Abstract
Strains of a novel anaerobic, Gram-negative coccus were isolated from the supra-gingival plaque of children. Independent strains from each of six subjects were shown, at a phenotypic level and based on 16S rRNA gene sequencing, to be members of the genus Veillonella. Analysis revealed that the six strains shared 99.7 % similarity in their 16S rRNA gene sequences and 99.0 % similarity in their rpoB gene sequences. The six novel strains formed a distinct group and could be clearly separated from recognized species of the genus Veillonella of human or animal origin. The novel strains exhibited 98 and 91 % similarity to partial 16S rRNA and rpoB gene sequences of Veillonella parvula ATCC 10790T, the most closely related member of the genus. The six novel strains could be differentiated from recognized species of the genus Veillonella based on partial 16S rRNA and rpoB gene sequencing. The six novel strains are thus considered to represent a single novel species of the genus Veillonella, for which the name Veillonella rogosae sp. nov. is proposed. The type strain is CF100T (=CCUG 54233T=DSM 18960T).
At the time of writing, the genus Veillonella comprised the following recognized species: Veillonella parvula, V. atypica, V. dispar, V. criceti, V. ratti, V. rodentium, V. caviae, V. montpellierensis and V. denticariosi (Mays et al., 1982; Rogosa, 1984; Jumas-Bilak et al., 2004; Byun et al., 2007). Members of the genus are non-fermentative, anaerobic, Gram-negative, small cocci isolated from the oral cavity and intestinal tract of humans and animals that gain energy from the utilization of short-chain organic acids (Delwiche et al., 1985). Species of the genus isolated from humans are V. parvula, V. atypica, V. dispar, V. montpellierensis and V. denticariosi, although a strain described as a member of the V. ratti –V. criceti group has been found in a semen sample from a patient attending for treatment of infertility at a urology unit (Marchandin et al., 2005). The above species may be associated with monomicrobial infections in humans (Marchandin et al., 2001).
Identification of taxa to the genus Veillonella is quite straightforward but the identification of members of the genus to the species level is difficult owing to the lack of discriminating phenotypic tests (Kolenbrander & Moore, 1992). Analysis of 16S rRNA gene sequences permits the identification of isolates as members of the genus Veillonella. However, very low levels of 16S rRNA gene sequence dissimilarity are found within the genus and, although V. atypica, V. montpellierensis and the species isolated from rodents are identifiable by this means, discrimination between V. dispar, V. parvula and V. denticariosi is problematic (Jumas-Bilak et al., 2004; Byun et al., 2007). Additionally, there have been frequent reports of the occurrence of intra-chromosomal heterogeneity between 16S rRNA genes in human isolates of Veillonella species, further complicating the identification of isolates on the basis of 16S rRNA gene sequencing (Marchandin et al., 2003). In order to overcome the limitations associated with using 16S rRNA gene sequences to identify Veillonella species, comparisons of partial dnaK, rpoB and gyrB gene sequences have been proposed as a reliable method to identify members of the genus. All recognized members of the genus can be discriminated by using dnaK gene sequence comparison and, in particular, V. parvula and V. dispar have been reliably identified in this way (Jumas-Bilak et al., 2004). In a more recent report, the ability of dnaK gene sequencing to differentiate between Veillonella species, including V. denticariosi, was confirmed, and comparisons of rpoB and gyrB gene sequences were also shown to enable the identification of species within the genus, with the rpoB gene being the more discriminatory (R. Byun, personal communication).
As part of a large study investigating the genotypic diversity of Veillonella species present in the dental plaque of caries-free children and in the infected dentine of carious lesions, we have frequently isolated strains that appeared based on 16S rRNA gene sequencing to be distinct from all recognized species of the genus. The aim of the present study was to determine the taxonomic status of these isolates by using a polyphasic approach.
Occlusal or buccal surface plaque was collected from nine caries-free children. The children or their carers gave consent to the collection of the samples and the study was approved by the local ethics committee (ref. 02/11/10). Veillonella-like isolates were recovered on agar consisting of (per litre deionized water) 5 g Bacto tryptone (Difco), 5 g Bacto yeast extract (Difco), 0.75 g sodium thioglycolate (Sigma), 0.002 g Bacto basic fuchsin (Difco), 21 ml 60 % sodium lactate (Sigma) and 15 g Bacto agar (Difco). The pH was adjusted to 7.5 prior to autoclaving; vancomycin (7.5 μg ml−1; Sigma) was added following autoclaving (Rogosa, 1956). Inoculated plates were incubated in an atmosphere consisting initially of 90 % (v/v) nitrogen, 5 % (v/v) hydrogen and 5 % (v/v) carbon dioxide at 37 °C for 4 days. Isolates were presumptively identified as members of the genus Veillonella on the basis of their ability to grow on the selective isolation medium and their typical colonial appearance: colonies were 2–4 mm in diameter, regular and slightly domed in shape with an entire edge.
To confirm the identity of the isolates as belonging to the genus Veillonella, genomic DNA was obtained from each isolate by suspending colonies in deionized water and heating the cells to 95 °C for 10 min. The 16S rRNA gene was amplified by using the primers 27f and 1492r (Lane, 1991) and sequencing was performed by using the 27f primer in conjunction with Big Dye Ready Reaction Termination Mix (ABI) to obtain short 16S rRNA gene sequences of 450–500 nt. These sequences were submitted to the Ribosomal Database Project (http://rdp.cme.msu.edu/) via the Sequence Match routine. The majority (>99 %) of isolates were identified as members of the genus Veillonella.
The partial 16S rRNA gene sequences (450–500 nt) of all the new isolates were aligned by using clustal w (Thompson et al., 1994) in BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) with partial 16S rRNA gene sequences of V. caviae DSM 20738T, V. rodentium ATCC 17743T, V. montpellierensis ADV 281.99T, V. parvula ATCC 10790T, V. dispar ATCC 17748T, V. atypica ATCC 17744T, V. ratti ATCC 17746T, V. criceti ATCC 17747T and V. denticariosi CIP 109448T. Phylogenetic relationships between taxa were analysed by using mega 3.1 (Kumar et al., 2004). Distances were calculated according to the Kimura two-parameter model and clustering was based on the neighbour-joining method of Saitou and Nei (1987) by using bootstrap values based on 1000 replicates. From these analyses, a group of isolates was identified that appeared to represent a novel taxon that was distinct from all other clinical isolates and from other type and reference strains investigated (data not shown).
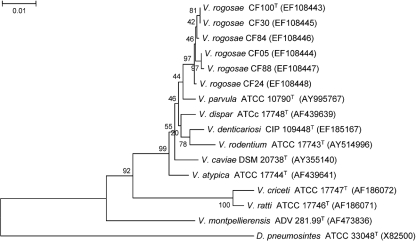
In order to understand the phylogenetic position of these isolates more clearly, more complete 16S rRNA gene sequences (1355 nt) were obtained from single strains (designated CF100T, CF05, CF30, CF84, CF88 and CF24), each isolated from a different, unrelated subject. The 16S rRNA gene sequences of these six strains, and also of V. parvula ATCC 10790T, exhibited a high level of sequence similarity with many cloned 16S rRNA gene sequences from taxa isolated from the human mouth or gut (data not shown). Alignment of the longer 16S rRNA gene sequences revealed similarity of >99.3 % (mean 99.7 %) among the six novel strains, demonstrating that they belonged to the same species. They showed the highest 16S rRNA gene sequences similarity to V. parvula ATCC 10790T (98 %). Phylogenetic relationships between the new isolates and recognized species of the genus Veillonella were investigated as described above with Dialister pneumosintes ATCC 33048T as the outgroup organism. The resulting neighbour-joining tree is shown in Fig. 1. When the 16S rRNA gene sequence data were analysed by using the minimum-evolution method in mega 3.1 (Kumar et al., 2004), the resultant tree had the same appearance with the novel strains on a distinct branch separated from other Veillonella species with a bootstrap value of 32 %. When the interior branch test of phylogeny (Kumar et al., 2004) was employed to test the significance of the branch containing the six novel strains, the confidence probability obtained with the neighbour-joining and minimum-evolution methods was 84 and 88 %, respectively, indicating that discrimination between V. parvula and the novel strains was approaching significance. However, 16S rRNA gene sequence analysis does not reliably distinguish between Veillonella species (Jumas-Bilak et al., 2004; Byun et al., 2007).
Fig. 1.
Neighbour-joining tree based on nearly complete 16S rRNA gene sequences (1309 nt), showing relationships between strains CF100T, CF05, CF30, CF84, CF88 and CF24 and the type strains of recognized members of the genus Veillonella. Dialister pneumosintes ATCC 33048T was used as the outgroup organism. Accession numbers for 16S rRNA gene sequences are given for each strain. Bootstrap values are indicated at corresponding nodes. Bar, 0.01 substitutions per site.
To overcome the above difficulty, comparisons of dnaK, gyrB and rpoB gene sequences have been used to differentiate between Veillonella species, including V. denticariosi (Jumas-Bilak et al., 2004; Byun et al., 2007). We used this approach in this study to determine the position of the six novel strains more accurately. No amplicons were produced when we used the primers previously described for the dnaK gene (Jumas-Bilak et al., 2004). We therefore used primers for the rpoB gene (Veill-rpoBF, GTAACAAAGGTGTCGTTTCTCG; Veill-rpoBR, GCACCRTCAAATACAGGTGTAGC) and were successful in obtaining and sequencing amplicons from all the strains investigated. The sequences (608 nt) were aligned, as described above, with the rpoB gene sequences of V. parvula strains ATCC 10790T, ATCC 17745, RBV162, RBV167, RBV180, RBV182, RBV175, RBV30, RBV123, RBV156, RBV173, RBV184, RBV54, RBV186, RBV188, RBV35, RBV136, RBV192, RBV183, RBV159, RBV146, RBV78, RBV179 and RBV185, V. atypica ATCC 17744T, V. dispar ATCC 17748T, V. denticariosi CIP 109448T, V. caviae DSM 20738T, V. criceti ATCC 17747T, V. ratti ATCC 17746T, V. rodentium ATCC 17743T and V. montpellierensis ADV 281.99T. Pseudoalteromonas atlantica T6c was used as the outgroup organism. The six novel strains formed a distinct taxon with robust bootstrap support (99 %) in the rpoB gene tree (Fig. 2). When the interior branch test of phylogeny was employed to test the significance of the branch containing the novel strains, the confidence probability obtained with the neighbour-joining and minimum-evolution methods was 99 %, indicating robust discrimination between V. parvula and the six novel strains. Mean rpoB gene sequence similarity among the six novel strains was 99.0 % and was 98.4 % among V. parvula strains, indicating homogeneity of the two taxa. Mean rpoB gene sequence similarity between the novel strains and strains belonging to V. parvula, the most closely related species, was 91 %, indicating that strains CF100T, CF05, CF30, CF84, CF88 and CF24 represent a novel species. We suggest that as 16S rRNA gene sequence comparisons are not suitable for differentiating between all members of the genus Veillonella, comparisons based on rpoB gene sequences represent a valid method for species characterization. This approach is validated given that V. dispar, V. parvula and V. denticariosi cannot be separated satisfactorily by using 16S rRNA gene sequence comparisons but can be defined clearly when rpoB gene sequence data are considered (Fig. 2).
Fig. 2.
Neighbour-joining tree based on partial rpoB gene sequences (608 nt), showing relationships between strains CF100T, CF05, CF30, CF84, CF88 and CF24 and recognized members of the genus Veillonella. Pseudoalteromonas atlantica T6c (gi:109698613 : 682317–686345) was used as the outgroup organism. The strains of V. parvula are not listed but were V. parvula ATCC 10790T (EF185158), V. parvula ATCC 17745 (EF185160) and an additional 22 rpoB gene sequences with accession numbers EU077574–EU077595. Bootstrap values are indicated at corresponding nodes. Bar, 0.1 substitutions per site.
As strains CF100T, CF05, CF30, CF84, CF88 and CF24 formed a phylogenetically distinct group based on rpoB gene sequencing, with robust bootstrap support, separate from V. parvula and all other recognized Veillonella species, we suggest that they represent a novel species of the genus Veillonella. We undertook a more complete phenotypic analysis of these strains.
Strains CF100T, CF05, CF30, CF84, CF88 and CF24 were tested, at least twice, by using the rapid ID 32A and API 20A identification kits (bioMérieux). Cells were small, Gram-negative coccoids, mainly appearing singly but with some short chains visible. Spores were not formed. No sugars were fermented in either of the identification kits. All strains were catalase-negative and did not hydrolyse aesculin, but one strain (CF84) weakly hydrolysed arginine. All isolates reduced nitrate. All were positive for pyroglutamic acid arylamidase and alkaline phosphatase was produced weakly by three of the six strains; all other test results were negative. No carbohydrates were fermented to acids in either test system. All strains fermented lactate, but not glucose, in peptone/yeast extract broth to produce major amounts of both propionic and acetic acids as the only acid end products when culture supernatants were analysed by using a GLC methodology (Delwiche et al., 1985). None of the six strains was positive for any of the following enzyme activities (Beighton et al., 1991) as measured by using 4-methyl umbelliferyl-linked fluorogenic substrates: α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, β-N-acetylgalactosaminidase, α-arabinosidase, β-N-acetylglucosaminidase, sialidase, α-fucosidase or β-fucosidase. The strains did not exhibit cytochrome oxidase activity. As with the recently described species V. montpellierensis (Jumas-Bilak et al., 2004), and other species of the genus (Kolenbrander & Moore, 1992), phenotypic characteristics were not sufficient to differentiate the novel isolates from other members of the genus.
Description of Veillonella rogosae sp. nov.
Veillonella rogosae (ro.go.sae. N.L. masc. gen. n. rogosae of Rogosa, named in honour of the late American microbiologist Morrison Rogosa, for his outstanding contributions to microbiology and to the taxonomy of the genus Veillonella).
Cells are Gram-negative, non-motile, non-sporulating coccoids (0.3–0.5 μm in diameter) that occur singly and in short chains. Colonies on Veillonella agar (Rogosa, 1956) are 2–4 mm in diameter and domed with an entire edge. Strictly anaerobic. Able to reduce nitrate, but unable to hydrolyse aesculin or arginine. Oxidase-negative. Unable to produce acids from carbohydrates and does not exhibit extracellular glycosidic enzyme activities. Exhibits pyroglutamic acid arylamidase activity and variable alkaline phosphatase activity. Major acid end products are acetic and propionic acids. Can be differentiated from other species of the genus Veillonella based on partial 16S rRNA and rpoB gene sequencing.
The type strain, CF100T (=CCUG 54233T=DSM 18960T), was isolated from supra-gingival plaque of a child. Strains CF05, CF30, CF84, CF88 and CF24, isolated from similar sources, are also included in the species.
Acknowledgments
This research was supported by The Dental Institute of King's College London and in part by the Wellcome Trust (grant number 076381). We thank Julie Downes for performing the acid end-product analysis.
Footnotes
References
- Beighton, D., Hardie, J. M. & Whiley, R. A. (1991). A scheme for the identification of viridans streptococci. J Med Microbiol 35, 367–372. [DOI] [PubMed] [Google Scholar]
- Byun, R., Carlier, J.-P., Jacques, N. A., Marchandin, H. & Hunter, N. (2007). Veillonella denticariosi sp. nov., isolated from human carious dentine. Int J Syst Evol Microbiol 57, 2844–2848. [DOI] [PubMed] [Google Scholar]
- Delwiche, E. A., Pestka, J. J. & Tortorello, M. L. (1985). The veillonellae: gram-negative cocci with a unique physiology. Annu Rev Microbiol 39, 175–193. [DOI] [PubMed] [Google Scholar]
- Jumas-Bilak, E., Carlier, J. P., Jean-Pierre, H., Teyssier, C., Gay, B., Campos, J. & Marchandin, H. (2004). Veillonella montpellierensis sp. nov., a novel, anaerobic, Gram-negative coccus isolated from human clinical samples. Int J Syst Evol Microbiol 54, 1311–1316. [DOI] [PubMed] [Google Scholar]
- Kolenbrander, P. E. & Moore, L. V. H. (1992). The genus Veillonella. In The Prokaryotes, 2nd edn, pp. 2034–2047. Edited by A. Balows, H. Trüper, M. Dworkin, W. Harder & K.-H. Schleifer. New York: Springer.
- Kumar, S., Tamura, K. & Nei, M. (2004). mega3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5, 150–163. [DOI] [PubMed] [Google Scholar]
- Lane, D. J. (1991). 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics, pp. 115–175. Edited by E. Stackebrandt & M. Goodfellow. Chichester: Wiley.
- Marchandin, H., Jean-Pierre, H., Carrière, C., Canovas, F., Darbas, H. & Jumas-Bilak, E. (2001). Prosthetic joint infection due to Veillonella dispar. Eur J Clin Microbiol Infect Dis 20, 340–342. [DOI] [PubMed] [Google Scholar]
- Marchandin, H., Teyssier, C., Simeon De Buochberg, M., Jean-Pierre, H., Carriere, C. & Jumas-Bilak, E. (2003). Intra-chromosomal heterogeneity between the four 16S rRNA gene copies in the genus Veillonella: implications for phylogeny and taxonomy. Microbiology 149, 1493–1501. [DOI] [PubMed] [Google Scholar]
- Marchandin, H., Teyssier, C., Jumas-Bilak, E., Robert, M., Artigues, A. C. & Jean-Pierre, H. (2005). Molecular identification of the first human isolate belonging to the Veillonella ratti–Veillonella criceti group based on 16S rRNA and dnaK gene sequencing. Res Microbiol 156, 603–607. [DOI] [PubMed] [Google Scholar]
- Mays, T. D., Holdeman, L. V., Moore, W. E. C., Rogosa, M. & Johnson, J. L. (1982). Taxonomy of the genus Veillonella Prévot. Int J Syst Bacteriol 32, 28–36. [Google Scholar]
- Rogosa, M. (1956). A selective medium for the isolation and enumeration of the veillonella from the oral cavity. J Bacteriol 72, 533–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogosa, M. (1984). Anaerobic Gram-negative cocci. In Bergey's Manual of Systematic Bacteriology, vol. 1, pp. 680–685. Edited by N. R. Krieg & J. G. Holt. Baltimore: Williams & Wilkins.
- Saitou, N. & Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994). clustal w: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]