Abstract
Antimicrobial-resistant pathogenic members of the Enterobacteriaceae are a well-defined global problem. We hypothesized that one of the main reservoirs of dissemination of antimicrobial resistance genes in Vietnam is non-pathogenic intestinal flora, and sought to isolate antimicrobial-resistant organisms from hospitalized patients and non-hospitalized healthy individuals in Ho Chi Minh City. The results identified substantial faecal carriage of gentamicin-, ceftazidime- and nalidixic acid-resistant members of the Enterobacteriaceae in both hospitalized patients and non-hospitalized healthy individuals. A high prevalence of quinolone resistance determinants was identified, particularly the qnrS gene, in both community- and hospital-associated strains. Furthermore, the results demonstrated that a combination of quinolone resistance determinants can confer resistance to nalidixic acid and ciprofloxacin, even in the apparent absence of additional chromosomal resistance mutations in wild-type strains and laboratory strains with transferred plasmids. These data suggest that intestinal commensal organisms are a significant reservoir for the dissemination of plasmid-mediated quinolone resistance in Ho Chi Minh City.
INTRODUCTION
Pathogenic enteric bacteria that exhibit antimicrobial resistance are a widespread phenomenon and arguably constitute a global epidemic. Whilst the depth of knowledge regarding antimicrobial-resistant organisms isolated from patients with infection or circulating in the hospital environment is broad, less is known about antimicrobial-resistant organisms that are disseminated in the community. Furthermore, little is known about the antimicrobial resistance patterns of community-acquired organisms that circulate in developing countries where antimicrobials are available without prior consultation with a physician.
Quinolones and fluoroquinolones are groups of antimicrobial compounds that are commonly used for the treatment of many bacterial infections. However, multiple studies have highlighted that, in recent years, resistance to fluoroquinolones has increased globally, particularly in members of the Enterobacteriaceae such as Salmonella and Klebsiella species (Chau et al., 2007; Strahilevitz et al., 2007; Wang et al., 2008). Quinolones target the DNA gyrase and topoisomerase IV enzymes of the bacterial cell, thus preventing DNA replication (Higgins et al., 2003). The main resistance mechanisms to quinolones are mutations in the gyrA and parC genes that alter the conformation of target amino acid residues within the protein (Jacoby, 2005). The more recent discovery and rapid dissemination of plasmid-mediated quinolone resistance (PMQR) genes has further highlighted the problem of quinolone and fluoroquinolone resistance and increased our understanding of resistance mechanisms associated with these antibacterial compounds (Robicsek et al., 2006a).
The first PMQR gene to be described was named qnrA and encodes a 218 aa pentapeptide repeat protein that is capable of protecting the DNA gyrase from the activity of quinolones (Robicsek et al., 2006a; Tran et al., 2005a, b). Since the discovery of QnrA, two related PMQR proteins have been described; these proteins are thought to act in a manner comparable to QnrA, as they share 40 and 59 % amino acid similarity and have been named QnrB and QnrS, respectively (Hata et al., 2005; Jacoby et al., 2006). In vitro acquisition of a Qnr determinant through conjugation confers a 16–125-fold increase in the MIC, depending on the nature of the donor strain and the quinolone tested (Robicsek et al., 2006a).
Two additional PMQR determinants have also been described, which act using two mechanisms distinct from that of the Qnr proteins. The first is the aac(6′)-Ib-cr gene, which encodes a variant aminoglycoside acetyltransferase (Melano et al., 2003; Park et al., 2006). This gene harbours two individual base pair substitutions, which result in the enzyme being able to acetylate ciprofloxacin and norfloxacin, reducing the activity of the fluoroquinolone and therefore increasing the MIC two- to fourfold (Robicsek et al., 2006b). The second is the qepA gene, which encodes a major facilitator subfamily quinolone-specific efflux pump (Perichon et al., 2007). Experimentally, the QepA protein does not alter the MICs of ampicillin, erythromycin, kanamycin or tetracycline, but it does decrease susceptibility to norfloxacin, enrofloxacin and ciprofloxacin by up to 64-fold (Yamane et al., 2007).
All of the Qnr determinants have been identified in clinical Enterobacteriaceae isolates; the majority of reported strains are isolates of Salmonella species and Klebsiella pneumoniae (Cattoir et al., 2007b; Gay et al., 2006; Mendes et al., 2008; Strahilevitz et al., 2007). The aac(6′)-Ib-cr gene has also been reported in pathogenic bacteria and, like the qnr genes, has been found worldwide. The qepA gene has been reported in members of the Enterobacteriaceae in Japan, China and France (Cattoir et al., 2008b; Ma et al., 2009; Yamane et al., 2008). In this study, we attempted to demonstrate the frequency of carriage of antimicrobial-resistant members of the Enterobacteriaceae in patients and healthy individuals living in Ho Chi Minh City, Vietnam. We determined the prevalence of PMQR genes in these strains and their effect on the MIC in wild-type strains with and without additional resistance-associated chromosomal mutations, as well as in laboratory strains transformed with isolated plasmid DNA.
METHODS
Bacterial isolation and identification.
The strains were collected from two distinct study populations comprising strains obtained from patients admitted to hospital (hospital strains) and those from healthy volunteers from the local community (community strains). Hospital strains were collected from 194 patients who were admitted to the Tetanus Intensive Care Ward at the Hospital for Tropical Diseases in Ho Chi Minh City over the periods of May–October 2004 and June–November 2005. Swabs for culture were taken from the axilla, nose, sputum and anus of all patients on admission and twice weekly.
The community strains were collected from stool samples from 27 healthy adults and 77 children (5–14 years) living in Ho Chi Minh City, who participated in a typhoid vaccine study in 2005 and 2006, and from nasal and rectal swabs from 100 consecutive 1–3-day-old healthy neonates, born after uncomplicated pregnancies on a general obstetrics ward in 2006. None of the individuals (including the mothers of the neonates) had had any known contact with antibiotics (prescribed or otherwise) for 4 weeks prior to sample collection.
Samples were cultured on MacConkey medium with and without supplementation of gentamicin (8 μg ml−1), ceftazidime (2 μg ml−1) or nalidixic acid (16 μg ml−1) (Sigma-Aldrich). All colony morphologies grown on MacConkey agar supplemented with antibiotics were Gram stained. All isolates confirmed to be Gram-negative and oxidase-negative were identified using an API 20E system (bioMérieux). For hospitalized patients, the first isolate growing on each of the selective agars, typically ranging from 2 days to 1 week after admission, was included in the study.
Antimicrobial susceptibility testing.
The susceptibility to gentamicin, amikacin, ceftazidime, piperacillin–tazobactam, imipenem and nalidixic acid of members of the Enterobacteriaceae growing on selective MacConkey agars was determined using a disc diffusion method on Mueller–Hinton agar plates, according to Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2007). Strains that were identified as resistant to ceftazidime were subjected to further phenotypic tests to confirm extended-spectrum β-lactamase (ESBL) production, using discs containing only cefotaxime (30 μg) and ceftazidime (30 μg) and both antimicrobials combined with clavulanic acid (10 μg), according to CLSI guidelines (CLSI, 2007). MICs were measured using E-test (AB Biodisk).
PCR and DNA sequencing.
Genomic DNA was isolated from all isolated members of the Enterobacteriaceae using a Wizard Genomic DNA Preparation kit (Promega), and PCR amplification of the gyrA, parC, qnrA, qnrB, qnrS, qepA and aac(6′)-Ib genes was performed using the primers outlined in Table 1.
Table 1.
Primers used in this study
na, Not applicable.
| Target gene | Gene size (bp) | Primer name | Predicted size of amplicon (bp) | Primer sequence (5′→ 3′) |
|---|---|---|---|---|
| qnrA | 657 | QnrA-F | 627 | TCAGCAAGAGGATTTCTCA |
| QnrA-R | GGCAGCACTATTACTCCCA | |||
| qnrB | 681 | QnrBm-F | 264 | GGMATHGAAATTCGCCACTG |
| QnrBm-R | TTTGCYGYYCGCCAGTCGAA | |||
| qnrS | 657 | QnrS-F | 491 | ATGGAAACCTACAATCATAC |
| QnrS-R | AAAAACACCTCGACTTAAGT | |||
| aac(6′)-Ib | 519 | Aac(6′)-Ib-F | 482 | TTGCGATGCTCTATGAGTGGCTA |
| Aac(6′)-Ib-R | CTCGAATGCCTGGCGTGTTT | |||
| qepA | 986 | QepA_F | 199 | GCAGGTCCAGCAGCGGGTAG |
| QepA_R | CTTCCTGCCCGAGTATCGTG | |||
| gyrA | 2628 | GyrA-F | 625 | CGACCTTGCGAGAGAAAT |
| GyrA-R | GTTCCATCAGCCCTTCAA | |||
| parC (E. coli) | 2258 | parC_E_F | 395 | AAACCTGTTCAGCGCCGCATT |
| parC_E_R | GTGGTGCCGTTAAGCAAA | |||
| parC (K. pneumoniae) | 2258 | parC_K_F1 | 389 | CTGAATGCCAGCGCCAAATT |
| parC_K_R1 | TGCGGTGGAATATCGGTCGC | |||
| RAPD | na | ERIC1 | na | ATGTAAGCTCCTGGGGATTCAC |
| ERIC2 | AAGTAAGTGACTGGGGTGAGCG | |||
| TT3 | GGCGAGGAGCG |
PCR amplification was carried out for 35 cycles at 94 °C for 30 s, 55 °C [gyrA, qnrB, parC, qepA and aac(6′)-Ib] or 48 °C (qnrA and qnrS) for 30 s, and 72 °C for 30 s. Amplification was performed using a DNA Engine Tetrad 2 (Bio-Rad) and BioTaq polymerase (Bioline). The resulting PCR amplicons were examined by electrophoresis and UV visualization on 2 % agarose gels containing ethidium bromide.
Amplicons produced from all strains specific for the gyrA, parC, qnrS, qepA and aac(6′)-Ib genes were sequenced with the same primers used for amplification. The forward and reverse strands of all PCR products were sequenced using BigDye terminators on a CEQ8000 DNA sequencer (Beckman Coulter).
The genetic environment of the qnrS gene was identified by cloning plasmid digestions in pUC18 and sequencing. DNA sequences were edited and analysed using Vector NTI software and compared with other sequences using blastn on the NCBI sequence database.
Molecular typing.
Strain differentiation was performed to identify clonally related strains isolated from different individuals. All hospital strains and all of the qnr- or aac(6′)-Ib-cr-positive community strains were typed by random amplified polymorphic DNA (RAPD) PCR using three different primers (Table 1) (Schultsz et al., 1997; Versalovic et al., 1991). All hospital strains from each 6-month period and all community strains were processed simultaneously for each primer, and electrophoresis was performed under identical conditions on the same day. RAPD patterns were clustered and interpreted by combining the resulting amplification patterns of all three primers and analysed using Bionumeric software (Applied Maths). Two isolates were considered to be clonally related when they had RAPD patterns that were identical. Isolates of a given species differing by one or two bands for all three primers combined were considered to be variants of a given type. This was based on similarities in patterns of multiple isolates obtained from individual patients, by visualization and computer-based analysis using Bionumerics.
Plasmid extraction and manipulation.
Plasmid DNA for electrotransformation was extracted using a Midiprep Plasmid DNA Extraction kit, following the manufacturer's recommendations (Qiagen). Escherichia coli TOP10 cells (Invitrogen) were transformed with isolated DNA using a Bio-Rad gene pulser, under conditions recommended by the manufacturer (Invitrogen). Transformants were selected on Luria–Bertani medium supplemented with 0.012 μg ciprofloxacin ml−1.
Conjugation was performed at a 1 : 1 ratio in liquid cultures by static incubation overnight at 37 °C. E. coli J53 (sodium azide resistant) was used as the recipient. Transconjugants were selected by plating onto Luria–Bertani medium supplemented with 0.03 μg ciprofloxacin ml−1 and 100 μg sodium azide ml−1.
Plasmid DNA for sizing and visualization was extracted using an alkaline lysis procedure, as described by Kado & Liu (1981). The resulting plasmid DNA was separated by electrophoresis in 0.7 % agarose gels made with 1× E buffer (Kado & Liu, 1981). Gels were run at 90 V for 3 h, stained with ethidium bromide and photographed. E. coli 39R861 containing plasmids of 7, 36, 63 and 147 kbp was used for sizing plasmid extractions on agarose gels.
RESULTS AND DISCUSSION
Isolation of antimicrobial-resistant commensal Enterobacteriaceae
A total of 194 hospitalized patients were tested over the study period. We isolated 70 E. coli strains, 123 K. pneumoniae strains and 29 other members of the Enterobacteriaceae, comprising Citrobacter species, Enterobacter cloacae, Proteus mirabilis, Klebsiella ornithinolytica, Pantoea species and a Vibrio fluvialis, resistant to gentamicin, ceftazidime or nalidixic acid, or a combination of these antimicrobials, from these patients. On the basis of RAPD results, we identified 53 unique E. coli strains, 62 unique K. pneumoniae strains and 16 other unique members of the Enterobacteriaceae.
The community group totalled 204 people and included 27 healthy adults, 77 healthy children and 100 healthy neonates. On the basis of culture on selective media containing gentamicin, ceftazidime or nalidixic acid, we isolated 340 resistant E. coli strains, 45 resistant K. pneumoniae strains and 28 other resistant members of the Enterobacteriaceae. Analysis of the RAPD patterns showed that all strains isolated from the different individuals were unique (data not shown). However, on the basis of typing and resistance patterns, we were able to isolate bacteria displaying a consistent RAPD pattern from consecutive isolates obtained over up to 14 days in 18 individuals, thus indicating carriage (data not shown).
We isolated organisms that were resistant to gentamicin, ceftazidime or nalidixic acid, or a combination of these, from 93 % (25/27) of the healthy adults, 92 % (71/77) of the healthy children, 64 % (64/100) of the healthy neonates and 68 % (132/194) of the tetanus patients. Combining the data for all 544 unique, resistant strains, 42 % of organisms were resistant to ceftazidime, 63 % were resistant to gentamicin and 74 % were resistant to nalidixic acid. All of the organisms that were resistant to ceftazidime were confirmed to be ESBL producers.
These results showed that the dissemination of resistant enteric bacteria is rife in the hospital and the community in Ho Chi Minh City. Antimicrobials are available without prescription in Vietnam; therefore, it is tempting to suggest that this is a primary source of selection for resistant organisms. However, none of the healthy individuals had had any antimicrobial therapy for at least 4 weeks prior to sample collection. The use of antimicrobials in the production of meat and vegetables is another potential major source of the ongoing selection of resistant organisms (Stobberingh & van den Bogaard, 2000; Teuber, 2001). In recent reports, investigators have shown intestinal colonization with fluoroquinolone-resistant and ESBL-producing E. coli in healthy schoolchildren in Latin America and in vegetarians in the USA (Pallecchi et al., 2007a, b; Sannes et al., 2008). These data suggest exposure to particular antimicrobials to such an extent as to maintain resistant organisms in the intestinal flora and/or transmission of resistant strains from individuals exposed to these agents followed by persistent carriage.
Amplification of plasmid-mediated quinolone resistance genes
All strains were subjected to PCR to amplify the individual qnrA, qnrB, qnrS, qepA and aac(6′)-Ib-cr genes. The majority of strains that generated a PMQR amplicon were positive for a single PMQR determinant (Table 2). Variability in PMQR determinant content was observed for a limited number of strains with identical RAPD patterns among the hospital strains. Based on a combination of RAPD pattern and PMQR content of strains, we identified 55 unique E. coli strains, 66 K. pneumoniae strains and 18 other members of the Enterobacteriaceae in this group.
Table 2.
Numbers of unique bacterial isolates from the hospital and community groups carrying a fluoroquinolone resistance determinant
| Source of bacterial isolate | Bacterial species (n) | PCR positive strains [n (%)] | |||||
|---|---|---|---|---|---|---|---|
| qnrA | qnrB | qnrS | aac(6′)-Ib-cr | qepA | >1 PMQR* | ||
| Hospital group | E. coli (55) | 5 (9.0) | 1 (1.8) | 5 (9.0) | 9 (16.4) | 1 (1.8) | 2 (3.6) |
| K. pneumoniae (66) | 4 (6.1) | 8 (12.1) | 52 (78.8) | 11 (16.7) | 0 (0) | 11 (16.7) | |
| Other† (18) | 6 (33.3) | 8 (44.4) | 6 (33.3) | 0 (0) | 0 (0) | 4 (22.2) | |
| Community group | E. coli (340) | 2 (0.6) | 0 (0) | 32 (9.4) | 9 (2.6) | 0 (0) | 1 (0.3) |
| K. pneumoniae (45) | 0 (0) | 0 (0) | 15 (33.3) | 1 (2.2) | 0 (0) | 1 (2.2) | |
| Other‡ (28) | 1 (3.6) | 2 (7.1) | 2 (7.1) | 1 (3.6) | 0 (0) | 1 (3.6) | |
*PCR positive for more than one PMQR gene inclusive of data in the rest of the table.
†Other bacterial species isolated from patients included three Enterobacter cloacae, three Citrobacter youngae, three Citrobacter freundii, two P. mirabilis, one Citrobacter species and one Citrobacter koseri.
‡Other bacterial species isolated from the community included two Citrobacter species, one Enterobacter cloacae and one Klebsiella terrigena.
Of the K. pneumoniae hospital strains, 78.8 % (52/66) produced amplicons for the qnrS gene (Table 2). The overall numbers of PMQR genes identified were lower in the community strains compared with the hospital strains; none the less, 9.4 % (32/340) of the E. coli community strains were PCR positive for the qnrS gene. We were able to detect aac(6′)-lb-cr-positive E. coli and K. pneumoniae from both hospital and community strains, albeit at a comparatively low frequency (Table 2). Of the 154 strains containing PMQR determinants, 98 (63.6 %) tested ESBL positive, including 59/112 (52.7 %) qnrS-positive strains.
The sequences for all of the amplicons of the qnrS fragment were indistinguishable and demonstrated 100 % sequence identity with qnrS1 from K. pneumoniae strain 052250 (GenBank accession no. EF683584). The sequences of the qnrA and qnrB amplicons and the single qepA PCR amplicon demonstrated 100 % identity to qnrA1 and qnrB1 from K. pneumoniae plasmid pMG252 (GenBank accession no. DQ831140) and K. pneumoniae plasmid pMG298 (GenBank accession no. DQ351241) and E. coli plasmid pHPA (GenBank accession no. AB263754), respectively.
These results indicated a very high prevalence of qnr genes, in particular the qnrS gene, in commensal isolates in Ho Chi Minh City. Recent reports showing qnr genes originating from waterborne bacteria may explain the dissemination of qnr genes in commensal bacteria (Cattoir et al., 2007a, 2008a). Faecal–oral transmission may facilitate exchange of qnr genes between waterborne and intestinal bacteria in the human host.
Effect of gyrA and parC mutations and PMQR genes on susceptibility to nalidixic acid and ciprofloxacin
Chromosomal quinolone resistance in E. coli and K. pneumoniae is determined predominantly by mutations at codons 83 and 87 in the gyrA gene. The combinatorial effect of harbouring one or more of the PMQR genes and the mutations in the gyrA gene on the susceptibility of the bacteria to nalidixic acid and ciprofloxacin was assessed in all unique hospital strains and all PMQR-positive community strains.
In the 55 unique hospital E. coli strains, 10 (18.2 %) had a single mutation at codon 83, none had a single mutation at codon 87, and 38 (69.1 %) had a double mutation in the gyrA gene. In 42 community E. coli strains, six (14.3 %) had a single mutation at codon 83 (Ser→Leu or Ala), none had a single mutation at codon 87 and 13 (31 .0%) had a double mutation (83Ser→Leu or Ala; 87Asp→Asn).
In the 66 hospital K. pneumoniae strains sequenced, Ser was mutated to Tyr, Ile and Phe at position 83 in eight (12.1 %), seven (10.6 %) and two (3.0 %) strains, respectively. Of the 15 community K. pneumoniae strains, Ser was mutated to Tyr at codon 83 in one strain (6.7 %). Asp was mutated to Ala at codon 87 in five (7.6 %) and one (6.7 %) hospital and community K. pneumoniae strains, respectively.
The combinatorial effects of the various PMQR genes and associated gyrA mutations on the MICs for nalidixic acid and ciprofloxacin are shown in Table 3 (hospital strains) and Table 4 (community strains). The biggest MIC increases associated with a single gene were associated with qnrS or aac(6′)-Ib-cr. We obtained two strains carrying the qnrS, qnrB and aac(6′)-Ib-cr genes with full resistance to nalidixic acid and ciprofloxacin, in the absence of known selective mutation in the gyrA gene (Table 3). The effect of PMQR genes on the resulting MIC to fluoroquinolones thus appeared to be reliant on the number and type of PMQR genes carried by the bacteria. The co-existence of qnrB and qnrS genes on two different plasmids within a strain has been shown previously. This combination did not have any additional effect on resistance to nalidixic acid (Hu et al., 2008), in contrast to our strains. We hypothesized a combinatorial effect, due to the proteins working in an independent manner: QnrB and QnrS protecting DNA gyrase and Aac(6′)-Ib-cr modifying the fluoroquinolone. However, as Aac(6′)-Ib-cr is thought not to modify quinolones such as nalidixic acid, we cannot currently rule out any possible effects of other quinolone resistance mechanisms, such as non-specific efflux pump activity.
Table 3.
Resulting MIC range (mg l−1) of E. coli and K. pneumoniae strains isolated from hospitalized patients, associated with a variety of combinations of PMQR genes and mutations in the gyrA gene
None of the strains contained mutations at codons 80 and 84 of the parC gene. NAL, Nalidixic acid; CIP, ciprofloxacin.
| PCR-positive gene | Strain (n) | Number of mutations in gyrA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | ||||||||
| n | NAL | CIP | n | NAL | CIP | n | NAL | CIP | ||
| None | E. coli (36) | 5 | 3–6 | 0.012–0.016 | 10 | >256 | 0.19–0.38 | 21 | >256 | >32 |
| K. pneumoniae (9) | 3 | 4–32 | 0.06–1 | 4 | >256 | 6–>32 | 2 | >256 | >32 | |
| qnrA | E. coli (5) | 0 | 0 | 5 | >256 | >32 | ||||
| K. pneumoniae (1) | 1 | 8 | 0.25 | 0 | 0 | |||||
| qnrB | E. coli (1) | 0 | 0 | 1 | >256 | >32 | ||||
| K. pneumoniae (1) | 1 | 12 | 0.38 | 0 | 0 | |||||
| qnrS | E. coli (4) | 2 | 24, 32 | 0.38 | 2 | >256 | >32 | |||
| K. pneumoniae (42) | 38* | 6–64 | 0.38–4 | 3 | >256 | 3–8 | 1 | >256 | >32 | |
| aac(6′)-Ib-cr | E. coli (8)† | 0 | 0 | 8 | >256 | >32 | ||||
| qnrA+qnrS | K. pneumoniae (2) | 2 | 24, 32 | 0.75, 1 | 0 | 0 | ||||
| qnrB+aac(6′)-Ib-cr | K. pneumoniae (3) | 0 | 1 | >256 | >32 | 2 | >256 | >32 | ||
| qnrS+aac(6′)-Ib-cr | E. coli (1) | 0 | 0 | 1 | >256 | >32 | ||||
| K. pneumoniae (3) | 2 | 16, 32 | 2 | 1 | >256 | >32 | 0 | |||
| qnrA+qnrS+aac(6′)-Ib-cr | K. pneumoniae (1) | 1 | 16 | 3 | 0 | 0 | ||||
| qnrB+qnrS+aac(6′)-Ib-cr | K. pneumoniae (4) | 2 | >256 | >32 | 0 | 2 | >256 | >32 | ||
*NAL: MIC50=16, MIC90=32; CIP: MIC50=0.75, MIC90=1.5.
†Included one qepA positive strain.
Table 4.
Resulting MIC range (mg l−1) of E. coli and K. pneumoniae strains isolated from healthy individuals, associated with a variety of combinations of PMQR genes and mutations in the gyrA gene
None of the strains contained mutations at codons 80 and 84 of the parC gene. NAL, Nalidixic acid; CIP, ciprofloxacin.
| PCR positive gene | Strains (n) | Number of mutations in gyrA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | ||||||||
| n | NAL | CIP | n | NAL | CIP | n | NAL | CIP | ||
| qnrA | E. coli (1) | 0 | 0 | 1 | >256 | 8 | ||||
| qnrS | E. coli (31) | 22* | 3–64 | 0.19–1.5 | 5 | >256 | 1.5–32 | 4 | >256 | >32 |
| K. pneumoniae (14) | 14† | 8–48 | 0.5–1.5 | 0 | 0 | |||||
| aac(6′)-Ib-cr | E. coli (9) | 0 | 1 | >256 | 2 | 8 | >256 | >32 | ||
| qnrA+qnrS | E. coli (1) | 1 | 6 | 0.5 | 0 | 0 | ||||
| qnrS+aac(6′)-Ib-cr | K. pneumoniae (1) | 0 | 0 | 1 | >256 | >32 | ||||
*NAL: MIC50=12, MIC90=48; CIP: MIC50=0.38, MIC90=0.75.
†NAL: MIC50=12, MIC90=16; CIP: MIC50=0.75, MIC90=1.5.
None of the PMQR-positive strains with a MIC of ≥8 mg nalidixic acid l−1 or ≥1 mg ciprofloxacin l−1 demonstrated any mutation at positions 80 and 84 of the parC gene, which have been shown to decrease the susceptibility of E. coli and K. pneumoniae to both nalidixic acid and ciprofloxacin (Brisse & Verhoef, 2001; Vila et al., 1996).
Characterization of qnrS-containing plasmids
We selected qnrS PCR-positive strains E. coli E66An (hospital), K. pneumoniae K1HV (community) and K. pneumoniae K18An (hospital) for further analysis (Table 5). We were unable to transfer nalidixic acid resistance by conjugation from either of the two K. pneumoniae strains into the E. coli recipient. However, we were able to transform E. coli TOP10 with purified plasmid DNA from both strains and select transformants on the basis of ciprofloxacin resistance. In contrast, we were able to transfer nalidixic acid resistance via both conjugation and transformation from E. coli E66An into another E. coli strain.
Table 5.
Resulting susceptibility patterns of wild-type, transconjugant and electrotransformant E. coli and K. pneumoniae strains
NAL, Nalidixic acid; CIP, ciprofloxacin.
| Strain | NAL MIC (mg l−1) | CIP MIC (mg l−1) | ESBL* | qnrS PCR |
|---|---|---|---|---|
| E. coli TOP10 | 1.5 | 0.006 | − | − |
| E. coli J53-Azi | 4 | 0.008 | − | − |
| K. pneumoniae K1HV | 8 | 0.75 | − | + |
| E. coli transformant 1 K1HV | 4 | 0.125 | − | + |
| E. coli transformant 2 K1HV | 4 | 0.125 | − | + |
| K. pneumoniae 18An | >256 | >32 | + | + |
| E. coli transformant 1 E18An | 4 | 0.125 | − | + |
| E. coli transformant 2 E18An | 4 | 0.094 | − | + |
| E. coli E66An | >256 | >32 | + | + |
| E. coli transformant E66An | 4 | 0.125 | + | + |
| E. coli transconjugant E66An | 16 | 0.75 | + | + |
*ESBL production was identified by resistance to ceftazidime (2 mg l−1) and confirmed by a double-disc method.
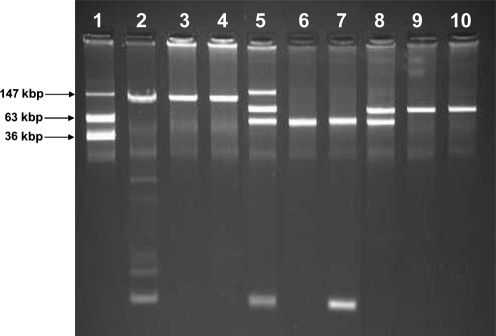
Comparison of the transformants derived from plasmid DNA isolated from both K. pneumoniae strains and the transformant and transconjugant derived from E. coli E66An indicated that sizes and resistance profiles varied across the different plasmids containing the qnrS gene (as confirmed by PCR) (Fig. 1, lanes 3, 4, 6 and 7; Table 5).
Fig. 1.
Agarose gel electrophoresis of alkaline plasmid lysis preparation from wild-type, transconjugant and electrotransformant E. coli and K. pneumoniae strains. Lanes: 1, E. coli 39R861; 2, K. pneumoniae 1HV; 3, E. coli transformant 1 1HV; 4, E. coli transformant 2 1HV; 5, K. pneumoniae 18An; 6, E. coli transformant 1 18An; 7, E. coli transformant 2 18An; 8, E. coli 66An; 9, E. coli transformant E66An; 10, E. coli transconjugant E66An.
The transconjugant strain derived from E. coli E66An, but not the transformants derived from the K. pneumoniae strains, additionally showed that ESBL production had been transferred alongside qnrS (Table 5). The correlation between qnrA or qnrB and ESBL genes is well known (Iabadene et al., 2008; Jiang et al., 2008; Oktem et al., 2008; Szabo et al., 2008; Wang et al., 2008), whilst the relationship between qnrS and other resistance genes is less well described. ESBL production was observed in only 52.7 % of the qnrS-positive strains in our study, and our transformation and conjugation experiments confirmed that qnrS genes can be located on plasmids that do not contain ESBL genes.
Sequence analysis of the region surrounding the qnrS gene in this E. coli strain demonstrated 100 % sequence identity to a previously sequenced transposon in plasmid pK245 in K. pneumoniae strain NK245, from a patient with hospital-acquired urinary tract infection in Taiwan (Chen et al., 2006). The qnrS gene appears to ‘piggy back’ on a mobile element, and selection may occur on the basis of the presence of other antimicrobial resistance genes (Chen et al., 2006). This is also suggested by the apparent redundant nature of the PMQR genes in those strains that have a double mutation in the gyrA gene. The variability in size of plasmids harbouring the qnrS gene, observed in our study, suggests that the genetic element carrying the qnrS gene has been mobilized onto numerous plasmids of different size.
Our study indicates that commensal organisms may represent the greatest reservoir and source of dissemination of plasmid-mediated antimicrobial resistance genes, such as qnrS, in Vietnam.
Acknowledgments
The authors wish to acknowledge the efforts of all of the nursing staff involved in sample collection. This work was supported by the Wellcome Trust, UK.
Abbreviations
ESBL, extended-spectrum β-lactamase
PMQR, plasmid-mediated quinolone resistance
RAPD, random amplified polymorphic DNA
References
- Brisse, S. & Verhoef, J. (2001). Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int J Syst Evol Microbiol 51, 915–924. [DOI] [PubMed] [Google Scholar]
- Cattoir, V., Poirel, L., Mazel, D., Soussy, C. J. & Nordmann, P. (2007a). Vibrio splendidus as the source of plasmid-mediated QnrS-like quinolone resistance determinants. Antimicrob Agents Chemother 51, 2650–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoir, V., Weill, F. X., Poirel, L., Fabre, L., Soussy, C. J. & Nordmann, P. (2007b). Prevalence of qnr genes in Salmonella in France. J Antimicrob Chemother 59, 751–754. [DOI] [PubMed] [Google Scholar]
- Cattoir, V., Poirel, L., Aubert, C., Soussy, C. J. & Nordmann, P. (2008a). Unexpected occurrence of plasmid-mediated quinolone resistance determinants in environmental Aeromonas spp. Emerg Infect Dis 14, 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoir, V., Poirel, L. & Nordmann, P. (2008b). Plasmid-mediated quinolone resistance pump QepA2 in an Escherichia coli isolate from France. Antimicrob Agents Chemother 52, 3801–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau, T. T., Campbell, J. I., Galindo, C. M., Van Minh Hoang, N., Diep, T. S., Nga, T. T., Van Vinh Chau, N., Tuan, P. Q., Page, A. L. & other authors (2007). Antimicrobial drug resistance of Salmonella enterica serovar Typhi in Asia and molecular mechanism of reduced susceptibility to the fluoroquinolones. Antimicrob Agents Chemother 51, 4315–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. T., Shu, H. Y., Li, L. H., Liao, T. L., Wu, K. M., Shiau, Y. R., Yan, J. J., Su, I. J., Tsai, S. F. & Lauderdale, T. L. (2006). Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-β-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 50, 3861–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2007). Performance Standards For Antimicrobial Susceptibility Testing, 17th Informational Supplement, M100-S17. Wayne, PA: Clinical and Laboratory Standards Institute.
- Gay, K., Robicsek, A., Strahilevitz, J., Park, C. H., Jacoby, G., Barrett, T. J., Medalla, F., Chiller, T. M. & Hooper, D. C. (2006). Plasmid-mediated quinolone resistance in non-Typhi serotypes of Salmonella enterica. Clin Infect Dis 43, 297–304. [DOI] [PubMed] [Google Scholar]
- Hata, M., Suzuki, M., Matsumoto, M., Takahashi, M., Sato, K., Ibe, S. & Sakae, K. (2005). Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob Agents Chemother 49, 801–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, P. G., Fluit, A. C. & Schmitz, F. J. (2003). Fluoroquinolones: structure and target sites. Curr Drug Targets 4, 181–190. [DOI] [PubMed] [Google Scholar]
- Hu, F. P., Xu, X. G., Zhu, D. M. & Wang, M. G. (2008). Coexistence of qnrB4 and qnrS1 in a clinical strain of Klebsiella pneumoniae. Acta Pharmacol Sin 29, 320–324. [DOI] [PubMed] [Google Scholar]
- Iabadene, H., Messai, Y., Ammari, H., Ramdani-Bouguessa, N., Lounes, S., Bakour, R. & Arlet, G. (2008). Dissemination of ESBL and Qnr determinants in Enterobacter cloacae in Algeria. J Antimicrob Chemother 62, 133–136. [DOI] [PubMed] [Google Scholar]
- Jacoby, G. A. (2005). Mechanisms of resistance to quinolones. Clin Infect Dis 41, S120–S126. [DOI] [PubMed] [Google Scholar]
- Jacoby, G. A., Walsh, K. E., Mills, D. M., Walker, V. J., Oh, H., Robicsek, A. & Hooper, D. C. (2006). qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob Agents Chemother 50, 1178–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y., Zhou, Z., Qian, Y., Wei, Z., Yu, Y., Hu, S. & Li, L. (2008). Plasmid-mediated quinolone resistance determinants qnr and aac(6')-Ib-cr in extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in China. J Antimicrob Chemother 61, 1003–1006. [DOI] [PubMed] [Google Scholar]
- Kado, C. I. & Liu, S. T. (1981). Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol 145, 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., Zeng, Z., Chen, Z., Xu, X., Wang, X., Deng, Y., Lü, D., Huang, L., Zhang, Y. & other authors (2009). High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac(6')-Ib-cr and qepA among ceftiofur-resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicrob Agents Chemother 53, 519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melano, R., Corso, A., Petroni, A., Centron, D., Orman, B., Pereyra, A., Moreno, N. & Galas, M. (2003). Multiple antibiotic-resistance mechanisms including a novel combination of extended-spectrum β-lactamases in a Klebsiella pneumoniae clinical strain isolated in Argentina. J Antimicrob Chemother 52, 36–42. [DOI] [PubMed] [Google Scholar]
- Mendes, R. E., Bell, J. M., Turnidge, J. D., Yang, Q., Yu, Y., Sun, Z. & Jones, R. N. (2008). Carbapenem-resistant isolates of Klebsiella pneumoniae in China and detection of a conjugative plasmid (blaKPC-2 plus qnrB4) and a blaIMP-4 gene. Antimicrob Agents Chemother 52, 798–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktem, I. M., Gulay, Z., Bicmen, M. & Gur, D. (2008). qnrA prevalence in extended-spectrum β-lactamase-positive Enterobacteriaceae isolates from Turkey. Jpn J Infect Dis 61, 13–17. [PubMed] [Google Scholar]
- Pallecchi, L., Bartoloni, A., Fiorelli, C., Mantella, A., Di Maggio, T., Gamboa, H., Gotuzzo, E., Kronvall, G., Paradisi, F. & Rossolini, G. M. (2007a). Rapid dissemination and diversity of CTX-M extended-spectrum β-lactamase genes in commensal Escherichia coli isolates from healthy children from low-resource settings in Latin America. Antimicrob Agents Chemother 51, 2720–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallecchi, L., Lucchetti, C., Bartoloni, A., Bartalesi, F., Mantella, A., Gamboa, H., Carattoli, A., Paradisi, F. & Rossolini, G. M. (2007b). Population structure and resistance genes in antibiotic-resistant bacteria from a remote community with minimal antibiotic exposure. Antimicrob Agents Chemother 51, 1179–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C. H., Robicsek, A., Jacoby, G. A., Sahm, D. & Hooper, D. C. (2006). Prevalence in the United States of aac(6')-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother 50, 3953–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perichon, B., Courvalin, P. & Galimand, M. (2007). Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA-mediated efflux in Escherichia coli. Antimicrob Agents Chemother 51, 2464–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robicsek, A., Jacoby, G. A. & Hooper, D. C. (2006a). The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis 6, 629–640. [DOI] [PubMed] [Google Scholar]
- Robicsek, A., Strahilevitz, J., Sahm, D. F., Jacoby, G. A. & Hooper, D. C. (2006b). qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob Agents Chemother 50, 2872–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannes, M. R., Belongia, E. A., Kieke, B., Smith, K., Kieke, A., Vandermause, M., Bender, J., Clabots, C., Winokur, P. & Johnson, J. R. (2008). Predictors of antimicrobial-resistant Escherichia coli in the feces of vegetarians and newly hospitalized adults in Minnesota and Wisconsin. J Infect Dis 197, 430–434. [DOI] [PubMed] [Google Scholar]
- Schultsz, C., Moussa, M., van Ketel, R., Tytgat, G. N. & Dankert, J. (1997). Frequency of pathogenic and enteroadherent Escherichia coli in patients with inflammatory bowel disease and controls. J Clin Pathol 50, 573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobberingh, E. E. & van den Bogaard, A. E. (2000). Spread of antibiotic resistance from food animals to man. Acta Vet Scand Suppl 93, 47–50. (Discussion 51–42) [PubMed] [Google Scholar]
- Strahilevitz, J., Engelstein, D., Adler, A., Temper, V., Moses, A. E., Block, C. & Robicsek, A. (2007). Changes in qnr prevalence and fluoroquinolone resistance in clinical isolates of Klebsiella pneumoniae and Enterobacter spp. collected from 1990 to 2005. Antimicrob Agents Chemother 51, 3001–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo, D., Kocsis, B., Rokusz, L., Szentandrassy, J., Katona, K., Kristof, K. & Nagy, K. (2008). First detection of plasmid-mediated, quinolone resistance determinants qnrA, qnrB, qnrS and aac(6')-Ib-cr in extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae in Budapest, Hungary. J Antimicrob Chemother 62, 630–632. [DOI] [PubMed] [Google Scholar]
- Teuber, M. (2001). Veterinary use and antibiotic resistance. Curr Opin Microbiol 4, 493–499. [DOI] [PubMed] [Google Scholar]
- Tran, J. H., Jacoby, G. A. & Hooper, D. C. (2005a). Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli topoisomerase IV. Antimicrob Agents Chemother 49, 3050–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, J. H., Jacoby, G. A. & Hooper, D. C. (2005b). Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob Agents Chemother 49, 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versalovic, J., Koeuth, T. & Lupski, J. R. (1991). Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 19, 6823–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila, J., Ruiz, J., Goni, P. & De Anta, M. T. (1996). Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother 40, 491–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, A., Yang, Y., Lu, Q., Wang, Y., Chen, Y., Deng, L., Ding, H., Deng, Q., Wang, L. & Shen, X. (2008). Occurrence of qnr-positive clinical isolates in Klebsiella pneumoniae producing ESBL or AmpC-type β-lactamase from five pediatric hospitals in China. FEMS Microbiol Lett 283, 112–116. [DOI] [PubMed] [Google Scholar]
- Yamane, K., Wachino, J., Suzuki, S., Kimura, K., Shibata, N., Kato, H., Shibayama, K., Konda, T. & Arakawa, Y. (2007). New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother 51, 3354–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane, K., Wachino, J., Suzuki, S. & Arakawa, Y. (2008). Plasmid-mediated qepA gene among Escherichia coli clinical isolates from Japan. Antimicrob Agents Chemother 52, 1564–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]