Short abstract
Identifying quantitative trait loci in Arabidopsis by population and pedigree studies.
Abstract
Two recent studies in Arabidopsis have identified quantitative trait loci (QTLs) by population- association and family-based studies, respectively, providing further data on the genetic architecture of complex-trait variation in plants.
Genetic variation for complex traits determines fitness in natural environments, as well as productivity of the crops that sustain all human populations [1]. Mapping and cloning of quantitative trait loci (QTLs) has begun to identify the genes responsible for this variation [2], as well as the evolutionary factors that maintain quantitative variation in populations [3]. Central to our understanding is to elucidate the genetic architecture of complex traits, which incorporates both the magnitude and the frequency of QTL alleles in a population.
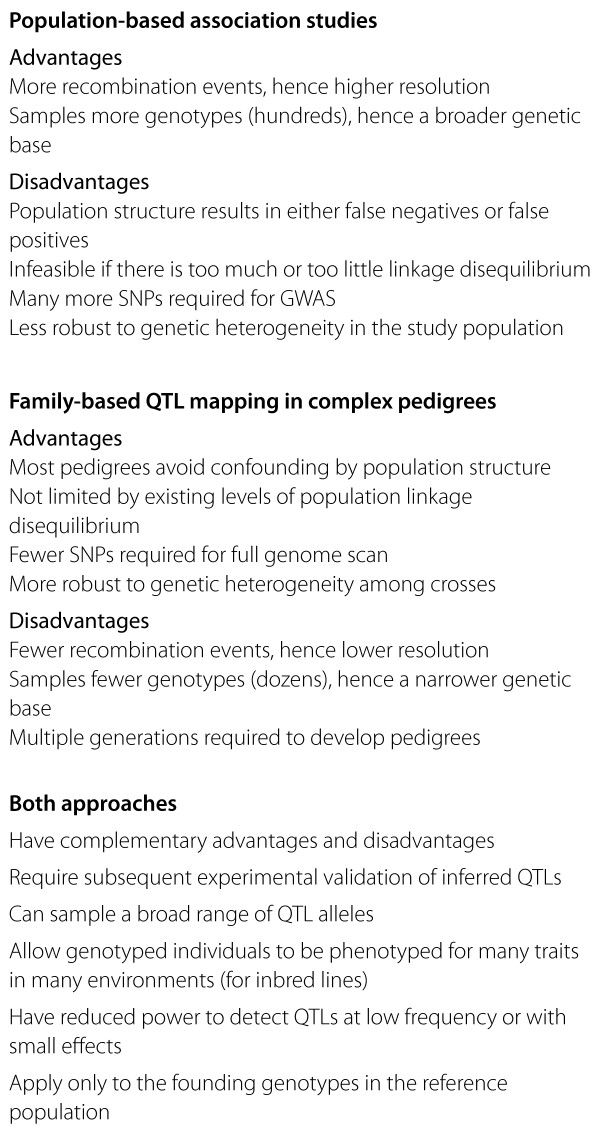
Two approaches have recently been applied to complex-trait analysis in plants, which both allow QTL identification in samples containing diverse genotypes. Population-based approaches such as genome-wide association studies (GWAS) use populations of unrelated individuals to examine genome-wide associations between single nucleotide polymorphisms (SNPs) and phenotypes. Alternatively, family-based QTL mapping can be applied to complex pedigrees from crosses among different founding genotypes. For Arabidopsis thaliana and most crop plants, inbred lines need be genotyped only once, enabling efficient and cost-effective phenotyping of many traits in multiple environments by a broad research community. Population- and family-based approaches have complementary advantages and disadvantages (Box 1), and together enable major advances in our understanding of quantitative trait variation. A recent paper in Nature by Atwell et al. [4] has taken a population-based approach to QTL association in a GWAS of some 200 inbred lines of Arabidopsis, while Kover et al. [5], writing in PLoS Genetics, take a family-based approach, describing a complex pedigree that can be used to fine-map QTLs in Arabidopsis.
Comparison of population-based and family-based approaches.
Population-based association studies
In plant populations, application of population-based association studies depends on the scale of linkage disequilibrium, which determines the degree to which molecular markers may be associated with the relevant phenotype. Optimal levels may allow resolution of QTLs to regions containing just a few genes. To resolve phenotypic effects among neighboring genes, GWAS take advantage of historical recombination events that have accumulated over thousands of generations in historical populations. However, it is difficult for association studies to identify QTLs that influence traits that are correlated with population structure, because many SNPs differ between populations. Failure to control for population structure results in false positives, whereas statistical methods to control for population structure, such as the mixed model, instead lead to false negatives.
The reasons for false positives and false negatives can be illustrated by a recent resequencing study [6] that examined nucleotide variation among 20 accessions of rice. Three historical lineages (indica, japonica, and aus) are differentiated by thousands of SNPs across the genome. Owing to their shared ancestry, members of each lineage share common SNP genotypes, that is, linkage disequilibrium among thousands of loci across the genome. This population structure occurs at neutral markers and at phenotypically important quantitative trait nucleotides (QTNs), which are shared by group members as a result of ecological and agricultural selection. Failure to correct for population structure causes false positives because many neutral SNPs are correlated with trait differences among groups. In contrast, correction for population structure adjusts for neutral SNP differences, but also causes false negatives by 'controlling away' the QTNs responsible for differences between structure groups. These complications of population structure can be avoided by more focused GWA studies that use a single historical population, as in most human studies. Alternatively, family-based complex pedigrees eliminate the confounding effects of population structure through controlled crosses.
Arabidopsis has excellent resources for population-based QTL studies. Atwell et al. [4] performed GWAS with around 200 lines scored for more than 200,000 SNPs, examining 107 phenotypes relating to flowering, development, plant defense, and physiological traits. Because of high levels of population structure they used mixed-model analyses [7], which control for relatedness among individuals at several levels, reducing spurious correlations between markers and phenotypes. Genetically simple traits such as pathogen resistance or ion concentrations were resolved clearly, showing the power of this approach. For quantitative traits the significant results are enriched near known candidate genes, but often give complex peaks encompassing many genes, without identifying a best candidate. In contrast to human association studies and results from family-based studies in maize (discussed below), individual QTLs with a large effect on phenotype (large-effect QTLs) are clearly evident in Arabidopsis. The authors also conclude that mixed-model analysis may not control for linkage disequilibrium arising from selection, as might be expected for ecologically and agriculturally important traits.
Genotyped populations for GWAS are being developed in plant species other than Arabidopsis, such as barley, maize and rice. In addition, targeted association studies in non-model organisms are able to combine sequence data from candidate genes with information on population structure based on a few thousand markers across the genome [8].
Family-based QTL mapping
Family-based QTL mapping in complex pedigrees has advantages and disadvantages that are complementary to those of population-based studies (see Box 1). Unlike GWAS, QTL resolution in family-based studies is unlikely to approach the single-gene level, as linkage analysis is based on recombinations accumulated over a few generations during pedigree development. However, most pedigrees avoid the confounding effects of population structure, and therefore escape the false positives and false negatives that can plague association studies.
In their family-based study, Kover et al. [5] used the Arabidopsis Multiparent Advanced Generation Inter-Cross (MAGIC) population. To develop this population, they crossed together 19 founding genotypes for four generations to increase the level of recombination, followed by six generations of self-pollination to develop 342 quasi-independent recombinant inbred lines. In comparison to population-based mapping, pedigree approaches can avoid complications of historical population structure, although QTLs cannot be resolved to regions of a few genes. Kover et al. [5] examined flowering time and other complex traits, and identified a number of QTLs near known candidate genes, including the flowering time genes FRIGIDA and FLOWERING LOCUS C, which also were evident in the GWAS of Atwell et al. [4].
In regard to crop plants, family-based complex pedigrees are particularly valuable in maize (Zea mays), which has high levels of outcrossing and a large effective population size. This results in very low linkage disequilibrium, which decays within hundreds of nucleotides in most populations. Using current technology, it is prohibitively expensive to score polymorphisms at this density, so GWAS remain challenging in maize. A different type of family breeding design has been used in maize compared with Arabidopsis to produce a complex pedigree known as the Nested Association Mapping (NAM) population, developed by a large collaboration among maize geneticists [9,10]. Twenty-five parents were each crossed to the fully sequenced B73 genotype, and 200 recombinant inbred lines were derived from each cross, giving 25 sets of lines, each set having a common parent.
A recent study [9,10] examining flowering time in nearly 1 million plants from around 5,000 NAM recombinant inbred lines found that the genetic architecture of flowering time was highly polygenic. Around 50 loci appeared to contribute to variation in flowering time, with many loci showing small, nearly additive effects. This is in striking contrast to Arabidopsis and rice, where large-effect QTLs have been found in many studies [2,4]. To some extent, this contrast may be less extreme than it initially seems. Large-effect flowering QTLs have been found in maize when researchers examine highly divergent parents, although QTL magnitude is sensitive to day length. Likewise, as sample sizes increase in Arabidopsis one anticipates that many small-effect flowering QTLs will be found. Nevertheless, these studies suggest that breeding system, effective population size, selective history, and population demography will influence the genetic architecture of complex traits. Combined population- and family-based QTL studies can begin to elucidate and explain these patterns of variation.
In summary, two complementary approaches to QTL identification are becoming available in model species and agriculturally important plants. Using genetically diverse founder populations, these approaches can elucidate the genetic architecture of complex traits, and estimate both the magnitude and frequency of QTL alleles.
Abbreviations
GWAS: genome-wide association study; NAM: Nested Association Mapping; QTL: quantitative trait locus; QTN: quantitative trait nucleotide; SNP: single nucleotide polymorphism.
Acknowledgements
I thank E Buckler, M Nordborg, and J Willis for comments on the manuscript. This work was supported by award R01-GM086496 from the National Institutes of Health and award EF-0723447 from the National Science Foundation.
References
- Mackay TFC, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nat Rev Genet. 2009;10:565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Aarts MGM, Bentsink L, Keurentjes JJB, Reymond M, Vreugdenhil D, Koornneef M. What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell. 2009;21:1877–1896. doi: 10.1105/tpc.109.068114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T, Willis JH, Goldstein DB. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat Rev Genet. 2007;8:845–856. doi: 10.1038/nrg2207. [DOI] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjalmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, Jiang R, Muliyati NW, Zhang X, Amer MA, Baxter I, Brachi B, Chory J, Dean C, Debieu M, de Meaux J, Ecker JR, Faure N, Kniskern JM, Jones JDG, Michael T, Nemri A, Roux F, Salt DE, Tang C. Genome-wide association study of 107 phenotypes in a common set of Arabidopsis thaliana inbred lines. Nature. 2010. doi:10.1038/nature08800. [DOI] [PMC free article] [PubMed]
- Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, Purugganan MD, Durrant C, Mott R. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000551. doi: 10.1371/journal.pgen.1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, Ulat VJ, Zeller G, Clark RM, Hoen DR, Bureau TE, Stokowski R, Ballinger DG, Frazer KA, Cox DR, Padhukasahasram B, Bustamante CD, Weigel D, Mackill DJ, Bruskiewich RM, Rätsch G, Buell CR, Leung H, Leach JE. Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc Natl Acad Sci USA. 2009;106:12273–12278. doi: 10.1073/pnas.0900992106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber AL, Briggs WH, Rucker J, Baltazar BM, de Jesus Sanchez-Gonzalez J, Feng P, Buckler ES, Doebley J. The genetic architecture of complex traits in teosinte (Zea mays ssp. parviglumis): New evidence from association mapping. Genetics. 2008;180:1221–1232. doi: 10.1534/genetics.108.090134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen MD, Kresovich S, Villeda HS, Bradbury P, Li H, Sun Q, Flint-Garcia S, Thornsberry J, Acharya C, Bottoms C, Brown P, Browne C, Eller M, Guill K, Harjes C, Kroon D, Lepak N, Mitchell SE, Peterson B, Pressoir G, Romero S, Oropeza Rosas M, Salvo S, Yates H, Hanson M, Jones E, Smith S, Glaubitz JC, Goodman M, Ware D. Genetic properties of the maize nested association mapping population. Science. 2009;325:737–740. doi: 10.1126/science.1174320. [DOI] [PubMed] [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, Goodman MM, Harjes C, Guill K, Kroon DE, Larsson S, Lepak NK, Li H, Mitchell SE, Pressoir G, Peiffer JA, Rosas MO, Rocheford TR, Romay MC, Romero S, Salvo S, Sanchez Villeda H, da Silva HS, Sun Q, Tian F, Upadyayula N. The genetic architecture of maize flowering time. Science. 2009;325:714–718. doi: 10.1126/science.1174276. [DOI] [PubMed] [Google Scholar]