Abstract
During the 2000s, a new clonal group with resistances to ampicillin, streptomycin, sulfonamides, and tetracycline (ASSuT) emerged in Italy among strains of Salmonella enterica serovar Typhimurium and its monophasic variant, Salmonella enterica subspecies enterica serovar 4,[5],12:i:−. The PulseNet Europe database allowed us to identify ASSuT strains of both S. Typhimurium and its monophasic variant, isolated in Denmark and the United Kingdom, with the same or very closely related pulsed-field gel electrophoresis (PFGE) patterns as the Italian strains, suggesting that the ASSuT clonal group is circulating in different European countries. With the aim of analyzing the molecular basis of antibiotic resistance, resistance genes were identified and their localization was investigated in 66 ASSuT strains and, as controls, in 11 strains with different resistance patterns and PFGE profiles, belonging both to S. Typhimurium and to its monophasic variant, isolated from humans in Italy, Denmark, and the United Kingdom. All the ASSuT strains were positive for the following resistance genes: blaTEM-1, strA-strB, sul2, and tet(B). A localization experiment demonstrated that the ASSuT resistance genes are chromosomally located. This study confirms that a multidrug-resistant clonal group, ASSuT, of S. Typhimurium and its monophasic variant has emerged and is circulating in Italy, Denmark, and the United Kingdom. Moreover, the results of this work demonstrate that the multidrug resistance in this clonal group of Salmonella strains is conferred by a new genomic island.
Salmonella enterica serovar Typhimurium is one of the most important food-borne zoonotic pathogens across the world, and the increasing occurrence and spread of multidrug-resistant (MDR) strains is a matter of concern. MDR S. Typhimurium definitive phage type 104 (DT104), resistant to ampicillin, chloramphenicol, streptomycin and spectinomycin, sulfonamides, and tetracyclines (resistance type [R type] ACSSpSuT), is a clear example of a clone that emerged in one country (the United Kingdom in the 1980s) and subsequently became widely distributed in at least four continents (41).
Recently strains of S. Typhimurium and its monophasic variant, Salmonella enterica serovar 4,[5],12:i:−, characterized by the four-drug resistance (tetraresistance) pattern ASSuT (with or without additional resistances but lacking resistance to chloramphenicol), has emerged in Italy (5). These strains accounted for more than 30% of isolates from cases of human infection during the last 5 years and were also observed among isolates from different farm animal species (20). Pulsed-field gel electrophoresis (PFGE) analysis showed that these tetraresistant strains belonged to a unique clonal group, different from the DT104 ACSSpSuT clone (13). Using the PulseNet Europe database (27), it has been possible to verify that ASSuT strains of S. Typhimurium and its monophasic variant isolated in Denmark and the United Kingdom exhibited PFGE patterns that were the same as or very closely related to those of the Italian strains, suggesting that an ASSuT clonal group was circulating in different European countries.
In Denmark, strains with the ASSuT R type accounted for 4.4% of S. Typhimurium strains from humans over the period from 1997 to 2002 (16); during the period from 2003 to 2008, the frequency of ASSuT strains from humans increased, varying from 10 to 22%. Moreover, a study of strains from pigs highlighted that from 2002 to 2006, the main phage type became DT120, and 50% of strains of this phage type belonged to R type ASSuT. Over the same period, the frequency of isolation of DT104 strains decreased (15).
In England and Wales, the ASSuT resistance pattern, with additional resistance to furazolidone (Fu), was first detected in 1964 in S. Typhimurium DT29. Such strains, which possessed resistance determinants on both conjugative and nonconjugative plasmids, caused substantive outbreaks in both cattle and humans until 1971 (41). During the 1980s there was an increase, from 1% in 1981 to 8% in 1990, of strains with the ASSuT R type, mainly belonging to DT193 (39). Since 1996, DT193 has been second only to DT104 in cases of human infection; of these strains, 56% were multiresistant, with strains of R type ASSuT predominating. Previous studies have shown that strains of DT193 of R type ASSuT have often been associated with pigs, and outbreaks associated with pigs have been reported (40). In addition, in 2006, strains of DT120 of R type ASSuT, with a unique PFGE profile (STYMXB.0083, according to PulseNet nomenclature), caused an outbreak in northeast England (3). From January 2004 to December 2006, there were 489 strains of R type ASSuT, with or without additional resistances, from a total of 4,426 isolates from human infections (11%) reported to the Health Protection Agency (HPA Salmonella data set 2009; www.HPA.org.uk).
In DT104 strains of R type ACSSuSpT, the resistance genes for ampicillin (antibiogram designation A) (blaPSE-1), chloramphenicol (antibiogram designation C) (floR), streptomycin-spectinomycin (antibiogram designation SSp) (aadA2), sulfonamides (antibiogram designation Su) (sul1), and tetracyclines (antibiogram designation T) [tet(G)] are chromosomally integrated within the 43-kb Salmonella genomic island 1 (SGI-1) (4). In contrast, the genes responsible for the tetraresistance in the ASSuT clonal group have not been identified so far.
The aim of this study was to analyze the molecular basis and location of ASSuT tetraresistance in S. Typhimurium and its monophasic variant strains from cases of human infection in Italy, Denmark, and the United Kingdom over the period from 2003 to 2006.
MATERIALS AND METHODS
Bacterial strains.
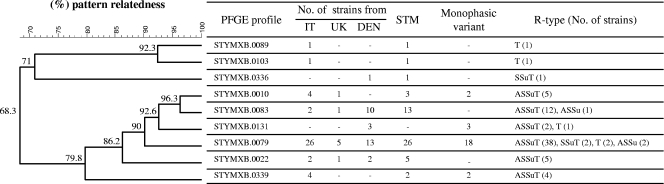
A total of 77 strains of S. Typhimurium and its monophasic variant were included in this study. Of these, 66 strains with the R type ASSuT showed a pattern relatedness of ≥79,8% to each other when analyzed by PFGE. Eleven strains with different R types and showing PFGE profiles related (8 strains) and unrelated (3 strains) to those of ASSuT strains were included as a control. A detailed description of the strains is given in Fig. 1 .
FIG. 1.
Dendrogram showing the pattern relatedness among the 77 strains of Salmonella Typhimurium (STM) and its monophasic variant included in this study. The dendrogram was generated by the BioNumerics software program. Similarity analysis was performed using the Dice coefficient (optimization [Opt], 1.00%; tolerance [Tol], 1.00%), and clustering was done by the unweighted pair group method with arithmetic mean (UPGMA). IT, Italy; UK, United Kingdom; DEN, Denmark. Antibiogram designations: A, ampicillin; S, streptomycin; Su, sulfonamides; T, tetracycline.
Strains were definitively assigned to Salmonella serovar Typhimurium or its monophasic variant on the basis of the presence or absence of the fljB gene (14). Antimicrobial susceptibility testing was performed for Italian and Danish strains using the disk diffusion method according to the CLSI recommendations (9, 10) and for United Kingdom strains by the method of Frost (18). The antimicrobials tested were those included in the Enternet reference panel (42). PFGE was performed according to the Pulsenet protocol (27) using XbaI as a restriction enzyme.
PCR amplification.
Standard PCR amplifications were performed with specific primers for the genes conferring resistance to ampicillin (blaSHV, blaOXA, blaTEM, and blaPSE), streptomycin (strA-strB and aadA2), sulfonamides (sul1 and sul2), and tetracyclines [tet(A), tet(B), tet(C), and tet(G)], using conditions described in Table 1. In addition, the presence of class 1 integron gene cassettes was investigated using specific primers for the 5′ and 3′ conserved segment. PCR amplicons of the blaTEM gene were fully sequenced in order to assess which subtype of the TEM gene was harbored.
TABLE 1.
Primers used in PCR amplification
| Target gene(s) | Name | DNA sequence (5′ to 3′) | Tm (°C) | Expected amplicon size (bp) | GenBank accession. no. | Reference |
|---|---|---|---|---|---|---|
| blaSHV | OS5 | TTATCTCCCTGTTAGCCACC | 60 | 796 | GQ463148 | 1 |
| OS6 | GATTTGCTGATTTCGCTCGG | |||||
| blaOXA | OO-1 | ACCAGATTCAACTTTCAA | 55 | 589 | GQ896560 | 22 |
| OO-2 | TCTTGGCTTTTATGCTTG | |||||
| blaTEM | TemA | ATAAAATTCTTGAAGAC | 50 | 1,073 | AY463797 | 11 |
| TemB | TTACCAATGCTTAATCA | |||||
| blaPSE | pse1-F | CGCTTCCCGTTAACAAGTAC | 60 | 419 | M69058 | 23 |
| pse1-R | CTGGTTCATTTCAGATAGCG | |||||
| strA-strB | SAF | AGCAGAGCGCGCCTTCGCTG | 62 | 703 | NC_001740 | 12 |
| SBR | CCAAAGCCCACTTCACCGAC | |||||
| aadA2 | AadA2F | TGTTGGTTACTGTGGCCGTA | 60 | 622 | AF071555 | 35 |
| AadA2R | GATCTCGCCTTTCACAAAGC | |||||
| sul1 | Sul1F | CTTCGATGAGAGCCGGCGGC | 65 | 436 | X12869 | 23 |
| Sul1b | GCAAGGCGGAAACCCGCGCC | |||||
| sul2 | Sul2F | TCAACATAACCTCGGACAGT | 60 | 707 | M36657 | 23 |
| Sul2R | GATGAAGTCAGCTCCACCT | |||||
| tet(A) | TetAF | GTAATTCTGAGCACTGTCGC | 60 | 956 | X61367 | 12 |
| TetAR | CTGCCTGGACAACATTGCTT | |||||
| tet(B) | TetBF | ACGTTACTCGATGCCAT | 50 | 1,169 | AM412236 | 12 |
| TetBR | AGCACTTGTCTCCTGTT | |||||
| tet(C) | TetCF | AACAATGCGCTCATCGT | 52 | 1,138 | Y19114 | 17 |
| TetCR | GGAGGCAGACAAGGTAT | |||||
| tet(G) | TetGF | CCGGTCTTATGGGTGCTCTA | 56 | 603 | F071555 | 35 |
| TetGR | CCAGAAGAACGAAGCCAGTC | |||||
| Integron variable region | 5′CS | GGCATCCAAGCAGCAAG | 56 | Variable | M73819 | 12 |
| 3′CS | AAGCAGACTTGACCTGA | |||||
| Integrase | Int1F | GCCTTGCTGTTCTTCTAC | 55 | 558 | X12870 | 22 |
| Int1B | GATGCCTGCTTGTTCTAC | |||||
| 16S rRNA gene | 16SF | TGTTGTGGTTAATAACCGCA | 56 | 571 | EU073022 | 44 |
| 16SR | CACAAATCCATCTCTGGA | |||||
| tet(B)a | TetB2 | CCAGTAGCTCCTGTGAT | 50 | 282 | AM412236 | This study |
| blaTEMa | TemC | AGCATCTTTTACTTTCA | 52 | 300 | AY463797 | This study |
Primers were used together with primers TetBF and TemA, respectively, only to produce probes.
Transfer experiment.
Conjugation experiments were performed using Escherichia coli CSH26NaR as the recipient strain, as described by Maimone et al. (28) with a few modifications. Overnight cultures of the donor and recipient strains were mixed in nutrient broth at a ratio of 1:6 and incubated overnight at three temperatures: 25°C, 28°C, and 37°C. Transconjugants were selected on Luria-Bertani (LB) agar plates containing nalidixic acid (40 μg/ml) together with tetracycline (3 μg/ml) or ampicillin (8 μg/ml).
Plasmid DNA was prepared with a PureLink HiPure plasmid filter purification kit (Invitrogen, Milan, Italy). Transformation experiments were performed using MAX Efficiency DH5α competent cells (Invitrogen, Milan, Italy), and transformants were selected on LB agar plates containing ampicillin (50 μg/ml) or tetracycline (8 μg/ml) (36).
PFGE with I-CeuI and preparation of total DNA.
PFGE with the I-CeuI enzyme was performed according to the PulseNet protocol (32), with a few modifications. Briefly, cell density for plug preparation was 2 at 600 nm, the final concentration of proteinase K in the plug was 1 mg/ml, and 1 additional wash (each) in H2O and in Tris-EDTA (TE) buffer was done. The plugs were digested with 0.04 U of I-CeuI enzyme (New England, Biolabs, Ipswich, MA) for 3 h at 37°C as described by Liu et al. (26). DNA fragments were separated on a 0.7% agarose gel, and lambda ladder concatemers (New England, Biolabs, Ipswich, MA) were used as a molecular marker.
The preparation of total DNAs was performed using the Wizard genomic DNA purification kit (Promega, Milan, Italy). Total DNAs were digested with the KpnI enzyme and electrophoresed in a 0.8% agarose gel at 16 V for 17 h.
Southern blot hybridization.
Restriction fragments obtained by PFGE with I-CeuI and total DNA digestion with KpnI were transferred onto positively charged nylon membranes (Roche Diagnostics, Monza, Italy) by standard methods (38) and hybridized with PCR-generated probes labeled with digoxigenin for specific resistance genes, the integrase gene, and ribosomal DNA (Table 1) under high-stringency conditions at 68°C without formamide, according to the manufacturer's procedure (DIG DNA labeling and detection kit; Roche Diagnostics, Monza, Italy).
RESULTS
Genetic characterization of the ASSuT resistance profile.
The investigation by PCR of the antimicrobial resistance genes showed that all 66 ASSuT strains were positive for blaTEM-1, strA-strB, sul2, and tet(B) and negative for all the other genes tested and for a class 1 integron(s) (Table 2). All strains of R type ASSu harbored the blaTEM-1, strA-strB, and sul2 genes, and two strains of R type SSuT were positive for strA-strB, sul2, and tet(B).
TABLE 2.
Antimicrobial resistance genes identified among Salmonella serovar Typhimurium and its monophasic variant strains
| No. of strains | R type | PFGE type | Presence of gene(s) |
|||||
|---|---|---|---|---|---|---|---|---|
| tet(A) | tet(B) | tet(C) | strA-strB | sul2 | tem1 | |||
| 38a | ASSuT | STYMXB.0079 | − | + | − | + | + | + |
| 4b | ASSuT | STYMXB.0339 | − | + | − | + | + | + |
| 5c | ASSuT | STYMXB.0010 | − | + | − | + | + | + |
| 12 | ASSuT | STYMXB.0083 | − | + | − | + | + | + |
| 5 | ASSuT | STYMXB.0022 | − | + | − | + | + | + |
| 2d | ASSuT | STYMXB.0131 | − | + | − | + | + | + |
| 2e | SSuT | STYMXB.0079 | − | + | − | + | + | − |
| 2f | ASSu | STYMXB.0079 | − | − | − | + | + | + |
| 1 | ASSu | STYMXB.0083 | − | − | − | + | + | + |
| 2 | T | STYMXB.0079 | − | + | − | − | − | − |
| 1g | T | STYMXB.0131 | − | + | − | − | − | − |
| 1 | T | STYMXB.0089 | − | − | + | − | − | − |
| 1 | T | STYMXB.0103 | + | − | − | − | − | − |
| 1 | SSuT | STYMXB.0336 | + | − | − | + | + | − |
Sixteen of these strains belong to the monophasic variant.
Two of these strains belong to the monophasic variant.
Two of these strains belong to the monophasic variant.
All the strains belong to the monophasic variant.
One of these strains belongs to the monophasic variant.
One of these strains belongs to the monophasic variant.
This strain belongs to the monophasic variant.
The strains not of R type ASSuT but positive for tet(B) showed PFGE profiles related to those of ASSuT strains. In particular, 2 strains with R type SSuT and 2 tetracycline-resistant strains exhibited the PFGE profile STYMXB.0079; one tetracycline-resistant strain showed the PFGE profile STYMXB.0131, a related profile observed only in some Danish ASSuT strains. The STYMXB.0079 PFGE profile and the related STYMXB.0083 were also shown by three strains of R type ASSu, which showed the same resistance gene profile as the ASSuT strains with the exception of tet(B) (Table 2). Tetracycline resistance genes different from tet(B) were found in 2 strains with the SSuT and T resistance patterns [tet(A)] and in one tetracycline-resistant strain [tet(C)].
Localization of resistance genes.
Conjugation and transformation experiments were performed on 2 representative strains of R type ASSuT belonging to 2 different PFGE profiles, STYMXB.0079 and STYMXB.0339. The experiments failed to transfer any genes encoding antimicrobial resistance from the donor to the recipient strain, suggesting an inability of horizontal transfer and a chromosomal localization of the resistance genes.
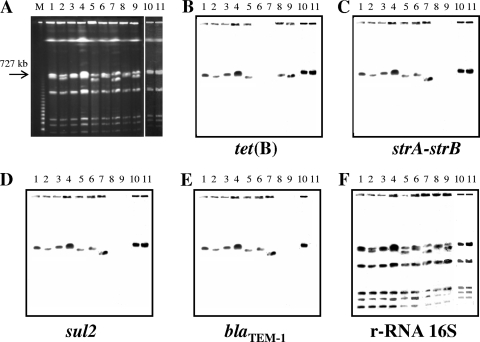
Chromosomal localization of resistance genes was investigated by PFGE using the I-CeuI restriction enzyme on a selection of 41 strains, including 18 strains from Italy, 15 from Denmark, and 8 from the United Kingdom. The strains were representative of each PFGE profile, and all harbored the tet(B) gene. The 3 Danish strains of R type ASSu were included as well.
Chromosomal digestion yielded seven PFGE fragments with sizes ranging from 40 to more than 3,000 kb. Southern blot hybridization was performed with specific probes for blaTEM-1, strA-strB, sul2, tet(B), 16S rRNA genes, and integrase of a class 1 integron (encoded by intI). All the probes bound to a fragment of approximately 750 kb in size, as did the ribosomal probe, thus demonstrating the chromosomal location of the resistance determinants (Fig. 2). Conversely, all the strains were negative for the presence of intI (data not shown).
FIG. 2.
(A) Pulsed-field gel electrophoresis with I-CeuI on a subset of strains with the following features. Lane 1, ASSuT STYMXB.0079; lane 2, ASSuT STYMXB.0339; lane 3, ASSuT STYMXB.0010; lane 4, ASSuT STYMXB.0083; lane 5, ASSuT STYMXB.0131; lane 6, ASSu STYMXB.0079; lane 7, ASSu STYMXB.0083; lane 8, T STYMXB.0079; lane 9, T STYMXB.0131; lane 10, ASSuT STYMXB.0022; lane 11, SSuT STYMXB.0079. Lane M is the molecular weight marker lane (Lambda ladder concatemers). (B to F) Southern blot hybridization was performed with tet(B), strA-strB, sul2, blaTEM-1, and 16S rRNA gene probes, respectively.
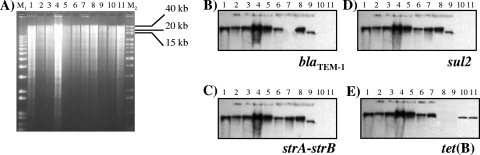
A southern blot experiment of total DNA, digested with KpnI, was performed on 11 strains with a PFGE pattern similarity of ≥79.8% (6 ASSuT strains, 1 SSuT strain, 2 ASSu strains, and 2 tetracycline-resistant strains). Hybridization with specific probes for the four resistance genes showed that all the genes colocalized on a fragment of about 40 kb (Fig. 3) for ASSuT strains, independent of their PFGE profile. An exception was represented by a strain with the PFGE profile STYMXB.0131, for which the resistance genes are localized in a slightly smaller fragment. A similar finding has been shown for an SSuT STYMXB.0079 strain and an ASSu strain with the PFGE profile STYMXB.0083. In addition, in one strain of ASSu STYMXB.0079 and in two strains resistant only to tetracycline, with the PFGE profiles STYMXB.0079 and STYMXB.0131, the resistance genes are localized on a KpnI fragment of approximately 20 kb (Fig. 3).
FIG. 3.
Total DNA digestion with KpnI enzyme on strains with the following features: Lane 1, ASSuT STYMXB.0079; lane 2, ASSuT STYMXB.0083; lane 3, ASSuT STYMXB.0339; lane 4, ASSuT STYMXB.0010; lane 5, ASSuT STYMXB.0022; lane 6, ASSuT STYMXB.0131; lane 7, SSuT STYMXB.0079; lane 8, ASSu STYMXB.0083; lane 9, ASSu STYMXB.0079; lane 10, T STYMXB.0079; lane 11, T STYMXB.0131. M1, 1 kb Plus DNA ladder; M2, 1 kb DNA extension ladder (Invitrogen, Milan, Italy). (B to E) Southern blot hybridization was performed with blaTEM-1, strA-strB, sul2, and tet(B) probes, respectively.
DISCUSSION
Resistance to antimicrobials in Salmonella is regarded as an important threat to public health. Results from a European collaborative study showed that 43% of strains were resistant to at least one antimicrobial and 18% were multiresistant (42). Among different serovars, S. Typhimurium consistently exhibited the highest percentage of strains with multiple resistances, belonging mainly to the ACSSpSuT DT104 clone (41). The dissemination of this multiresistant clone is still a matter of concern despite a marked reduction of its frequency in Europe since 2002 (29).
In Italy since 2000, strains with R type ASSuT have become increasingly common, both in S. Typhimurium and among its monophasic variant strains (13, 20). In addition, an increase in the frequency of these strains has been observed in both Denmark and the United Kingdom.
Until recently, comparison of strains from different countries had been difficult, mainly because of the lack of standardization in typing methodology. Standardization of PFGE coupled with the establishment of a central database and a common nomenclature system facilitated by the international network PulseNet Europe has made the identification of clones circulating within and between countries easier. In this study, the PulseNet database assisted in ascertaining that strains of R type ASSuT isolated in Denmark and the United Kingdom exhibited the same PFGE profiles as Italian strains (13), with the unique exception of STYMXB.0131, a PFGE profile shown only by Danish monophasic variant strains but which has a 79.8% homology with the other PFGE profiles.
The aim of this study was to analyze the molecular basis of antibiotic resistance in strains of R type ASSuT, both S. Typhimurium and its monophasic variant, isolated from humans in Italy, Denmark, and the United Kingdom. All the ASSuT strains, regardless of their origin, harbored the same resistance genes: blaTEM-1, strA-strB, sul2, and tet(B). These resistance genes are common in Salmonella spp., and the association of these genes has been frequently described. In particular, the strA-strB genes are usually associated with sul2. Both genes are frequently located on small broad-host-range plasmids, such as RSF1010 or pPB1, but have also been detected on the chromosome of an S. Typhimurium isolate (12). Moreover, a similar association of the four resistance genes in ASSuT strains has been observed in various plasmids, for example, pHCM1, typical of Salmonella enterica serovar Typhi (30), and pU302L, found in an S. Typhimurium isolate (8). In relation to tetracycline resistance, in all ASSuT strains the only gene coding for resistance to tetracycline was tet(B). This gene has previously been identified in S. Typhimurium strains at a frequency of 48% but always in combination with tet(A), tet(G), or both (31). In addition, tet(B) was the unique gene also encoding tetracycline resistance in strains showing an R type different from ASSuT but with a PFGE profile related to the main profile STYMXB.0079.
The localization experiments with ASSuT strains demonstrated that the resistance genes were localized on the bacterial chromosome, in particular on a KpnI fragment of approximately 40 kb. Only in the Danish strains with PFGE profile STYMXB.0131, the resistance genes are localized on a fragment with a slightly smaller size, probably due to a deletion(s) not affecting resistances genes.
Overall these findings are indicative of the existence of a new resistance island, different from SGI-1 of the ACSSpSuT DT104 clone not only in terms of resistance gene content but also in the absence of class 1 integrons. In this respect, SGI-1 possesses two class 1 integrons, located in the chromosome and carrying the aadA2, blaPSE-1, and sul1 genes, conferring resistance to streptomycin, β-lactams, and sulfonamides, in addition to the floR and tet(G) genes, conferring resistance to chloramphenicol and tetracyclines, respectively (4).
The chromosomal localization of resistance genes was also demonstrated in strains with R types different from ASSuT but with related PFGE profiles. However, in strains with the R types SSuT, ASSu, and resistance only to tetracycline, the genes are localized on a KpnI fragment smaller than the fragment detected in ASSuT strains. It is conceivable that rearrangements or deletions of the resistance island may have occurred, thereby generating partial resistance patterns. A similar finding has been described for DT104 strains showing partial R types but the same PFGE profile of ACSSpSuT strains and in which deleted SGI-1 was observed (2, 6, 7, 25, 33).
Despite S. Typhimurium ASSuT strains having been found in the United States (19, 34), Spain (37), France (43), and the Czech Republic (24), to our knowledge no molecular analyses have been undertaken in relation to the resistance genes or their genetic background. However, a partial characterization performed with French ASSuT strains concerning the presence of integrons and β-lactam resistance genes demonstrated that these strains were negative for class 1 integrons and harbored the blaTEM-1 gene (43). Similarly, in the Czech Republic, ASSuT strains of S. Typhimurium were negative for the presence of SGI-1 and class 1 integrons (24). Although not conclusive, the results of these studies are in agreement with our results and seem to suggest that the strains of R type ASSuT from the respective countries may belong to the same clonal group as described above, suggesting its wide dissemination within Europe.
Finally, this report demonstrates that strains of S. Typhimurium and its monophasic variant with the R type ASSuT belong to the same clonal group. The monophasic variant strains studied exhibited the same chromosomally located resistance genes as the ASSuT S. Typhimurium strains. In contrast, Spanish monophasic variant strains of R type ACSSuT and with additional resistance to gentamicin and trimethoprim exhibited DT104-related PFGE profiles but differed from the DT104 clone in terms of the resistance gene profile and localization (21, 22).
In conclusion, this study demonstrates the existence of a second genomic island carrying antibiotic resistance genes distinct from those previously detected in SGI-1. This new genomic island confers resistance to ampicillin, streptomycin, sulfonamides, and tetracyclines in a clonal group of S. Typhimurium strains and its monophasic variant circulating in Italy, Denmark, and the United Kingdom. Further studies are needed to complete the characterization of this new resistance island and to better understand its organization.
Acknowledgments
This study was partially funded by the European Community Network of Excellence Med-Vet-Net Workpackages 21 (contract no. FOOD-CT-2004-506122).
Footnotes
Published ahead of print on 21 April 2010.
REFERENCES
- 1.Arlet, G., M. Rouveau, and A. Philippon. 1997. Substitution of alanine for aspartate at position 179 in the SHV-6 extended-spectrum beta-lactamase. FEMS Microbiol. Lett. 152:163-167. [DOI] [PubMed] [Google Scholar]
- 2.Baggesen, D. L., D. Sandvang, and F. M. Aarestrup. 2000. Characterization of Salmonella enterica serovar Typhimurium DT104 isolated from Denmark and comparison with isolates from Europe and the United States. J. Clin. Microbiol. 38:1581-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Best, E. L., M. D. Hampton, S. Ethelberg, E. Liebana, F. A. Clifton-Hadley, and E. J. Threlfall. 2009. Drug-resistant Salmonella Typhimurium DT 120: use of PFGE and MLVA in a putative international outbreak investigation. Microb. Drug Resist. 15:133-138. [DOI] [PubMed] [Google Scholar]
- 4.Briggs, C. E., and P. M. Fratamico. 1999. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob. Agents Chemother. 43:846-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busani, L., C. Graziani, A. Battisti, A. Franco, A. Ricci, D. Vio, E. Digiannatale, F. Paterlini, M. D'Incau, S. Owczarek, A. Caprioli, and I. Luzzi. 2004. Antibiotic resistance in Salmonella enterica serotypes Typhimurium, Enteritidis and Infantis from human infections, foodstuffs and farm animals in Italy. Epidemiol. Infect. 132:245-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carattoli, A., E. Filetici, L. Villa, A. M. Dionisi, A. Ricci, and I. Luzzi. 2002. Antibiotic resistance genes and Salmonella genomic island 1 in Salmonella enterica serovar Typhimurium isolated in Italy. Antimicrob. Agents Chemother. 46:2821-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casin, I., J. Breuil, A. Brisabois, F. Moury, F. Grimont, and E. Collatz. 1999. Multidrug-resistant human and animal Salmonella typhimurium isolates in France belong predominantly to a DT104 clone with the chromosome- and integron-encoded beta-lactamase PSE-1. J. Infect. Dis. 179:1173-1182. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. Y., G. W. Nace, B. Solow, and P. Fratamico. 2007. Complete nucleotide sequences of 84.5- and 3.2-kb plasmids in the multi-antibiotic resistant Salmonella enterica serovar Typhimurium U302 strain G8430. Plasmid 57:29-43. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2006. Institute performance standards for antimicrobial susceptibility testing; 26th informational supplement. CLSI/NCCLS document M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10.CLSI/NCCLS. 2002. Institute performance standards for antimicrobial performance standard for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals, approved standard. CLSI/NCCLS document M31-A2. CLSI/NCCLS, Wayne, PA.
- 11.Coque, T. M., A. Oliver, J. C. Perez-Diaz, F. Baquero, and R. Canton. 2002. Genes encoding TEM-4, SHV-2, and CTX-M-10 extended-spectrum beta-lactamases are carried by multiple Klebsiella pneumoniae clones in a single hospital (Madrid, 1989 to 2000). Antimicrob. Agents Chemother. 46:500-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daly, M., L. Villa, C. Pezzella, S. Fanning, and A. Carattoli. 2005. Comparison of multidrug resistance gene regions between two geographically unrelated Salmonella serotypes. J. Antimicrob. Chemother. 55:558-561. [DOI] [PubMed] [Google Scholar]
- 13.Dionisi, A. M., C. Graziani, C. Lucarelli, E. Filetici, L. Villa, S. Owczarek, A. Caprioli, and I. Luzzi. 2009. Molecular characterization of multidrug-resistant strains of Salmonella enterica serotype Typhimurium and monophasic variant (S. 4,[5],12:i:-) isolated from human infections in Italy. Foodborne Pathog. Dis. 6:711-717. [DOI] [PubMed] [Google Scholar]
- 14.Echeita, M. A., S. Herrera, J. Garaizar, and M. A. Usera. 2002. Multiplex PCR-based detection and identification of the most common Salmonella second-phase flagellar antigens. Res. Microbiol. 153:107-113. [DOI] [PubMed] [Google Scholar]
- 15.Emborg, H. D., D. L. Baggesen, and F. M. Aarestrup. 2008. Ten years of antimicrobial susceptibility testing of Salmonella from Danish pig farms. J. Antimicrob. Chemother. 62:360-363. [DOI] [PubMed] [Google Scholar]
- 16.Ethelberg, S., M. Lisby, M. Torpdahl, G. Sorensen, J. Neimann, P. Rasmussen, S. Bang, U. Stamer, H. B. Hansson, K. Nygard, D. L. Baggesen, E. M. Nielsen, K. Molbak, and M. Helms. 2004. Prolonged restaurant-associated outbreak of multidrug-resistant Salmonella Typhimurium among patients from several European countries. Clin. Microbiol. Infect. 10:904-910. [DOI] [PubMed] [Google Scholar]
- 17.Frech, G., and S. Schwarz. 2000. Molecular analysis of tetracycline resistance in Salmonella enterica subsp. enterica serovars Typhimurium, Enteritidis, Dublin, Choleraesuis, Hadar and Saintpaul: construction and application of specific gene probes. J. Appl. Microbiol. 89:633-641. [DOI] [PubMed] [Google Scholar]
- 18.Frost, J. A. 1994. Testing for resistance to antibacterial drugs, p. 73-82. In H. Chart (ed.), Methods in Practical Laboratory Bacteriology. CRC Press, New York, NY.
- 19.Gebreyes, W. A., and C. Altier. 2002. Molecular characterization of multidrug-resistant Salmonella enterica subsp. enterica serovar Typhimurium isolates from swine. J. Clin. Microbiol. 40:2813-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graziani, C., L. Busani, A. M. Dionisi, C. Lucarelli, S. Owczarek, A. Ricci, M. Mancin, A. Caprioli, and I. Luzzi. 2008. Antimicrobial resistance in Salmonella enterica serovar Typhimurium from human and animal sources in Italy. Vet. Microbiol. 128:414-418. [DOI] [PubMed] [Google Scholar]
- 21.Guerra, B., I. Laconcha, S. M. Soto, M. A. Gonzalez-Hevia, and M. C. Mendoza. 2000. Molecular characterisation of emergent multiresistant Salmonella enterica serotype [4,5,12:i:-] organisms causing human salmonellosis. FEMS Microbiol. Lett. 190:341-347. [DOI] [PubMed] [Google Scholar]
- 22.Guerra, B., S. M. Soto, J. M. Arguelles, and M. C. Mendoza. 2001. Multidrug resistance is mediated by large plasmids carrying a class 1 integron in the emergent Salmonella enterica serotype [4,5,12:i:-]. Antimicrob. Agents Chemother. 45:1305-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerra, B., E. Junker, A. Miko, R. Helmuth, and M. C. Mendoza. 2004. Characterization and localization of drug resistance determinants in multidrug-resistant, integron-carrying Salmonella enterica serotype Typhimurium strains. Microb. Drug Resist. 10:83-91. [DOI] [PubMed] [Google Scholar]
- 24.Havlickova, H., H. Hradecka, I. Bernardyova, and I. Rychlik. 2009. Distribution of integrons and SGI1 among antibiotic-resistant Salmonella enterica isolates of animal origin. Vet. Microbiol. 133:193-198. [DOI] [PubMed] [Google Scholar]
- 25.Heir, E., B. A. Lindstedt, I. Nygard, T. Vardund, V. Hasseltvedt, and G. Kapperud. 2002. Molecular epidemiology of Salmonella typhimurium isolates from human sporadic and outbreak cases. Epidemiol. Infect. 128:373-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, S. L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukinmaa, S., U. M. Nakari, A. Liimatainen, and A. Siitonen. 2006. Genomic diversity within phage types of Salmonella enterica ssp. enterica serotypes Enteritidis and Typhimurium. Foodborne Pathog. Dis. 3:97-105. [DOI] [PubMed] [Google Scholar]
- 28.Maimone, F., B. Colonna, P. Bazzicalupo, B. Oliva, M. Nicoletti, and M. Casalino. 1979. Plasmids and transposable elements in Salmonella wien. J. Bacteriol. 139:369-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meakins, S., I. S. Fisher, C. Berghold, P. Gerner-Smidt, H. Tschape, M. Cormican, I. Luzzi, F. Schneider, W. Wannett, J. Coia, A. Echeita, and E. J. Threlfall. 2008. Antimicrobial drug resistance in human nontyphoidal Salmonella isolates in Europe 2000-2004: a report from the Enter-net International Surveillance Network. Microb. Drug Resist. 14:31-35. [DOI] [PubMed] [Google Scholar]
- 30.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 31.Pasquali, F., A. De Cesare, A. Ricci, C. Kehrenberg, S. Schwarz, and G. Manfreda. 2004. Phage types, ribotypes and tetracycline resistance genes of Salmonella enterica subsp. enterica serovar Typhimurium strains isolated from different origins in Italy. Vet. Microbiol. 103:71-76. [DOI] [PubMed] [Google Scholar]
- 32.Peters, T. M., C. Maguire, E. J. Threlfall, I. S. Fisher, N. Gill, and A. J. Gatto. 2003. The Salm-gene project—a European collaboration for DNA fingerprinting for food-related salmonellosis. Euro Surveill. 8:46-50. [DOI] [PubMed] [Google Scholar]
- 33.Poppe, C., K. Ziebell, L. Martin, and K. Allen. 2002. Diversity in antimicrobial resistance and other characteristics among Salmonella typhimurium DT104 isolates. Microb. Drug Resist. 8:107-122. [DOI] [PubMed] [Google Scholar]
- 34.Rabatsky-Ehr, T., J. Whichard, S. Rossiter, B. Holland, K. Stamey, M. Headrick, T. Barrett, F. Angulo, and the NARMS Working Group. 2004. Multidrug-resistant strains of Salmonella enterica Typhimurium, United States, 1997-1998. Emerg. Infect. Dis. 10:795-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Randall, L. P., S. W. Cooles, M. K. Osborn, L. J. Piddock, and M. J. Woodward. 2004. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J. Antimicrob. Chemother. 53:208-216. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J. E., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Soler, P., R. Gonzalez-Sanz, M. J. Bleda, G. Hernandez, A. Echeita, and M. A. Usera. 2006. Antimicrobial resistance in non-typhoidal Salmonella from human sources, Spain, 2001-2003. J. Antimicrob. Chemother. 58:310-314. [DOI] [PubMed] [Google Scholar]
- 38.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 39.Threlfall, E. J., B. Rowe, and L. R. Ward. 1993. A comparison of multiple drug resistance in salmonellas from humans and food animals in England and Wales, 1981 and 1990. Epidemiol. Infect. 111:189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Threlfall, E. J., L. R. Ward, J. A. Skinner, and A. Graham. 2000. Antimicrobial drug resistance in non-typhoidal salmonellas from humans in England and Wales in 1999: decrease in multiple resistance in Salmonella enterica serotypes Typhimurium, Virchow, and Hadar. Microb. Drug Resist. 6:319-325. [DOI] [PubMed] [Google Scholar]
- 41.Threlfall, E. J. 2002. Antimicrobial drug resistance in Salmonella: problems and perspectives in food- and water-borne infections. FEMS Microbiol. Rev. 26:141-148. [DOI] [PubMed] [Google Scholar]
- 42.Threlfall, E. J., I. S. Fisher, C. Berghold, P. Gerner-Smidt, H. Tschape, M. Cormican, I. Luzzi, F. Schnieder, W. Wannet, J. Machado, and G. Edwards. 2003. Antimicrobial drug resistance in isolates of Salmonella enterica from cases of salmonellosis in humans in Europe in 2000: results of international multi-centre surveillance. Euro Surveill. 8:41-45. [DOI] [PubMed] [Google Scholar]
- 43.Weill, F. X., F. Guesnier, V. Guibert, M. Timinouni, M. Demartin, L. Polomack, and P. A. Grimont. 2006. Multidrug resistance in Salmonella enterica serotype Typhimurium from humans in France (1993 to 2003). J. Clin. Microbiol. 44:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ziemer, C. J., and S. R. Steadham. 2003. Evaluation of the specificity of Salmonella PCR primers using various intestinal bacterial species. Lett. Appl. Microbiol. 37:463-469. [DOI] [PubMed] [Google Scholar]