Abstract
Mycobacterium kansasii carrying IS1245, a highly prevalent insertion sequence among Mycobacterium avium isolates, was detected in a mixed culture of M. avium and M. kansasii. The insertion sequence was stable and able to transpose by a replicative mechanism in M. kansasii. These findings may have significant implications for molecular diagnosis and treatment outcome.
Mycobacterium avium and Mycobacterium kansasii are major human mycobacterial pathogens, and both can cause pulmonary infections in immunocompetent individuals, as well as disseminated infections in the immunocompromised (6). Coinfection with M. avium and M. kansasii has also been documented in HIV-positive patients (8, 15). Current treatment of M. avium and M. kansasii infections is based on long-term antibiotic therapy and relies on different drug combinations, justifying all efforts directed to the correct identification of clinical isolates. Insertion sequences (IS) are mobile genetic elements within mycobacteria that are often species specific, a property that can be exploited for diagnosis and epidemiological studies (4). The host range of the IS1245 element was considered to be limited to M. avium subsp. avium, M. avium subsp. hominissuis, and M. avium subsp. silvaticum (3, 9, 11-13, 17).
Detection of IS1245 by PCR was used for confirmation of the presence of M. avium in a mixed culture from a bone marrow specimen from an HIV-positive patient. Fifteen slow-growing colonies were recovered from the bone marrow primary culture by plating dilutions on Middlebrook 7H10 solid medium supplemented with oleic acid, albumin, catalase, and dextrose (7H10-OADC). Four colonies (88.1 to 88.4) were nonpigmented, slow-growing mycobacteria identified as M. avium by PCR-restriction enzyme analysis (PRA) using the 16S-23S rRNA internal transcribed spacer (ITS) sequence as the target (14). Eleven colonies (88.5 to 88.15) produced yellow pigment after exposure to light and were identified as M. kansasii by PRA-ITS (Fig. 1 A). For further characterization of these colonies, a 427-bp fragment from IS1245 was amplified by PCR (3). Unexpectedly, IS1245 amplicons were detected not only in the nonpigmented M. avium colonies but also in 8 out of 11 M. kansasii colonies (Fig. 1B). Amplicons generated with M. kansasii DNA were sequenced and showed 100% identity with the deposited IS1245 sequence (accession number L33879) (data not shown). Final identification of the 15 colonies to the species level was obtained by sequencing a 440-bp fragment of the 5′ 16S rRNA gene (Escherichia coli positions 54 to 510) (16). Colonies 88.1 through 88.4 showed 100% sequence similarity to the corresponding sequence of M. avium type strain ATCC 25291 (accession number EF521895). Colonies 88.5 through 88.15 showed 100% similarity to the corresponding sequence of M. kansasii type strain CIP 104589 (accession number AF547940). The two sequences differed at 12 positions (97.3% similarity).
FIG. 1.
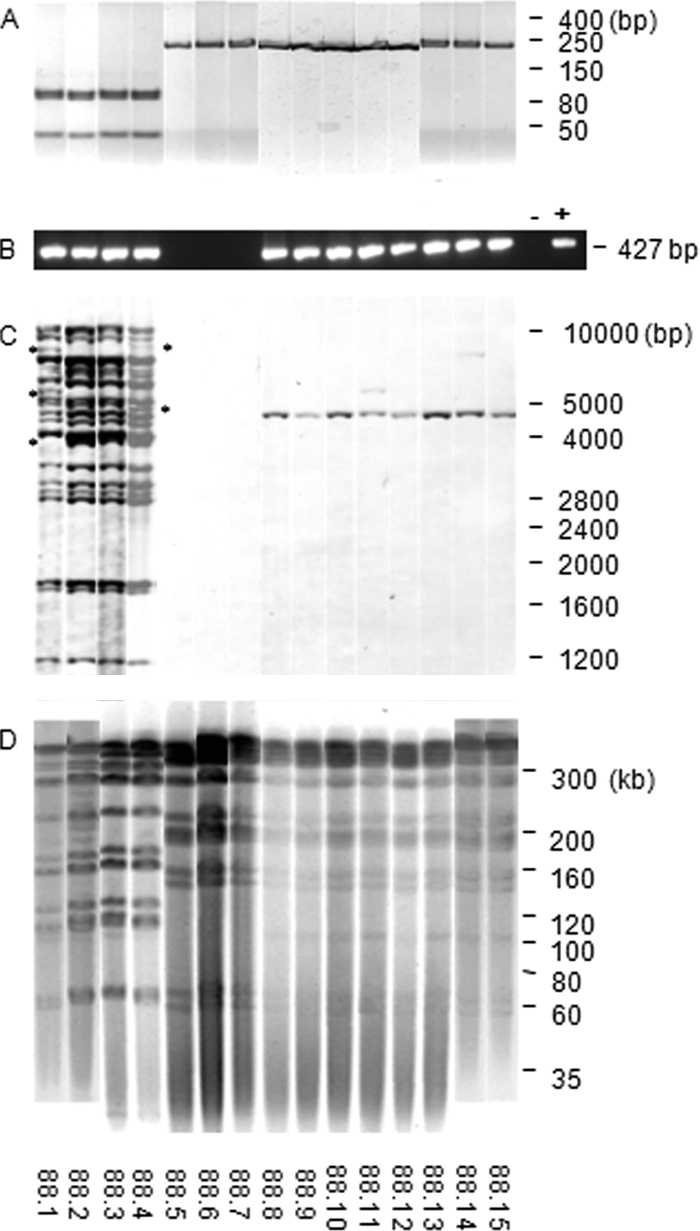
Molecular characterization of 15 single colonies isolated from the original M. avium-M. kansasii mixed culture by PRA-ITS (A), PCR-IS1245 (B), RFLP-IS1245 (C), and pulsed-field gel electrophoresis (D). Lanes: colonies 88.1 to 88.4, M. avium; colonies 88.5 to 88.15, M. kansasii; −, PCR negative control (water); +, PCR positive control (DNA from M. avium ATCC 25291T). Asterisks indicate RFLP-IS1245 band polymorphisms in M. avium colonies. On the right are molecular size markers.
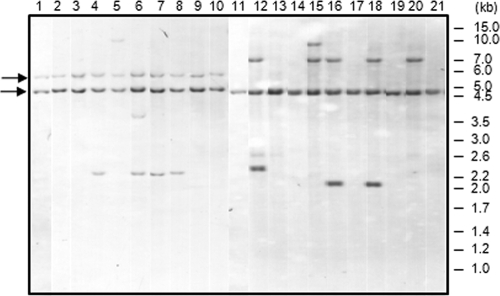
To confirm the presence of IS1245 copies in the eight colonies of M. kansasii, restriction fragment length polymorphism (RFLP) experiments were performed by using IS1245 as a probe (17). While the M. avium colonies (88.1 to 88.4) produced multiband RFLP patterns characteristic of M. avium subsp. hominissuis, the eight M. kansasii colonies that had produced amplicons by PCR-IS1425 (88.8 to 88.15) showed a single IS1245-hybridizing band of approximately 4,750 bp. Moreover, a second IS1245-hybridizing band was detected in two of these colonies (88.11 and 88.14). Furthermore, the three PCR-IS1245-negative colonies of M. kansasii (88.5 to 88.7) lacked IS1245 hybridization bands (Fig. 1C). Except for the presence of IS1245, pulsed-field gel electrophoresis patterns indicated that all of the 11 M. kansasii colonies were, in fact, the same strain (Fig. 1D).
The results obtained strongly suggest that M. kansasii acquired the IS1245 element from the M. avium strain in a horizontal DNA transfer event which occurred either within the patient or during specimen or culture storage. This hypothesis was reinforced by the fact that both species were isolated from a unique specimen collected from one patient and by the finding of several M. kansasii colonies (88.5 to 88.7) of the same strain devoid of this insertion sequence element in that specimen (Fig. 1B, C, and D). The presence of the IS1245 element in M. kansasii was not detected by the analysis of five additional M. avium-M. kansasii mixed cultures from different patients (data not shown), which agrees with the hypothesis that the event described here was the result of a particular horizontal DNA transfer episode.
Antimicrobial susceptibility testing was performed with three isolates using a microdilution method previously described (10). The drugs tested included streptomycin, isoniazid, ethambutol, rifampin, rifabutin, ciprofloxacin, amikacin, azithromycin, and clarithromycin. Interpretative criteria followed NCCLS recommendations (10). M. avium colony 88.3 was resistant to isoniazid (MIC, 1 μg/ml), ethambutol (MIC, 8 μg/ml), azithromycin (MIC, >8 μg/ml), and clarithromycin (MIC, >32 μg/ml). M. kansasii colonies 88.5 (IS1245 negative) and 88.8 (IS1245 positive) were susceptible to all of the drugs tested, showing that the acquisition of IS1245 did not change the pattern of susceptibility of this M. kansasii strain to the drugs tested.
In order to examine the stability of IS1245 in M. kansasii, two colonies (88.11 and 88.12) were grown in liquid Middlebrook 7H9-OADC on a shaker at 37°C until the optical density at 600 nm reached 0.6 to 0.8. Ten serial passages were then performed by diluting the cultures 1:10 in the same medium and incubating them again under the same conditions. After 10 passages, the cultures were added to solid 7H10-OADC by the spread plate method and 19 isolated colonies were analyzed by RFLP-IS1245. The original IS1245 hybridization bands were maintained in all of the colonies isolated after 10 passages, demonstrating the stability of this insertion sequence in M. kansasii, at least during short-term (4 months) in vitro processing. However, one to three new hybridization bands were visualized in 11 out of 19 M. kansasii colonies analyzed, pointing to the occurrence of insertion sequence transposition by a replicative mechanism (Fig. 2).
FIG. 2.
RFLP-IS1245 of colonies 88.11 and 88.12 and individual colonies isolated after 10 serial passages in 7H9-OADC liquid medium. Lanes: 1, original colony 88.11; 2 to 10, isolated colonies after subculture of colony 88.11; 11, original colony 88.12; 12 to 21, isolated colonies after subculture of colony 88.12. Black arrows indicate the hybridization bands present in the original colonies. On the right are molecular size markers.
Colonies of both species showed variations in RFLP-IS1245 patterns during this study. Besides the IS1245 transposition detected in M. kansasii, two or three RFLP-IS1245 band polymorphisms were also detected in the M. avium colonies (Fig. 1C). Recent IS1245 transposition events in M. avium have been observed in other studies with single colonies of the same isolate or isolates collected from individual patients over time (1, 3, 11, 12).
The IS1245 insertion sequence was initially considered to be species specific for M. avium, but additional studies have shown that this element is sporadically present in M. intracellulare, M. malmoense, M. scrofulaceum, and M. nonchromogenicum (2, 7, 9). To our knowledge, the results obtained in this study confirm, for the first time, the presence of one or more copies of IS1245 in a strain of M. kansasii by PCR, RFLP, and DNA sequencing.
As a consequence, the utilization of IS1245 as a genetic marker for the identification of M. avium and its distinction from M. kansasii should be done with caution and combined with other genetic markers. The absence of IS1245 copies in some M. avium strains must also be taken into account (5, 11, 13).
IS1245 transposition events such as those described here can produce genome plasticity by the interruption or deletion of genes, affecting biological properties, which deserves further investigation.
Acknowledgments
This study received financial support from the Foundation for Research Support of the State of Sao Paulo (FAPESP-06/01533-9). M.C.D.S.R. was the recipient of a fellowship from the National Council for Scientific and Technological Development (CNPq).
A. Leyva provided help with English editing.
Footnotes
Published ahead of print on 14 April 2010.
REFERENCES
- 1.Bauer, J., and A. B. Andersen. 1999. Stability of insertion sequence IS1245, a marker for differentiation of Mycobacterium avium strains. J. Clin. Microbiol. 37:442-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beggs, M. L., R. Stevanova, and K. D. Eisenach. 2000. Species identification of Mycobacterium avium complex isolates by a variety of molecular techniques. J. Clin. Microbiol. 38:508-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guerrero, C., C. Bernasconi, D. Burki, T. Bodmer, and A. Telenti. 1995. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J. Clin. Microbiol. 33:304-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guilhot, C., M. Jackson, and B. Gicquel. 1999. Mobile genetic elements and plasmids: tools for genetic studies, p. 17-37. In C. Ratledge and J. Dale (ed.), Mycobacteria: molecular biology and virulence. Blackwell Science Ltd., Oxford, United Kingdom.
- 5.Inagaki, T., K. Nishimori, T. Yagi, K. Ichikawa, M. Moriyama, T. Nakagawa, T. Shibayama, K. Uchiya, T. Nikai, and K. Ogawa. 2009. Comparison of a variable-number tandem-repeat (VNTR) method for typing Mycobacterium avium with mycobacterial interspersed repetitive-unit-VNTR and IS1245 restriction fragment length polymorphism typing. J. Clin. Microbiol. 47:2156-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katoch, V. M. 2004. Infections due to non-tuberculous mycobacteria (NTM). Indian J. Med. Res. 120:290-304. [PubMed] [Google Scholar]
- 7.Keller, A. P., M. L. Beggs, B. Amthor, F. Bruns, P. Meissner, and W. H. Haas. 2002. Evidence of the presence of IS1245 and IS1311 or closely related insertion elements in nontuberculous mycobacteria outside of the Mycobacterium avium complex. J. Clin. Microbiol. 40:1869-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massenkeil, G., M. Opravil, M. Salfinger, A. von Graevenitz, and R. Lüthy. 1992. Disseminated coinfection with Mycobacterium avium complex and Mycobacterium kansasii in a patient with AIDS and liver abscess. Clin. Infect. Dis. 14:618-619. [DOI] [PubMed] [Google Scholar]
- 9.Mijs, W., P. de Haas, R. Rossau, T. Van der Laan, L. Rigouts, F. Portaels, and D. van Soolingen. 2002. Molecular evidence to support a proposal to reserve the designation Mycobacterium avium subsp. avium for bird-type isolates and ‘M. avium subsp. hominissuis’ for the human/porcine type of M. avium. Int. J. Syst. Evol. Microbiol. 52(Pt. 5):1505-1518. [DOI] [PubMed] [Google Scholar]
- 10.National Committee for Clinical Laboratory Standards. 2003. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard, NCCLS document M24-A. National Committee for Clinical Laboratory Standards, Wayne, PA. [PubMed]
- 11.Oliveira, R. S., M. P. Sircili, E. M. D. Oliveira, S. C. Balian, J. S. Ferreira-Neto, and S. C. Leão. 2003. Identification of Mycobacterium avium genotypes with distinctive traits by combination of IS1245-RFLP and restriction analysis of hsp65. J. Clin. Microbiol. 41:44-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pestel-Caron, M., and R. D. Arbeit. 1998. Characterization of IS1245 for strain typing of Mycobacterium avium. J. Clin. Microbiol. 36:1859-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritacco, V., K. Kremer, T. van der Laan, J. E. Pijnenburg, P. E. de Haas, and D. van Soolingen. 1998. Use of IS901 and IS1245 in RFLP typing of Mycobacterium avium complex: relatedness among serovar reference strains, human and animal isolates. Int. J. Tuberc. Lung Dis. 2:242-251. [PubMed] [Google Scholar]
- 14.Roth, A., U. Reischl, A. Streubel, L. Naumann, R. M. Kroppenstedt, M. Habicht, M. Fischer, and H. Mauch. 2000. Novel diagnostic algorithm for identification of mycobacteria using genus-specific amplification of the 16S-23S rRNA gene spacer and restriction endonucleases. J. Clin. Microbiol. 38:1094-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasaki, Y., F. Yamagishi, K. Suzuki, M. Saitoh, and M. Izumizaki. 1997. A case of AIDS with disseminated Mycobacterium kansasii infection in which Mycobacterium avium complex was also detected from his sputum repeatedly. Kekkaku 72:573-577. [PubMed] [Google Scholar]
- 16.Tortoli, E. 2003. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clin. Microbiol. Rev. 16:319-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Soolingen, D., J. Bauer, V. Ritacco, S. C. Leao, I. Pavlik, V. Vincent, N. Rastogi, A. Gori, T. Bodmer, C. Garzelli, and M. J. Garcia. 1998. IS1245 restriction fragment length polymorphism typing of Mycobacterium avium isolates: proposal for standardization. J. Clin. Microbiol. 36:3051-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]