Abstract
Polymorphic variability in Helicobacter pylori factors CagA and VacA contributes to bacterial virulence. The presence of one CagA EPIYA-C site is an independent risk factor for gastroduodenal ulceration (odds ratio [OR], 4.647; 95% confidence interval [CI], 2.037 to 10.602), while the presence of the vacA i1 allele is a risk factor for increased activity (OR, 5.310; 95% CI, 2.295 to 12.287) and severity of gastritis (OR, 3.862; 95% CI, 1.728 to 8.632).
Helicobacter pylori, colonizing the gastric mucosa of 35 to 70% of people worldwide, is the etiologic factor for peptic ulcer development and increases the risk for gastric cancer. H. pylori pathogenesis is exerted via distinct virulent factors such as the secreted cytotoxin VacA (vacuolating cytotoxin A), the cag pathogenicity island (cagPAI) encoding the type IV secretion system (T4SS), and the cytotoxin-associated gene A (CagA) protein (6). We analyzed H. pylori clinical isolates from the antrum of 144 Greek adults (mean age ± standard deviation [SD], 52.6 ± 13.7 years; 78 male) diagnosed with peptic ulcer (gastric, n = 21; duodenal, n = 44) and non-peptic ulcer disease (nonulcer dyspepsia, n = 61; esophagitis, n = 18) on the basis of functional CagA EPIYA motifs as well as vacA alleles for signal, intermediate, and middle regions, as described previously (13, 15), and assessed putative associations with disease parameters and gastric inflammatory response.
Approximately 27% of the strains were found to be cagA negative with complete absence of the cagPAI. Among the 96 cagA-positive isolates, 15 (10.4%) lacked a functional T4SS as they induced minimal interleukin-8 (IL-8) levels (Fig. 1A ), and no phosphorylated CagA was detected (Fig. 1B) following infection of gastric epithelial AGS cells (15). Infection with strains possessing a functional T4SS led to significantly higher IL-8 secretion, irrespective of the number of EPIYA-C sites, and to CagA phosphorylation (Fig. 1A and B). Hence, for univariate and multivariate logistic regression analysis, cagA-positive isolates with a nonfunctional T4SS were grouped together with cagA-negative cases, comprising the “None” category. In single H. pylori strain infections, the majority of isolates (n = 59, 41.0%) were of the ABC EPIYA type (15), with a second EPIYA-C repeat observed in 19 (13.2%) strains, while ABCCC strains were also identified (n = 2, 1.4%). In 11 cases (7.7%), the presence of mixed infection by isogenic strains differing solely with regard to the number of EPIYA-C repeats was identified as shown before (13).
FIG. 1.
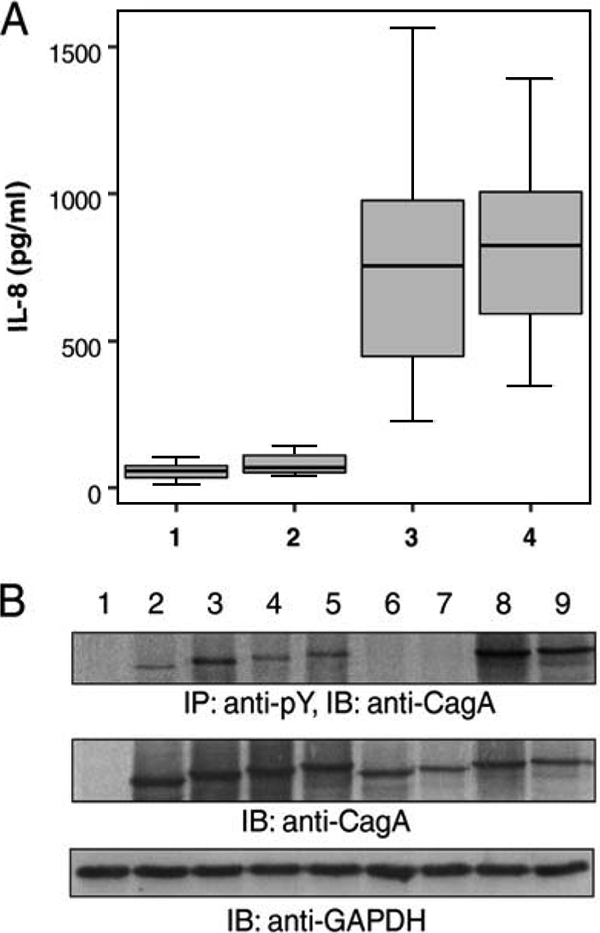
(A) Levels of secreted IL-8 following infection of gastric epithelial AGS cells with H. pylori clinical strains (1, cagA negative; 2, cagPAI defective; 3, 1 EPIYA-C repeat; 4, ≥2 EPIYA-C repeats). No difference was observed between cagA-negative and cagPAI-defective strains (U = 32.500 and P = 0.201 by the Mann-Whitney U test). cagA- and cagPAI-positive strains induced higher levels of IL-8 than cagPAI-defective strains (U = 0.000 and P < 0.0001 by the Mann-Whitney U test), irrespective of the number of EPIYA-C sites (U = 224.500 and P = 0.627 by the Mann-Whitney U test). (B) Tyrosine phosphorylation and expression patterns of CagA protein following infection of AGS cells with representative H. pylori clinical strains. CagA tyrosine phosphorylation was detected by immunoblotting (IB) following immunoprecipitation (IP) with PY20 antiphosphotyrosine antibody. The expression of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was utilized as a total protein loading control. Lanes: 1, CagA-negative clinical isolate; 2 to 5, CagA-positive isolates with functional cagPAI harboring 2 (AB), 3 (ABC), 4 (ABCC), and 5 (ABCCC) motifs in CagA, respectively; 6 and 7, CagA-positive H. pylori strains carrying 3 (ABC) and 4 (ABCC) EPIYA motifs with defective cagPAI, respectively, as depicted by the absence of phosphorylated CagA; 8 and 9, CagA-positive strains with 5 (ABCCC) EPIYA motifs.
The dominant vacA polymorphisms for the signal, intermediate, and middle regions were s1, i1, and m2, respectively, as reported for Western-type H. pylori strains (5, 17). More specifically, 102 (70.8%) isolates were identified as vacA s1, with 57 (39.6%) carrying the vacA m1 allele simultaneously. No strain with vacA s2/m1 was recorded. Of the 91 vacA i1 strains, 54 (59.3%) were also typed as vacA s1/m1, whereas 38/53 (71.7%) vacA i2 strains were s2/m2 (P < 0.001). Depending on the vacA genotype, strains were further classified into three categories (7, 14), namely, nonvacuolation (s2/i2/m2, s1/i2/m1, s1/i2/m2, and s2/i1/m2), low vacuolation (s1/i1/m2), and high vacuolation (s1/i1/m1). Vacuolating vacA s1/i1/m1 or s1/i1/m2 types were present in strains harboring functional CagA variants with more EPIYA-C phosphorylation repeats, with frequencies reaching approximately 90% in cases of multiple infections, whereas cagPAI-defective strains were almost exclusively related to a nonvacuolating vacA genotype (P < 0.001).
vacA s1 and i1 polymorphisms were found to be associated with marked chronic inflammatory infiltration and activity of chronic gastritis in the antrum (Table 1). No association was observed with the density of H. pylori colonization or the presence of gastric atrophy and intestinal metaplasia (IM), even though in 41/57 (71.9%) of recorded IM the strain carried the vacA i1 allele (P = 0.111). However, the risk for IM development increased 2-fold upon infection with vacA m1 strains (odds ratio [OR], 2.182; 95% confidence interval [CI], 1.098 to 4.338; P = 0.026). Heavy H. pylori colonization (OR of 6.866 and 95% CI of 3.072 to 15.344 [P < 0.001] and OR of 8.476 and 95% CI of 3.633 to 19.777 [P < 0.001], respectively) and infection with vacA i1 strains (OR of 3.862 and 95% CI of 1.728 to 8.632 [P = 0.001] and OR of 5.310 and 95% CI of 2.295 to 12.287 [P = 0.001], respectively) were recognized as independent risk factors for the development of severe chronic inflammatory infiltration and marked activity of chronic gastritis in the antrum. This is the first report associating vacA intermediate region polymorphisms with increased activity of antral gastritis. vacA s1 strains of Western origin have previously been associated with more-severe gastric inflammation (5, 11, 16). The only reports relating specific cagA polymorphisms with histological lesions involve strains of East Asian origin carrying the ABD EPIYA sites, which present distinct biological properties compared to Western-type ABC EPIYA motifs (4).
TABLE 1.
Univariate logistic regression analysis showing association of vacA and cagA polymorphisms with severity and activity of chronic gastritis in the antrum of 144 Greek adults
| Risk factor | Chronic inflammatory infiltration |
Activity of chronic gastritis |
||||||
|---|---|---|---|---|---|---|---|---|
| No. (%) of isolates with mild/moderate severity | No. (%) of isolates with marked severity | OR (95% CI)a | P | No. (%) of isolates with mild/moderate activity | No. (%) of isolates with marked activity | OR (95% CI)a | P | |
| vacA alleles | ||||||||
| s2 | 25 (17.4) | 17 (11.8) | Reference | 27 (18.8) | 15 (10.4) | Reference | ||
| s1 | 34 (23.6) | 68 (47.2) | 2.941 (1.402-6.171) | 0.004 | 38 (26.4) | 64 (44.4) | 3.032 (1.435-6.405) | 0.004 |
| i2 | 30 (20.8) | 23 (16.0) | Reference | 34 (23.6) | 19 (13.2) | Reference | ||
| i1 | 29 (20.1) | 62 (43.1) | 2.789 (1.385-5.614) | 0.004 | 31 (21.5) | 60 (41.7) | 3.463 (1.704-7.040) | 0.001 |
| cagA EPIYA status | ||||||||
| None | 32 (22.2) | 21 (14.6) | Reference | 33 (22.9) | 20 (13.9) | Reference | ||
| 1 EPIYA-C repeat | 18 (12.5) | 41 (28.5) | 3.471 (1.589-7.58) | 0.002 | 23 (16.0) | 36 (25.0) | 2.583 (1.204-5.539) | 0.015 |
| ≥2 EPIYA-C repeat | 6 (4.2) | 15 (10.4) | 3.810 (1.274-11.389) | 0.017 | 6 (4.2) | 15 (10.4) | 4.125 (1.376-12.363) | 0.011 |
| Mixed infections | 3 (2.1) | 8 (5.6) | 4.063 (0.966-17.091) | 0.056 | 3 (2.1) | 8 (5.6) | 4.4 (1.044-18.542) | 0.044 |
| Vacuolation potential | ||||||||
| None | 32 (22.2) | 24 (16.7) | Reference | 35 (24.3) | 21 (14.6) | Reference | ||
| Low | 12 (8.3) | 22 (15.3) | 2.444 (1.014-5.895) | 0.047 | 11 (7.6) | 23 (16.0) | 3.485 (1.418-8.566) | 0.007 |
| High | 15 (10.4) | 39 (27.1) | 3.467 (1.563-7.690) | 0.002 | 19 (13.2) | 35 (24.3) | 3.070 (1.411-6.681) | 0.005 |
Reference, used as the reference category for the calculation of risk in each case.
The development of gastric ulcers (GU) or duodenal ulcers (DU) was associated with the occurrence of H. pylori strains harboring functional EPIYA-C repeats in CagA (P = 0.001) as well as with the vacA s1 allele (P = 0.004) and marked activity of chronic gastritis (P = 0.014). Despite that, CagA EPIYA polymorphisms were found to be the only independent risk factor for ulcer disease (Table 2), with over 50% of strains with 1 (57.6%) or 2 or more (52.4%) EPIYA-C repeats found to be isolated from ulcer cases (8 GU/26 DU and 4 GU/7 DU, respectively) and the majority (8/11, 72.8%) of multiple infections with isogenic strains (4 GU and 4 DU) (Table 2). To date, infection with cagA-positive H. pylori has been well associated with gastroduodenal ulcers (2, 6, 12), whereas variability in the EPIYA phosphorylation sites and in particular CagA variants with an increased number of EPIYA-C repeats or of East Asian origin have been reported to augment the risk for gastric adenocarcinoma (1, 4, 8-10, 20). In our study, we observed that the presence of single or multiple infecting strains rather than the number of EPIYA-C sites in CagA per se is probably crucial in determining the type of gastric disease, since the majority of mixed infections with isogenic strains expressing CagA with various numbers of EPIYA-C repeats were isolated from peptic ulcer patients. Previous reports relate ulcer lesions with the presence of vacA s1 and i1 types (3, 5, 14), although in our sample, the association of VacA determinants with peptic ulcer disease was not sustained through multivariate analysis, possibly reflecting geographical differences in the prevalence of the various genotypes (17-19).
TABLE 2.
Multivariate logistic regression model depicting parameters relating to the development of peptic ulcers
| Risk factor | No. (%) of cases with non-peptic ulcers | No. (%) of cases with peptic ulcers | OR (95% CI)a | P |
|---|---|---|---|---|
| cagA EPIYA status | ||||
| Defective cagPAI | 41 (28.5) | 12 (8.3) | Reference | <0.001 |
| 1 EPIYA-C repeat | 25 (17.4) | 34 (23.6) | 4.647 (2.037-10.602) | 0.015 |
| ≥2 EPIYA-C repeat | 10 (6.9) | 11 (7.6) | 3.758 (1.288-10.969) | 0.003 |
| Mixed infections | 3 (2.1) | 8 (5.6) | 9.111 (2.085-39.810) | <0.001 |
| vacA alleles | ||||
| s2 | 30 (20.8) | 11 (7.6) | Reference | |
| s1 | 49 (34.0) | 54 (37.5) | 0.24 | |
| i2 | 34 (23.6) | 19 (13.2) | Reference | |
| i1 | 45 (31.3) | 46 (31.9) | 0.76 | |
| Activity of chronic gastritis | ||||
| Mild | 11 (7.6) | 1 (0.7) | Reference | |
| Moderate | 32 (22.2) | 21 (14.6) | 0.383 | |
| Marked | 36 (25.0) | 43 (29.9) | 0.129 |
Reference, used as the reference category for the calculation of risk in each case.
Collectively, our data indicate a distinct yet coordinated activity of virulence factors associated with H. pylori pathogenesis, with CagA contributing to the development of particular disease phenotypes, such as peptic ulcer, and VacA differentially affecting the inflammatory process. Our findings emphasize the necessity to meticulously assess the functionality of virulence factors in H. pylori clinical strains so as to discern the true biological significance that lies beneath the plasticity of the H. pylori genome.
Nucleotide sequence accession numbers.
Partial cagA nucleotide sequences were submitted to the GenBank/EMBL/DDBJ databases under accession numbers FN561978 to FN562025.
Acknowledgments
This study was supported by a Hellenic Pasteur Institute grant (HPI-922616) and in part by an Actions Concertées Inter Pasteuriennes (ACIP) program entitled “Etude de la fréquence de l'infection par Helicobacter pylori (Hp) des patients présentant une pathologie gastro-duodénale, de la sensibilité des souches aux antibiotiques et de la diversité de la région 3′ du gène cagA.”
Footnotes
Published ahead of print on 14 April 2010.
REFERENCES
- 1.Argent, R. H., M. Kidd, R. J. Owen, R. J. Thomas, M. C. Limb, and J. C. Atherton. 2004. Determinants and consequences of different levels of CagA phosphorylation for clinical isolates of Helicobacter pylori. Gastroenterology 127:514-523. [DOI] [PubMed] [Google Scholar]
- 2.Atherton, J. C. 2006. The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Annu. Rev. Pathol. 1:63-96. [DOI] [PubMed] [Google Scholar]
- 3.Atherton, J. C., P. Cao, R. M. Peek, Jr., M. K. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 270:17771-17777. [DOI] [PubMed] [Google Scholar]
- 4.Azuma, T., S. Yamazaki, A. Yamakawa, M. Ohtani, A. Muramatsu, H. Suto, Y. Ito, M. Dojo, Y. Yamazaki, M. Kuriyama, Y. Keida, H. Higashi, and M. Hatakeyama. 2004. Association between diversity in the Src homology 2 domain-containing tyrosine phosphatase binding site of Helicobacter pylori CagA protein and gastric atrophy and cancer. J. Infect. Dis. 189:820-827. [DOI] [PubMed] [Google Scholar]
- 5.Basso, D., C. F. Zambon, D. P. Letley, A. Stranges, A. Marchet, J. L. Rhead, S. Schiavon, G. Guariso, M. Ceroti, D. Nitti, M. Rugge, M. Plebani, and J. C. Atherton. 2008. Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology 135:91-99. [DOI] [PubMed] [Google Scholar]
- 6.Blaser, M. J., and J. C. Atherton. 2004. Helicobacter pylori persistence: biology and disease. J. Clin. Invest. 113:321-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cover, T. L., and S. R. Blanke. 2005. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat. Rev. Microbiol. 3:320-332. [DOI] [PubMed] [Google Scholar]
- 8.Higashi, H., R. Tsutsumi, A. Fujita, S. Yamazaki, M. Asaka, T. Azuma, and M. Hatakeyama. 2002. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc. Natl. Acad. Sci. U. S. A. 99:14428-14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones, K. R., Y. M. Joo, S. Jang, Y. J. Yoo, H. S. Lee, I. S. Chung, C. H. Olsen, J. M. Whitmire, D. S. Merrell, and J. H. Cha. 2009. Polymorphism in the CagA EPIYA motif impacts development of gastric cancer. J. Clin. Microbiol. 47:959-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naito, M., T. Yamazaki, R. Tsutsumi, H. Higashi, K. Onoe, S. Yamazaki, T. Azuma, and M. Hatakeyama. 2006. Influence of EPIYA-repeat polymorphism on the phosphorylation-dependent biological activity of Helicobacter pylori CagA. Gastroenterology 130:1181-1190. [DOI] [PubMed] [Google Scholar]
- 11.Nogueira, C., C. Figueiredo, F. Carneiro, A. T. Gomes, R. Barreira, P. Figueira, C. Salgado, L. Belo, A. Peixoto, J. C. Bravo, L. E. Bravo, J. L. Realpe, A. P. Plaisier, W. G. Quint, B. Ruiz, P. Correa, and L. J. van Doorn. 2001. Helicobacter pylori genotypes may determine gastric histopathology. Am. J. Pathol. 158:647-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nomura, A. M., G. I. Perez-Perez, J. Lee, G. Stemmermann, and M. J. Blaser. 2002. Relation between Helicobacter pylori cagA status and risk of peptic ulcer disease. Am. J. Epidemiol. 155:1054-1059. [DOI] [PubMed] [Google Scholar]
- 13.Panayotopoulou, E. G., D. N. Sgouras, K. Papadakos, A. Kalliaropoulos, G. Papatheodoridis, A. F. Mentis, and A. J. Archimandritis. 2007. Strategy to characterize the number and type of repeating EPIYA phosphorylation motifs in the carboxyl terminus of CagA protein in Helicobacter pylori clinical isolates. J. Clin. Microbiol. 45:488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhead, J. L., D. P. Letley, M. Mohammadi, N. Hussein, M. A. Mohagheghi, M. Eshagh Hosseini, and J. C. Atherton. 2007. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology 133:926-936. [DOI] [PubMed] [Google Scholar]
- 15.Sgouras, D. N., E. G. Panayotopoulou, K. Papadakos, B. Martinez-Gonzalez, A. Roumbani, J. Panayiotou, C. vanVliet-Constantinidou, A. F. Mentis, and E. Roma-Giannikou. 2009. CagA and VacA polymorphisms do not correlate with severity of histopathological lesions in Helicobacter pylori-infected Greek children. J. Clin. Microbiol. 47:2426-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soltermann, A., S. Koetzer, F. Eigenmann, and P. Komminoth. 2007. Correlation of Helicobacter pylori virulence genotypes vacA and cagA with histological parameters of gastritis and patient's age. Mod. Pathol. 20:878-883. [DOI] [PubMed] [Google Scholar]
- 17.Van Doorn, L. J., C. Figueiredo, F. Megraud, S. Pena, P. Midolo, D. M. Queiroz, F. Carneiro, B. Vanderborght, M. D. Pegado, R. Sanna, W. De Boer, P. M. Schneeberger, P. Correa, E. K. Ng, J. Atherton, M. J. Blaser, and W. G. Quint. 1999. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology 116:823-830. [DOI] [PubMed] [Google Scholar]
- 18.van Doorn, L.-J., C. Figueiredo, R. Sanna, M. J. Blaser, and W. G. Quint. 1999. Distinct variants of Helicobacter pylori cagA are associated with vacA subtypes. J. Clin. Microbiol. 37:2306-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaoka, Y., T. Kodama, O. Gutierrez, J. G. Kim, K. Kashima, and D. Y. Graham. 1999. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J. Clin. Microbiol. 37:2274-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamaoka, Y., T. Kodama, K. Kashima, D. Y. Graham, and A. R. Sepulveda. 1998. Variants of the 3′ region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J. Clin. Microbiol. 36:2258-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]


