Abstract
Background
A decrease in retinoic acid levels due to alcohol consumption has been proposed as a contributor to such conditions as fetal alcohol spectrum diseases and ethanol-induced cancers. One molecular mechanism, competitive inhibition by ethanol of the catalytic activity of human alcohol dehydrogenase (EC 1.1.1.1) (ADH) on all-trans retinol oxidation has been shown for the ADH7 isoform. Ethanol metabolism also causes an increase in the free NADH in cells, which might reasonably be expected to decrease the retinol oxidation rate by product inhibition of ADH isoforms.
Method
To understand the relative importance of these two mechanisms by which ethanol decreases the retinol oxidation in vivo we need to assess them quantitatively. We have built a model system of four reactions: (1) ADH oxidation of ethanol and NAD+ (2) ADH oxidation of retinol and NAD+ (3) oxidation of ethanol by a generalized Ethanoloxidase that uses NAD+ (4) NADHoxidase which carries out NADH turnover.
Results
Using the metabolic modeling package SCRUMPY, we have shown that the ethanol-induced increase in NADH contributes from 0–90% of the inhibition by ethanol, depending on [ethanol] and ADH isoform. Furthermore, while the majority of flux control of retinaldehyde production is exerted by ADH, Ethanoloxidase and the NADHoxidase contribute as well.
Discussion
Our results show that the ethanol-induced increase in NADH makes a contribution of comparable importance to the ethanol competitive inhibition throughout the range of conditions likely to occur in vivo, and must be considered in the assessment of the in vivo mechanism of ethanol interference with fetal development and other diseases.
Keywords: alcohol dehydrogenase, simulation, retinol, ethanol
Introduction
Retinoic acid (RA), a carboxylic acid metabolite of retinol (vitamin A), plays essential roles in embryogenesis, cellular differentiation and proliferation. An insufficient level of RA in the tissues resulting from ethanol consumption contributes to alcohol-related diseases such as fetal alcohol syndrome, liver cirrhosis, and gastrointestinal cancers. The causal connection between RA and cancer is exemplified in a study by the upregulation of several hepatic cancer markers in ethanol-fed rats; the wild-type phenotype was restored with retinoic acid feeding (Chung et al., 2001). In chronic alcoholics, the level of RA is decreased due to poor nutrition, increased mobilization/breakdown of RA (Leo and Lieber, 1999), and to decreased synthesis from retinol.
The primary pathway of RA synthesis from retinol is via the NAD+-dependent alcohol dehydrogenases (EC 1.1.1.1, ADHi) and aldehyde dehydrogenases (E.C. 1.2.1.5, ALDH) (Deltour et al., 1996; Napoli, 1996) with ADH generally considered to be the flux controlling step in RA synthesis. All ADH isoforms oxidize ethanol, and all but the ADH3 form have been shown to oxidize retinol. Several studies support a role for ADH7 in RA production, as it is present in embryonic tissues at the time and locations where RA is produced, with ADH1 also involved in RA production in development (Molotkov and Duester, 2002) and in adult brain (Martinez et al., 2001). ADH1 and its various isoforms and polymorphisms (1A, 1B1, 1B2, 1B3, 1C1, 1C2) may play similar roles in the liver.
First shown in 1971, ethanol inhibits retinol oxidation (Mezey and Holt, 1971) by human liver ADH as an alternate substrate with retinol for ADH. NAD+-dependent RA synthesis was shown to be inhibitable by ethanol in rat esophagus (Shiraishi-Yokoyama et al., 2003) or rat liver/colon (Parlesak et al., 2000). It was hypothesized in 1991 that competitive inhibition of retinol oxidation by ADH by ethanol was a cause of fetal alcohol syndrome (Deltour et al., 1996; Duester, 1991; Pullarkat, 1991), and that it may play a similar role in ethanol-related cancers (reviewed by (Wang, 2003). This viewpoint on the mechanism of the ethanol effect on retinol oxidation has dominated subsequent thinking, and we will return to this issue in more detail in the Discussion.
The extra cytosolic NADH production from ethanol oxidation by ADHs forces the NADH concentration higher until the rate of NADH reoxidation rises to match the production rate. Ethanol oxidation thus has the broad effect of shifting the redox state of cells, with liver undergoing, in rat, approximately a 4-fold increase in NADH at high levels of ethanol (Christensen and Higgins, 1979). Because each of the ADH isoforms has a product inhibition constant, Kiq, for NADH just above the physiological range of 0.5–2µM (see table 1), increases in NADH would be expected to decrease the apparent rate of oxidation of alcohols, including retinol to retinaldehyde, by ADH.
Table 1.
Kinetic constants used for each ADH isoform for ADHethanol (Eq. A1). Ki values in parenthesis were not available from the literature. In that case, the assumption was made that Ki=Km.
| Isoform | ADH1A | ADH1B1 | ADH1B2 | ADH1B3 | ADH1C1 | ADH4 | ADH7 |
| Vf (/min) | 21.12 | 61.16 | 378.7 | 222 | 115.7 | 13.12 | 718 |
| Vr (/min) | 350 | 83.27 | 2639 | 2560 | 2100 | 337 | 14470 |
| Km Ethanol (mM) |
4.58 | 0.273 | 1.103 | 44.5 | 1.33 | 23.93 | 36.4 |
| Km Acetaldehyde (mM) | 3.15 | 0.0828 | 0.24 | 3.4 | 0.33 | 30 | 20 |
| Km NAD (mM) | 0.013 | 0.0074 | 0.180 | 0.614 | 0.0079 | 0.00471 | 0.210 |
| Km NADH (mM) | 0.011 | 0.0064 | 0.105 | 0.260 | 0.007 | 0.016 | 0.096 |
| Ki NAD (mM) | 0.032 | 0.09 | 0.287 | 2.3 | (0.0079) | (0.00471) | 0.575 |
| Ki NADH (mM) | 0.0004 | 0.00019 | 0.0097 | 0.00665 | 0.00098 | (0.016) | 0.0057 |
| Ki Acetaldehyde (mM) |
(0.315) | 0.0015 | 0.022 | 0.210 | (0.033) | (3) | 0.36 |
Because of its significance to fetal development, this effect of NADH increases has been modeled computationally for ADH7 Using only the kinetic equation for human ADH7 oxidation of retinol given by Chou et al. (2002), Yin et al (2003) showed that an increase in NADH from 0.5 to 10µM could decrease the retinol oxidation rate by ADH4 by 43% at high levels of retinol, though ethanol competition was not considered. Plapp & Berst (2006) simulated the effects of ethanol on human ADH7 oxidation of retinol to retinoic acid through both ADH and ALDH at 1 and 50mM ethanol (0.5 and 5µM NADH, respectively). They concluded that ethanol inhibited retinol oxidation by the human ADH7. Their model appears to predict a 50% decrease in the rate of RA accumulation through ADH7 in the presence of ethanol due to the competition by both ethanol and NADH. The interplay between the competition for ADH7 by ethanol and the inhibition of the enzyme by NADH may act in concert to decrease retinol oxidation in vivo.
While ADH7 is a significant contributor to ethanol oxidation in the stomach and intestines, as well as in embryos, it is not present in human liver tissue. Thus, it has not yet been determined if this same two-pronged inhibition by ethanol (direct and indirectly through NADH levels) is also a property of the retinol oxidation by other ADH isoforms found in liver.
The amount of NADH is a property of the cellular system of reactions because it represents the balance of all NAD+ and NADH-consuming reactions in a cell. Previous computational models have parameterized NADH (e.g., =2.0uM when ethanol =50mM, (Plapp and Berst, 2006)). This neglects to consider the dynamic impact of enzymes involved in NADH oxidation or synthesis, enzymes which would (a) be contributing to the NADH pool and (b) which would be affected by the level of NADH.
When considering a single isoform of ADH in its oxidation of retinol, there remain other ADH forms that are able to oxidize ethanol and produce NADH. The activity of these other enzymes affects the flux of retinol oxidation by ADH isoforms inasmuch as they contribute to the production of NAD+ or NADH in the cell. To more fully understand the impact of ethanol on retinol oxidation, and the genetic/enzymatic basis for disease, we must account for the control of retinol flux by these other enzymes.
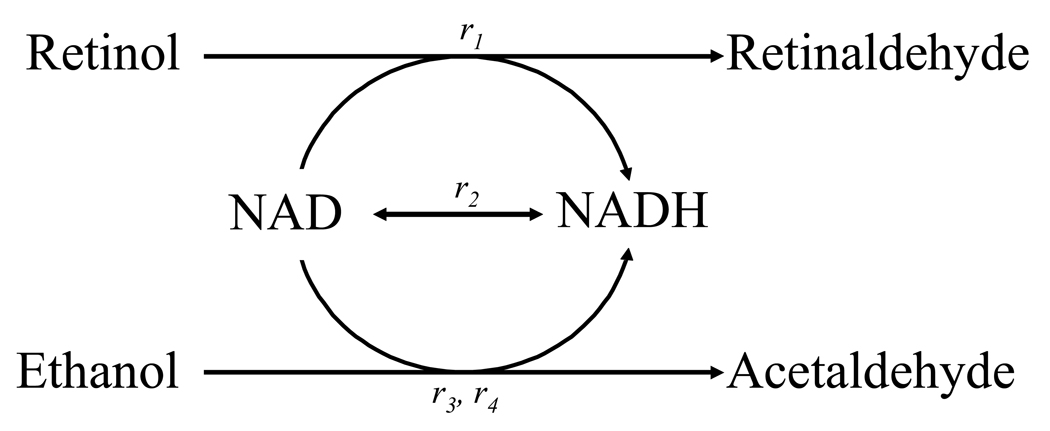
In this study we use a metabolic model of retinol oxidation to retinaldehyde that has properties that mimic those in vivo (e.g., metabolite changes, relative fluxes). This model system consists of (1) ADH oxidation of ethanol and NAD+ (2) ADH oxidation of retinol and NAD+ (3) oxidation of ethanol by a generalized Ethanoloxidase that uses NAD+ (4) NADHoxidase which carries out NADH turnover (Fig 1). Each ADH isoform has been considered separately. The model described here has been submitted to the Online Cellular Systems Modeling Database and can be freely accessed at: http://jjj.biochem.sun.ac.za/database/ and at PubMed Central.
Figure 1.
The system as modeled:
r1 Oxidation of retinol by the specific ADH isoform
r2 General NAD/H turnover.
r3 Oxidation of ethanol by the specific ADH isoform
r4 Other ethanol oxidation activity.
NAD+ and NADH are treated as internal, all other reactants are external. Details of reaction kinetics can be found in the Appendix. Parameter values and reactant concentrations are given in Tables 1–3.
Though we evaluated models with all available ADH isoforms that oxidize retinol, we have paid special attention to ADH7, because of its apparent significance in the development of fetal alcohol syndrome, and the ADH1B forms because of the genetic polymorphisms that have been evaluated for the effects of alcohol and alcoholism.
We are able to quantify, for the first time, the distribution of inhibition of retinol oxidation by ADH isoforms, distinguishing between NADH increases vs. direct ethanol competition when ethanol increases from 1–50mM. Also, we have determined the distribution of control of retinol flux to retinaldehyde for isoforms of ADH relative to other ethanol and NADH oxidizing enzymes in a cell. Using published enzyme contents of liver and gastric mucosal tissues has enabled us to estimate the effect of retinol oxidation pathways by elevated ethanol.
Methods
The retinol oxidation system was modeled as shown in Figure 1. In addition to the oxidation of retinol and ethanol by the ADH isoform, background ethanol and NAD/H turnover reactions were included. The latter of these was assumed to have reversible Michaelis-Menten kinetics (Appendix, Eq. A2) The background Ethanoloxidase was assumed to have Ordered Bi-Bi kinetics, which is the mechanism for ADH isoforms which make up the majority of the ethanol oxidation (Segal, 1975), (Appendix, Eq. A4). For the retinol/retinaldehyde-inhibited and the ethanol-inhibited oxidation of retinol by ADH isoforms, full kinetic equations were derived as described in the Appendix (Eq. A1 & A3). Parameter values for the background Ethanoloxidase and turnover reactions were selected to maintain NAD/H ratios at a 4-fold decrease (Christensen and Higgins, 1979) used elsewhere (Plapp and Berst, 2006; Plapp et al., 2001) in the presence of varying ethanol concentrations with ADH isoforms and background Ethanoloxidase activity also present. Parameter values for ADH isoforms for both retinol and ethanol oxidation, and for the background Ethanoloxidase were taken or estimated from the literature (Tables 1, 2, & 3). The Ki for acetal for ADH7 (Plapp and Berst, 2006) and those for the ADH1B1, B2, and B3 have been measured as 1.5 ± 0.1, 22 ± 14, and 210 ± 5 µM, respectively (unpublished data, Matsumoto & Crabb 2008). For the remaining isoforms, the Ki for acetal is estimated to be 1/10th the Km of acetal, similar to the ratio for the reported forms. The ADH4 isoform values of Ki for NAD+ and NADH have not been determined and were estimated to be equal to the Km values. The Ki for retinol was assumed to be equal to the Km for retinol, except for ADH7 for which the constant has been determined.
Table 2.
Kinetic Constants used for particular ADH isoforms for r1 (Eq. A1) oxidizing retinol. Ki values in parenthesis were not available from the literature. In those cases, the assumption was made that KiX=KmX. References are the sources of the kinetic constants in tables 1 and 2.
Table 3.
The kinetics constants used for the Ethanoloxidase (Eq. A4) for each isoform model.
| Model | ADH1A | ADH1B1 | ADH1B2 | ADH1B3 | ADH1C1 | ADH4 | ADH7 |
| Vf (/min) | 1530 | 1321 | 764 | 1319 | 1380 | 1551 | 1008 |
| Km Ethanol (mM) | 2.1 | 1.85 | 3.46 | 3.93 | 2.64 | 1.4 | 1.3 |
| Km NAD (mM) | 0.1 | 0.056 | 0.008 | 0.0687 | 0.0988 | 0.1 | 0.05 |
| Km NADH (mM) | 3.5 | 0.004 | 2.3 | 0.59 | 3.21 | 3.76 | 0.01 |
| Kia (mM) | 0.1 | 0.665 | 0.001 | 0.00214 | 0.0796 | 0.327 | 0.577 |
The steady-state fluxes, steady-state NADH, and flux control coefficients of retinol for the model were evaluated using the ScrumPy modeling package (Poolman, 2006). (Available from http://mudshark.brookes.ac.uk/ScrumPy). The model can be downloaded from http://jjj.biochem.sun.ac.za/ in ScrumPy and SBML format. ScrumPy was optimized for the Ubuntu LINUX operating system.
The flux control of steady-state flux (J) across a particular reaction by enzyme activities and concentrations of external substrates are quantified by flux control coefficients (Eq. 1) described extensively elsewhere (Fell, 1997). Briefly, a flux control coefficient quantifies the relative change in flux across a given reaction, resulting from a change in enzyme activity.
| Eq. 1 |
Control coefficients were determined for ADH isoforms, the Ethanoloxidase and NADHoxidase activities on retinol oxidation.
Characteristics of the model of oxidation by ADH isoforms were calculated at ethanol concentrations ranging from 1–50mM for each of the isoforms in the presence (Case 1) and absence (Case 2) of terms for ethanol inhibition of retinol oxidation, that is, inhibition by ethanol was included as:
(denominator term 3, Eq. A1). The term Ethanol/KmEthanol was eliminated from the kinetic equation for retinol oxidation to determine the rate of retinol oxidation in the absence of ethanol competitive inhibition. The calculations were performed at 1nM, and 0.5, 1, and 2µM retinol. The lower level (1nM) was used to approximate the free retinol in the presence of the cellular retinol binding protein (Kd= 16nM, 5µM binding protein in liver (Kedishvili et al., 1998). The retinol/retinaldehyde ratio was set at 10:1. Acetaldehyde was set to 1µM. For ADH7, four additional computation sets were generated to assess the impact of our method of choosing kinetic constants. Cases 3& 4: The effect of the activity of the background Ethanoloxidase (r4, Fig 1) was investigated for ADH7 by repeating the calculations over a 100-fold variation of the activity of this reaction with competitive inhibition terms (Case 3) present and absent (Case 4) from the r1 kinetic equation. Cases 5&6: The impact of using the kinetic constants determined by a single vs. variation of the in the presence (Case 5) and absence (case 6) of the competitive inhibition term from the r1 kinetic equation with all kinetic constants available from (Plapp and Berst, 2006) the remainder from Table 2, at 1µM retinol and 0.1µM retinaldehyde to evaluate the impact of a specific set of kinetic constants rather than averages because that report offered the most complete set of kinetic constants measured in one study. They paper reported a lower Km of ADH7 for ethanol, which would lead to a higher estimate of inhibition by ethanol of ADH7 and lower contribution of NADH inhibition.
In all cases, the total NAD/H pool was held at constant levels 0.5005mM because it has been reported (Veech et al., 1972) that the cytosolic level of NAD+ in rat hepatocytes was 0.5mM with 0.0005 mM NADH reflecting the free NAD/NADH ratios in this tissue (Christensen and Higgins, 1979). This value has been used in previous simulations for ADH (Lee et al., 2006; Plapp and Berst, 2006),
Since this is a steady-state rather than a dynamic model, only the relative velocities are analyzed for reactions, such that flux values were normalized to the velocity for Case 2 at 1mM ethanol. The relative contributions of competitive inhibition and alteration in NAD/H ratios to the net inhibition of retinol oxidation flux were calculated relative to the reference state (0.5µM NADH and no ethanol effect). This would be equivalent to the rate of retinol oxidation in the absence of ethanol at 0.5µM NADH.
Results
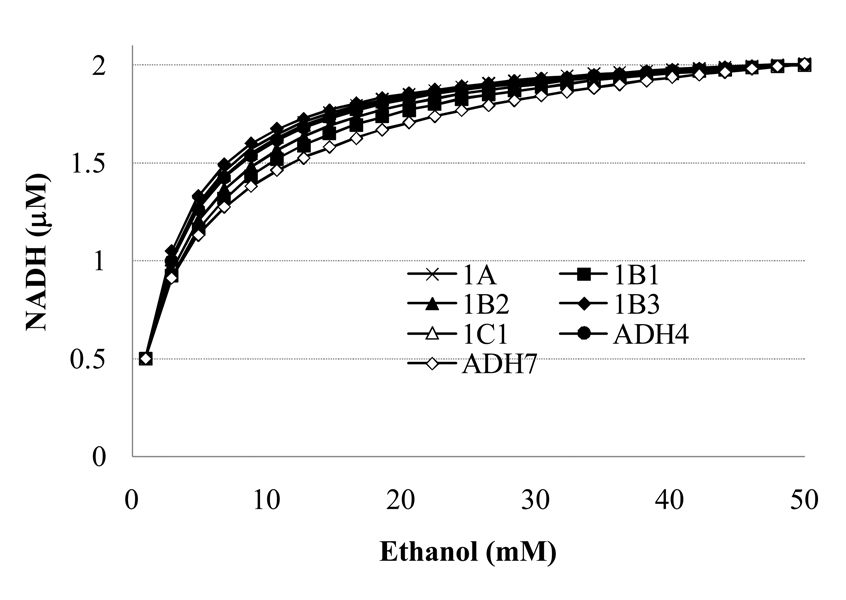
All of the 4-enzyme systems had an increase from 0.5 to 2µM NADH for the range of ethanol from 1 to 50mM (Fig. 2) at 1µM retinol over range of ethanol. NADH varied by at most 0.002 µM across the range of retinol concentrations. The identical patterns are found for all variables at 1nM retinol (data not shown). Because the NADH is slightly lower over most of the range of ethanol levels for ADH7, the inhibition of retinol oxidation by NADH may be considered a minimal estimate for ADH7 relative to the other isoforms. The exact shape of this relationship in cells is not known, thus these are reasonable approximations of the NADH level as a function of ethanol.
Figure 2.
Steady-state NADH as a function of 1–50mM ethanol with the kinetics of r1 and r3 parameterized for each of the isoforms in turn. The graphs for 1nM and 1µM retinol are indistinguishable for each isoform because the NADH at a given ethanol varies by less than 1%.
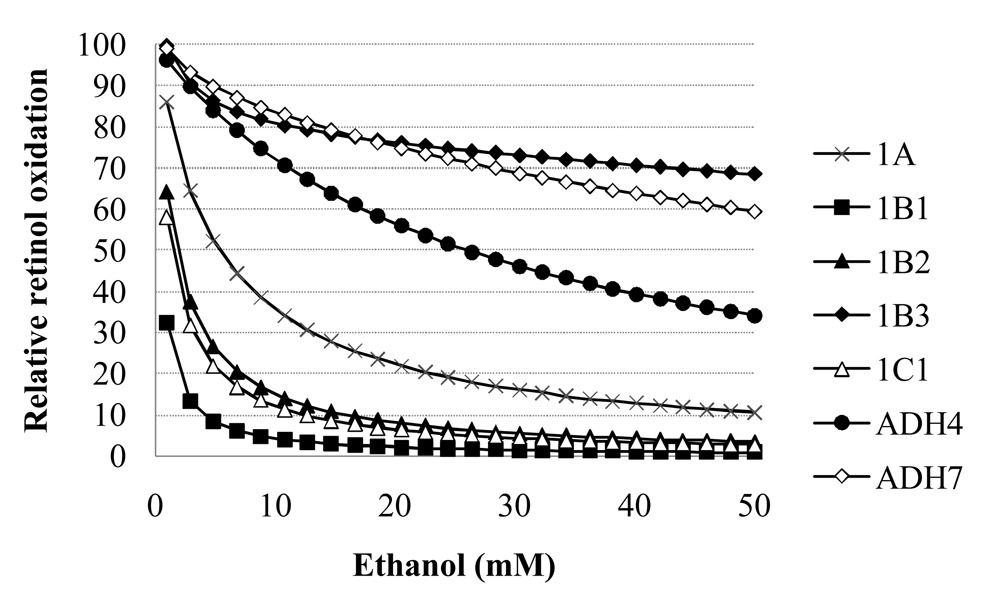
Ethanol increases lead to a decreased rate of retinol oxidation for systems with each ADH isoform (Fig 3). Sensitivity to the effects of ethanol on the system varied greatly across the ADH isoforms. Relative to the 1mM ethanol case, the rate of retinol oxidation at 50mM ethanol decreased by 30–99% (Fig. 3). The % inhibition at 50mM vs. 1mM ethanol does not correlate with the Km for ethanol for each isoform (R2 =0.25).The most inhibited and least inhibited by the increased ethanol levels (and accompanying NADH increases) were both types of the ADH1B isoforms (ADH1B1 and ADH1B3, respectively.)
Figure 3.
Relative retinol oxidation rate for each ADH isoform as a function of ethanol level for Case 1 (both ethanol and NADH inhibition operative).
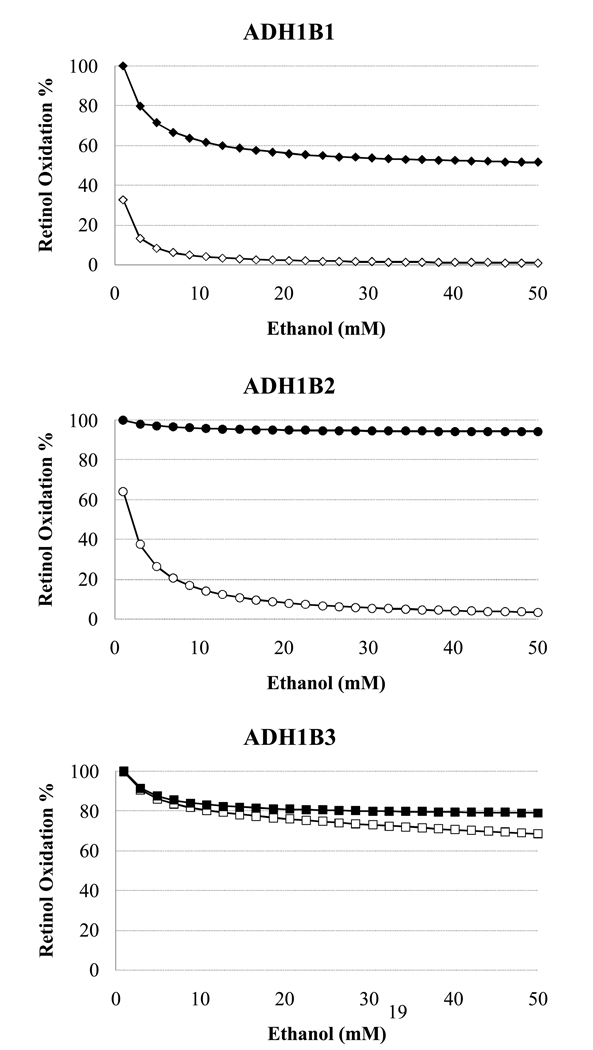
For the ADH1B isoforms, the amount of inhibition accounted for solely by the change in NADH varies widely (solid symbols, Fig. 4 i, ii, iii). Though the total inhibition is similar for ADH1B1 and ADH1B2, the amount resulting from the redox shift is much greater for ADH1B1 (Fig. 4i vs. ii), with a reduction of nearly 50% in retinol oxidation rate by ADH1B1 when only the ethanol-induced increase in NADH level inhibits the reaction, compared with less than 10% for ADH1B2. In contrast, for the ADH1B3 formis inhibited 31% by 50mM ethanol, with two-thirds of this inhibition accounted for by the NADH increase alone.
Figure 4.
Relative retinol oxidation rate for the three ADH1B isoforms (1B1, 1B2, 1B3) as a function of ethanol level. Rates are relative to the NADH-inhibited rate at 1mM ethanol. solid=NADH inhibition alone (Case 2) ; open= NADH and ethanol competitive inhibition of the ADH isoform (Case 1).
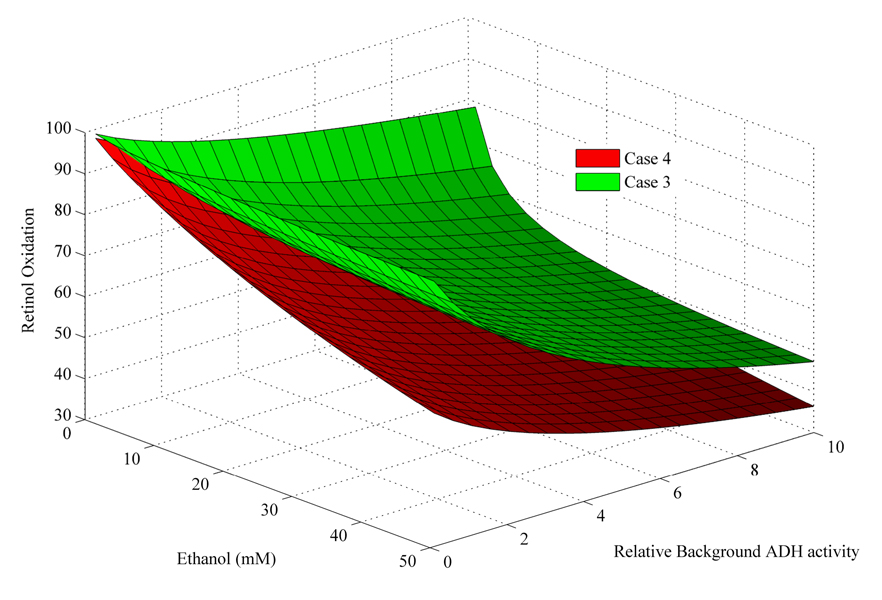
To ensure that the pattern of inhibition by NADH changes was not a result of the parameters chosen for the pooled enzymes, we evaluated the effect of a 100-fold range of activity for the Ethanoloxidase, 0.1–10X the activity for the ADH7 isoform model (Cases 3 & 4). This variation has the direct impact of changing the NADH level in the model. The pattern of inhibition by NADH (red points, Fig. 5) vs. total inhibition (blue points, Fig. 5) is consistent with the original case; that is, retinol oxidation is inhibited partially by NADH changes and partially by ethanol competition. The effect of ethanol on retinol oxidation was greater for ADH7 in cases 5 & 6 than for the averaged kinetic constants: NADH inhibited by, on average, an additional 2.5% and ethanol and NADH by an additional 14% more than in cases 1 & 2. For cases 5 & 6, ethanol accounted for 13% more of the total inhibition than in cases 1 & 2.
Figure 5.
Plot of cases 3&4 from above to show the impact of variation in ADH on the rate of retinol oxidation rate as a function of ethanol.
Red= case 3, competitive inhibition by ethanol and NADH at ADH7
Blue= case 4, no ethanol competition at ADH7.
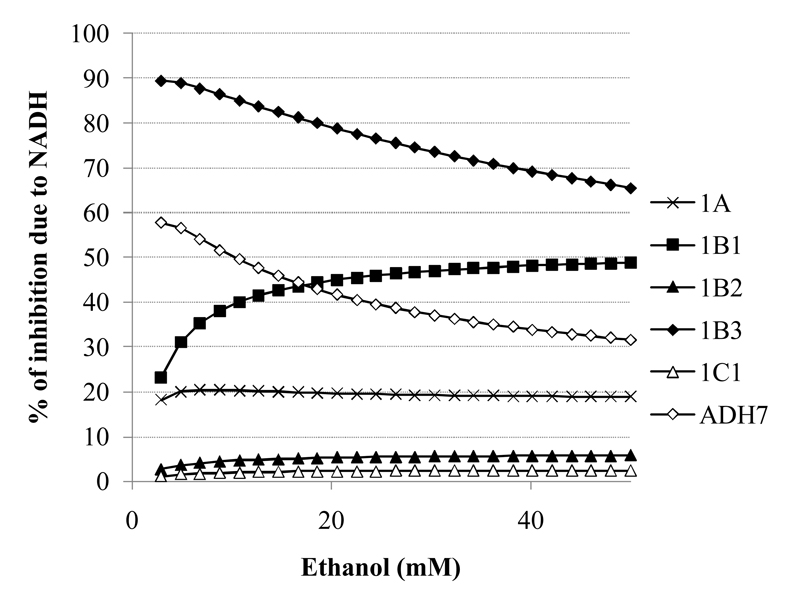
The % of total inhibition accounted for by changes in NADH varies greatly across isoforms and with ethanol levels (Fig. 6). Only for ADH4 is less than 1% of the inhibition due to the NADH increases. In contrast, for ADH1B3 and ADH7, the majority of inhibition of retinol oxidation is due to NADH levels, with the role of ethanol increasing as the ethanol levels are raised. The pattern is the opposite for ADH1B1, where the impact of NADH levels increases with ethanol levels, approaching a 50/50 distribution of direct inhibition by ethanol vs. the elevated NADH levels (Fig. 6).
Figure 6.
The fraction of inhibition of retinol oxidation that is accounted for by the ethanol-induced increase in NADH varies greatly between ADH isoforms. Total inhibition: Uninhibited rate – rate for case 1 at each ethano levell. Inhibition by NADH: Uninhibited rate – rate for case 2 at each ethanol. % inhibition by NADH: Inhibition by NADH/ Total inhibition * 100.
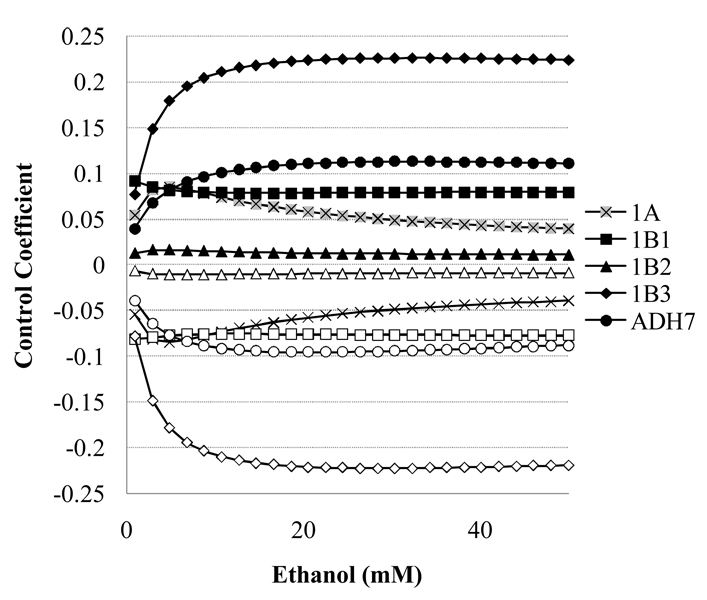
The control coefficients (Eq. 1) of the retinol oxidation rate by the ADH isoforms, and by pooled enzymes of the NADH turnover (NADHoxidase and Ethanoloxidase) follow different patterns. For all the ADH isoforms, the control coefficients are very high and constant (>0.98, data not shown). In contrast, the control by the enzymes of NADH turnover varies and is sensitive to changes in ethanol (Fig. 7.) A property of the design of the model is that the control by NADHoxidase and Ethanoloxidase are nearly identical in magnitude but with opposite signs.. There is no correlation between control at 50mM ethanol and turnover number, Km for ethanol nor Km for NADH for Ethanoloxidase (r2<0.5) and the kinetics of the NADHoxidase are held constant across models.. Therefore, the choice of parameters for the pooled NADHoxidase or Ethanoloxidase does not dictate the magnitude of the control coefficients, but rather these values are a property of the system. The levels of NADH have the greatest impact on retinol oxidation by ADH1B3, with control coefficients for Ethanoloxidase and NADHoxidase increasing to more than ± 0.2. Patterns and magnitudes of the control coefficients show some isoform variability.
Figure 7.
The control of the rate of retinol oxidation flux (Eq. 1) by the Ethanoloxidase (open symbols) and NADHoxidase (solid symbols). Control coefficients for y either “enzyme” in the ADH1C1, and 4 models are <0.005 (data not shown).
Each of the above results is affected by the choice of kinetic constants in the model. For Cases 5 and 6, which uses a lower Km for Ethanol for ADH7, the impact of changes in ethanol alone increases and that of NADH changes decreases. Retinol oxidation is inhibited by (on average) 8% more by ethanol 8% less by NADH across the range of ethanol (data not shown). The control coefficients for both Ethanoloxidase and NADHoxidase are >20% higher for Cases 5& 6 than 1&2 (data not shown). Using either set of kinetic constants, changes in NADH are seen to be significant contributors to retinol inhibition.
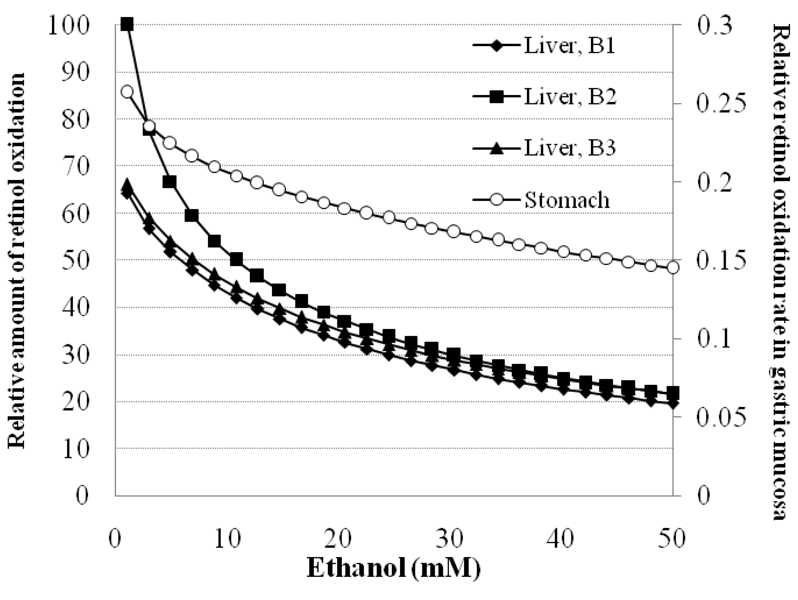
Ethanol inhibition of retinol oxidation in specific tissues is affected by genetic polymorphisms in ADH1B (Fig. 8). Ethanol increases result in a 40–80% decrease in retinol oxidation. Stomach mucosa has the smallest absolute and smallest percentage inhibition (open circles) because of the small amount of retinol oxidizing capacity relative to liver (0.0235g/stomach vs. 9.3g per liver). At 7mM ethanol, liver retinol oxidation for 1A/1B2/1C1/4 genotype individuals is still higher than oxidation in uninhibited livers of other genotypes.he retinol oxidation rate in liver decreases to below 50% of uninhibited rate by less than 15mM ethanol, below intoxicated levels.
Figure 8.
Retinol oxidation in liver (solid symbols; ADH1A, ADH1B, ADH1C, ADH4) or gastric mucosa (open symbols; ADH1C & ADH7) as affected by ethanol concentrations and polymorphisms at ADH1B. Retinol oxidation is scaled to the uninhibited rate of liver expressing ADH7. Amount of each isoform from Lee, et al (2006) at rates predicted by the model for cases 1 & 2. (Polymorphisms at ADH1B ■=B2♦=liver with B1 ▲=liver with B3 ○=Gastric mucosa, expressing ADH1C1 and ADH7)
Discussion
Pullarkat (1991) and Duester (1991) separately hypothesized that the mechanism by which ethanol leads to fetal alcohol syndrome is via a decrease in retinoic acid production. Duester (1991), in a paper that has been cited more than 60 times, specifically hypothesized that “many of the abnormalities observed in fetal alcohol syndrome may be caused by high levels of ethanol acting as a competitive inhibitor of ADH-catalyzed retinol oxidation in the embryo or fetus.” Since then, all but a few reports on ADH and retinol in the literature have focused on this competitive inhibition as the mechanism by which elevated ethanol decreases ADH oxidation of retinol.
Because ethanol ingestion can lead to a 10-fold increase in liver NADH, the inhibition of ADH by NADH must be considered as a potential mechanism. Recent reviews of the impact of ethanol on retinoid metabolism did not mention the potential inhibition of retinol oxidation by ADH from ethanol-induced redox shifts (Crabb et al., 2001; Wang, 2005). The impact of these NADH shifts have been simulated for ADH7 (Plapp and Berst, 2006; Yin et al., 2003), with NADH shifts alone shown to account for approximately a 50% decrease in retinol oxidation rate, the size of the effect being dependent on retinol and retinaldehyde levels (Yin et al., 2003). We have extended this analysis to the other retinol-oxidizing ADH isoforms and show that they are inhibited from 30 to 97% by the combination of increases in ethanol and NADH (Fig. 3). We have shown here that at low levels of ethanol, the NADH elevation inhibits ADH7 at least as much as the oft-cited direct ethanol competition (Fig. 6). For ADH1B3, NADH inhibition is 1.5-to-9 times greater than the direct ethanol inhibition, but is only a small fraction of the inhibition by ethanol for ADH1B2 (Fig 4). NADH inhibition is both significant in magnitude and is genotype-dependent, warranting its consideration in the overall mechanism of ethanol’s effect on retinoic acid synthesis and the resulting physiological effects (e,g., FAS).
Liver injury from ischemia is enhanced by ethanol in perfused rat liver (Younes et al., 1989). Khan and O’Brien (1999) showed that hepatocytes were more susceptible to ethanol-induced cytotoxicity in low oxygen (correlating with the cellular redox state) or with additions of sorbitol or xylitol, which further decreased the NAD+/NADH. This NADH-sensitive, ethanol-induced cytotoxicity was at least partially mediated by ADH, since it was inhibited by methylpyrazole or dimethylsulfoxide. Although there is no indication that the mechanism of cytotoxicity in these short term (<2hrs) hepatocyte studies is the result of diminished retinoic acid levels, it does highlight the ability of NADH reoxidation to mediate ethanol effects on the cell. This is consistent with our calculation that the flux control by the enzymes that recycle NAD+/NADH (NADHoxidases) had control coefficients that are up to 1/5th that of the ADH isoform oxidizing retinol (Fig. 7).
We have shown that ethanol, directly and through increases in NADH, is capable of significantly inhibiting retinol oxidation in human tissues (gastric mucosa, liver) by each ADH isoform. In liver and gastric mucosa retinol oxidation by ADH is significantly inhibited by ethanol (Fig. 8.) The small difference in effect of polymorphisms of ADH1B1 and ADH1B2 (Fig. 8) do not account for increased relative risk for head and neck cancer with the ADH1B*1/1 isoform (Brennan et al., 2004). The ADH1B2 isoform is believed to be protective against alcoholism; here we show that this polymorphism leads to greater retinol oxidation at all ethanol levels as well as a greater amount of inhibition by ethanol/NADH. Measurements of the amounts of ADH isoforms in other human tissues and specific cells (e.g., hepatic stellate cells, fetal tissues) would make it possible to make precise predictions of the relative retinol oxidation rates in the presence of ethanol.
The significance of all this is that the NAD/H recycling potential of a cell has a discernable impact on the rate of retinol oxidation. There is a broad range in this relative potential across tissue types, with liver among the highest in ability to turn over NAD/H. The ingestion of ethanol along with other pharmacological agents that alter the redox state (e.g., metformin) or a compromised liver function might further diminish the ability of the liver to oxidize retinol. Tissues with higher NAD/H turnover capacity would be protected from some of the detrimental effects of ethanol on retinol oxidation. The presence of the ADH1C1, more active in oxidizing ethanol than ADH1C2, is associated with increased risk of cancer (Visapaa et al., 2004). The authors suggested that the mediator of this increased risk might be acetaldehyde. Because rapid ethanol oxidation would lead not only to increased acetaldehyde but also to increased cytosolic NADH, Visapaa’s results on cancer rates are also consistent with increased inhibition of retinol oxidation by inhibition of ADH isoforms by the redox shift. All other factors being equal, forms of ADH that are more active toward ethanol may remove ethanol as a competitive retinol oxidation inhibitor, but thereby generate inhibitory NADH.
Though it is not unexpected that NADH increases would inhibit ADH as a product, the degree of inhibition by NADH is not a simple function of the product inhibition constant. The enzyme with the lowest Kiq (ADH1B1) is the most inhibited, and the highest Kiq (ADH4) is least inhibited, but the Kiq values and inhibition for the other three enzymes for which Kiq has been determined do not show correlation (overall R2 = 0.49). It is therefore necessary to use a simulation of the system to assess the relationship between elevated NADH and the inhibition of retinol oxidation.
The limitations of the current report result from incomplete data from which to make the models. We also used averaged kinetic constants for the other ADH isoforms; because some reported values varied widely, it is possible that in specific cases the exact impact on retinol oxidation by ethanol would be affected- especially if the Kms for ethanol and NADH are significantly different from their averaged values. However, it was not our intention to predict a specific quantitative contribution of the NADH inhibition, but to illustrate that under a wide range of conditions that are likely to cover those arising in cells in vivo, the effect makes a significant contribution to the reduction in retinol oxidation. In other words, the uncertainties in the relative activities of the enzymes involved, and in their kinetics, may vary the degree to which elevated NADH affects retinol oxidation, but not the fact that it does inhibit retinol oxidation by human ADH isoforms to an extent that needs consideration.
Retinol in tissues may be oxidized by three distinct classes of enzymes: microsomal or cytosolic NADP-dependent retinol dehydrogenases (RoDH), or ADH isoforms. The contribution of each to retinol oxidation in various tissues, throughout development, and in the presence of ethanol is a matter of some disagreement. The potential of other enzymes to take over the synthesis of retinol if production by ADH is blocked, e.g., in ADH1 or ADH7 knockout mice, (Deltour et al., 1999) might limit the application of the current results. Nevertheless, the broadly accepted mechanism by which the presence of ethanol interferes with retinol oxidation is interference with ADH isoforms, partially because the NADP+/NADPH ratio in liver is 0.01 vs. 1000 for NAD+/NADH. Under such conditions, NAD+-dependent ADH would act in the direction of retinol oxidation and the NADP+-dependent cytosolic RoDH would function in the direction of reduction of retinal to retinol. In human embryonic hepatocyte cytosolic extracts, NAD+ supported retinol oxidation was six-fold greater than NADP+ -supported retinol oxidation (Khalighi et al., 1999). A side-by-side comparison of these enzymes in detergent-free assays and cell culture concluded that ADH isoforms were the enzymes that oxidized retinol and the RoDHs in reduction of retinal (Gallego et al., 2006).
The majority of retinol is bound to the cellular retinol binding protein (CRBP), decreasing free retinol to less than 0.1µM (Kedishvili et al., 1998). It has variously been reported that CRBP-bound retinol is a substrate for the RoDHs (for example (Kedishvili et al., 1998; Napoli, 1999) or that only free retinol is the substrate for both RoDH and ADHs (Gallego et al., 2006). We did not see any difference in the impact of ethanol and NADH on the pattern of retinol oxidation nor its flux control over a 1000-fold range of free retinol (1nM—1µM). If the RoDHs can oxidize CRBP-bound retinol, these enzymes might be a dominant producer of retinal and, thereby, retinoic acids in tissues in which they are present. However, in the absence of a hypothetical mechanism by which these enzymes are inhibited by ethanol, the importance of these enzymes to mediation of alcohol-related diseases cannot be assessed.
Leaving considerations of this particular system to one side, this study also demonstrates the fact that even apparently simple systems can have quite subtle properties that can only be properly understood via a metabolic modeling approach.
Conclusion
The ethanol-induced increase in NADH is of comparable significance in decreasing retinoic acid production to the broadly accepted role of ethanol competition with retinol at alcohol dehydrogenases (Duester, 1991). This potential mechanism for retinoic acid inhibition by ethanol has not previously been quantified and may provide an additional site for prevention and treatment of ethanol-induced, retinoic acid-related diseases. In further consideration of the mechanisms underlying ethanol’s cause of FASD and cancers in alcoholics, shifts in the NAD/H ratio and the other ADH isoforms of fetal and adult tissues must be considered potentially to contribute substantially to the inhibition of the retinol oxidation rate.
Table 4.
Comparison of retinol oxidation rates with a published simulation ((Plapp and Berst, 2006)). In their simulation, they evaluated ethanol effects at 0.1mM (with 0.5µM NADH) and 10 and 50mM ethanol (with 2µM NADH). In our ADH7 model, the lowest ethanol was 1mM at 0.5µM NADH; NADH at 10 and 50mM ethanol is as shown in Figure 2.
| Condition | Rate estimated from Figure 6 in (Plapp and Berst, 2006) | Rate relative to low ethanol | Rate relative to low ethanol in ADH7 model |
| Low ethanol | 0.15µM/min | 50% | 81% |
| 10mM ethanol | 0.075µM/min | ||
| 50mM Ethanol | 0.02µM/min | 13% | 57% |
Acknowledgments
This work was supported by National Institutes of Health Grant # PO RR016454 from the INBRE Program for the National Center for Research Resources and by the UK BBSRC.
Appendix
Chou (Chou et al., 2002) determined that the retinol dehydrogenase function “conforms to an asymmetric mechanism, random for the oxidation of all-trans-retinol and ordered for the reduction of all-trans-retinal under rapid equilibrium for binding the substrate and coenzyme for both reactions.” Ethanol and acetaldehyde, which bind to the enzyme-NAD+ and enzyme-NADH forms, respectively, were treated as dead-end inhibitors of the enzyme-coenzyme complexes (Segal, 1975). Terms (1+Ethanol/KmEthanol) and (1+Acetaldehyde/KmAcetaldehyde) were added to the appropriate terms in the denominator for these complexes (Eq. A2). A similar approach was taken for the formulation of the equation for ethanol oxidation by ADH (Eq. A3). Retinol and retinaldehyde were treated as dead-end inhibitors of the oxidation. Because retinol can bind in random fashion, the term (1+Retinol/KmRetinol) was multiplied by both the constant term and the NAD+ term in the denominator. Retinaldehyde, like acetaldehyde, binds to the enzyme-NADH complex and so the (1+Retinaldehyde/KmRetnialdehyde) was added only to the NADH term in the denominator of Eq. A3 (Segal, 1975).
| Eq.A1 |
NADHoxidase velocity was calculated from the following equation for reversible Michaelis-Menten reactions (Eq. A2).
| Eq. A2 |
| Eq.A3 |
| Eq. A4 |
where Kia and Kiq are the dissociation constants for NAD+ and NADH, respectively, from the free enzyme.
Footnotes
HUGO recommended designations for the various classes and isoforms of ADH are used. Names in this paper (ADH7, ADH4) have common synonyms (ADH4 and ADH2, respectively) from Duester G, Farres J, Felder MR, Holmes RS, Hoog J-O, Pares X, Plapp BV, Yin S-J, Jornvall H (1999) Recommended nomenclature for the vertebrate alcohol dehydrogenase gene family. Biochemical Pharmacology 58(3):389–395.
References
- Allali-Hassani A, Peralba JM, Martras S, Farres J, Pares X. Retinoids, w-hydroxyfatty acids and cytotoxic aldehydes as physiological substrates, and H2-receptor antagonists as pharmacological inhibitors of human class IV alcohol dehydrogenase. FEBS Letters. 1998;326:362–366. doi: 10.1016/s0014-5793(98)00374-3. [DOI] [PubMed] [Google Scholar]
- Bosron WF, Magnes LJ, Li T-K. Kinetic and electrophoretic properties of native and recombined isoenzymes of human liver alcohol dehydrogenase. Biochemistry. 1983;22:1852–1857. doi: 10.1021/bi00277a017. [DOI] [PubMed] [Google Scholar]
- Brennan P, Lewis S, Hashibe M, Bell DA, Boffetta P, Bouchardy C, Caporaso N, Chen C, Coutelle C, Diehl SR, Hayes RB, Olshan AF, Schwartz SM, Sturgis EM, Wei Q, Zavras AI, Benhamou S. Pooled Analysis of Alcohol Dehydrogenase Genotypes and Head and Neck Cancer: A HuGE Review. Am. J. Epidemiol. 2004;159(1):1–16. doi: 10.1093/aje/kwh003. [DOI] [PubMed] [Google Scholar]
- Burnell JC, Carr LG, Dwulet FE, Edenberg HJ, Li TK, Bosron WF. The human beta 3 alcohol dehydrogenase subunit differs from beta 1 by a Cys for Arg-369 substitution which decreases NAD(H) binding. Biochemical and Biophysical Research Communications. 1987;146(3):1127–1133. doi: 10.1016/0006-291x(87)90779-0. [DOI] [PubMed] [Google Scholar]
- Burnell JC, Li T-K, Bosron WF. Purification and steady-state kinetic characerization of human liver β3β3 alcohol dehydrogenase. Biochemistry. 1989;28:6810–6815. doi: 10.1021/bi00443a005. [DOI] [PubMed] [Google Scholar]
- Chou C-F, Lai C-L, Chang Y-C, Duester G, Yin S-J. Kinetic mechanism of human class IV alcohol dehydrogenase functioning as retinol dehydrogenase. Journal of Biological Chemistry. 2002;277:25209–25016. doi: 10.1074/jbc.M201947200. [DOI] [PubMed] [Google Scholar]
- Christensen EL, Higgins JJ. In: Effect of acute and chronic administration of ethanol on the redox states of brain and liver, in Biochemistry and Pharmacology of Ethanol. Majchrowicz E, Noble EP, editors. vol 1. New York: Plenum Press; 1979. pp. 191–244. [Google Scholar]
- Chung J, Liu C, Smith DE, Seitz HK, Russell RM, Wang X-D. Restoration of retinoic acid concentration supresses ethanol-enhanced c-Jun expression and hepatocyte proliferation in rat liver. Carcinogenesis. 2001;22(8):1213–1219. doi: 10.1093/carcin/22.8.1213. [DOI] [PubMed] [Google Scholar]
- Crabb DW, Pinairs J, Hasanadka R, Fang M, Leo MA, Lieber CS, Tsukamoto H, Motomura K, Miyahara T, Ohata M, Bosron WF, Sanghani S, Kedishvili NY, Shiraishi H, Yokoyama H, Miyagi M, Ishii H, Bergheim I, Menzl I, Parlesak A, Bode C. Alcohol and Retinoids. Alcoholism: Clinical and Experimental Research. 2001;25(5):207S–217S. doi: 10.1097/00000374-200105051-00034. [DOI] [PubMed] [Google Scholar]
- Deetz JS, Leuhr CA, Vallee BL. Human Liver Alcohol Dehydrogenase Isozymes: Reduction of Aldehydes and Ketones. Biochemistry. 1984;23(26):6822–6828. doi: 10.1021/bi00321a084. [DOI] [PubMed] [Google Scholar]
- Deltour L, Ang HL, Duester G. Ethanol inhibition of retinoic acid synthesis as a potential mechanism for fetal alcohol syndrome. FASEB Journal. 1996;10(9):1050–1057. [PubMed] [Google Scholar]
- Deltour L, Foglio MH, Duester G. Metabolic deficiencies in alcohol dehydrogenase Adh1, Adh3, and Adh4 null mutant mice. Journal of Biological Chemistry. 1999;274:16796–16801. doi: 10.1074/jbc.274.24.16796. [DOI] [PubMed] [Google Scholar]
- Duester G. A hypothetical mechanism for fetal alcohol syndrome involving ethanol inhibition of retinoic acid synthesis at the alcohol dehydrogenase step. Alcoholism: Clinical and Experimental Research. 1991;15(3):568–572. doi: 10.1111/j.1530-0277.1991.tb00562.x. [DOI] [PubMed] [Google Scholar]
- Duester G, Farres J, Felder MR, Holmes RS, Hoog J-O, Pares X, Plapp BV, Yin S-J, Jornvall H. Recommended nomenclature for the vertebrate alcohol dehydrogenase gene family. Biochemical Pharmacology. 1999;58(3):389–395. doi: 10.1016/s0006-2952(99)00065-9. [DOI] [PubMed] [Google Scholar]
- Fell DA. Understanding the control of metabolism. Miami: Portland Press; 1997. [Google Scholar]
- Gallego O, Belyaeva O, Porte S, Ruiz F, Stetsenko A, Shabrova E, Kostereva N, Farres J, Pares X, Kedishvili NY. Comparative functional analysis of human medium-chain dehydrogenases, short-chain dehydrogenases/reductases and aldo-keto reductases with retinoids. Biochemical Journal. 2006;399(1):101–109. doi: 10.1042/BJ20051988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C-L, Liao C-S, Wu C-W, Hwong C-L, Lee A-R, Yin S-J. Contribution to first-pass metabolism of ethanol and inhibition by ethanol for retinol oxidation in human alcohol dehydrogenase family. Implications for etiology of fetal alcohol syndrome and alcohol-related diseases. European Journal of Biochemistry. 1998;254(1):25–31. doi: 10.1046/j.1432-1327.1998.2540025.x. [DOI] [PubMed] [Google Scholar]
- Hellgren M, Strömberg P, Gallego O, Martras S, Farrés J, Persson B, Parés X, Höög J. Alcohol dehydrogenase 2 is a major hepatic enzyme for human retinol metabolism. Cellular and Molecular Life Sciences (CMLS) 2007;64(4):498–505. doi: 10.1007/s00018-007-6449-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedishvili NY, Bosron WF, Stone CL, Hurley TD, Peggs CF, Thomasson HR, Popov KM, Carr LG, Edenberg HJ, Li T-K. Expression and kinetic characterization of recombinant human stomach alcohol dehydrogenase. Journal of Biological Chemistry. 1995;270(8):3625–3630. doi: 10.1074/jbc.270.8.3625. [DOI] [PubMed] [Google Scholar]
- Kedishvili NY, Gough WH, Davis WI, Parsons S, Li T-K, Bosron WF. Effect of cellular retinol-binding protein on retinol oxidation by human class IV retinol/alcohol dehydrogenase and inhibition by ethanol. Biochemical and Biophysical Research Communications. 1998;249:191–196. doi: 10.1006/bbrc.1998.9105. [DOI] [PubMed] [Google Scholar]
- Khalighi M, Brzezinski MR, Chen H, Juchau MR. Inhibition of human prenatal biosynthesis of all-trans-retinoic acid by ethanol, ethanol metabolites, and products of lipid peroxidation reactions: A possible role for CYP2E1. Biochemical Pharmacology. 1999;57(7):811–821. doi: 10.1016/s0006-2952(98)00362-1. [DOI] [PubMed] [Google Scholar]
- Khan S, O’Brien PJ. Role of the cellular redox state in modulating acute ethanol toxicity in isolated hepatocytes. Clinical Biochemistry. 1999;32(7):585–589. doi: 10.1016/s0009-9120(99)00059-4. [DOI] [PubMed] [Google Scholar]
- Lands WE. A review of alcohol clearance in humans. Alcohol. 1998;15(2):147–160. doi: 10.1016/s0741-8329(97)00110-9. [DOI] [PubMed] [Google Scholar]
- Lee S-L, Chau G-Y, Yao C-T, Wu C-W, Yin S-J. Functional assessment of human alcohol dehydrogenase family in ethanol metabolism: Significance of first-pass metabolism. Alcoholism: Clinical and Experimental Research. 2006;30(7):1132–1142. doi: 10.1111/j.1530-0277.2006.00139.x. [DOI] [PubMed] [Google Scholar]
- Leo MA, Lieber CS. Alcohol, vitamin A, and beta-carotene: adverse interactions, including hepatotoxicity, and carcinogenicity. American Journal of Clinical Nutrition. 1999;69:1071–1085. doi: 10.1093/ajcn/69.6.1071. [DOI] [PubMed] [Google Scholar]
- Mardh G, Luehr CA, Vallee BL. Human Class I Alcohol Dehydrogenases Catalyze the Oxidation of Glycols in the Metabolism of Norepinephrine. PNAS. 1985;82(15):4979–4982. doi: 10.1073/pnas.82.15.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez SE, Vaglenova J, Sabria J, Martinez MC, Farres J, Pares X. Distribution of alcohol dehydrogenase mRNA in the rat central nervous system: Consequences for brain ethanol and retinoid metabolism. European Journal of Biochemistry. 2001;268:5045–5056. [PubMed] [Google Scholar]
- Martras S, Alvarez R, Gallego O, Dominguez M, de Lera AR, Farres J, Pares X. Kinetics of human alcohol dehydrogenase with ring-oxidized retinoids: effect of Tween 80. Archives of Biochemistry and Biophysics. 2004a;430(2):210–217. doi: 10.1016/j.abb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Martras S, Alvarez R, Martinez SE, Torres D, Gallego O, Duester G, Farres J, de Lera AR, Pares X. The specificity of alcohol dehydrogenase with cis-retinoids: Activity with 11-cis-retinol and localization in retina. European Journal of Biochemistry. 2004b;271:1660–1670. doi: 10.1111/j.1432-1033.2004.04058.x. [DOI] [PubMed] [Google Scholar]
- Mezey E, Holt PR. The inhibitory effect of ethanol on retinol oxidation by human liver and cattle retina. Experimental and Molecular Pathology. 1971;15:148–156. doi: 10.1016/0014-4800(71)90095-5. [DOI] [PubMed] [Google Scholar]
- Molotkov A, Duester G. Retinol/Ethanol drug interaction during acute alcohol intoxication in mice involves inhibition of retinol metabolism to retinoic acid by alcohol dehydrogenase. Journal of Biological Chemistry. 2002;277(25):22553–22557. doi: 10.1074/jbc.M201603200. [DOI] [PubMed] [Google Scholar]
- Moreno A, Pares X. Purification and characterization of a new alcohol dehydrogenase from human stomach. Journal of Biological Chemistry. 1991;266(2):1128–1133. [PubMed] [Google Scholar]
- Napoli JL. Retinoic acid biosynthesis and metabolism. FASEB Journal. 1996;10:993–1001. doi: 10.1096/fasebj.10.9.8801182. [DOI] [PubMed] [Google Scholar]
- Napoli JL. Interactions of retinoid binding proteins and enzymes in retinoid metabolism. Biochemica et Biophysica Acta. 1999;1440(2–3):139–162. doi: 10.1016/s1388-1981(99)00117-1. [DOI] [PubMed] [Google Scholar]
- Parlesak A, Menzl I, Feuchter A, Bode JC, Bode C. Inhibition of retinol oxidation by ethanol in the rat liver and colon. Gut. 2000;47:825–831. doi: 10.1136/gut.47.6.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plapp BV, Berst KB. In: Human Alcohol Dehydrogenase 4: Mechanism, Specificity and Effects of Ethanol on Retinoid Metabolism, in Enzymology and Molecular Biology of Carbonyl Metabolism: 12. Weiner H, Plapp BV, Lindahl R, Maser E, editors. West Lafayette, Indiana: Purdue University Press; 2006. pp. 190–199. [Google Scholar]
- Plapp BV, Mitchell JL, Berst KB. Mouse alcohol dehydrogenase 4: kinetic mechanism, substrate specificity and simulation of effects of ethanol on retinoid metabolism. Chemico-Biological Interactions. 2001;130–132:445–456. doi: 10.1016/s0009-2797(00)00284-2. [DOI] [PubMed] [Google Scholar]
- Poolman MG. ScrumPy: metabolic modelling with python. Systems Biology. 2006;153(5):375–378. doi: 10.1049/ip-syb:20060010. [DOI] [PubMed] [Google Scholar]
- Pullarkat R. Hypothesis: prenatal ethanol-induced birth defects and retinoic acid. Alcoholism: Clinical and Experimental Research. 1991;15(3):565–567. doi: 10.1111/j.1530-0277.1991.tb00561.x. [DOI] [PubMed] [Google Scholar]
- Segal IH. Wiley Classics Library Edition 1993 ed. New York: John Wiley & Sons, Inc.; 1975. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-state Enzyme Systems. [Google Scholar]
- Shiraishi-Yokoyama H, Yokoyama H, Matsumoto M, Ishii H. The existence of an NAD-dependent pathway for retinoic acid formation from vitamin A (retinol) in rat esophagus and its inhibition by ethanol and histamine 2 (H2) receptor antagonists. Medical Science Monitor. 2003;9(12):BR403–BR406. [PubMed] [Google Scholar]
- Strömberg P, Svensson S, Hedberg JJ, Nordling E, Höög JO. Identification and characterisation of two allelic forms of human alcohol dehydrogenase 2. Cellular and Molecular Life Sciences (CMLS) 2002;59(3):552–559. doi: 10.1007/s00018-002-8447-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veech RL, Guynn R, Veloso D. The time-course of the effects of ethanol on the redox and phosphorylation states of rat liver. Biochemical Journal. 1972;127:387–397. doi: 10.1042/bj1270387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner FW, Burger AR, Vallee BL. Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes. Biochemistry. 1983;22:1857–1863. doi: 10.1021/bi00277a018. [DOI] [PubMed] [Google Scholar]
- Wang X-D. Retinoids and Alcohol-related Carcinogenesis. Journal of Nutrition. 2003;133:287S–290S. doi: 10.1093/jn/133.1.287S. [DOI] [PubMed] [Google Scholar]
- Wang X-D. Alcohol, vitamin A, and cancer. Alcohol. 2005;35:251–258. doi: 10.1016/j.alcohol.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Yang Z-N, Davis GJ, Hurley TD, Stone CL, Li T-K, Bosron WF. Catalytic Efficiency of Human Alcohol Dehydrogenases for Retinol Oxidation and Retinal Reduction. Alcoholism: Clinical and Experimental Research. 1994;18(3):587–591. doi: 10.1111/j.1530-0277.1994.tb00914.x. [DOI] [PubMed] [Google Scholar]
- Yin S-J, Bosron WF, Magnes LJ, Li T-K. Human liver alcohol dehydrogenase: Purification and kinetic characterization of the b2b2, b2b1, and b2g1 “Oriental” isoenzymes. Biochemistry. 1984;23:5847–5853. doi: 10.1021/bi00319a026. [DOI] [PubMed] [Google Scholar]
- Yin S-J, Chou C-F, Lai C-L, Lee S-L, Han C-L. Human class IV alcohol dehydrogenase: kinetic mechanism, functional roles, and medical relevance. Chemico-Biological Interactions. 2003;143–144:219–227. doi: 10.1016/s0009-2797(02)00167-9. [DOI] [PubMed] [Google Scholar]
- Younes M, Wagner H, Strubelt O. Enhancement of acute ethanol hepatotoxicity under conditions of low oxygen supply and ischemia/reperfusion. The role of oxygen radicals. Biochemical Pharmacology. 1989;38(20):3573–3581. doi: 10.1016/0006-2952(89)90130-5. [DOI] [PubMed] [Google Scholar]