Abstract
It has proven challenging to investigate the molecular epidemiology of Mycobacterium leprae, the causative agent of leprosy, due to difficulties with culturing of the organism and a lack of genetic heterogeneity between strains. Recently, a cost-effective panel of variable-number tandem-repeat (VNTR) markers has been developed. Use of this panel allows some of those limitations to be overcome and has allowed the genotyping of 475 M. leprae strains from six different countries. In the present report, we provide a comprehensive analysis of the relationships among the strains in order to investigate the patterns of transmission and migration of M. leprae. We find phylogenetic analysis to be inadequate and have developed an alternative method, structure-neighbor clustering, which assigns isolates with the most similar genotypes to the same groups and, subsequently, subgroups, without inferring how the strains descended from a common ancestor. We validate the approach by using simulated data and detecting expected epidemiological relationships from experimental data. Our results suggest that most M. leprae strains from a given country cluster together and that the occasional isolates assigned to different clusters are a consequence of migration. We found three genetically distinguishable populations among isolates from the Philippines, as well as evidence for the significant influx of strains to that nation from India. We also report that reference strain TN originated from the Philippines and not from India, as was previously believed. Lastly, analysis of isolates from the same families and villages suggests that most community infections originate from a common source or person-to-person transmission but that infection from independent sources does occur with measurable frequency.
Leprosy is a chronic disease caused by the bacterium Mycobacterium leprae and is characterized by disfiguring bacterial inclusions in the skin, peripheral nerves, and respiratory mucosa. Despite the success of multidrug therapy, leprosy remains prevalent in several developing countries, and in some regions, the incidence of new cases remains relatively unchanged (3). The World Health Organization (WHO) presently estimates the global prevalence to be over 210,000 cases, but it also estimates that more than 240,000 new cases are detected annually, underlining a considerable deficit in the understanding of basic aspects of disease contraction and prevention (8). Fundamental information about the epidemiology of M. leprae remains unknown, including the potential sources of infection, the organism's exact mode of transmission, and the potential importance of person-to-person contact (8). A basic understanding of the pathogenesis of M. leprae is also wanting, including the number of organisms required to produce a successful infection and the time between infection and the first onset of symptoms. This lack of knowledge is a consequence of two primary factors: (i) the difficulty in obtaining samples of M. leprae in sufficient quantities for even the most basic molecular studies and (ii) the failure of national and international granting agencies to provide support for basic research on M. leprae.
M. leprae cannot be cultured in vitro on artificial media and must be grown either in armadillos or in the footpads of mice. Because the doubling time of the organism is on the order of 14 days, weeks or even months are required to grow material in amounts sufficient for genomic studies. Samples for molecular analyses have largely been restricted to those obtained directly from patients by biopsy or as slit skin smears, in which sufficient numbers of bacteria can be ensured. Regardless of the approach used to obtain the sample, however, it cannot be ensured that the end material represents a pure culture.
Throughout the 1970s and 1980s, the WHO provided materials, research leadership, and financial support for investigating basic aspects of M. leprae biology and for the development of vaccines against that organism. Under that initiative, significant advances were made in generating monoclonal antibody banks, developing seroepidemiological tools, and investigating the cellular basis of immunity and the role of T cells in leprosy lesions (2). Yet, because of the success of multidrug therapy, in the 1990s, the WHO gradually turned its attention away from basic leprosy research and toward the goal of eradicating leprosy through drug treatment, with the stated goal of reducing leprosy to <1 case per 10,000 individuals (2). Although the NIH has continued to provide grants for basic leprosy research, many leprosy researchers have transitioned to more highly funded studies of the related organism Mycobacterium tuberculosis, the causative agent of tuberculosis (2).
Understanding of the fundamental aspects of leprosy, such as transmission patterns, infectivity, and incubation time, remains important and increasingly depends upon the existence of robust molecular tools for investigating the epidemiology of M. leprae. Microbial epidemiology, in general, requires both the ability to distinguish isolates from each other and the ability to estimate the relationships among those isolates. Recent resequencing of four M. leprae strains has revealed that the level of genetic variation for that organism is unusually low, about one single nucleotide polymorphism (SNP) per 28 kb (14). The paucity of SNPs does not provide sufficient genetic heterogeneity to permit inferences of the population structure to be made at a resolution sufficient to fully explore the epidemiological relationships between strains and, additionally, requires methods which incur high costs and a high level of technical expertise (14). This has led investigators to instead utilize cost-effective variable-number tandem repeats (VNTRs; also known as microsatellite and minisatellite loci), which are short tandemly repeated DNA motifs that are useful as molecular markers because they are highly polymorphic in populations and much more polymorphic than SNPs (12). Polymorphisms consist of differences in the numbers of repeat sequences contained by a VNTR and arise when subunits are inserted or deleted during mitosis (26). The availability of complete M. leprae genome sequences has permitted the identification of over 50 potentially useful VNTR loci, from which a panel of 16 was recently characterized in great detail (8). To date, those 16 VNTR loci have been used in six independent survey studies in Brazil, China, Colombia, India, the Philippines, and Thailand (3, 7, 18, 21, 25, 28). However, because the data were reported independently, there has been no analysis of the results taken as a whole.
Here, we integrate the VNTR genotype data from all six studies, plus those from two additional studies of isolates from the Philippines (13, 19), in order to provide a comprehensive study of the microbial epidemiology of Mycobacterium leprae. We find that conventional phylogenetic analysis is inadequate for estimation of the relationships among those strains. Therefore, we have developed a novel two-step approach, structure-neighbor clustering, to reliably assign M. leprae isolates to a hierarchy of closely related groups and subgroups. We use data simulation studies to evaluate the accuracy of our approach and find that isolates are arranged into appropriate clusters and subclusters with an accuracy exceeding 90% under a variety of different conditions.
MATERIALS AND METHODS
Phylogenetic analysis.
Neighbor-joining (NJ) trees were estimated by using the PAUP* (version 4.10-beta) program (27). A Perl script, FormatVNTR2, was used to encode repeat numbers as standard characters. The source code and documentation for all programs used in the study are available in the supplemental material. Majority-rule bootstrap trees were estimated from 1,000 bootstrap pseudoreplicates. Trees were visualized by using the FigTree (version 1.2.3) program (http://tree.bio.ed.ac.uk/software/figtree/).
Data simulation of VNTR evolution.
VNTR data sets were simulated by using the EvolveVNTR3 program, a modification of the EvolveAGene3 sequence evolution simulation program (10). Briefly, the user specifies (i) the number of taxa, (ii) whether the tree will be symmetric or random (in the sense that each branch has an equal probability of leading to a terminal or an interior node), and (iii) the average number of changes per site along each branch (branch length). The EvolveVNTR3 program generates a tree topology for the specified number of taxa and then assigns to each branch a length that is randomly drawn from a uniform distribution between zero and twice the mean branch length. The user provides a VNTR profile (the repeat number at each locus) that serves as the root sequence that initiates the simulation. At each change, a random locus is chosen, and the repeat number at that locus is changed by 1 unit 75% of the time and by 2 units 25% of the time. Changes increase and decrease the repeat number with equal probabilities. The VNTR profile at each node is recorded, and the true tree is saved in the Newick format. The output data file includes the VNTR profile of each tip of the tree, and in addition, the VNTR profile of the interior nodes with a probability of 0.2 is included. The inclusion of some interior nodes mimics the situation in most natural microbial populations, some ancestors of which survive to the present.
Structure-neighbor clustering.
All steps of the structure-neighbor clustering approach have been automated by using a Perl script, RunStructure, and ancillary scripts, FormatVNTR2, MissingData, and StructureBack.
The FormatVNTR2 Perl script formats input files for use by the Structure and PAUP* programs and writes a file consisting of the pairwise distances among all individuals. For each pair of isolates, that distance is calculated as the mean number of repeat differences per site, averaged over all of the loci for which neither individual had missing data.
(i) Stage 1: clustering by the structure program.
The Structure (version 2.3.1) Bayesian population structure inference program (17) was used to cluster strains according to their VNTR profiles, as described in the Structure program manual. The no-admixture model was used for simulated data because the simulation does not involve any recombination. For the M. leprae data, the no-admixture model was also used because (i) there is no evidence of recombination within M. leprae populations and (ii) the log likelihood (lnL) of the data was higher under the no-admixture model than under the admixture model (data not shown). Each run included a burn-in of 30,000 generations and proceeded for 100,000 generations after the burn-in. The execution of the Structure program for structure-neighbor clustering is mediated by the script RunStructure. Prior to analysis, the script excludes isolates that contain 40% or greater missing data. The user next specifies the maximum number of clusters (K) to be considered. The RunStructure program progressively executes the Structure program for values of K ranging from 1 to the maximum value chosen by the user and for each run considers the log likelihood of the data given that number of clusters. The RunStructure program determines which value of K achieves the highest log likelihood corresponding to the most likely number of clusters and uses that run for all subsequent operations. The Structure program reports for each individual the posterior probability that it belongs to each of the K populations. In order to assign an individual to a cluster, the RunStructure program requires that the posterior probability that the individual belongs to its most likely population be ≥0.8; otherwise, the individual is not assigned to a cluster and is reported to be “unclustered.” The RunStructure program produces a summary file that lists the composition of each cluster and of the unclustered set and reports for each individual the probability that it belongs to its assigned cluster.
(ii) Stage 2: subclustering by nearest neighbors.
The RunStructure program uses the distance matrix file generated by the FormatVNTR2 script and the summary file written by the RunStructure program to calculate for each isolate in a cluster the nearest neighbor(s) within the cluster. An isolate may have more than one nearest neighbor if there are equidistant nearest neighbors. This process defines the nearest-neighbor networks that join subsets of the isolates within a larger cluster.
(iii) Data visualization.
The RunStructure program produces output files that diagram the groups defined in stages 1 and 2, which can be visualized with the GraphViz (version 2.24) program. The GraphViz program is authored by AT&T and is made freely available under the common public license at http://www.graphviz.org/About.php.
RESULTS
Phylogenetic analysis.
The VNTR profiles of 475 M. leprae isolates were considered for our analysis (3, 7, 13, 18, 19, 21, 25, 28), but 14 of those isolates were excluded because missing or ambiguous data represented 7 or more of the 16 loci (∼44%). We initially estimated a neighbor-joining phylogeny for those isolates and estimated the confidence in that tree by the standard approach of bootstrapping with 1,000 pseudoreplicates. The resulting tree is essentially a “star phylogeny”: only 1.5% of the clades had bootstrap confidences of >70%, the generally accepted cutoff for taking an inference seriously, and the vast majority of isolates emanated directly from a common central node. Thus, there is little internal structure to the phylogeny that defines relationships among various isolates, and nearly all of the few relationships inferred are unreliable. This finding suggests that there is insufficient information contained in the 16 VNTR loci to permit the confident estimation of relationships among isolates by the conventional method of phylogenetic analysis.
Structure-neighbor clustering.
Given the limitations of phylogenetic analysis, we developed a more generalized clustering approach in order to identify functional groups of genetically similar M. leprae strains, without determining the precise hierarchy of how individual isolates are related to each other. This two-stage method, structure-neighbor clustering, first assigns isolates to generalized populations sharing similar genotypes, and in the second stage, the individuals in each cluster are further subdivided into subclusters of the most genetically similar isolates by using nearest-neighbor networks. We have automated this process using a Perl script, RunStructure.
The first stage of structure-neighbor clustering is carried out with the Structure program (17), a Bayesian model-based algorithm designed to probabilistically assign individuals to different populations on the basis of allele frequencies at multiple SNP or VNTR loci. The Structure program has found widespread use in inferring population structures from genome-wide SNP genotype analyses (16). Given a number of assumed groups, K, the Structure program attempts to assign individuals to each of the groups. The true value of K is typically not known, so to estimate that number, the Structure program is run multiple times by using different values of K. The log likelihood of the data for each run is evaluated: the log likelihood generally increases as K increases, reaches a maximum, and then declines as K increases further (Fig. 1). The value of K for which the highest log likelihood is achieved represents the most likely number of clusters into which the data can be partitioned, and that run is selected for further analysis.
FIG. 1.
Representative graph of log likelihood as a function of the number of clusters. The graph was obtained by using a simulated data set in which the mean branch length was 5. In this case, the most likely number of clusters for which the highest log likelihood is achieved is 5.
For each individual, the Structure program estimates a posterior probability that it belongs to each of the K groups. The individual would normally be assigned to the population for which that probability p is the greatest. However, our approach imposes an additional, more stringent, criterion for assignment: an individual is assigned to its most likely cluster only if p is ≥0.8; otherwise, it is reported as unassigned.
In our experience, the populations defined by the Structure program tend to be large and thus provide insufficient resolution for inferring epidemiological relationships. We therefore further divided the inferred populations into subgroups of the most genetically similar individuals, using nearest-neighbors networks. We performed a pairwise comparison between each of the individuals in the same population, calculating the mean number of repeat number differences averaged over the loci at which both individuals have data. That matrix of pairwise distances was used to determine for each individual within the population its “nearest neighbor(s),” which is the isolate or isolates that have the least number of genetic differences with that individual. The connections between nearest neighbors define networks of genetically similar isolates. These networks can be as small as two individuals, when both isolates are reciprocally each other's nearest neighbors, or they can theoretically contain all of the members of a population, if the connections define a chain or conglomeration that links each of those isolates. Regardless of the size of the nearest-neighbor networks, we equate the term “subgroups” with the isolated networks, since the boundaries of those networks define groups of individuals that share closer relations to other members of the network than they do to individuals in other networks.
Data simulation studies.
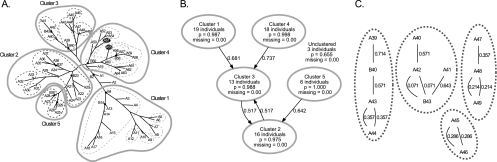
To evaluate the accuracy with which structure-neighbor clustering assigns groups and subgroups of individuals, we first evaluated that approach using simulated data in which the actual order of descent for each isolate and the relationships between individuals are known. We simulated bacterial phylogenies (Fig. 2A) and the VNTR profiles for each bacterial isolate by the use of different parameters for the amount of divergence. We produced a total of five data sets for each of the following mean branch lengths: 1, 2, 3, 5, 7, and 10. Our simulation program retained the VNTR profiles of the internal nodes with a low probability, which mimics the case for real bacterial populations, which typically include a number of ancestral strains that have persisted to the present day (6). The simulated data were subjected to structure-neighbor clustering (Fig. 2B and C), and the results of those analyses were compared to the topology of the true phylogenies (Fig. 2A).
FIG. 2.
Simulated data and structure-neighbor clustering results. (A) Representative phylogeny of simulated data for an average branch length of 5 and for 75 individuals. The labels for terminal nodes begin with “A,” and those of ancestral nodes begin with “B.” The locations of ancestral nodes are indicated by a dot. For comparison, the clusters (solid lines) and subclusters (dashed lines) defined by structure-neighbor clustering are indicated. Two isolates are misassigned to subclusters (A51 and A55 of cluster 3) and are shown in white text with a black background. (B) Stage 1 clustering of the simulated data by use of the Structure program. Each cluster reports the number of individuals in the cluster, the mean probability that the individuals belong to that cluster (p), and the mean number of loci with missing data (here, this number is 0). Arrows point from each cluster to its nearest neighbor and are labeled with the distances between those clusters estimated by the Structure program. Three individuals are left unassigned to clusters, indicating that they cannot be assigned with a probability of ≥0.8; the mean probability that those individuals could have been assigned to their most likely clusters is 0.655. Sixty-two terminal nodes and 10 ancestral nodes were successfully clustered. (C) Stage 2 clustering of simulated data by use of nearest-neighbor networks. The subclusters of cluster 3 are depicted. Individuals are connected to their nearest neighbors by lines that are labeled with the calculated distance between those individuals. The boundaries of the subclusters (dashed lines) have been drawn for clarity.
We evaluated the success of structure-neighbor clustering using two different metrics. First, we defined the “efficiency” of clustering as the fraction of individuals that could confidently (p ≥ 0.8) be assigned to clusters. We also evaluated the “accuracy” of structure-neighbor clustering, defined as the fraction of the individuals that have been correctly assigned to the proper clusters and subclusters. Members of a cluster were deemed correctly assigned if they constituted a clade, in other words, if they could be traced back to a single ancestral node without passing through another cluster. Likewise, the members of a subcluster were considered properly assigned if they could be traced back to a single ancestral node without passing through another subcluster (Fig. 2A). Although subclustering analysis provides valuable information about how the most genetically similar strains are related to one another, there are limitations to the inferences that can be drawn from those networks. For example, it is tempting to assume that ancestral nodes are those that have the most nearest-neighbor connections with presumed descendants. However, our simulation studies do not support a correlation between the topology of subclusters and the strains' order of descent, and we caution that no such inferences should be derived from the topology of the nearest-neighbor networks.
We evaluated each of the five simulations, spanning a total of six mean branch lengths, for both efficiency and accuracy (Table 1). While efficiency decreases sharply as the mean branch lengths increase, accuracy remains well above 0.9 under all conditions. In an analogous fashion, we evaluated the accuracy of neighbor-joining phylogenetic reconstructions of the simulated data by computing the degree of topological agreement between those estimations and the simulated phylogenies that they are meant to reflect (15). (It is not necessary to compute an efficiency score for the neighbor-joining trees, since all isolates are necessarily included in the reconstruction.) The accuracy for neighbor-joining reconstructions averaged about 0.73 for all branch lengths considered, considerably less than that obtained by structure-neighbor clustering. These results suggest that relationships among isolates can be estimated from VNTR profiles much more reliably by structure-neighbor clustering than is possible by estimating those relationships from neighbor-joining phylogenetic trees.
TABLE 1.
Reconstruction of simulated data by structure-neighbor clustering and neighbor-joining phylogenetic analysis
| Method and run no. | Result for BLa of: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
2 |
3 |
5 |
7 |
10 |
|||||||
| Effb | Accc | Eff | Acc | Eff | Acc | Eff | Acc | Eff | Acc | Eff | Acc | |
| Structure-neighbor clustering | ||||||||||||
| 1 | 0.96 | 0.97 | 0.95 | 1.00 | 0.96 | 0.99 | 0.96 | 0.99 | 0.80 | 0.91 | 0.52 | 0.95 |
| 2 | 0.94 | 1.00 | 0.94 | 1.00 | 0.90 | 0.97 | 0.88 | 0.96 | 0.82 | 0.93 | 0.85 | 0.82 |
| 3 | 0.99 | 1.00 | 0.96 | 0.99 | 0.95 | 0.99 | 0.95 | 0.99 | 0.89 | 0.91 | 0.90 | 0.95 |
| 4 | 0.99 | 1.00 | 0.96 | 0.99 | 0.94 | 0.97 | 0.80 | 1.00 | 0.94 | 0.88 | 0.62 | 1.00 |
| 5 | 0.94 | 1.00 | 0.97 | 0.99 | 0.96 | 0.90 | 0.62 | 0.85 | 0.96 | 0.93 | 0.82 | 0.97 |
| Mean | 0.96 | 0.99 | 0.96 | 0.99 | 0.94 | 0.96 | 0.84 | 0.96 | 0.88 | 0.91 | 0.74 | 0.94 |
| SEMd | 0.01 | 0.01 | 0.01 | 0.00 | 0.01 | 0.02 | 0.06 | 0.03 | 0.03 | 0.01 | 0.07 | 0.03 |
| Neighbor-joining phylogenetic reconstruction | ||||||||||||
| 1 | 0.79 | 0.55 | 0.57 | 0.83 | 0.75 | 0.69 | ||||||
| 2 | 0.83 | 0.87 | 0.82 | 0.75 | 0.76 | 0.74 | ||||||
| 3 | 0.80 | 0.54 | 0.76 | 0.83 | 0.74 | 0.75 | ||||||
| 4 | 0.82 | 0.56 | 0.81 | 0.76 | 0.77 | 0.80 | ||||||
| 5 | 0.81 | 0.60 | 0.82 | 0.74 | 0.72 | 0.75 | ||||||
| Mean | 0.81 | 0.62 | 0.75 | 0.78 | 0.75 | 0.75 | ||||||
| SEM | 0.01 | 0.06 | 0.05 | 0.02 | 0.01 | 0.02 | ||||||
BL, mean branch length.
Eff, efficiency.
Acc, accuracy.
SEM, standard error of the mean.
Because the experimental data for M. leprae do not include information about the length of every VNTR for every isolate, we modified the simulated data in order to examine the effects of missing data on structure-neighbor clustering. For simulations with branch lengths of 5 and 10, we randomly replaced simulated VNTR loci with missing data either with a frequency equal to that observed for the experimental data set (“average missing data”) or at twice that probability (“high missing data”). Once again, we performed structure-neighbor clustering and neighbor-joining phylogenetic reconstruction of those data and evaluated the accuracy and efficiency of the results compared to the known phylogeny (Table 2). Missing data somewhat decreased the efficiency of structure-neighbor clustering but did not influence the accuracy of either neighbor-joining phylogenetic reconstruction or structure-neighbor clustering. For the results typically obtained by M. leprae genotyping, it appears that the amount of missing data should not be a limiting factor with respect to the accuracy of structure-neighbor clustering.
TABLE 2.
Effect of missing data on reconstruction by structure-neighbor clustering and neighbor-joining phylogenetic analysis
| Method and run no. | Result for BLa of: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 |
10 |
|||||||||||
| No missing data |
Avg missing data |
High missing data |
No missing data |
Average missing data |
High missing data |
|||||||
| Effb | Accc | Eff | Acc | Eff | Acc | Eff | Acc | Eff | Acc | Eff | Acc | |
| Structure-neighbor clustering | ||||||||||||
| 1 | 0.96 | 0.99 | 0.92 | 0.97 | 0.29 | 1.00 | 0.52 | 0.95 | 0.51 | 0.97 | 0.12 | 1.00 |
| 2 | 0.88 | 0.96 | 0.88 | 1.00 | 0.47 | 1.00 | 0.85 | 0.82 | 0.66 | 0.85 | 0.64 | 0.85 |
| 3 | 0.95 | 0.99 | 0.97 | 0.97 | 0.93 | 0.93 | 0.90 | 0.95 | 0.67 | 1.00 | 0.90 | 0.94 |
| 4 | 0.80 | 1.00 | 0.84 | 0.95 | 0.80 | 0.97 | 0.62 | 1.00 | 0.83 | 0.99 | 0.50 | 0.97 |
| 5 | 0.62 | 0.85 | 0.62 | 0.96 | 0.93 | 0.94 | 0.82 | 0.97 | 0.86 | 0.91 | 0.73 | 0.96 |
| Mean | 0.84 | 0.96 | 0.85 | 0.97 | 0.69 | 0.97 | 0.74 | 0.94 | 0.71 | 0.94 | 0.58 | 0.95 |
| SEMd | 0.06 | 0.03 | 0.06 | 0.01 | 0.13 | 0.01 | 0.07 | 0.03 | 0.06 | 0.03 | 0.13 | 0.03 |
| Neighbor-joining phylogenetic reconstruction | ||||||||||||
| 1 | 0.83 | 0.80 | 0.78 | 0.69 | 0.70 | 0.71 | ||||||
| 2 | 0.75 | 0.73 | 0.73 | 0.74 | 0.70 | 0.70 | ||||||
| 3 | 0.83 | 0.50 | 0.77 | 0.75 | 0.72 | 0.74 | ||||||
| 4 | 0.76 | 0.78 | 0.75 | 0.80 | 0.78 | 0.79 | ||||||
| 5 | 0.74 | 0.68 | 0.65 | 0.75 | 0.73 | 0.74 | ||||||
| Mean | 0.78 | 0.70 | 0.74 | 0.75 | 0.72 | 0.74 | ||||||
| SEM | 0.02 | 0.05 | 0.02 | 0.02 | 0.02 | 0.02 | ||||||
BL, mean branch length.
Eff, efficiency.
Acc, accuracy.
SEM, standard error of the mean.
Structure-neighbor analysis of M. leprae VNTR data.
The complete VNTR profiles for all 475 M. leprae strains were subjected to structure-neighbor clustering. The most probable number of clusters (K) was 10, for which the log likelihood of the data was −7,705.
(i) Identifying which loci should be included.
The study that originally characterized the M. leprae VNTR loci utilized here (8) considered the reliability of each locus by requiring that, for as many as three experimental replicates, the number of repeats was read independently by two different individuals. Concordance was calculated as the fraction of instances in which the two readers agreed on the repeat number, with concordance being considered directly proportional to reliability. By this metric, two loci, AT(15) and TA(18), ranked unusually poorly, demonstrating concordances of 0% and 50%, respectively. Similarly, it was noted that locus 18-8 produced a large degree of nonspecific amplification products that complicated genotype interpretation, especially when the locus was tested by using higher concentrations of the DNA template. We therefore analyzed the M. leprae VNTR data set after excluding those three potentially unreliable loci singly, in pairs, or all together (Table 3). The effects of those changes were monitored by determining the lnL of the data given for the most likely number of clusters, in which a higher lnL represents a more probable interpretation of the data. The absolute value of the log likelihood is affected by the amount of data, i.e., the number of loci used. The appropriate comparisons are therefore between data sets that include the same number of loci. Since the stochastic nature of the Bayesian Structure program means that independent runs of the same data set do not yield identical lnL values, Table 3 reports the mean ± standard error of the lnL from three independent analyses, unless otherwise indicated.
TABLE 3.
Evaluation of potentially unreliable loci
| No. of VNTR loci used | Locus or loci excluded | Comment | lnL ± SEMb | Efficiency ± SEM |
|---|---|---|---|---|
| 16 | None | Full data set | −7,842 ± 69 | 0.71 ± 0.05 |
| 15 | (AC)8a | Randomly selected locus | −7,314 ± 60 | 0.69 ± 0.03 |
| 15 | (GTA)9 | Randomly selected locus | −7,155 ± 3 | 0.66 ± 0.02 |
| 15 | 12-5 | Randomly selected locus | −7,721 ± 5 | 0.73 ± 0.02 |
| 15 | Average of (AC)8a, (GTA)9, and 12-5 | Mean of one randomly selected locus (n = 9) | −7,397 ± 86 | 0.69 ± 0.02 |
| 15 | (AT)15 | Worst concordance, 22.3% missing dataa | −7,032 ± 67 | 0.68 ± 0.002 |
| 15 | (TA)18 | Second-worst concordance, 18.2% missing dataa | −6,951 ± 68 | 0.78 ± 0.04 |
| 15 | 18-8 | Multiple stutter bands, 30.2% missing dataa | −7,675 ± 72 | 0.62 ± 0.02 |
| 15 | (TA)10 | Most missing data (45.1%) | −7,361 ± 69 | 0.71 ± 0.02 |
| 14 | (AT)17, 12-5 | Two random loci | −6,655 ± 45 | 0.73 ± 0.01 |
| 14 | (AT)15, (TA)18 | Combination of two suspect loci | −6,126 ± 9 | 0.61 ± 0.01 |
| 14 | (AT)15, 18-8 | Combination of two suspect loci | −6,822 ± 67 | 0.66 ± 0.02 |
| 14 | (TA)18 18-8 | Combination of two suspect loci | −6,721 ± 66 | 0.59 ± 0.02 |
| 13 | (AT)17, 6-7, 12-5 | Three randomly selected loci | −6,268 ± 37 | 0.76 ± 0.02 |
| 13 | (AT)15, (TA)18, 18-8 | All three suspect loci | −5,897 ± 68 | 0.70 ± 0.05 |
| 13 | (AT)15, (TA)18, (GGT)5 | Two suspect loci plus a randomly selected locus | −6,038 ± 2 | 0.66 ± 0.01 |
| 13 | (AT)15, (TA)18, (AC)8a | Two suspect loci plus a randomly selected locus | −5,580 ± 6 | 0.62 ± 0.01 |
| 13 | Average of two trials | Mean of two suspect loci plus one randomly selected locus (n = 6) | −5,809 ± 84 | 0.64 ± 0.01 |
Comment from reference 8.
SEM, standard error of the mean.
When each of three randomly chosen loci [(AC)8a, (GRA)9, and 12-5] were excluded, the log likelihoods were somewhat reduced and ranged from −7,155 to −7,721 with a mean of −7,397 ± 86 (average ± standard error of the mean). Excluding either the (AT)15 or the (TA)18 locus increased lnL by factors of e365 and e446, respectively, relative to the value obtained by excluding a randomly selected locus, while excluding the 18-8 locus reduced lnL by a factor of e278. The gains in lnL suggest that removing either (AT)15 or (TA)18 does result in the more reliable interpretation of the data by structure-neighbor clustering. Data for both (AT)15 and (TA)18 were missing for a significant fraction of the M. leprae isolates, so we considered the possibility that the improvement in lnL is the result of excluding loci with missing data. Locus (TA)10 is considered reliable, but a higher fraction of isolates were missing data for that locus than for any of the suspect loci. However, excluding (TA)10 did not improve the lnL relative to that obtained by excluding a randomly selected locus (Table 3). We conclude that the gains in lnL achieved by removing the unreliable loci are not solely the effect of eliminating missing data from the analysis.
When (AT)15 and (TA)18 were simultaneously excluded, lnL was increased by a factor of e529 relative to that achieved by excluding two randomly selected loci. Excluding either of those loci in conjunction with locus 18-8 again resulted in a reduced lnL.
We next considered the possibility that the exclusion of locus 18-8 might increase reliability only when (AT)15 and (TA)18 were also excluded. Although the exclusion of all three loci increased lnL by e371 relative to the value obtained by excluding three random loci, the increase was no greater that that achieved by excluding (AT)15 and (TA)18 in conjunction with a randomly chosen locus.
lnL is not, however, the only factor worth considering in deciding which loci to include in a structure-neighbor analysis. When (AT)15 and (TA)18 were excluded but 18-8 was included, the efficiency was only 0.61, whereas it was 0.70 when 18-8 was additionally excluded. That reduction in clustering efficiency reflects a decrease in the probability with which isolates can be assigned to a cluster. Indeed, when 18-8 was included, the mean probability with which clusterable isolates were assigned was 0.924 ± 0.001, whereas the mean probability was 0.936 ± 0.002 when 18-8 was excluded. On that basis, we judge that locus 18-8 contributes little but noise to the analysis and that it may be resource effective to exclude it in future studies.
We conclude not only that loci (AT)15 and (TA)18 are unreliable in the laboratory, as reported by Gillis et al. (8), but also, with consideration of the comprehensive data set, that those loci are unreliable in the field. Those VNTR markers are confounding and should not be used in future studies of M. leprae.
(ii) Population structure of M. leprae.
We repeated structure-neighbor clustering of the 475 M. leprae strains using only the 13 loci deemed reliable (locus 18-8 was excluded) (Fig. 3). Ten isolates were automatically excluded because of excessive amounts of missing data, leaving 465 strains in the final analysis. During the first step of clustering, performed by the Structure program, 101 of those isolates could not be assigned to a cluster with a probability of ≥0.8 and were excluded from further analysis (see Table S1 in the supplemental material). The efficiency of clustering was therefore 0.782, similar to that observed in data simulation studies when the mean branch length of the simulated trees was 10. Although the cutoff probability for assigning strains to clusters is 0.8, the mean probabilities for all clusters in this instance exceeded 0.92. We therefore have considerable confidence in the assignment of strains to their respective populations.
FIG. 3.
Stage 1 clustering of M. leprae data by use of the Structure program. The mean probability that isolates belong to the cluster (p) and the mean number of loci per individual for which data are missing (indicated by “missing”) are reported for each cluster. For example, the 42 individuals in cluster 1 had missing data at an average of 0.60 loci. Individuals with an insufficient probability of being assigned to any cluster are indicated. Arrows are drawn from each cluster to its nearest neighbor.
The eight populations defined at this step reflected the source countries reasonably well. For example, all but one of the strains from China belonged to cluster 8, 85% of the strains from Brazil belonged to cluster 5, and 88% of the strains from India belonged to cluster 6. A few minority stains, typically one or two from any given country, that were isolated from geographically separate regions existed within most clusters and therefore probably reflect immigration. Notable exceptions to those trends are the strains isolated from the Philippines, the majority of which constituted three major populations, clusters 1, 3, and 4. It appears that the strains from the Philippines are genetically diverse enough that they can be divided into two closely related but distinct groups (clusters 1 and 3), plus a third minority group (cluster 4) that is more closely related to strains from India. This finding suggests a significant influx of strains to that nation from India. Clusters 2 and 7 each primarily consisted of strains isolated in Thailand, suggesting the existence of two distinct populations from Thailand, but each also included a significant number of strains isolated in the Philippines, suggesting immigrations from Thailand to the Philippines.
Surprisingly, we also found that strain TN, the first M. leprae strain to be sequenced (4), does not originate from India, as reported previously (14). The present analysis infers with a high degree of confidence (p = 1.0) that TN belongs to a cluster of isolates almost exclusively from the Philippines (cluster 3). This result suggests that although TN was isolated from a source in India, it was genetically very similar to strains endemic to the Philippines, suggesting the migration of the TN strain.
Higher resolution is afforded by the second stage of structure-neighbor clustering, which defines subgroups within larger populations. Figure 4 shows typical results of stage 2 structure-neighbor analyses. Depicted is cluster 1, in which six subclusters have been defined for the 42 strains by using nearest-neighbor networks. Although subcluster analysis provides valuable insight into the genetic relationships between individual strains, we again emphasize that no inferences about the order of descent can be derived from the nearest-neighbor topologies of the subclusters. Diagrams of stage 2 structure-neighbor clustering for clusters 1 to 8 are available in the supplemental material.
FIG. 4.
Stage 2 clustering of the M. leprae strains within cluster 1. Nearest neighbors are connected by lines, and the genetic distance between isolates is indicated. Subclusters are equivalent to the individual nearest-neighbor networks within a cluster.
Recent studies conducted in the Philippines examined pairs (and one trio) of M. leprae strains isolated from each of eight different families (18, 19) or, similarly, pairs of strains taken from each of eight different villages (19) (Table 4). Because of their tight geographical distribution, those paired strains may be expected to be closely related and would thus fall within the same clusters and subclusters defined by the structure-neighbor approach. Of the total of the 16 sets of paired strains, only 1 had an unassigned member that precluded further analysis. Of the remaining 15 pairs of isolates, 14 pairs were within the same cluster and 13 grouped both within the same cluster and within the same subcluster. For the 13 pairs contained in the same subcluster, all but 1 pair of isolates was directly linked by a nearest-neighbor connection. These inferences suggest that the groups and subgroups identified by structure-neighbor clustering, as well as the relationships between strains inferred through nearest-neighbor networks, correlate well with expected epidemiological relationships determined with real-world data. At the same time, we found that for one multicase family (family F3), the isolates were confidently assigned to entirely different populations. We cannot rule out the possibility that the results from family F3 reflect reporting errors. Nevertheless, caution is warranted in assuming that multiple infections within the same family always represent direct transmission from one family member to another, as it appears that independent infections from genetically dissimilar sources occur with measurable frequency.
TABLE 4.
Multicase families and pairs from the same village
| Group and case designation | Strain | Same cluster? | Same subcluster? | Nearest neighbors? |
|---|---|---|---|---|
| Multicase family | ||||
| MCF-1a | IDEAL50 | Yes | Yes | Yes |
| MCF-1 | IDEAL53 | Cluster 1 | ||
| MCF-2a | IDEAL68 | (IDEAL68 unassigned) | Yes | Yes |
| MCF-2 | IDEAL69 | Yes | ||
| MCF-2 | IDEAL70 | Cluster 3 | ||
| F1b | F1-L42 | Yes | Yes | Yes |
| F1 | F1-M20 | Cluster 1 | ||
| F2b | F2-M90 | Yes | No | No |
| F2 | F2-M91 | Cluster 2 | ||
| F3b | F3-L14 | No | No | No |
| F3 | F3-L29 | |||
| F4b | F4-M26 | (F4-M26 unassigned) | ?d | ?d |
| F4 | F4-M85 | |||
| F5b | F5-M46 | Yes | Yes | Yes |
| F5 | F5-M71 | Cluster 3 | ||
| F6b | F6-L55 | Yes | Yes | Yes |
| F6 | F6-L64 | Cluster 4 | ||
| Pairs from the same village | ||||
| B1c | B1-M11 | Yes | Yes | Yes |
| B1 | B1-M59 | Cluster 1 | ||
| B2c | B2-M50 | Yes | Yes | Yes |
| B2 | B2-M61 | Cluster 1 | ||
| B3c | B3-R34 | Yes | Yes | Yes |
| B3 | B3-L36 | Cluster 3 | ||
| B4c | B4-M71 | Yes | Yes | No |
| B4 | B4-M72 | Cluster 3 | ||
| B5c | B5-L03 | Yes | Yes | Yes |
| B5 | B5-L57 | Cluster 2 | ||
| B6c | B6-R19 | Yes | Yes | Yes |
| B6 | B6-M69 | Cluster 4 | ||
| B7c | B7-M81 | Yes | Yes | Yes |
| B7 | B7-M82 | Cluster 4 | ||
| B8c | B8-L60 | Yes | Yes | Yes |
| B8 | B8-L91 | Cluster 7 |
Finally, the structure-neighbor method identified 417 unique VNTR types (equivalent to genotypes) among the 465 strains that were clustered. This is in contrast to the 16 SNP types, again equivalent to genotypes, found for the 400 strains that were subjected to SNP analysis at 84 informative polymorphic sites (14). There were 31 sets of strains, ranging from 2 to 6 members per set, in which the members within a set had indistinguishable VNTR types. In some cases, the strains were identical at all 13 VNTR loci, and in others, some members contained missing data and were therefore identical only at the loci for which data were available. The most interesting pair was CD236 from India and I40 from the Philippines, which were identical at all 13 VNTR loci. Except for reference strain TN (also supposedly from India), CD236 was the only member of cluster 1 (Fig. 3) that was not from the Philippines. This example represents another case of the very recent immigration of a strain from the Philippines into India.
DISCUSSION
Understanding the most basic epidemiology of leprosy infection has been greatly hampered by the absence of inexpensive molecular strain typing methods capable of revealing enough genetic variation to, in turn, allow estimation of the relationships among isolates with a reasonable degree of confidence and accuracy. The method that can be used to type Mycobacterium leprae isolates by the use of 16 VNTR loci and as few as 10 cells as the starting material, which has recently been developed and validated (8), largely meets the need for an inexpensive typing method. There are several reasons why VNTR typing is superior to SNP typing for investigating the epidemiology of M. leprae. First, VNTR loci evolve much faster than SNPs, with the consequence being that VNTR typing reveals considerably more genetic heterogeneity than examination of an equivalent number of SNPs. For instance, in their survey of 400 M. leprae strains using 84 informative SNP sites, Monot et al. (14) identified only 16 unique SNP types, whereas in the present study, 465 strains included 417 unique VNTR types. The higher resolution of VNTR typing is the result of having an average of 9.5 alleles per locus, whereas none of the SNPs had more than 2 alleles per site. Second, the number of possible alleles per locus is another factor distinguishing SNPs and VNTRs. The VNTRs used in this study had an average of 9.5 alleles per locus, but none of the SNPs had more than two alleles, of a maximum of only four possible alleles, per site (14). It is therefore easier to distinguish between isolates on the basis VNTR typing. Lastly, the cost of typing the full VNTR panel is currently about 6.5 times lower than that of amplifying and sequencing the 84 SNP loci identified by Monot et al. (14). This cost difference is especially significant for researchers in the countries where leprosy is endemic. A total of eight studies involving six countries have now used VNTR loci to type hundreds of M. leprae strains, making it clear that use of the VNTR panel for epidemiological surveys is practical. The present study brings together the results of those surveys into a single, comprehensive analysis of the molecular epidemiology of M. leprae.
Phylogenetic analysis, the study of evolutionary relatedness among species, is typically used to infer how bacterial strains are related to each other through a common ancestry (5, 11). Although such analyses can be enlightening, it has been recognized that phylogenetic reconstructions can be inadequate or misleading when the amount of genetic diversity distinguishing individuals is small (6, 20). Our initial phylogenetic analysis of the M. leprae strains indicates that there is insufficient genetic polymorphism at the available VNTR loci to infer relationships between isolates in any meaningful or reliable way. In light of this finding, caution should be used when inferences from published phylogenies of M. leprae (3, 19) are drawn, unless metrics for those estimations can be provided with a high degree of confidence. Because of the practical constraints posed by working with M. leprae, it is unlikely that significantly more VNTR loci will become available in the near future, and thus, an alternative method to phylogenetic analysis is required to estimate such relationships.
An alternative method to phylogenetic analysis for inferring relationships from limited VNTR loci, eBURST (6), has been proposed, and we have previously suggested the use of that approach for the examination of M. leprae (9). Unfortunately, eBURST cannot be applied to samples for which data are missing for any of the loci. For the available M. leprae strains, only 178 of the 461 VNTR profiles (38.6%) contained data for all 16 loci. Additionally, eBURST requires that strains differ at no more than a single locus in order to form connections between them, which is the case for only a small minority of M. leprae strains. Clearly, the constraints of eBURST make it an impractical tool for use for estimation of the relationships between M. leprae isolates.
We therefore developed a more generalized approach to enable the clustering of genetically similar strains into groups and subgroups. We refer to this approach as structure-neighbor clustering. The approach requires two separate stages: in the first stage, isolates are clustered into large populations of genetically similar strains by using the Structure population structure inference program (17), and in the second stage, the individuals in each population are further grouped into subclusters, on the basis of nearest-neighbor networks. Using simulated data for which the true relationships can be known, we have found that the fraction of individuals that can reliably be assigned to clusters is a function of the genetic distances between them and, to a lesser extent, the average amount of missing data. Under the conditions under which the isolates were examined in the present study, 74% or more of the isolates could be successfully assigned to clusters (Table 1). The accuracy with which individuals are assigned to clusters and further assigned to subclusters averaged 95% across a wide variety of conditions, whereas the accuracy of neighbor-joining phylogenetic reconstructions by use of the same data was 73% (Table 1). We found that the inclusion of missing data somewhat decreased the number of individuals that could be assigned to clusters, but for those individuals that could be assigned, accuracy was not affected (Table 2).
Structure-neighbor clustering outperforms phylogenetic analysis only because it provides fewer, but more reliable, inferences about the population structure from the data. Phylogenetic analysis attempts to infer the order of descent of each individual in the data set from hypothetical common ancestors. In contrast, structure-neighbor clustering merely assigns isolates with the most similar genotypes to the same groups and/or subgroups, without making any attempt to infer how they have descended from a common progenitor. Of course, it is assumed that the relatedness within any group (either a cluster or a subcluster) is the consequence of descent from a common ancestor, but our approach does not identify those ancestors: indeed, we caution that no inferences about the order of descent should be drawn from the topology of connections within subclusters. A secondary factor contributing to the robustness of structure-neighbor clustering is that it does not attempt to analyze all of the individuals provided, whereas phylogenetic analysis does. Instead, it imposes requirements for the maximal amount of missing data permitted for an individual (less than 40%) as well as minimum standards of confidence (Bayesian posterior probability ≥ 0.8) for the clustering of individuals, the result of which is to include only those individuals for which the data permit the strongest inferences to be made.
Given that structure-neighbor clustering provides fewer inferences than phylogenetic analysis, is it actually useful for epidemiological purposes? We believe that it is. First, for most epidemiological purposes, it is sufficient to be able to identify what strains are closely related without concern about the precise order of descent from common ancestors. Structure-neighbor clustering reliably and accurately meets that need, whereas phylogenetic methods attempt to provide additional information with inaccurate and misleading results. Although the structure-neighbor approach cannot estimate relationships for every isolate, those for which it can estimate relationships are assigned to groups with a high degree of reliability. Second, as more individuals are added to the available pool of data (especially if measures are taken to minimize missing data), progressively more reliable connections are likely to emerge and the efficiency of the structure-neighbor approach is likely to increase beyond its present 77%. Additionally, future improvements to the structure-neighbor program may improve accuracy and resolution. The Structure program currently considers VNTR alleles either to be the same length or to have different lengths. VNTR loci tend to mutate in units of one tandem repeat at a time (1), so the Structure program's binary treatment of data ignores the fact that alleles which have more similar numbers of repeats are more genetically similar than alleles which have more pronounced differences in the number of repeats. The inclusion of a parameter for ordered character states in the Structure program may therefore improve future analyses but was outside the scope of the present work.
In this study, the application of the structure-neighbor method to the available M. leprae VNTR data pooled from eight recent studies yields a number of valuable insights that could not have been obtained by considering each data set independently. We have described a strategy to determine what VNTR loci are unreliable and have the potential to confound analyses and have found that 13 of the 16 available VNTR markers are suitable for use in molecular epidemiology. Examining relationships at the resolution of countries through stage 1 analysis of those data (Fig. 3) reveals a general agreement between clusters and the country of origin for the samples assigned within. However, strains from the Philippines group into three major populations, suggesting an unusual amount of genetic heterogeneity among the isolates from that nation. Two of those populations are closely related but distinct groups, while the third, smaller group is more closely related to strains from India, suggesting a significant degree of strain migration from India to the Philippines. Another surprising result is the inferred origin of M. leprae strain TN, the first M. leprae strain to be sequenced (4). That isolate is presumed to have come from India (14), but our analysis demonstrates with a high degree of confidence that it belongs to a cluster otherwise composed entirely of strains from the Philippines. TN may have been isolated in India, but if so, it was clearly a recent immigrant from the Philippines. With that in mind, it is unfortunate that strain TN was used as an outgroup to root a phylogenetic tree of strains from the Philippines (19), because although the authors could not have known it at the time, it now seems likely that TN is a member of the in-group.
Two recent studies (18, 19) isolated multiple M. leprae strains from the same families or villages, permitting examination of that organism's epidemiology at a more local level. As expected, we found that most of the strains from the same villages and families grouped closely together in the same cluster and/or subcluster in structure-neighbor analysis. However, we identified one multicase family (family F3) in which the separate isolates were confidently assigned to genetically dissimilar populations. This finding not only suggests that the majority of leprosy cases in those communities and families are either laterally transferred between individuals or obtained from the same natural source but also suggests that the independent infection of family members from different sources occurs at a nonnegligible frequency.
It is clear that the quality of structure-neighbor clustering is dependent on the quality of the underlying data used for those analyses, and we encourage all M. leprae investigators to employ only the 14 reliable VNTR loci that we have identified, using appropriate quality checks, as described by Gillis et al. (8). Although locus 18-8 is reliable, it appears to contribute little but noise to the clustering of the global sets of isolates that we have considered here; but it is possible that it may contribute positively to strain resolution in more local comparisons, e.g., those limited to a single country or a single region within a country. A future study will specifically address that issue and will further consider how best loci should be selected for inclusion in data sets. Furthermore, the value of the molecular epidemiology of M. leprae increases with an increase in the number of available strains, which improves the ability to make connections among those data. With that in mind, the research community would benefit from a formally funded database for M. leprae VNTR data, developed and curated at a university or other institution. Until that time, the authors will provide a Web-based, read-only version of that database, diagrams of the clusters and subclusters based on the most current M. leprae database, and detailed instructions for adding high-quality data to the database. That database can be accessed at http://web.me.com/barryghall/Leprosy/Database/Database.html.
We also point out that structure-neighbor clustering may have application for inferring the population structures of other organisms. For example, VNTR data from 12 loci have been used, in conjunction with spoligotyping data, to estimate phylogenetic trees for Mycobacterium tuberculosis (22-24). Bootstrap percentiles or other measures of clade confidence have not been reported for those reconstructions, so it is not possible to assess the accuracies of the phylogenies. However, a simulation study has suggested that at least 25 VNTR loci are required to produce NJ trees for bacteria that are 90% accurate (B. G. Hall, unpublished results). Given these observations, structure-neighbor clustering might be considered an alternative means to estimate the relationships among M. tuberculosis isolates.
Supplementary Material
Footnotes
Published ahead of print on 29 March 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Boyer, J. C., N. A. Yamada, C. N. Roques, S. B. Hatch, K. Riess, and R. A. Farber. 2002. Sequence dependent instability of mononucleotide microsatellites in cultured mismatch repair proficient and deficient mammalian cells. Hum. Mol. Genet. 11:707-713. [DOI] [PubMed] [Google Scholar]
- 2.Brenan, P. J. 2009. IDEAL: in the footsteps of IMMLEP and THELEP. Lepr. Rev. 80:236-245. [PubMed] [Google Scholar]
- 3.Cardona-Castro, N., J. C. Beltran-Alzate, I. M. Romero-Montoya, E. Melendez, F. Torres, R. M. Sakamuri, W. Li, and V. Vissa. 2009. Identification and comparison of Mycobacterium leprae genotypes in two geographical regions of Colombia. Lepr. Rev. 80:316-321. [PubMed] [Google Scholar]
- 4.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. [DOI] [PubMed] [Google Scholar]
- 5.Diamant, E., Y. Palti, R. Gur-Arie, H. Cohen, E. M. Hallerman, and Y. Kashi. 2004. Phylogeny and strain typing of Escherichia coli, inferred from variation at mononucleotide repeat loci. Appl. Environ. Microbiol. 70:2464-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fontes, A. N., R. M. Sakamuri, I. M. Baptista, S. Ura, M. O. Moraes, A. N. Martinez, E. N. Sarno, P. J. Brennan, V. D. Vissa, and P. N. Suffys. 2009. Genetic diversity of Mycobacterium leprae isolates from Brazilian leprosy patients. Lepr. Rev. 80:302-315. [PubMed] [Google Scholar]
- 8.Gillis, T., V. Vissa, M. Matsuoka, J. H. Richardus, R. Truman, B. G. Hall, and P. J. Brenan. 2009. Characterisation of short tandem repeats for genotyping Mycobacterium leprae. Lepr. Rev. 80:250-260. [PubMed] [Google Scholar]
- 9.Hall, B. G. 2009. Molecular epidemiology of Mycobacterium leprae: a solid beginning. Lepr. Rev. 80:246-249. [PubMed] [Google Scholar]
- 10.Hall, B. G. 2008. Simulating DNA coding sequence evolution with EvolveAGene 3. Mol. Biol. Evol. 25:688-695. [DOI] [PubMed] [Google Scholar]
- 11.Hall, B. G., and M. Barlow. 2006. Phylogenetic analysis as a tool in molecular epidemiology of infectious diseases. Ann. Epidemiol. 16:157-169. [DOI] [PubMed] [Google Scholar]
- 12.Hughes, C. R., and D. C. Queller. 1993. Detection of highly polymorphic microsatellite loci in a species with little allozyme polymorphism. Mol. Ecol. 2:131-137. [DOI] [PubMed] [Google Scholar]
- 13.Kimura, M., R. M. Sakamuri, N. A. Groathouse, B. L. Rivoire, D. Gingrich, S. Krueger-Koplin, S. N. Cho, P. J. Brennan, and V. Vissa. 2009. Rapid variable-number tandem-repeat genotyping for Mycobacterium leprae clinical specimens. J. Clin. Microbiol. 47:1757-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monot, M., N. Honore, T. Garnier, N. Zidane, D. Sherafi, A. Paniz-Mondolfi, M. Matsuoka, G. M. Taylor, H. D. Donoghue, A. Bouwman, S. Mays, C. Watson, D. Lockwood, A. Khamispour, Y. Dowlati, S. Jianping, T. H. Rea, L. Vera-Cabrera, M. M. Stefani, S. Banu, M. Macdonald, B. R. Sapkota, J. S. Spencer, J. Thomas, K. Harshman, P. Singh, P. Busso, A. Gattiker, J. Rougemont, P. J. Brennan, and S. T. Cole. 2009. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat. Genet. 41:1282-1289. [DOI] [PubMed] [Google Scholar]
- 15.Nye, T. M., P. Lio, and W. R. Gilks. 2006. A novel algorithm and web-based tool for comparing two alternative phylogenetic trees. Bioinformatics 22:117-119. [DOI] [PubMed] [Google Scholar]
- 16.Parker, H. G., L. V. Kim, N. B. Sutter, S. Carlson, T. D. Lorentzen, T. B. Malek, G. S. Johnson, H. B. DeFrance, E. A. Ostrander, and L. Kruglyak. 2004. Genetic structure of the purebred domestic dog. Science 304:1160-1164. [DOI] [PubMed] [Google Scholar]
- 17.Pritchard, J. K., M. Stephens, and P. Donnelly. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakamuri, R. M., J. Harrison, R. Gelber, P. Saunderson, P. J. Brennan, M. Balagon, and V. Vissa. 2009. A continuation: study and characterisation of Mycobacterium leprae short tandem repeat genotypes and transmission of leprosy in Cebu, Philippines. Lepr. Rev. 80:272-279. [PubMed] [Google Scholar]
- 19.Sakamuri, R. M., M. Kimura, W. Li, H. C. Kim, H. Lee, M. D. Kiran, W. C. Black IV, M. Balagon, R. Gelber, S. N. Cho, P. J. Brennan, and V. Vissa. 2009. Population-based molecular epidemiology of leprosy in Cebu, Philippines. J. Clin. Microbiol. 47:2844-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salipante, S. J., J. M. Thompson, and M. S. Horwitz. 2008. Phylogenetic fate mapping: theoretical and experimental studies applied to the development of mouse fibroblasts. Genetics 178:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinde, V., H. Newton, R. M. Sakamuri, V. Reddy, S. Jain, A. Joseph, T. Gillis, I. Nath, G. Norman, and V. Vissa. 2009. VNTR typing of Mycobacterium leprae in South Indian leprosy patients. Lepr. Rev. 80:290-301. [PubMed] [Google Scholar]
- 22.Sola, C., I. Filliol, M. C. Gutierrez, I. Mokrousov, V. Vincent, and N. Rastogi. 2001. Spoligotype database of Mycobacterium tuberculosis: biogeographic distribution of shared types and epidemiologic and phylogenetic perspectives. Emerg. Infect. Dis. 7:390-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sola, C., I. Filliol, E. Legrand, S. Lesjean, C. Locht, P. Supply, and N. Rastogi. 2003. Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect. Genet. Evol. 3:125-133. [DOI] [PubMed] [Google Scholar]
- 24.Sola, C., I. Filliol, E. Legrand, I. Mokrousov, and N. Rastogi. 2001. Mycobacterium tuberculosis phylogeny reconstruction based on combined numerical analysis with IS1081, IS6110, VNTR, and DR-based spoligotyping suggests the existence of two new phylogeographical clades. J. Mol. Evol. 53:680-689. [DOI] [PubMed] [Google Scholar]
- 25.Srisungnam, S., J. Rudeeaneksin, A. Lukebua, S. Wattanapokayakit, S. Pasadorn, K. Mahotarn, Ajincholapan, R. M. Sakamuri, M. Kimura, P. J. Brennan, B. Phetsuksiri, and V. Vissa. 2009. Molecular epidemiology of leprosy based on VNTR typing in Thailand. Lepr. Rev. 80:280-289. [PubMed] [Google Scholar]
- 26.Streisinger, G., Y. Okada, J. Emrich, J. Newton, A. Tsugita, E. Terzaghi, and M. Inouye. 1966. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb. Symp. Quant. Biol. 31:77-84. [DOI] [PubMed] [Google Scholar]
- 27.Swofford, D. L. 2000. PAUP*. Phylogenetic analysis using parsimony (*and other methods). 4.0 ed. Sinauer Associates, Sunderland, MA.
- 28.Xing, Y., J. Liu, R. M. Sakamuri, Z. Wang, Y. Wen, V. Vissa, and X. Weng. 2009. VNTR typing studies of Mycobacterium leprae in China: assessment of methods and stability of markers during treatment. Lepr. Rev. 80:261-271. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.