Abstract
DNA extraction from formalin-fixed paraffin-embedded (FFPE) tissues is difficult and requires special protocols in order to extract small amounts of DNA suitable for amplification. Most described methods report an amplification success rate between 60 and 80%; therefore, there is a need to improve molecular detection and identification of fungi in FFPE tissue. Eighty-one archived FFPE tissues with a positive Gomori methenamine silver (GMS) stain were evaluated using five different commercial DNA extraction kits with some modifications. Three different panfungal PCR assays were used to detect fungal DNA, and two housekeeping genes were used to assess the presence of amplifiable DNA and to detect PCR inhibitors. The sensitivities of the five extraction protocols were compared, and the quality of DNA detection (calculated for each kit as the number of housekeeping gene PCR-positive samples divided by the total number of samples) was 60 to 91% among the five protocols. The efficiencies of the three different panfungals used (calculated as the number of panfungal-PCR-positive samples divided by the number of housekeeping gene PCR-positive samples) were 58 to 93%. The panfungal PCR using internal transcribed spacer 3 (ITS3) and ITS4 primers yielded a product in most FFPE tissues. Two of the five DNA extraction kits (from TaKaRa and Qiagen) showed similar and promising results. However, one method (TaKaRa) could extract fungal DNA from 69 of the 74 FFPE tissues from which a housekeeping gene could be amplified and was also cost-effective, with a nonlaborious protocol. Factors such as sensitivity, cost, and labor will help guide the selection of the most appropriate method for the needs of each laboratory.
Given the rise in the incidence of invasive fungal infections (IFIs) and the expanding spectrum of fungal pathogens, early and accurate identification of the causative microorganisms in formalin-fixed paraffin-embedded (FFPE) tissue is essential (20). Tissue samples collected and processed for pathological diagnosis represent a unique source of archived and morphologically defined disease-specific biological material (24). Histopathologic examination remains one of the major diagnostic tools in mycology because it permits rapid, presumptive identification of fungal infections. In recent years, however, there have been cases with discrepant histologic and culture results at final diagnosis; such discrepancies could lead to unnecessary pharmaceutical exposure and/or inappropriate treatment (17, 24).
Recent efforts to improve the sensitivity and specificity of diagnostic tests have focused on culture-independent methods, in particular, nucleic acid-based methods, such as PCR assays. PCR-based detection of fungal DNA sequences can be rapid, sensitive, and specific and can be applied to fresh and FFPE tissues (16). The majority of fungal assays target multicopy loci, in particular, the ribosomal DNA (rDNA) genes (18S, 28S, and 5.8S) and the intervening internal transcribed spacer (ITS) regions (ITS1 and ITS2) in order to maximize the yield of amplified DNA and allow high specificity (9).
Several protocols have been described for the extraction of DNA from fresh tissue, blood, and cells in cultures, but extraction from FFPE tissues is difficult because the material is frequently scarce and degraded and often contains remnants of either substances that inhibit the amplification reaction or chemicals, such as formalin, that inhibit the proteinase K used in the DNA extraction procedure. In general, FFPE tissue requires special protocols in order to extract small amounts of DNA suitable for amplification (6, 7, 10, 18).
In this work, we evaluated five commercial kits for the extraction of high-quality DNA from FFPE tissues that can be applied in molecular studies. To the best of our knowledge, three of the five protocols have not been previously evaluated in the context of extracting fungal DNA. After DNA extraction, the subsequent molecular analyses included two housekeeping gene PCR assays and three different panfungal PCR assays, followed by sequencing of the DNA fragments obtained. The protocols were assessed for time spent in performing the procedure, quality of DNA detection, and efficiency of fungal-DNA detection.
MATERIALS AND METHODS
Eighty-one archived FFPE tissue samples were examined. The samples came from the collections of the Mycotic Diseases Branch (n = 46) and the Infectious Diseases Pathology Branch (n = 29), Centers for Disease Control and Prevention (CDC), and from the Department of Pathology, University of Alabama at Birmingham (n = 6). The specimens included 51 human cases (Table 1), 24 human mock tissues (Table 2), and 6 animal cases.
TABLE 1.
Results of histopathology, PCR (DNA extracted with Takara Dexpat), and DNA sequence analysis of 51 FFPE human tissues
| Sample | Tissue site (type of specimen) | Clinical diagnosis | Histopathology (GMS)a | PCR resultsb |
Result of sequencing | |
|---|---|---|---|---|---|---|
| HKG | ITS3-4 | |||||
| 1 | Lung | Aspergillosis | Sparse | + | + | Aspergillus flavus |
| 2 | Lung | Blastomycosis | Abundant | + | + | B. dermatitidis |
| 3 | Sinus | Aspergillosis | Sparse | − | − | |
| 4 | Prostate | Aspergillosis | Medium | + | + | A. flavus |
| 5 | Unknown | Zygomycosis/Aspergillosis | Sparse | + | + | Aspergillus niger |
| 6 | Nasal cartilage | Zygomycosis | Sparse | − | − | |
| 7 | Sinus | Aspergillosis | Medium | + | + | Aspergillus fumigatus |
| 8 | Sinus | Aspergillosis | Abundant | + | + | A. fumigatus |
| 9 | Nasal septum | Aspergillosis | Abundant | − | − | |
| 10 | Orbital tissue | Aspergillosis | Abundant | + | + | A. niger |
| 11 | Turbinate | Aspergillosis | Abundant | + | + | A. flavus |
| 12 | Sinus | Aspergillosis | Sparse | − | − | |
| 13 | Sinus | Aspergillosis | Abundant | + | + | A. flavus |
| 14 | Colon | Histoplasmosis | Abundant | + | + | H. capsulatum |
| 15 | Unknown | Aspergillosis | Abundant | + | + | A. flavus |
| 16 | Unknown | Aspergillosis | Abundant | + | + | A. flavus |
| 17 | Lung | Blastomycosis | Abundant | + | + | B. dermatitidis |
| 18 | Bone marrow | Histoplasmosis | Abundant | + | + | H. capsulatum |
| 19 | Bone marrow | Histoplasmosis | Abundant | + | + | H. capsulatum |
| 20 | Bone marrow | Histoplasmosis | Abundant | + | + | H. capsulatum |
| 21 | Bone marrow | Histoplasmosis | Abundant | + | + | H. capsulatum |
| 22 | Bone marrow | Histoplasmosis | Abundant | + | + | H. capsulatum |
| 23 | Lung | Aspergillosis | Sparse | − | − | |
| 24 | Cardiac muscle | Zygomycosis | Sparse | − | − | |
| 25 | Nasal | Aspergillosis | Medium | + | + | A. flavus |
| 26 | Skin | Zygomycosis | Abundant | + | + | Apophysomyces elegans |
| 27 | Colon | Aspergillosis | Abundant | + | + | A. fumigatus |
| 28 | Colon | Aspergillosis | Abundant | + | + | A. fumigatus |
| 29 | Lung | Histoplasmosis | Abundant | + | + | H. capsulatum |
| 30 | Skin | Zygomycosis | Medium | + | − | |
| 31 | Lung | Zygomycosis | Abundant | + | + | Rhizopus oryzae/Amylomyces rouxii |
| 32 | Lung | Zygomycosis | Sparse | + | − | |
| 33 | Kidney | Zygomycosis | Abundant | + | + | Rhizopus oryzae/A. rouxii |
| 34 | Lung | Zygomycosis | Sparse | + | + | R. oryzae/R. microsporus/R. azygosporus |
| 35 | Lung | Aspergillosis | Sparse | + | − | |
| 36 | Lung | Aspergillosis | Sparse | + | − | |
| 37 | Kidney | Aspergillosis | Medium | + | − | |
| 38 | Unknown | Zygomycosis | Sparse | + | + | R. oryzae/A. rouxii |
| 39 | Unknown | Zygomycosis | Sparse | + | + | R. oryzae/A. rouxii |
| 40 | Unknown | Zygomycosis | Sparse | + | + | Saksenaea vasiformis |
| 41 | Nasal | Zygomycosis | Medium | + | + | C. echinulata/C. septata |
| 42 | Forearm | Zygomycosis. | Medium | + | + | R. oryzae/R. azygosporus/R. microsporus |
| 43 | Lung | Zygomycosis | Sparse | + | + | R. microsporus/R. azygosporus |
| 44 | Abdominal | Zygomycosis | Medium | + | + | R. oryzae/A. rouxii |
| 45 | Lung | Zygomycosis | Sparse | + | − | |
| 46 | Skin | Zygomycosis | Abundant | + | + | R. microsporus/R. azygosporus |
| 47 | Spleen | Zygomycosis | Medium | + | + | R.s oryzae/A. rouxii |
| 48 | Skin | Zygomycosis | Medium | + | + | R. oryzae/A. rouxii |
| 49 | Axilla | Zygomycosis | Medium | + | + | R. oryzae/A. rouxii |
| 50 | Eye | Histoplasmosis | Abundant | + | + | H. capsulatum |
| 51 | Lung | Aspergillosis | Medium | + | + | A. fumigatus |
Fungal structures are reported as abundant, medium and sparse.
HKG, housekeeping gene (human β-globin gene). +, positive; −, negative.
TABLE 2.
Results of PCR (DNA extracted with TaKaRa Dexpat) and DNA sequence analysis of 24 FFPE human mock tissues
| Organisma | Strain sourceb | PCR resultc | Result of sequencing |
|---|---|---|---|
| Mucor ramosissimus | NRRL 3042 | + | M. ramosissimus |
| Rhizopus oryzae | NRRL 28631 | + | R. oryzae/Amylomyces rouxii |
| Rhizopus azygosporus | NRRL 28627 | + | R. oryzae/R. microsporus/R. azygosporus/ |
| Rhizopus microspores | NRRL 28630 | + | R. oryzae/R. microsporus/R. azygosporus/ |
| Cunninghamella bertholletiae | NRRL 6436 | + | C. bertholletiae |
| Cunninghamella elegans | NRRL 28624 | + | C. elegans/C. bertholletiae |
| Absidia corymbifera | NRRL 28639 | + | A. corymbifera |
| Apophysomyces elegans | NRRL 28632 | + | A. elegans |
| Saksenaea vasiformis | NRRL 5251 | + | S. vasiformis |
| Conidiobolus incongruus | NRRL 28636 | + | C. incongruus |
| Cokeromyces recurvatus | NRRL A-18802 | + | C. recurvatus |
| Basidiobolus sp. | MDB B5743 | + | Basidiobolus sp. |
| Syncephalastrum racemosum | MDB B6101 | + | S. racemosum |
| Rhizomucor pusillus | MDB B5448 | + | R. pusillus/R. tauricus |
| Scedosporium apiospermum | MDB B5400 | + | S. apiospermum |
| Fusarium oxysporum | MDB B6908 | + | Fusarium sp. |
| Aspergillus fumigatus | MDB IFI 03-0127 | + | A. fumigatus |
| Aspergillus flavus | MDB IFI 01-0074 | + | A. flavus |
| Penicillium sp. | MDB/CDC | + | P. funiculosum/P. minioluteum |
| Penicillium sp. | MDB/CDC | + | P. meleagrinum/P. sumatrense |
| Penicillium sp. | MDB/CDC | + | P. purpurogenum/P. funiculosum P. minioluteum |
| H. capsulatum (yeast) | MDB/CDC | + | H. capsulatum |
| Candida albicans | MDB/CDC | + | C. albicans |
| B. dermatitidis (yeast) | MDB/CDC | + | B. dermatitidis |
All of the species listed have been reported to cause human and other animal infections.
NRRL numbers designate strains from the ARS Culture Collection, National Center for Agricultural Utilization Research, U.S. Department of Agriculture. MDB/CDC, Mycotic Diseases Branch, Centers for Disease Control and Prevention.
+, positive.
Twenty-four human mock tissues were generated from formalin-fixed and paraffin-embedded pellets of minced normal human tissues mixed with yeast or mold cells from isolates obtained from the Mycotic Diseases Branch and the U.S. Department of Agriculture ARS culture collections (Table 2). Briefly, yeasts were grown in Sabouraud dextrose agar (SDA) (Oxoid) for 48 h at 36°C, and molds were cultured for 5 to 7 days in SDA (Oxoid) at room temperature. Isolates were subcultured three times, and then cells were collected and suspended in 70% ethanol overnight. The ethanol was removed, and the cells were then resuspended in 10% formalin until preparation was performed (less than 24 h). The cells suspended in formalin were centrifuged at 3,000 rpm for 10 min, and the protocol for embedding the cells in paraffin was then performed using HistoGel, following the manufacturer's instructions (Richard-Allan Scientific [subsidiary of Thermo Fisher Scientific, Kalamazoo, MI]).
For each tissue block, 5-μm sections were cut from each specimen; a new sterile blade was used for each one (outer sections were discarded). Four or five scrolls from serial sections were placed in each of five Eppendorf tubes for the different DNA extraction protocols. A corresponding section from each tissue was used for Gomori methenamine silver (GMS) staining. The microtome (Leica RM2145; Leica Microsystems Inc., IL) was cleaned using DNAZap (Applied Biosystems/Ambion Inc., TX) each time before the paraffin-embedded tissue was cut and after the procedure.
DNA extraction protocols. (i) Protocol 1: QIAamp DNA FFPE Tissue Kit (Qiagen; catalog no. 56404).
Nucleic acids were extracted according to the manufacturer's instructions with three modifications: (a) After incubation and washing with xylene and ethanol, the tube was incubated at room temperature (15 to 25°C) for 1 h. (b) The pellet was digested with ATL buffer (Qiagen) and proteinase K at 56°C for 2 h. (c) After proteinase K treatment, the pellet was incubated with recombinant lyticase (L4276; Sigma-Aldrich Corporation, St. Louis, MO; 2 U/100 μl solution) for 45 min at 37°C.
(ii) Protocol 2: TaKaRa Dexpat (Takara Bio Inc.; catalog no. TAK 9091).
DNA extraction using TaKaRa Dexpat was performed as described by Paterson et al. (19) using recombinant lyticase (L4276; Sigma-Aldrich Corporation, St. Louis, MO; 2 U/100 μl solution). We omitted the step using 28 mM β-mercaptoethanol.
(iii) Protocol 3: PureLink Genomic DNA Mini Kit (Invitrogen; catalog no. K1820-00).
DNA was extracted according to the manufacturer's instructions with the following modifications: (a) One milliliter of xylene was added to an Eppendorf tube containing 4 or 5 scrolls, which was then centrifuged in an Eppendorf centrifuge at 14,000 rpm for 2 min. The supernatant was removed, and 1 ml of absolute ethanol was added, followed by centrifugation at 14,000 rpm for 3 min. The supernatant was discarded, and the samples were air dried for 1 h. (b) PureLink Genomic Digestion Buffer (180 μl) from the PureLink Genomic DNA Mini Kit (Invitrogen) and proteinase K (final concentration, 2 mg/ml; Invitrogen) were added and incubated at 55°C for 2 h and then at 90°C for 1 h. (c) Recombinant lyticase (L4276; Sigma-Aldrich Corporation, St. Louis, MO; 2 U/100 μl solution) was added and incubated for 45 min at 37°C. After the DNA was cooled at room temperature, it was extracted using the PureLink Genomic DNA Mini Kit (Invitrogen) based on binding of the DNA to silica columns, in accordance with the manufacturer's instructions.
(iv) Protocol 4: WaxFree DNA (TrimGen Genetic Diagnostics; catalog no. WF.50).
DNA was extracted according to the manufacturer's instructions with the following modifications: (a) Incubation with recombinant lyticase (L4276; Sigma-Aldrich Corporation, St. Louis, MO; 2-U/100 μl solution) for 45 min at 37°C. (b) Ethanol precipitation of the supernatant after centrifugation at 425 × g for 2 min in the Waxfree DNA Kit's WR filter.
(v) Protocol 5: QuickExtract FFPE DNA Extraction Kit (Epicenter Biotechnologies; catalog no. QEF81805).
DNA was extracted according to the manufacturer's instructions with the following modifications: (a) QuickExtract FFPE DNA Extraction Solution (150 μl) was used. (b) Incubation with recombinant lyticase (L4276; Sigma-Aldrich Corporation, St. Louis, MO; 2 U/100 μl solution) for 45 min at 37°C. (c) Centrifugation in an Eppendorf centrifuge at 14,000 rpm for 10 min and ethanol precipitation of the supernatant.
PCR amplification and DNA sequencing.
In order to confirm the presence or absence of amplifiable DNA or the possible presence of PCR inhibitors in the human and animal samples, fragments of two housekeeping genes, the human β-globin gene for human tissue (260 bp) and the mouse actin gene for mouse tissue (450 bp), were amplified by PCR as described by Bialek et al. (references 5 and 6, respectively), the only modification being the use of MgCl2 at 2.5 mM.
DNA extracted from fungal elements was detected with panfungal PCRs using ITS1, ITS3, or ITS5 forward primer and ITS4 universal reverse primer, which amplify the ITS1 and/or ITS2 regions of fungal rDNA genes (27). The PCR mixture (50 μl) included 10 μl of DNA, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 1.5 U of Taq polymerase (Roche Diagnostics, Indianapolis, IN), and 0.2 μM each primer. The amplification program for the ITS3 and ITS4 primers included an initial denaturation at 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 51°C for 1 min, and extension at 72°C for 1 min and then once at 72°C for 10 min. The primers ITS3 and ITS4 amplify a fragment of between 300 and 400 bp. For PCR using the ITS1/ITS5 and ITS4 primers, the conditions included an initial denaturation at 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 53°C for 30 s, and extension at 72°C for 90 s and then once at 72°C for 10 min. A fragment between 400 and 600 bp was amplified when primers ITS1/ITS5 and ITS4 were used. All PCRs were run in a PCR Peltier Thermal Cycler PT100 (MJ Research, Waltham, MA). The PCR products were visualized on a UV transilluminator after electrophoresis on 2% agarose gels (Sigma) using ethidium bromide at 0.5 μg/ml.
All PCR products were cleaned with ExoSap-It (USB Corporation, Cleveland, OH) following the manufacturer's instructions. The DNA sequencing was done at the DFBMD Genomics Unit, CDC, using an ABI Prism 3730 Genetic Analyzer (Applied Biosystems). The sequences obtained were edited and aligned using Sequencher version 4.8 software (Gene Codes Corp., Ann Arbor, MI). A homology search of all sequences was carried out using BLASTn (National Center for Biotechnology Information, Washington, DC).
RESULTS
Comparison of the five protocols.
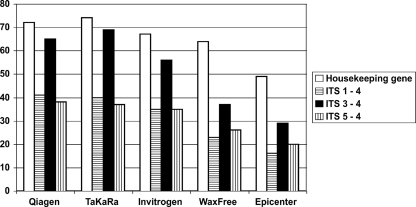
Of the 81 DNAs extracted from FFPE tissue samples, the numbers that were positive after the human β-globin or mouse actin assay (housekeeping gene PCR) were 72 (89%), 74 (91%), 67 (83%), 64 (79%), and 49 (60%), respectively, for the QIAamp DNA FFPE Tissue Kit, TaKaRa Dexpat Kit, Invitrogen PureLink Genomic DNA Mini Kit, TrimGen WaxFree DNA Kit, and Epicenter QuickExtract FFPE DNA Extraction Kit protocols (Fig. 1, white bars); the numbers of samples in which the housekeeping gene could not be amplified (demonstrating inhibition of PCR) were 9 (11%), 7 (8.6%), 14 (17.3%), 17 (21%), and 32 (40.8%), respectively. The proportions of DNA samples with positive housekeeping gene PCR for which a panfungal PCR was also positive were 65/72 (90.3%), 69/74 (93.2%), 56/67 (83.6%), 37/64 (57.8%), and 29/49 (59.2%) when the ITS3-ITS4 primers were used (Fig. 1, black bars); the proportions when the other two primer pairs were used were consistently lower, namely, 41/72, 40/74, 35/67, 23/64, and 16/49 for the ITS1-ITS4 primers and 38/72, 37/74, 35/67, 26/64, and 20/49 for the ITS5-ITS4 primers (Fig. 1, hatched bars). Of the 51 samples from human patients, 43 (84.3%), 44 (86.3%), 38 (74.5%), 37 (72.5%), and 27 (53%) were positive by PCR for the human β-globin locus, and of those, 36 (83.7%), 39 (88.6%), 28 (73.7%), 20 (54%), and 15 (55%) were also positive in the panfungal PCR using primers ITS3-ITS4. Of the 30 additional samples, i.e., of the 24 mock human and 6 animal tissues, 29 (including all animal tissues), 30 (all tissues), 29 (including all animal tissues), 27, and 22 were positive for the housekeeping gene PCR when Qiagen, Takara, Invitrogen, TrimGen, and Epicenter kits were used, respectively; of those, 29 (all), 30 (all), 28, 16, and 14 were also positive for the panfungal PCR using primers ITS3-ITS4.
FIG. 1.
Comparison of DNA amplifications using 5 extraction protocols and 3 panfungal PCR primer pairs in 81 FFPE tissues.
Results for FFPE tissues found to have molds or yeasts by histopathology.
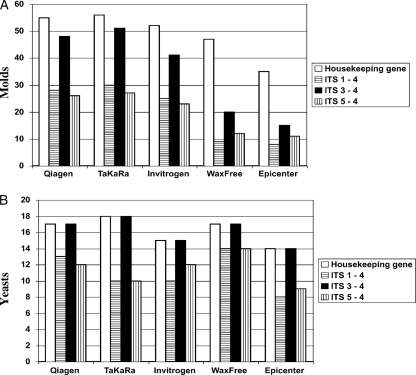
We separated the 81 FFPE tissues used in this study into those containing molds (n = 63) and yeasts (n = 18) and compared the results of housekeeping gene PCR and all panfungal PCRs with the 5 different DNA extraction protocols (Fig. 2A and B). When only FFPE tissues that contained molds (n = 63) were considered, the results were very similar to the full set of results shown in Fig. 1. The housekeeping gene PCR was positive in 55 DNAs extracted with the QIAamp DNA FFPE Tissue Kit, 56 for TaKaRa Dexpat, 52 for the PureLink Genomic DNA Mini Kit, 47 for WaxFree DNA, and 35 for the QuickExtract FFPE DNA Extraction Kit (Fig. 2A). In contrast, for FFPE tissues containing yeast (n = 18), all DNA extraction protocols gave similar results, ranging from 15 of 18 to 18 of 18 samples demonstrating an ITS amplicon (Fig. 2B). The panfungal PCR with primers ITS3 and ITS4 again gave the best amplification of DNA from FFPE tissues.
FIG. 2.
Comparison of 5 protocols for DNA extraction in 63 FFPE tissues containing molds (A) and in 18 FFPE tissues containing yeast (B). Protocols and primers are as in Fig. 1.
Identification of fungi.
In all FFPE tissues in this study from which fungal DNA could be amplified by a panfungal PCR, molecular identification by direct ITS sequencing was possible (Tables 1 and 2). In FFPE tissues that contained mucormycetes, the identification was to the genus level.
Other considerations.
We also compared the five protocols in terms of time spent on the procedure, quality of DNA detection (calculated as the number of housekeeping gene PCR positives divided by the total number of samples tested), and efficiency of fungal-DNA detection (calculated as the number of PCR positives reported by a panfungal PCR assay divided by the number of housekeeping gene PCR positives) (Table 3). The time spent on the procedure was 2.5 to 6 h, the price (kit only) ranged from $42 to $180, the quality of DNA detection ranged from 60% to 92%, and the efficiency of fungal-DNA detection ranged from 58% to 94% for the different protocols evaluated (Table 3). The efficiencies of fungal-DNA detection were clearly better for tissues that contained yeasts than for those containing molds (Fig. 2A and B). In fact, for yeasts, the fungal detection efficiency was consistently 100% for the panfungal PCR using the ITS3-ITS4 primers in all 5 protocols, and the DNA detection quality was best for the TaKaRa Dexpat protocol.
TABLE 3.
Comparison of 5 protocols for DNA extraction in 81 FFPE tissues in terms of time, quality of DNA detection, and efficiency of PCR
| Method | Time (h:s) | DNA quality (%)a | PCR efficiency (%)b |
|---|---|---|---|
| QIAamp DNA FFPE Tissue Kit (Qiagen) | 6:00 | 89 (72/81) | 90.3 (65/72) |
| TaKaRa Dexpat (Takara Bio Inc.) | 2:30 | 91.4 (74/81) | 93.2 (69/74) |
| PureLink Genomic DNA Mini Kit (Invitrogen) | 6:00 | 83 (67/81) | 83.6 (56/67) |
| WaxFree DNA (TrimGen Genetic Diagnostics) | 5:00 | 79 (64/81) | 57.8 (37/64) |
| QuickExtract FFPE DNA Extraction Kit (Epicenter Biotechnologies) | 3:50 | 60.5 (49/81) | 59.2 (29/49) |
Quality of DNA detection (calculated as the number of housekeeping genes PCR positive divided by the total number of samples tested).
Efficiency of detection of fungal DNA (calculated as the number of samples positive by any panfungal PCR assay divided by the number of samples housekeeping gene PCR positive).
DISCUSSION
Rapid and accurate identification to the genus and species levels of fungal pathogens in infected tissues is crucial for correct management of fungal infections. In many cases, a fungal infection is diagnosed only retrospectively in FFPE material that was never submitted for fungus culture. In other cases, viable fungi cannot be recovered from tissue submitted for mycology culture. The advent of novel antifungal therapies that have varying effects among different fungal agents has necessitated further identification in tissue beyond a simple determination that fungal elements are present. Several immunohistochemistry protocols and reagents are available to detect and identify certain groups of fungi, such as aspergilli and mucormycetes, but not to identify them to the genus and/or species level (14, 15). The ability to extract, detect, and identify fungal DNA in FFPE tissue has represented a major advance in fungal diagnostics to fill this gap, and a number of research studies in this area have been reported (4, 5, 6, 18).
It has become very important to have protocols for DNA extraction from FFPE tissues that are efficient and reproducible and that also yield DNA of high molecular weight with low levels of fragmentation and high quality. Isolating high-quality fungal DNA from FFPE tissue can be difficult, because only minimal quantities of intact DNA may be present in the sample. While routine formalin fixation preserves the tissue morphology, the process can cause the formation of protein-DNA cross-links, limiting the analysis of nucleic acids by reducing the quantity and size of amplified products compared to those obtained from fresh or frozen tissues (12, 21, 28). Furthermore, the success of PCR from preserved tissue can vary with the type of fixative, fixation or storage time, temperature, and PCR conditions. Isolating DNA from FFPE tissues can also be technically challenging, because PCR inhibitors may be present (11). In this study, we evaluated 81 FFPE tissues in which fungal elements could be detected with the GMS stain, using five different commercial DNA extraction kits with some modifications and three panfungal PCR assays, followed by DNA sequencing. Three of the five commercially available kits tested (Invitrogen, TrimGen, and Epicenter) have not been used before for extracting fungal DNA from FFPE tissues. Our results indicate that two of the five DNA extraction kits (TaKaRa and Qiagen) showed similar and promising results. However, DNA extraction with the TaKaRa kit followed by amplification of the DNA using panfungal PCR with ITS3 and ITS4 primers provide a highly sensitive and useful tool for the detection of a wide range of fungi. The validity and clinical applicability of the assays were confirmed by testing 24 human mock tissues infected with a wide variety of pathogens. We correctly identified all of them using the same methodology (Table 2). Our 5 modified DNA extraction protocols allowed amplification of a housekeeping gene (human β-globin or mouse actin) in different numbers of the 81 FFPE tissue samples, ranging from 49 (Epicenter) to 74 (TaKaRa). The TaKaRa and Qiagen methods proved to be the most efficient of the five protocols (Fig. 1).
Bialek and his group described a GAPDH (glyceraldehyde-3-phosphate dehydrogenase) nested PCR for amplification of human DNA in FFPE tissues and reported the quality of DNA as 62% (4) and 79.4% (3) using the QIAamp tissue kit (Qiagen) with their modifications for DNA extraction (3, 4). Two years later, the same group reported an improvement in the quality of the human DNA extracted (92.3%) using the human β-globin gene as a housekeeping gene for the PCR (4). Similarly, Paterson et al. (18), using the TaKaRa Dexpat kit (Takara Bio Inc.), reported the quality of DNA extracted as up to 93%. We found similar results when using the TaKaRa kit (91.4%) and the QIAamp kit (89%) with our modifications and amplifying a human β-globin housekeeping gene fragment. There was still some failure in amplifying human DNA from some FFPE tissues, and this is a well-documented effect of the formalin fixation process (11, 12).
The ITS3-ITS4 primer pair, which amplifies a 300- to 400-bp fragment of the ITS2 region of the rDNA gene, provided the best result of the three panfungal assays tested in this study. In samples where a housekeeping gene could be amplified, we could obtain a PCR product in up to 69 of 74 samples (93.2%; Takara) or 65 of 72 samples (90.3%; Qiagen). In contrast, the panfungal PCR using either ITS1-ITS4 or ITS5-ITS4 primers, which amplify a larger fragment (400 to 600 bp) of the ITS1 and ITS2 rDNA region, was never positive in more than 57% of the samples with a positive housekeeping gene PCR (Fig. 1). A likely reason for the lower yield is the length of intact DNA that is needed for amplification, since the longer amplicons are more difficult to achieve when DNA is highly fragmented or cross-linked, e.g., during formalin fixation. Other short DNA targets, such as the ∼300-bp D2 region of the large ribosomal subunit, may also be suitable targets for identification of fungal DNA in tissue, although we did not test alternative targets in this study. We found that in some samples with a report of fungal elements by GMS staining and that were housekeeping gene PCR positive, all PCR assays failed to amplify fungal DNA (Fig. 1). This might have been due to undetectable amounts of fungal DNA in the total volume extracted or to mutations in the ribosomal ITS region(s) leading to a lack of primer binding sites. Amplifiable human DNA detected by a housekeeping gene PCR does not necessarily indicate the presence of sufficient amounts of amplifiable fungal DNA. Other authors have also reported failure in amplifying fungal DNA in spite of a positive histopathological report and positive amplification of a human housekeeping gene (2, 4, 6, 18).
The final identification of the pathogens was performed using BLAST searches of the GenBank database. Only the nucleotide sequences of type or reference strains in the GenBank database were considered for identification purposes. When we used the BLAST algorithm to align and compare the sequences obtained via ITS3-ITS4 primers with the reference sequences, we found that the maximal level of identity (MLI) was equal to or higher than 98%, but we were not able to identify fungi to the species level in FFPE tissues that contained mucormycetes, Fusarium spp., and some Penicillium spp. Similar problems have been reported in the literature, but this limitation is more common in the molecular identification of molds than in yeasts, even when specific genes and specific PCRs are used (1, 6, 8, 13, 16, 22, 23, 25, 26). We had no cases of FFPE tissues containing black molds. Further studies are needed to see how well the extraction methods perform for this group of fungi.
The recovery of DNA from FFPE tissue that contained yeasts provided similar results with all five DNA extraction protocols (Fig. 2B). However, the efficiency in amplifying a housekeeping gene was best with the TaKaRa kit, followed by Qiagen and TrimGen. We conclude that extracting DNA from FFPE tissue containing yeasts, including the yeast phase of the dimorphic pathogens Blastomyces dermatitidis and Histoplasma capsulatum, is not particularly difficult, and laboratories can choose among the different techniques with more confidence.
Bialek and coauthors, using specific nested PCR, showed an efficiency for the detection of fungal DNA of up to 90% when FFPE tissues contained yeasts and up to 58% when tissues contained molds (4, 6). Our results obtained with the 63 FFPE tissues that contained molds (Fig. 2A) suggest that three protocols, Qiagen, TaKaRa, and Invitrogen, provided better recovery of mold DNA when ITS3 and ITS4 primers were used for the PCR. It is important to note that all of our protocols included a requirement for the use of recombinant lyticase. This step is mandatory in tissues containing molds to ensure that the hyphal mat is dissolved and fungal DNA is released. When lyticase is omitted, no fungal DNA can be recovered (unpublished observation). The use of recombinant lyticase ensures that no exogenous fungal DNA is inadvertently added during the extraction procedure. Paterson et al. (19), using Aspergillus conidia, earlier reported that recombinant lyticase improved DNA extraction when the TaKaRa Dexpat kit (Takara Bio Inc.) and the QIAamp DNA Mini Kit (Qiagen) were used.
In conclusion, although molecular identification from FFPE tissues remains difficult, this study has demonstrated that fungal-DNA extraction with protocols including the use of recombinant lyticase was possible for up to 91% of cases and that ITS2 sequencing can be a useful tool in the identification of a wide variety of clinically significant pathogens. After comparing the quality of DNA detection, the efficiency of fungal DNA detection, and the time spent in the procedure, we found that the best of the five DNA extraction protocols were TaKaRa and Qiagen, and we recommend a panfungal PCR using ITS3 and ITS4 primers.
Acknowledgments
C.M.-C. was supported in part by an International Travel Fellowship from the American Society for Microbiology (ASM) and the Oak Ridge Institute for Science and Education (ORISE) during his time at the CDC. This research was supported in part by an appointment (S.R.) to the Emerging Infectious Diseases (EID) Fellowship Program administered by the Association of Public Health Laboratories (APHL) and funded by the CDC.
We thank the DFBMD Genomics Unit for performing the DNA sequencing. We thank Invitrogen USA for donating kits for evaluation, Mark Lindsley for technical assistance, and David Warnock for supporting this project.
The use of product names does not imply their endorsement by the U.S. Department of Health and Human Services. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 14 April 2010.
REFERENCES
- 1.Alvarez, E., D. A. Sutton, J. Cano, A. W. Fothergill, A. Stchgel, M. G. Rinaldi, and J. Guarro. 2009. Spectrum of Zygomycetes species identified in clinically significant specimens in the United States. J. Clin. Microbiol. 47:1650-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aviel-Ronen, S., C. Q. Zhu, B. P. Coe, N. Liu, S. K. Watson, W. L. Lam, and M. S. Tsao. 2006. Large fragment Bst DNA polymerase for whole genome amplification of DNA from formalin-fixed paraffin-embedded tissues. BMC Genomics 7:312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bialek, R., A. Cascante, T. Hermann, C. Aepinus, V. Shearn-Bochsler, and A. Legendre. 2003. Nested PCR assay for detection of Blastomyces dermatitidis DNA in paraffin-embedded canine tissue. J. Clin. Microbiol. 41:205-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bialek, R., A. Feucht, C. Aepinus, G. Just-Nübling, V. J. Robertson, J. Knobloch, and R. Hohle. 2002. Evaluation of two nested PCR assays for detection of Histoplasma capsulatum DNA in human tissue. J. Clin. Microbiol. 40:1644-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bialek, R., J. Fischer, A. Feucht, L. Najvar, K. Dietz, J. Knobloch, and J. R. Graybill. 2001. Diagnosis and monitoring of murine histoplasmosis by a nested PCR assay. J. Clin. Microbiol. 39:1506-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bialek, R., F. Konrad, J. Kern, C. Aepinus, L. Cecenas, G. M. Gonzalez, G. Just-Nubling, B. Willinger, E. Presterl, C. Lass-Florl, and V. Rickerts. 2005. PCR based identification and discrimination of agents of mucormycosis and aspergillosis in paraffin wax embedded tissue. J. Clin. Pathol. 58:1180-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, W., M. Hashibe, J. Y. Rao, H. Morgenstern, and Z. F. Zhang. 2003. Comparison of methods for DNA extraction from paraffin-embedded tissues and buccal cells. Cancer Detect. Prev. 27:397-404. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Y. C., J. D. Eisner, M. M. Kattar, L. Rassoulian-Barrett, K. Lafe, S. L. Yarfitz, A. P. Limaye, and B. T. Cookson. 2000. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of rRNA genes. J. Clin. Microbiol. 38:2302-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2008. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing: approved guideline. MM18-A. Clinical and Laboratory Standards Institute. Wayne, PA.
- 10.Coura, R., J. C. Prolla, L. Meurer, and P. Ashton-Prolla. 2005. An alternative protocol for DNA extraction from formalin fixed and paraffin wax embedded tissue. J. Clin. Pathol. 58:894-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank, T., S. M. Svoboda-Newman, and E. D. His. 1996. Comparison of methods for extracting DNA from formalin-fixed paraffin sections for nonisotopic PCR. Diagn. Mol. Pathol. 5:220-224. [DOI] [PubMed] [Google Scholar]
- 12.Greer, S. E., S. L. Paterson, N. B. Kiviat, and M. M. Manos. 1991. PCR amplification from paraffin-embedded tissues. Effects of fixative and fixation time. Am. J. Clin. Pathol. 95:117-124. [DOI] [PubMed] [Google Scholar]
- 13.Hata, D. J., S. P. Buckwalter, B. S. Pritt, G. D. Roberts, and N. L. Wengenack. 2008. Real-time PCR method for detection of Zygomycetes. J. Clin. Microbiol. 46:2353-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen, H. E., B. Aalbaek, P. Lind, and H. V. Krogh. 1996. Immunohistochemical diagnosis of systemic bovine zygomycosis by murine monoclonal antibodies. Vet. Pathol. 33:176-183. [DOI] [PubMed] [Google Scholar]
- 15.Jensen, H. E., B. Halbaek, P. Lind, H. V. Krogh, and P. L. Frandsen. 1996. Development of murine monoclonal antibodies for the immunohistochemical diagnosis of systemic bovine aspergillosis. J. Vet. Diagn. Invest. 8:68-75. [DOI] [PubMed] [Google Scholar]
- 16.Lau, A., S. Chen, T. Sorrell, D. Carter, R. Malik, P. Martin, and C. Halliday. 2007. Development and clinical application of a panfungal PCR assay to detect and identify fungal DNA in tissue specimens. J. Clin. Microbiol. 45:380-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez, F. A., R. S. Crowley, L. Wastila, H. A. Valantine, and J. S. Remington. 1998. Scedosporium apiospermum (Pseudallescheria boydii) infection in a heart transplant recipient: a case of mistaken identity. J. Heart Lung Transplant. 17:321-324. [PubMed] [Google Scholar]
- 18.Paterson, P. J., S. Seaton, T. D. McHugh, J. McLaughlin, M. Potter, H. G. Prentice, and C. C. Kibbler. 2006. Validation and clinical application of molecular methods for the identification of molds in tissue. Clin. Infect. Dis. 42:51-56. [DOI] [PubMed] [Google Scholar]
- 19.Paterson, P. J., S. Seaton, J. McLaughlin, and C. C. Kibbler. 2003. Development of molecular methods for the identification of aspergillus and emerging moulds in paraffin wax embedded tissue sections. J. Clin. Pathol. Mol. Pathol. 56:368-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perfect, J. R., and W. A. Schell. 1996. The new fungal opportunists are coming. Clin. Infect. Dis. 22:S112-S118. [DOI] [PubMed] [Google Scholar]
- 21.Quach, N., M. F. Goodman, and D. Shibata. 2004. In vitro mutation artifacts after formalin fixation and error prone translesion synthesis during PCR. BCM Clin. Pathol. 12:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rickerts, V., G. Just-Nubling, F. Konrad, J. Kern, E. Lambrecht, A. Bohme, V. Jacobi, and R. Bialek. 2006. Diagnosis of invasive aspergillosis and mucormycosis in immunocompromised patients by seminested PCR assay of tissue samples. Eur. J. Clin. Microbiol. Infect. Dis. 25:8-13. [DOI] [PubMed] [Google Scholar]
- 23.Rickerts, V., S. Mousset, E. Lambrecht, K. Tintelnot, R. Schwerdtfeger, E. Presterl, V. Jacobi, G. Just-Nubling, and R. Bialek. 2007. Comparison of histopathological analysis, culture, and polymerase chain reaction assay to detect invasive mold infections from biopsy specimens. Clin. Infect. Dis. 44:1078-1083. [DOI] [PubMed] [Google Scholar]
- 24.Sangoi, A. R., W. M. Rogers, T. A. Longacre, J. G. Montoya, E. J. Baron, and N. Banaei. 2009. Challenges and pitfalls of morphologic identification of fungal infections in histologic and cytologic specimens: a ten-year retrospective review at a single institution. Am. J. Clin. Pathol. 131:364-375. [DOI] [PubMed] [Google Scholar]
- 25.Schwarz, P., S. Bretagne, J. C. Gantier, D. Garcia-Hermoso, O. Lortholary, F. Dromer, and E. Dannaoui. 2006. Molecular identification of Zygomycetes from culture and experimentally infected tissues. J. Clin. Microbiol. 44:340-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voigt, K., E. Cigelnik, and K. O'Donnell. 1999. Phylogeny and PCR identification of clinically important Zygomycetes based on nuclear ribosomal-DNA sequence data. J. Clin. Microbiol. 37:3957-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplication and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, CA.
- 28.Wu, L., N. Patten, C. T. Yamashiro, and B. Chui. 2002. Extraction and amplification of DNA from formalin-fixed, paraffin-embedded tissues. Appl. Immunohistochem. Mol. Morphol. 10:269-274. [DOI] [PubMed] [Google Scholar]