Abstract
Between 2000 and 2004, dengue virus type 1 (DENV-1) genotypes I and II from Asia were introduced into the Pacific region and co-circulated in some localities. Envelope protein gene sequences of DENV-1 from 12 patients infected on the island of New Caledonia were obtained, five of which carried genotype I viruses and six, genotype II viruses. One patient harboured a mixed infection, containing viruses assigned to both genotypes I and II, as well as a number of inter-genotypic recombinants. This is the first report of a population of dengue viruses isolated from a patient containing both parental and recombinant viruses.
Dengue is a major cause of mortality and morbidity in the tropics (Gubler & Meltzer, 1999), and the ability of positive-sense RNA viruses, such as dengue virus (DENV), to evolve rapidly (Craig et al., 2003; Sittisombut et al., 1997; Thu et al., 2004; Wang et al., 2002; Wittke et al., 2002) poses a serious challenge to the development and efficacy of vaccines. The principal source of the genetic diversity in DENV is error-prone replication with RNA-dependent RNA polymerase (Steinhauer et al., 1992), such that one genomic mutation occurs in nearly every cycle of virus replication. DENV populations at a particular locality may also change rapidly due to periodic selective sweeps (Bennett et al., 2003), to the introduction of exotic strains of virus (A-Nuegoonpipat et al., 2004; Messer et al., 2003; Rico Hesse et al., 1997; Thu et al., 2004), to random genetic bottlenecks (Sittisombut et al., 1997; Thu et al., 2004; Wittke et al., 2002) or to intra-serotypic recombination (AbuBakar et al., 2002; Domingo et al., 2006; Holmes et al., 1999; Tolou et al., 2001; Worobey et al., 1999). Whilst it is widely accepted that another positive-strand RNA virus, polio virus, undergoes both intra-serotypic (Ledinko, 1963) and inter-serotypic (Minor et al., 1986; Tolskaya et al., 1983) recombination, as well as inter-species recombination (Kew et al., 2002), there has been considerable debate about whether recombination occurs in DENV (Rico-Hesse, 2003) and the relevance of any recombination to the deployment of live-attenuated flavivirus vaccines (Monath et al., 2005; Seligman & Gould, 2004). However, there is no mechanistic reason why recombination cannot occur in DENV and this process is being described with increasing frequency in hepatitis C virus, another member of the family Flaviviridae (Bernardin et al., 2006; Gao et al., 2007; Kageyama et al., 2006; Legrand-Abravanel et al., 2007; Moreno et al., 2006; Noppornpanth et al., 2006).
The ecological conditions most likely to facilitate recombination in DENV involve the co-circulation of multiple viral populations, including distinct genotypes, so that there are opportunities for a mosquito vector to ingest multiple variants by feeding on a number of infected hosts, or for a host to be infected almost simultaneously by vectors infected with different viruses. These opportunities exist throughout most of south-east Asia. Further, as there are numerous reports of multiple serotypes of DENV recovered from single hosts (Gubler et al., 1985; Laille et al., 1991; Twiddy et al., 2002; Wang et al., 2003), it is likely that mixed infections with different genotypes of the same serotype occur where they co-circulate, but that current diagnostic procedures are unable to detect such mixtures. An opportunity for frequent mixed infections with different strains of DENV arose after 2000, when multiple genotypes of DENV-1 were introduced into the Pacific from distant Asian localities, including the Philippines, Myanmar/Thailand and Malaysia (A-Nuegoonpipat et al., 2004).
To determine the origin of the DENV-1 circulating in New Caledonia during an outbreak in 2002 and 2003, virus was first recovered from 12 patients by culturing 100 μl serum on 25 cm2 monolayers of C6-36 Aedes albopictus cells in 7–8 ml RPMI 1640 tissue-culture medium at 30 °C for 7 days. RNA was recovered from virus in supernate from the infected cultures of C6-36 cells and from the serum of patient D1.New Caledonia.416/03 by using QIAamp viral RNA mini columns (Qiagen) according to the manufacturer's instructions. The envelope (E) protein gene was amplified by RT-PCR (A-Nuegoonpipat et al., 2004) using oligonucleotide primers D1-764F and D1-2467R (see Supplementary Table S1, available in JGV Online), corresponding to regions of the pre-membrane and non-structural 1 proteins of DENV-1, and a mixture of Taq and Pwo polymerases (Expand Long Template DNA polymerase; Roche). The PCR product was analysed on 1.5 % (w/v) agarose–Tris/acetate EDTA gels and the band of cDNA of interest was excised and purified by using a High Pure PCR Purification kit (Roche) according to the manufacturer's instructions. Approximately 100 ng cDNA or 600 ng plasmid (see below) was sequenced, using 6.4 pmol oligonucleotide primer and an ABI PRISM Dye Terminator cycle sequencing ready reaction kit (Perkin Elmer) according to the manufacturer's instructions. The product was purified by using Dye-EX spin columns (Qiagen) and analysed at the Australian Genome Research Facility on a 3730XL nucleotide sequencer (Applied Biosystems). Sequences are identified as DENV serotype, country of origin, sample number (and clone number)/year sample obtained.
To determine the DENV-1 genotype of each of 12 New Caledonian viruses, we conducted a maximum-likelihood (ML) phylogenetic analysis of the complete E gene (1485 bp) by using the paup* package (Swofford, 2003) with the best-fit GTR+Γ4 model of nucleotide substitution, determined by using modeltest (Posada & Crandall, 1998), and 1000 bootstrap neighbour-joining replications under the same ML model (the sequence alignment is provided as a nexus file in JGV Online). This analysis revealed that five patients were infected with genotype I viruses, whilst six patients were infected with viruses of genotype II (tree not shown; available from authors on request). However, it was not possible to obtain an unambiguous consensus nucleotide sequence, using cDNA as a template, for the E gene of a single virus isolate – D1.New Caledonia.416/03 – taken from a patient who developed dengue in New Caledonia in February 2003 (chromatogram available in JGV Online as Supplementary Fig. S1; all other isolates gave unambiguous sequences). Therefore, cDNA corresponding to the E genes of viruses from this patient was derived by RT-PCR using primers D1-764F and D1-2467R or D1-938F and D1-2386R (used to prepare clones D1.New Caledonia.416.3/03, 10/03, 14/03 and 91/03) and purified from 1.5 % (w/v) agarose–Tris/acetate EDTA gels by using a High Pure PCR purification kit (Roche) according to the manufacturer's instructions. 5′-ATP extensions were added to the cDNA (Craig et al., 2003) and it was ligated into pGEM-T Easy plasmids (Promega) and used to transform Escherichia coli JM109 or DH5α bacteria (Sambrook & Russell, 2001). Plasmids were purified from individual bacterial colonies, which grew on Luria–Bertani agar supplemented with 100 μg ampicillin ml−1, 0.5 mM IPTG and 80 μg 5-bromo-4-chloro-indoyl-β-d-galactosidase ml−1 (Sambrook & Russell, 2001), using a commercial plasmid purification kit (Qiagen).
The nucleotide sequences of 24 clones derived from the virus population of D1.New Caledonia.416/03 (including seven obtained directly from serum and denoted ‘416S’ in all figures) varied at 266 sites. Clones 19 and 32 had single-nucleotide deletions at E250 and E308, respectively, resulting in frame-shifts and the production of downstream stop codons. The deduced amino acid sequences of the clones varied at 110 sites (after replacement of the nucleotide deletions in the sequences of clones 19 and 32). Clone 32 also had a stop codon (UAG) at amino acid position E248, as did clone 7 at E391.
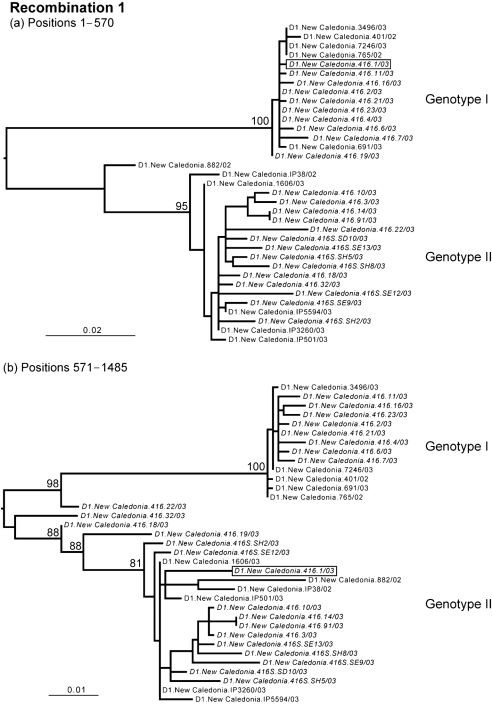
Notably, some sequences of the clones from patient D1.New Caledonia.416/03 segregated with both genotype I and genotype II DENV-1, which were circulating in New Caledonia during the same outbreak, whereas others occupied ‘intermediate’ phylogenetic positions between the two genotypes, suggestive of recombination (Figs 1–3). To determine whether recombination had occurred among the viruses sampled from patient D1.New Caledonia.416/03, we employed the rdp2 program (Martin et al., 2005), which employs six different algorithms to identify recombinants and their break points: rdp, SiScan, bootscan, chimeric, MaxChi and geneconv. Default settings were used in all cases. Strikingly, all six methods identified frequent recombination events; for example, geneconv, which is considered one of the most robust methods for detecting recombination (Posada, 2002), identified 1609 possible recombination events. Among all six methods in rdp2, there was particularly strong evidence for recombination break points at nucleotide positions 570, 620 and 1100.
Fig. 1.
The changing phylogenetic position of clone D1.New Caledonia.416.1/03 (boxed), indicative of inter-genotypic recombination in DENV-1. The maximum-likelihood phylogenetic tree was inferred by using all (n=24) clones from patient D1.New Caledonia.416/03 (in italics), as well as the consensus sequences from 11 other patients sampled from New Caledonia, inferred for the entire E gene (1485 bp). Clones derived from serum are denoted ‘416S’. Bootstrap values are shown for key nodes only and all branches are scaled according to the number of nucleotide substitutions per site.
Fig. 3.
The changing phylogenetic position of clone D1.New Caledonia.416.22/03 (boxed), indicative of inter-genotypic recombination in DENV-1 (sequence labels as defined in the legend to Fig. 1). Bootstrap values are shown for key nodes only and all branches are scaled according to the number of nucleotide substitutions per site.
To test further for recombination, ML phylogenetic trees were inferred for the regions of nucleotide sequence on either side of the break points detected by rdp2 using the methods described above (Figs 1–3). This analysis provided compelling evidence for recombination among the genotype I and II viruses that co-infected patient D1.New Caledonia.416/03, involving clones 1 (recombination event 1), 19 (recombination event 2) and 22 (recombination event 3). For each of these three recombinants, there was strong evidence for the phylogenetic incongruence indicative of recombination; the viruses occupied different phylogenetic (i.e. genotypic) positions on either side of specific break points that were supported by high numbers of bootstrap replications. Although clones 1, 19 and 22 represent clear instances of inter-genotype recombination, the extremely high frequency of recombination identified by rdp2, as well as the changing phylogenetic positions of other sequences (for example, clones 18 and 32 in Fig. 3), suggests that this process has occurred very frequently in this patient. Crucially, none of the break points were in regions containing sequences the same as, or complementary to, the sequences of oligonucleotide primers used for sequencing or for PCR, indicating that they are bona fide recombinants rather than laboratory artefacts.
As less than 50 μl (badly degraded) serum was available from patient D1.New Caledonia.416/03 after virus isolation had been performed, only seven clones of the E protein gene could be obtained from this source. The sequences of these clones served to confirm the presence of genotype 1 DENV-1 in this patient, but as none were recombinant, we were unable to distinguish whether the recombinant genomes that we detected in this virus isolate arose in the patient or during the isolation of the virus in cultures of A. albopictus mosquito cells. Indeed, it is highly likely that C6-36 cells amplify some subpopulations of virus selectively. Nonetheless, our results provide unambiguous evidence of intra-serotypic recombination in populations of DENV-1, if not in a patient, then at least in cells derived from a species of mosquito involved in dengue transmission. However, we are unable to determine whether these recombinants are viable, as it not possible to plaque-purify DENV to homogeneity. Even if the recombinant E protein is functional, nucleotide insertions and deletions elsewhere in the genome could render the virus non-functional.
This is only the second observation of both parental and recombinant DENV in a population within a single host (Craig et al., 2003). Whilst there are several reports of infections of humans with multiple DENV serotypes, most techniques employed to identify virus in patient tissues would not detect infections with multiple genotypes of the same DENV serotype, as is clearly necessary to give rise to the recombinant viral populations described here. We therefore propose that recombination events in DENV may occur more frequently than has been reported, but that the practice of determining consensus nucleotide sequences from cDNA greatly limits the chances of their detection; only if the recombinant represents the dominant viral population will it be sampled by consensus sequencing. Further, nucleotide deletions, which also may be the footprints of recombination events, have been observed in a number of DENV populations (Aaskov et al., 2006; Craig et al., 2003; Lin et al., 2004; Wang et al., 2002). Hence, the examination of a much larger number of recombinant viruses and their parents may enable ‘hot spots’ of recombination to be identified and permit a systematic analysis of the factors mediating recombination in these viruses (Nagy & Simon, 1997).
By continuing to monitor the epidemiology of the ongoing outbreak of DENV-1 virus infection in the Pacific, it may be possible to determine whether the recombination events described here have conferred any fitness benefit on these viruses, although it is likely that most recombinants are deleterious and so removed by purifying selection. This notwithstanding, it is remarkable that both parental and different recombinant viral genomes were observed in an isolate from a single patient, particularly as the recombination appeared to have involved two genotypes of DENV-1 imported into New Caledonia (A-Nuegoonpipat et al., 2004). This suggests that recombination in DENV may be more common than thought previously, but that the process will remain largely undetected until more studies of clonal diversity are undertaken.
Supplementary Material
Fig. 2.
The changing phylogenetic position of clone D1.New Caledonia.416.19/03 (boxed), indicative of inter-genotypic recombination in DENV-1 (sequence labels as defined in the legend to Fig. 1). Bootstrap values are shown for key nodes only and all branches are scaled according to the number of nucleotide substitutions per site.
Acknowledgments
This study was supported by grants from the Lee Foundation, the Wellcome Trust and the Royal Society.
Footnotes
A table showing oligonucleotide primers used for PCR or sequencing of the DENV-1 E genes, a chromatogram of the sequence of complementary-strand cDNA obtained following RT-PCR and a sequence alignment (nexus format) are available as supplementary material with the online version of this paper.
References
- Aaskov, J., Buzacott, K., Thu, H. M., Lowry, K. & Holmes, E. C. (2006). Long-term transmission of defective RNA viruses in humans and Aedes mosquitoes. Science 311, 236–238. [DOI] [PubMed] [Google Scholar]
- AbuBakar, S., Wong, P.-F. & Chan, Y.-F. (2002). Emergence of dengue virus type 4 genotype IIA in Malaysia. J Gen Virol 83, 2437–2442. [DOI] [PubMed] [Google Scholar]
- A-Nuegoonpipat, A., Berlioz-Arthaud, A., Chow, V., Endy, T., Lowry, K., Mai, L. Q., Ninh, T. U., Pyke, A., Reid, M. & other authors (2004). Sustained transmission of dengue virus type 1 in the Pacific due to repeated introductions of different Asian strains. Virology 329, 505–512. [DOI] [PubMed] [Google Scholar]
- Bennett, S. N., Holmes, E. C., Chirivella, M., Rodriguez, D. M., Beltran, M., Vorndam, V., Gubler, D. J. & McMillan, W. O. (2003). Selection-driven evolution of emergent dengue virus. Mol Biol Evol 20, 1650–1658. [DOI] [PubMed] [Google Scholar]
- Bernardin, F., Herring, B., Page-Shafer, K., Kuiken, C. & Delwart, E. (2006). Absence of HCV viral recombination following superinfection. J Viral Hepat 13, 532–537. [DOI] [PubMed] [Google Scholar]
- Craig, S., Thu, H. M., Lowry, K., Wang, X.-F., Holmes, E. C. & Aaskov, J. G. (2003). Diverse dengue type 2 virus populations contain recombinant and both parental viruses in a single mosquito host. J Virol 77, 4463–4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo, C., Palacios, G., Jabado, O., Reyes, N., Niedrig, M., Gascon, J., Cabrerizo, M., Lipkin, W. I. & Tenorio, A. (2006). Use of a short fragment of the C-terminal E gene for detection and characterization of two new lineages of dengue virus 1 in India. J Clin Microbiol 44, 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, F., Nainan, O. V., Khudyakov, Y., Li, J., Hong, Y., Gonzales, A. C., Spelbring, J. & Margolis, H. S. (2007). Recombinant hepatitis C virus in experimentally infected chimpanzees. J Gen Virol 88, 143–147. [DOI] [PubMed] [Google Scholar]
- Gubler, D. J. & Meltzer, M. (1999). Impact of dengue/dengue haemorrhagic fever on the developing world. Adv Virus Res 53, 35–70. [DOI] [PubMed] [Google Scholar]
- Gubler, D. J., Kuno, G., Sather, G. E. & Waterman, S. H. (1985). A case of natural concurrent human infection with two dengue viruses. Am J Trop Med Hyg 34, 170–173. [DOI] [PubMed] [Google Scholar]
- Holmes, E. C., Worobey, M. & Rambaut, A. (1999). Phylogenetic evidence for recombination in dengue virus. Mol Biol Evol 16, 405–409. [DOI] [PubMed] [Google Scholar]
- Kageyama, S., Agdamag, D. M., Alesna, E. T., Leano, P. S., Heredia, A. M., Abellanosa-Tac-An, I. P., Jereza, L. D., Tanimoto, T., Yamamura, J. & Ichimura, H. (2006). A natural inter-genotypic (2b/1b) recombinant of hepatitis C virus in the Philippines. J Med Virol 78, 1423–1428. [DOI] [PubMed] [Google Scholar]
- Kew, O., Morris-Glasgow, V., Landaverde, M., Burns, C., Shaw, J., Garib, Z., Andre, J., Blackman, E., Freeman, C. J. & other authors (2002). Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science 296, 356–359. [DOI] [PubMed] [Google Scholar]
- Laille, M., Deubel, V. & Sainte-Marie, F. F. (1991). Demonstration of concurrent dengue 1 and dengue 3 infections in six patients by the polymerase chain reaction. J Med Virol 34, 51–54. [DOI] [PubMed] [Google Scholar]
- Ledinko, N. (1963). Genetic recombination with poliovirus type 1. Studies of crosses between a normal horse serum-resistant mutant and several guanidine-resistant mutants of the same strain. Virology 20, 107–119. [DOI] [PubMed] [Google Scholar]
- Legrand-Abravanel, F., Claudinon, J., Nicot, F., Dubois, M., Chapuy-Regaud, S., Sandres-Saune, K., Pasquier, C. & Izopet, J. (2007). New natural intergenotypic (2/5) recombinant of hepatitis C virus. J Virol 81, 4357–4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S.-R., Hsieh, S.-C., Yueh, Y.-Y., Lin, T.-H., Chao, D.-Y., Chen, W.-J., King, C.-C. & Wang, W.-K. (2004). Study of sequence variation of dengue type 3 virus in naturally infected mosquitoes and human hosts: implications for transmission and evolution. J Virol 78, 12717–12721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, D. P., Williamson, C. & Posada, D. (2005). rdp2: recombination detection and analysis from sequence alignments. Bioinformatics 21, 260–262. [DOI] [PubMed] [Google Scholar]
- Messer, W. B., Gubler, D. J., Harris, E., Sivananthan, K. & de Silva, A. M. (2003). Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg Infect Dis 9, 800–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor, P. D., John, A., Ferguson, M. & Icenogle, J. P. (1986). Antigenic and molecular evolution of the vaccine strain of type 3 poliovirus during the period of excretion by a primary vaccinee. J Gen Virol 67, 693–706. [DOI] [PubMed] [Google Scholar]
- Monath, T. P., Kanesa-Thasan, N., Guirakhoo, F., Pugachev, K., Almond, J., Lang, J., Quentin-Millet, M.-J., Barrett, A. D. T., Brinton, M. A. & other authors (2005). Recombination and flavivirus vaccines: a commentary. Vaccine 23, 2956–2958. [DOI] [PubMed] [Google Scholar]
- Moreno, M. P., Casane, D., Lopez, L. & Cristina, J. (2006). Evidence of recombination in quasispecies populations of a hepatitis C virus patient undergoing anti-viral therapy. Virol J 3, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, P. D. & Simon, A. E. (1997). New insights into the mechanism of RNA recombination. Virology 235, 1–9. [DOI] [PubMed] [Google Scholar]
- Noppornpanth, S., Lien, T. X., Poovorawan, Y., Smits, S. L., Osterhaus, A. D. & Haagmans, B. L. (2006). Identification of a naturally occurring recombinant genotype 2/6 hepatitis C virus. J Virol 80, 7569–7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada, D. (2002). Evaluation of methods for detecting recombination from DNA sequences: empirical data. Mol Biol Evol 19, 708–717. [DOI] [PubMed] [Google Scholar]
- Posada, D. & Crandall, K. A. (1998). modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818. [DOI] [PubMed] [Google Scholar]
- Rico-Hesse, R. (2003). Microevolution and virulence of dengue viruses. Adv Virus Res 59, 315–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico-Hesse, R., Harrison, L. M., Salas, R. A., Tovar, D., Nisalak, A., Ramos, C., Boshell, J., de Mesa, M. T. R., Nogueira, R. M. R. & da Rosa, A. T. (1997). Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology 230, 244–251. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. & Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Seligman, S. J. & Gould, E. A. (2004). Live flavivirus vaccines: reasons for caution. Lancet 363, 2073–2075. [DOI] [PubMed] [Google Scholar]
- Sittisombut, N., Sistayanarain, A., Cardosa, M. J., Salminen, M., Damrongdachakul, S., Kalayanarooj, S., Rojanasuphot, S., Supawadee, J. & Maneekarn, N. (1997). Possible occurrence of a genetic bottleneck in dengue serotype 2 viruses between the 1980 and 1987 epidemic seasons in Bangkok, Thailand. Am J Trop Med Hyg 57, 100–108. [DOI] [PubMed] [Google Scholar]
- Steinhauer, D. A., Domingo, E. & Holland, J. J. (1992). Lack of evidence for proofreading mechanisms associated with an RNA polymerase. Gene 122, 281–288. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L. (2003). paup*: phylogenetic analysis using parsimony (*and other methods), version 4. Sunderland, MA: Sinauer Associates.
- Thu, H. M., Lowry, K., Myint, T. T., Shwe, T. N., Han, A. M., Khin, K. K., Thant, K. Z., Thein, S. & Aaskov, J. G. (2004). Myanmar dengue outbreak associated with displacement of serotypes 2, 3 and 4 by dengue 1. Emerg Infect Dis 10, 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolou, H. J. G., Couissinier-Paris, G. P., Durand, J.-P., Mercier, V., de Pina, J. J., de Micco, P., Billoir, F., Charrel, R. N. & de Lamballerie, X. (2001). Evidence for recombination in natural populations of dengue virus type 1 based on the analysis of complete genome sequences. J Gen Virol 82, 1283–1290. [DOI] [PubMed] [Google Scholar]
- Tolskaya, E. A., Romanova, L. A., Kolesnikova, M. S. & Agol, V. I. (1983). Intertypic recombination in poliovirus: genetic and biochemical studies. Virology 124, 121–132. [DOI] [PubMed] [Google Scholar]
- Twiddy, S. S., Farrar, J. F., Chau, N. V., Wills, B., Gould, E. A., Gritsun, T., Lloyd, G. & Holmes, E. C. (2002). Phylogenetic relationships and differential selection pressures among genotypes of dengue-2 virus. Virology 298, 63–72. [DOI] [PubMed] [Google Scholar]
- Wang, W.-K., Sung, T.-L., Lee, C.-N., Lin, T.-Y. & King, C.-C. (2002). Dengue type 3 virus in plasma is a population of closely related genomes: quasispecies. J Virol 76, 4662–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W.-K., Chao, D.-Y., Lin, S.-R., King, C.-C. & Chang, S. C. (2003). Concurrent infections by two dengue virus serotypes among dengue patients in Taiwan. J Microbiol Immunol Infect 36, 89–95. [PubMed] [Google Scholar]
- Wittke, V., Robb, T. E., Thu, H. M., Nisalak, A., Nimmannitya, S., Kalayanrooj, S., Vaughn, D. W., Endy, T. P., Holmes, E. C. & Aaskov, J. G. (2002). Extinction and rapid emergence of strains of dengue 3 virus during an interepidemic period. Virology 301, 148–156. [DOI] [PubMed] [Google Scholar]
- Worobey, M., Rambaut, A. & Holmes, E. C. (1999). Widespread intra-serotypic recombination in natural populations of dengue virus. Proc Natl Acad Sci U S A 96, 7352–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.