Sulaiman, I. M., P. R. Hira, L. Zhou, F. M. Al-Ali, F. A. Al-Shelahi, H. M. Shweiki, J. Iqbal, N. Khalid, and L. Xiao. 2005. Unique endemicity of cryptosporidiosis in children in Kuwait. J. Clin. Microbiol. 43:2805-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tumwine, J. K., A. Kekitiinwa, N. Nabukeera, D. E. Akiyoshi, S. M. Rich, G. Widmer, X. Feng, and S. Tzipori. 2003. Cryptosporidium parvum in children with diarrhea in Mulago Hospital, Kampala, Uganda. Am. J. Trop. Med. Hyg. 68:710-715. [PubMed] [Google Scholar]
- 30.Xiao, L. 2010. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 124:80-89. [DOI] [PubMed] [Google Scholar]
- 31.Xiao, L., C. Bern, J. Limor, I. Sulaiman, J. Roberts, W. Checkley, L. Cabrera, R. H. Gilman, and A. A. Lal. 2001. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis. 183:492-497. [DOI] [PubMed] [Google Scholar]
- 32.Xiao, L., L. Escalante, C. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]