Abstract
An outbreak of Legionnaires' disease at a long-term care facility in Ontario, Canada from September to October 2005 resulted in the death of 23 residents and the illness of 112 other people. In response, molecular methods were developed to detect Legionella pneumophila in clinical lung samples and to subtype isolates from clinical and environmental samples. The targeted genetic loci included Legionella-specific virulence determinants (mip, icmO, sidA and lidA) and core bacterial determinants (ftsZ, trpS and dnaX). An established amplified fragment length polymorphism typing method provided the first indication of genetic relatedness between strains recovered from clinical samples and strains cultured from environmental samples taken from the outbreak site. These associations were verified using the European Working Group for Legionella Infections sequence-based typing protocol targeting the flaA, pilE, asd, mip, mompS and proA loci. These molecular typing methods confirmed the outbreak source as a contaminated air conditioning cooling tower.
INTRODUCTION
Legionella pneumophila is a ubiquitous bacterium in natural aquatic environments that can also persist in human-controlled systems containing water, such as air conditioning and plumbing infrastructures. Intracellular growth in protozoa can permit this pathogen to survive chlorination, and the generation of aerosols in these systems contributes to the transmission to humans where infection of alveolar macrophages results in respiratory illness (Steinert et al., 2002; McDade et al., 1977). The first recognized outbreak of Legionnaires' disease occurred in 1976 in Philadelphia, USA, and Legionella is now accepted as a significant cause of community- and nosocomial-acquired pneumonia, including in long-term care facilities (Seenivasan et al., 2005).
During outbreaks of Legionnaires' disease the recovery or detection of L. pneumophila in clinical specimens can be sufficient for detection of the aetiological agent. However, due to the ubiquitous nature of this bacterium in the environment, molecular typing methods are needed both to determine the relatedness of outbreak strains and for definitive identification of the outbreak source. Culture of clinical specimens (e.g. from the lower respiratory tract) is the gold standard for diagnosis of Legionnaires' disease but rapid detection of L. pneumophila serogroup 1, the most common subtype associated with human disease, can be achieved with immunofluorescence and serological assays targeting L. pneumophila antigen and L. pneumophila-specific antibodies, respectively (Fields et al., 2002). Molecular detection of genus and species is through assays targeting characteristic sites in the rRNA and mip genes, respectively (Fields et al., 2002; Ballard et al., 2000; Wilson et al., 2003; Rantakokko-Jalava & Jalava, 2001). The mip locus is also one of six loci used in a sequence-based typing (SBT) scheme for the molecular subtyping of L. pneumophila strains (Gaia et al., 2005). The mip locus therefore encodes conserved species-specific polymorphisms useful for molecular diagnostics and also strain-specific polymorphisms that can distinguish amongst L. pneumophila isolates. The effectiveness of the SBT scheme for determining the genetic relatedness of L. pneumophila outbreak-associated strains, in comparison to other subtyping methods such as PFGE, has been previously demonstrated (Scaturro et al., 2005). In this study, molecular reagents were developed during an outbreak of Legionnaires' disease for the detection of L. pneumophila in clinical specimens and typing of clinical and environmental isolates.
METHODS
The case definition of Legionnaires' disease was chest X-ray confirmed pneumonia, clinical diagnosis or pathological evidence of pneumonia at autopsy, and living or working within 3 kilometres (1.9 miles) of the outbreak site since a month prior to the outbreak. Each case of Legionnaires' disease was definitively confirmed after a positive culture for L. pneumophila serogroup 1, a positive urine antigen test (Binax) or positive immunofluorescence antibody serology (standing titre of ⩾1 : 256 or fourfold seroconversion of ⩾1 : 128).
Legionella strains were recovered from clinical specimens and environmental samples as described by Wilkinson (1988) and Fields (1994). Briefly, Legionella spp. from environmental samples were first selected using buffered charcoal yeast agar (BCYE) plates supplemented with polymyxin B, anisomycin and vancomycin (PAV). Colonies resembling Legionella spp. on the BCYE-PAV plates were then differentiated using BCYE, BCYE (lacking cysteine) and blood agar plates. Isolation of Legionella spp. from lung samples was performed using solely BCYE plates. All incubations were performed at 37 °C in candle jars or CO2 incubators. Lung autopsy materials were transported in viral transport media, swabs and water samples were collected in sterile containers, and air samples were collected with a XMX-CV aerosol collection system (Dycor). Direct fluorescent antibody assay (DFA) analysis on clinical specimens were done using monoclonal or polyclonal DFA products directed towards L. pneumophila serogroups (Prolab and Monofluo L. pneumophila kit; Bio-Rad). Additionally, total DNA was prepared from 19 lung autopsy samples from 7 patients using the DNeasy tissue extraction kit (Qiagen).
Oligonucleotides designed for molecular detection and typing are described in Table 1. PCR was performed using these oligonucleotides (1 μM), dNTPs (0.2 mM), magnesium sulfate (2 mM), Tris/sulfate (60 mM; pH 8.9), ammonium sulfate (18 mM) and 0.5 U High Fidelity Platinum Taq polymerase (Invitrogen) per 25 ml reaction, and the following thermocycling conditions: 94 °C for 5 min; 35 cycles of 94 °C for 30 s, 55 °C for 30 s and 68 °C for 30 s (except with sidA, lidA and icmO 1 min); and a one-round final extension at 68 °C for 5 min. DNA sequencing was performed with the oligonucleotides used to generate the amplicon. SBT using targets designed by the European Working Group for Legionella Infections (EWGLI) and amplified fragment length polymorphism (AFLP) analysis were performed as described by Gaia et al. (2003, 2005) and Valsangiacomo et al. (1995).
Table 1.
Oligonucleotides developed in this study for the detection and subtyping of L. pneumophila
| Target | Oligonucleotide | Sequence (5′ to 3′) | Product size (bp) | Purpose* |
|---|---|---|---|---|
| dnaX | GIL296 | ATACCTGTGTTGCCATTGAGC | 255 | PCR-Lpn ID |
| GIL297 | ATTTTTGTGGGTCAGTCGTTG | PCR-Lpn ID | ||
| dnaX | GIL313 | TTTGCGCTGGTGTATTATCATC | 936 (with GIL296) | PCR-sequencing |
| ftsZ | GIL298 | GCGATTGCATCTCCTTTATTG | 351 | PCR-Lpn ID |
| GIL299 | CTGAGCCTGCTTACGAATCAC | PCR-Lpn ID | ||
| ftsZ | GIL312 | CGATGCACAAGCTTTAAGAGG | 946 (with GIL299) | PCR-sequencing |
| trpS | GIL300 | TCAGCCTAATTTGTCGATTGG | 274 | PCR-Lpn ID |
| GIL301 | TTGCCAAACAGGACATTTTTC | PCR-Lpn ID | ||
| trpS | GIL308 | TTGATTGGCTAGCTTGTGGAG | 999 | PCR-sequencing |
| GIL309 | AATCAAGCCCCATGACTTCTC | PCR-sequencing | ||
| mip | GIL306 | GCTTTAACCGAACAGCAAATG | 267 | PCR-Lpn ID |
| GIL307 | AACGGTACCATCAATCAGACG | PCR-Lpn ID | ||
| lidA | GIL314 | TCACATCAAGTTAAAACATCAG | 882 | PCR-Lpn ID |
| GIL315 | ATGCTCACGCTGTAAGGATTG | PCR-Lpn ID | ||
| icmO | GIL316 | AATTTTCGGTTCAACGGGTAG | 763 | PCR-Lpn ID |
| GIL317 | CAGTGCGGGTAATAAAACCAC | PCR-Lpn ID | ||
| sidA | GIL318 | GAAATTCGTGCCATAGAACTCC | 330 | PCR-Lpn ID |
| GIL319 | CGGGGCTATGATTTCTTCTATG | PCR-Lpn ID |
*Lpn ID, L. pneumophila-specific detection.
RESULTS AND DISCUSSION
An outbreak of Legionnaires' disease at an extended care facility in Scarborough, Ontario, Canada, from September to October, 2005 resulted in 23 deaths. All mortalities occurred with residents of the care facility (hereafter, ‘outbreak site’) and additional cases were identified in 47 residents, 39 staff and 21 visitors to the outbreak site, as well as in 5 occupants and staff of a neighbouring care facility and residence (hereafter, ‘adjacent site’). A total of 82 people were affected with symptoms resembling Legionnaires' disease and 53 people presented with the milder form of illness cause by Legionella, Pontiac fever. A total of 79 people were hospitalized, of which 54 were residents at the outbreak site. This study describes the molecular detection of L. pneumophila in the lung autopsy material from the first mortalities, and the subtyping of L. pneumophila strains subsequently isolated from clinical and environmental samples.
Molecular detection of L. pneumophila in lung tissue DNA extractions and subtyping of pure cultures was achieved using species-specific oligonucleotides (Table 1). These were designed for various genetic loci after comparative analyses of the complete sequence data from three L. pneumophila group 1 strains (Lens, Paris and Philadelphia: GenBank accession numbers NC_006369, NC_006368 and NC_002942, respectively) and with the other publicly available bacterial sequences. Oligonucleotides corresponded to conserved regions amongst the three L. pneumophila genomes, but were unique compared to other genera and/or species (data not shown). The target loci included Legionella-specific virulence determinants (mip, icmO, sidA and lidA) and core bacterial determinants (ftsZ, trpS and dnaX). These core bacterial determinants have been predicted to contain species-specific polymorphisms (Zeigler, 2003), and we have previously utilized ftsZ and trpS for speciation of Vibrio spp. and detection of Salmonella enterica serovar Typhi, respectively (Tracz et al., 2006, 2007). Published primer sets specific for the Legionella genus (16S rRNA) and human β-globin locus were used as additional control reactions (Raggam et al., 2005; Keller & Mannack, 1993).
PCR-based detection assays for L. pneumophila were conducted on 19 lung tissue DNA extractions (from 7 different patients) using ftsZ, trpS, dnaX and mip L. pneumophila-specific primer sets; control reactions included 293T tissue culture cells spiked with Legionella micdadei ATCC 33218 or spiked with L. pneumophila ATCC 33153 (Table 2). Using this approach, all clinical lung samples were positive for the presence of L. pneumophila, except for a single lung tissue extract (05-L-025, patient 3). The two other lung specimens from patient 3 were positive for L. pneumophila and the control PCR reactions for specimen 05-L-025 failed, indicating that DNA extraction was likely unsuccessful for this individual specimen. DFA testing on one lung autopsy specimen from each patient was positive for L. pneumophila serogroup 1, whereas L. pneumophila was cultured from lung specimens for six of the seven patients.
Table 2.
PCR-based detection of L. pneumophila
Templates included clinical lung autopsy material and tissue culture spiked with live bacterial culture (prepared using a Qiagen tissue extraction kit) or pure bacterial culture (prepared from boiled cell lysates).
| Sample* | Patient | Description | dnaX | ftsZ | trpS | mip | 16S† | β-globin‡ |
|---|---|---|---|---|---|---|---|---|
| L. pneumophila ATCC 33153 | na | Pure culture | + | + | + | + | + | − |
| L. micdadei ATCC 33218 | na | Pure culture | − | − | − | − | + | − |
| 293T tissue culture | na | Tissue culture | − | − | − | − | − | + |
| 293T+L. pneumophila | na | Spiked tissue | + | + | + | + | + | + |
| 293T+L. micdadei | na | Spiked tissue | − | − | − | − | + | + |
| 05-L-011 | 1 | Autopsy tissue | + | + | + | + | + | + |
| 05-L-012 (DFA+) | 1 | Autopsy tissue | + | + | + | + | + | + |
| 05-L-013 | 1 | Autopsy tissue | + | + | + | + | + | + |
| 05-L-014 | 2 | Autopsy tissue | + | + | + | + | + | + |
| 05-L-015 | 2 | Autopsy tissue | + | + | + | + | + | + |
| 05-L-024 (DFA+) | 2 | Autopsy tissue | + | + | + | + | + | + |
| 05-L-016 | 3 | Autopsy tissue | + | + | + | + | + | + |
| 05-L-017 | 3 | Autopsy tissue | + | + | + | + | + | + |
| 05-L-025 (DFA+) | 3 | Autopsy tissue | − | − | − | − | − | − |
| 05-L-018 | 4 | Autopsy tissue | + | + | + | + | + | + |
| 05-L-019 | 4 | Autopsy tissue | + | + | + | + | + | + |
| 05-L-027 (DFA+) | 4 | Autopsy tissue | + | + | + | + | + | + |
| 05-L-020 | 5 | Autopsy tissue | + | + | + | + | + | + |
| 05-L-021 | 5 | Autopsy tissue | + | + | + | + | + | + |
| 05-L-026 (DFA+) | 5 | Autopsy tissue | + | + | + | + | + | + |
| 05-L-022 | 6 | Autopsy tissue | + | + | + | + | + | + |
| 05-L-023 | 6 | Autopsy tissue | + | + | + | + | + | + |
| 05-L-028 (DFA+) | 6 | Autopsy tissue | + | + | + | + | + | + |
| 05-L-029 (DFA+) | 7 | Autopsy tissue | + | + | + | + | + | + |
na, Not applicable.
*DFA+, DFA detected L. pneumophila serogroup 1 in the clinical specimen.
†Legionella genus-specific primers (Raggam et al., 2005).
‡Human-specific primers (Keller & Mannack, 1993).
Environmental sampling was performed throughout the outbreak site and the adjacent site, including cooling towers, air filtration systems and internal plumbing systems. Samples included sediments and biofilms in these infrastructure, collection of standing and free-flowing water, and collection of air-borne particulate matter. Additionally, sampling was performed of the soil and natural water systems in the surrounding area, which included a ravine and creek directly behind the outbreak site. Notably, PCR-based detection of L. pneumophila directly in environmental samples failed, including those that ultimately were cultured for this organism. The amount of L. pneumophila DNA extracted from these samples was likely below the detection threshold for these methods, and therefore these methods might be limited to analysis of pure culture and clinical samples. The only environmental samples that yielded L. pneumophila isolates were associated with the cooling towers at both the outbreak and adjacent sites (Table 3). Isolates 05-L-77, -79, -192 and -193 were typed as serogroup 1, corresponding to the same serogroup observed in the lung autopsy material by DFA analysis.
Table 3.
SBT of L. pneumophila isolates cultured from lung autopsy tissue or from environmental samples
The allele profile number for each locus is indicated. AFLP analysis was performed on selected isolates and the patterns correlated to those illustrated in Fig. 1.
| Strain | Site of collection; AFLP result | flaA | pilE | asd | mip | mompS | proA |
|---|---|---|---|---|---|---|---|
| 05-L-013 | Lung, patient 1 | 2 | 19 | 5 | 10 | 18 | 1 |
| 05-L-025 | Lung, patient 3 | 2 | 19 | 5 | 10 | 18 | 1 |
| 05-L-026 | Lung, patient 5 | 2 | 19 | 5 | 10 | 18 | 1 |
| 05-L-028 | Lung, patient 6 | 2 | 19 | 5 | 10 | 18 | 1 |
| 05-L-029 | Lung, patient 7 | 2 | 19 | 5 | 10 | 18 | 1 |
| 060017 | Lung, patient 4; pattern A | 2 | 19 | 5 | 10 | 18 | 1 |
| 060018 | Lung, patient 1; pattern B | 2 | 19 | 5 | 10 | 18 | 1 |
| 060019 | Outbreak site, cooling tower; pattern A | 2 | 19 | 5 | 10 | 18 | 1 |
| 060020 | Outbreak site, cooling tower middle cavity; pattern B | 2 | 19 | 5 | 10 | 18 | 1 |
| 05-L-0192-1 | Outbreak site, cooling tower bottom cavity | 2 | 19 | 5 | 10 | 18 | 1 |
| 05-L-0192-2 | Outbreak site, cooling tower bottom cavity | 2 | 19 | 5 | 10 | 18 | 1 |
| 05-L-0192-3 | Outbreak site, cooling tower bottom cavity | 2 | 19 | 5 | 10 | 18 | 1 |
| 05-L-0192-4 | Outbreak site, cooling tower bottom cavity | 2 | 19 | 5 | 10 | 18 | 1 |
| 05-L-0192-5 | Outbreak site, cooling tower bottom cavity | 2 | 19 | 5 | 10 | 18 | 1 |
| 05-L-0192-6 | Outbreak site, cooling tower bottom cavity | 2 | 19 | 5 | 10 | 18 | 1 |
| 05-L-0192-7 | Outbreak site, cooling tower bottom cavity | 2 | 19 | 5 | 10 | 18 | 1 |
| 05-L-0193 | Outbreak site, cooling tower | 2 | 19 | 5 | 10 | 18 | 1 |
| CPHL-3 | Adjacent site; pattern C | 1 | 4 | 3 | 1 | 1 | 1 |
| CPHL-5 | Adjacent site; pattern C | 1 | 4 | 3 | 1 | 1 | 1 |
| CPHL-7 | Adjacent site; pattern C | 1 | 4 | 3 | 1 | 1 | 1 |
| 05-L-077-1 | Adjacent site, west cooling tower | 1 | 4 | 3 | 1 | 1 | 1 |
| 05-L-079-2 | Adjacent site, east cooling tower | 1 | 4 | 3 | 1 | 1 | 1 |
| ATCC 33153 | L. pneumophila group 1 (Knoxville) reference strain | 7 | 6 | 17 | 3 | 24 | 11 |
To discriminate the environmental isolates beyond the level of serogroup, and to determine genetic relatedness to the lung isolates, AFLP analysis was used as a rapid molecular typing method (Fig. 1). The AFLP pattern produced from adjacent site isolates was different than the patterns obtained for both the outbreak site cooling tower and lung isolates, wherein two similar (but non-identical) patterns were observed amongst both groups of these outbreak-associated isolates. These data provided the first indication that the cooling tower at the outbreak site was responsible for the event.
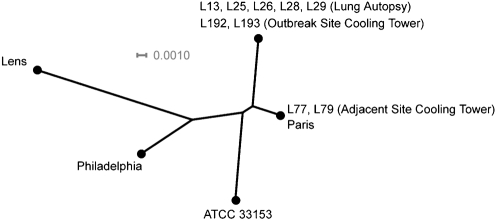
Fig. 1.
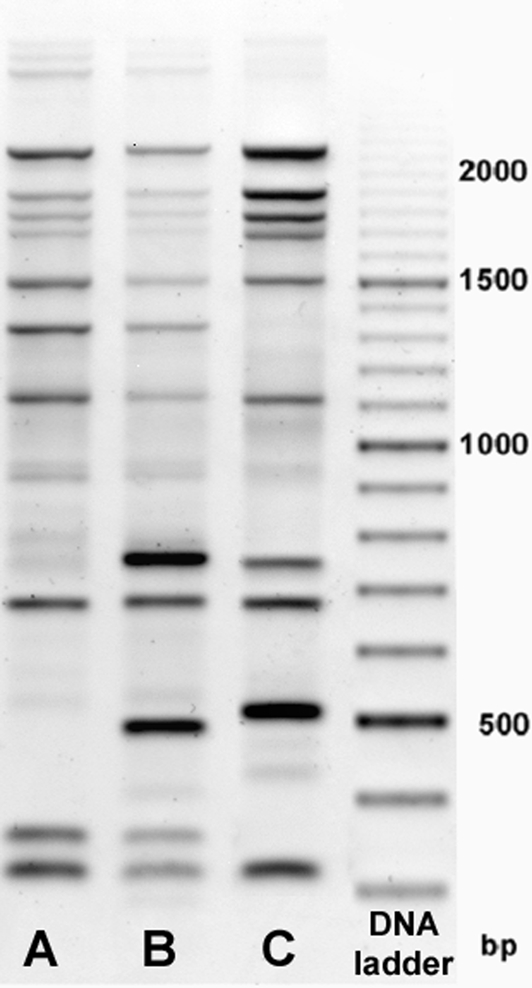
AFLP analysis of outbreak-associated (A, B) and adjacent site (C) isolates. Molecular mass markers are indicated.
The clinical and environmental L. pneumophila isolates were also prepared as template for DNA sequencing using the ftsZ, trpS, dnaX, icmO, sidA and lidA loci. Phylogenetic comparisons indicated that each of the lung isolates and strains recovered from the cooling tower at the outbreak site had identical genotypes at all loci (Fig. 2). Furthermore, the outbreak site and lung isolates had a different genotype at all loci in comparison to reference strains and strains isolated at the adjacent site, whereas the adjacent site isolates had identical genotypes as the endemic ‘Paris' strain. SBT using an established protocol (Gaia et al., 2005, 2003) was also utilized to determine the genetic relatedness of the isolates (Table 3), and the allelic profile of the lung-associated strains was again identical to the strains recovered from the outbreak site cooling tower, but different from the profile of isolates from the adjacent site. Notably, none of the individual SBT alleles reported in this study were novel to the database maintained by EWGLI, but the cumulative profile of the outbreak strain had not been reported before and therefore no associations to previously examined strains were evident. However, the isolates from the cooling tower of the adjacent site encoded the most frequently recorded profile in the EWGLI database (N. Fry, personal communication).
Fig. 2.
Genetic relatedness of outbreak, environmental and reference L. pneumophila strains. Phylogeny is based upon a neighbour-joining tree of the concatenated segments of core bacterial loci (trpS, ftsZ and trpS) and Legionella-specific virulence determinants (lidA, icmO, sidA, total 4243 nucleotides per entry). GenBank accession numbers for previously sequenced loci are presented in the text, and strains from the current study are abbreviated from the form 05-L-0XX to LXX. The source is indicated in parentheses. Bar, scale of the distance score.
Molecular reagents were developed during this outbreak of Legionnaires' disease to supplement bacterial culture and immunological detection methods for L. pneumophila in clinical lung specimens. Additional validation will be required to confirm the specificity and sensitivity of the primer sets in other clinical samples such as bronchial alveolar lavage fluid and urine. Molecular typing analyses (both SBT and AFLP) demonstrated that isolates from the cooling tower at the outbreak site were related to isolates from lung autopsy specimens, but divergent from isolates recovered from an adjacent building and reference strains. Notably, AFLP analyses distinguished two clones amongst the human and environmental outbreak-associated isolates suggesting increased sensitivity for this method; however, the nature of the observed genetic diversity was undefined. The genetic loci developed in this study for SBT effectively determined the relatedness of clinical and environmental samples, but the established EWGLI SBT protocol was of increased utility because it similarly differentiated the L. pneumophila strains but additionally allowed for comparison to a global collection of SBT data deposited in a web-accessible database.
Acknowledgments
We would like to thank Toronto Public Health for collecting clinical and environmental samples, PulseNet Canada and the DNA Core Facility at the National Microbiology Laboratory for DNA sequencing and oligonucleotide synthesis, Elsie Grudeski for technical assistance, Norman Fry of the Health Protection Agency Centre for Infections for assistance with SBT analyses, and Donald Low and Lai-King Ng for their support.
Abbreviations
AFLP, amplified fragment length polymorphism
DFA, direct fluorescent antibody assay
EWGLI, European Working Group for Legionella Infections
SBT, sequence-based typing
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the sequences of the new isolates are DQ451531–DQ451547.
References
- Ballard, A. L., Fry, N. K., Chan, L., Surman, S. B., Lee, J. V., Harrison, T. G. & Towner, K. J. (2000). Detection of Legionella pneumophila using a real-time PCR hybridization assay. J Clin Microbiol 38, 4215–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields, B. W. (1994). Procedures for the Recovery of Legionella from the Environment. US Department of Health and Human Services. Atlanta, GA: Centers for Disease Control and Prevention.
- Fields, B. S., Benson, R. F. & Besser, R. E. (2002). Legionella and Legionnaires' disease: 25 years of investigation. Clin Microbiol Rev 15, 506–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaia, V., Fry, N. K., Harrison, T. G. & Peduzzi, R. (2003). Sequence-based typing of Legionella pneumophila serogroup 1 offers the potential for true portability in legionellosis outbreak investigation. J Clin Microbiol 41, 2932–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaia, V., Fry, N. K., Afshar, B., Luck, P. C., Meugnier, H., Etienne, J., Peduzzi, R. & Harrison, T. G. (2005). Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J Clin Microbiol 43, 2047–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, G. H. & Mannack, M. M. (1993). DNA Probes, pp. 396–397. New York: Stockton Press.
- McDade, J. E., Shepard, C. C., Fraser, D. W., Tsai, T. R., Redus, M. A. & Dowdle, W. R. (1977). Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med 297, 1197–1203. [DOI] [PubMed] [Google Scholar]
- Raggam, R. B., Leitner, E., Berg, J., Muhlbauer, G., Marth, E. & Kessler, H. H. (2005). Single-run, parallel detection of DNA from three pneumonia-producing bacteria by real-time polymerase chain reaction. J Mol Diagn 7, 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantakokko-Jalava, K. & Jalava, J. (2001). Development of conventional and real-time PCR assays for detection of Legionella DNA in respiratory specimens. J Clin Microbiol 39, 2904–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaturro, M., Losardo, M., De Ponte, G. & Ricci, M. L. (2005). Comparison of three molecular methods used for subtyping of Legionella pneumophila strains isolated during an epidemic of Legionellosis in Rome. J Clin Microbiol 43, 5348–5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seenivasan, M. H., Yu, V. L. & Muder, R. R. (2005). Legionnaires' disease in long-term care facilities: overview and proposed solutions. J Am Geriatr Soc 53, 875–880. [DOI] [PubMed] [Google Scholar]
- Steinert, M., Hentschel, U. & Hacker, J. (2002). Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol Rev 26, 149–162. [DOI] [PubMed] [Google Scholar]
- Tracz, D. M., Tabor, H., Jerome, M., Ng, L.-K. & Gilmour, M. W. (2006). Genetic determinants and polymorphisms specific for human-adapted serovars of Salmonella enterica that cause enteric fever. J Clin Microbiol 44, 2007–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracz, D. M., Backhouse, P. G., Olson, A. B., McCrea, J. K., Walsh, J. A., Ng, L.-K. & Gilmour, M. W. (2007). Rapid detection of Vibrio species using liquid microsphere arrays and real-time PCR targeting the ftsZ locus. J Med Micriobiol 56, 56–65. [DOI] [PubMed] [Google Scholar]
- Valsangiacomo, C., Baggi, F., Gaia, V., Balmelli, T., Peduzzi, R. & Piffaretti, J. C. (1995). Use of amplified fragment length polymorphism in molecular typing of Legionella pneumophila and application to epidemiological studies. J Clin Microbiol 33, 1716–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, H. W. (1988). Hospital-Laboratory Diagnosis of Legionella pneumophila Infections. Atlanta, GA: Centers for Disease Control and Prevention.
- Wilson, D. A., Yen-Lieberman, B., Reischl, U., Gordon, S. M. & Procop, G. W. (2003). Detection of Legionella pneumophila by real-time PCR for the mip gene. J Clin Microbiol 41, 3327–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler, D. R. (2003). Gene sequences useful for predicting relatedness of whole genomes in bacteria. Int J Syst Evol Microbiol 53, 1893–1900. [DOI] [PubMed] [Google Scholar]