Abstract
Luciferase-based imaging allows a global view of microbial pathogenesis. We applied this technique to gammaherpesvirus infection by inserting a luciferase expression cassette into the genome of murine herpesvirus-4 (MuHV-4). The recombinant virus strongly expressed luciferase in lytically infected cells without significant attenuation. We used it to compare different routes of virus inoculation. After intranasal infection of anaesthetized mice, luciferase was expressed in the nose and lungs for 7–10 days and in lymphoid tissue, most consistently the superficial cervical lymph nodes, for up to 30 days. Gastrointestinal infection was not observed. Intraperitoneal infection was very different to intranasal, with strong luciferase expression in the liver, kidneys, intestines, reproductive tract and spleen, but none in the nose or lungs. The nose has not previously been identified as a site of MuHV-4 infection. After intranasal infection of non-anaesthetized mice, it was the only site of non-lymphoid luciferase expression. Nevertheless, lymphoid colonization and persistence were still established, even at low inoculation doses. In contrast, virus delivered orally was very poorly infectious. Inoculation route therefore had a major impact on pathogenesis. Low dose intranasal infection without anaesthesia seems most likely to mimic natural transmission, and may therefore be particularly informative about normal viral gene functions.
INTRODUCTION
Gammaherpesviruses are highly prevalent and cause considerable disease. A major challenge in combating this disease is to understand natural infection, for example how gammaherpesviruses first enter their hosts. Neither Epstein–Barr virus (EBV) nor Kaposi's sarcoma-associated herpesvirus (KSHV) are easy to analyse, because their infections are largely limited to humans. Gammaherpesviruses that allow experimental in vivo analysis can therefore tell us a great deal. Murine herpesvirus-4 (MuHV-4) (Nash et al., 2001; Stevenson & Efstathiou, 2005) currently provides the most accessible model. The archetypal MHV-68 strain came from a bank vole (Myodes glareolus) (Blaskovic et al., 1980). However, MuHV-4 has also been isolated from yellow-necked mice (Apodemus flavicollis) (Kozuch et al., 1993), and closely related viruses have been isolated from a shrew (Crocidura russula) (Chastel et al., 1994) and from wood mice (Apodemus sylvaticus) (Blasdell et al., 2003), suggesting that MuHV-4-like viruses have wide host ranges. Although MuHV-4 has not been isolated from house mice (Mus musculus), the natural correlate of inbred laboratory strains, its benign persistence in laboratory mice contrasts with the high pathogenicity or marked attenuation generally shown by herpesviruses in xenogenic hosts (Stevenson et al., 2002a). There is serological evidence that MuHV-4 (or a close relative) naturally infects house mice (Mistrikova et al., 2000), and MuHV-4 major histocompatibility complex (MHC) class I degradation (Boname & Stevenson, 2001; Lybarger et al., 2003), transporter associated with antigen processing (TAP) degradation (Boname et al., 2004), complement inhibition (Kapadia et al., 1999) and chemokine binding (Parry et al., 2000; van Berkel et al., 2000) all work in laboratory mice. Such immune evasion functions are typically species-specific, for example MuHV-4 K3 downregulates MHC class I expression poorly in rat cells (our unpublished data). MuHV-4 infection of laboratory mice therefore seems to provide a reasonable pathogenesis model.
Experimental MuHV-4 infection typically employs intranasal virus inoculation under general anaesthesia. This leads to a lytic infection of lung alveolar epithelial cells that is controlled within 2 weeks (Nash & Sunil-Chandra, 1994). Virus meanwhile seeds to lymphoid tissue and drives the proliferation of latently infected B cells. This peaks at 2 weeks post-infection (p.i.) and is controlled by 4 weeks. A predominantly latent infection of memory B cells (Flano et al., 2002) then persists. In addition to intranasal infection, MuHV-4 has been given intraperitoneally (Weck et al., 1996), subcutaneously (Raslova et al., 2001), intravenously (Sunil-Chandra et al., 1992), orally (Blaskovic et al., 1984), intracerebrally (Terry et al., 2000) and by gavage (Peacock & Bost, 2000). Its capacity to infect many different anatomical sites corresponds to a broad tropism for different fibroblast and epithelial cell lines (Gillet et al., 2007a).
All inoculation routes lead to B-cell infection, and latently infected B cells can in theory transport MuHV-4 from any one site to any other. It is therefore often assumed that the different modes of infection are fairly equivalent. However, the peak of B-cell colonization coincides with a strong CD8+ T-cell response (Stevenson et al., 1999a) that limits lytic spread (Stevenson et al., 1999b), and there is evidence from EBV that latently infected B cells recirculate mainly through their site of initial infection (Laichalk et al., 2002). The exposure of different epithelial and fibroblast populations to MuHV-4 infection may therefore depend strongly on inoculation route. Even without a direct comparison, it is clear that inoculation route can affect experimental outcomes. For example, intraperitoneal MuHV-4 infection led to the conclusion that B cells support acute lytic replication and macrophages support long-term latency (Weck et al., 1996), whereas after intranasal infection, macrophage colonization is transient and B cells provide the long-term latent reservoir (Sunil-Chandra et al., 1992).
A major impetus to establishing a realistic form of experimental MuHV-4 infection has been the finding that in vitro and in vivo virus neutralization are quite different (Gillet et al., 2007b). The implication is that to understand neutralization, we must also understand host entry. Global imaging provides one way to compare different infection routes. Here we imaged MuHV-4 lytic gene expression by luciferase expression and charge-coupled-device camera scanning (Hutchens & Luker, 2007). After intranasal virus inoculation without anaesthesia, luciferase expression was limited to the nose and superficial cervical lymph nodes (SCLN), but still established a persistent infection. In contrast, orally delivered virus was poorly infectious. The nose therefore seems a likely physiological route of host entry. Studies of infection by this route may give new insights into MuHV-4 gene functions.
METHODS
Mice.
Female BALB/c mice were infected with MuHV-4 when 6–12 weeks old. Intranasal infections with anaesthesia were in 30 μl aliquots, those without were in 5 μl. All experiments conformed to local animal ethics regulations; those in Cambridge also followed Home Office Project Licence 80/1992. For luciferase imaging, mice were injected intraperitoneally with luciferin, anaesthetized with ketamine/xylazine or isoflurane, then scanned with an IVIS Lumina (Caliper Life Sciences). In preliminary experiments we used the manufacturer's recommended dose of 150 μg luciferin g−1. In later experiments this was reduced to 2 mg/mouse without noticeably less signal. Signal intensity was fairly constant between 3 and 10 min after injection. Mice were routinely imaged after 5 min. For quantitative comparisons, we used Living Image software (Caliper Life Sciences) to obtain the maximum radiance (photons per s per cm2 per steradian, i.e. photons s−1 cm−2 sr−1) over each region of interest, relative to a negative control region.
Cells.
Baby hamster kidney (BHK-21) cells, NIH-3T3 cells, NIH-3T3-CRE cells (Stevenson et al., 2002b) and NIH-3T3-TET50 cells were propagated in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 2 mM glutamine, 100 U penicillin ml−1, 100 μg streptomycin ml−1 and 10 % fetal calf serum. NIH-3T3-TET50 cells were made by serially transducing NIH-3T3 cells with three retroviruses: one expressed ORF50 from a promoter with doxycycline-responsive (TRE) promoter; one expressed constitutively a transcriptional suppressor with doxycycline-inactivated TRE-binding; and one expressed constitutively a transcriptional activator with doxycycline-activated TRE-binding. All together allowed doxycycline-inducible ORF50 expression. The ORF50 coding sequence was amplified by PCR from infected cell cDNA and cloned into pREV-TRE (Clontech). The TRE-binding transcriptional suppressor was excised from pTET-tTS (Clontech) with EcoRI/ClaI, the ClaI site was blunted with T4 DNA polymerase and the fragment was ligated into the EcoRI-blunted XhoI sites of pMSCV-IRES-PURO. The TRE-binding transcriptional activator was from pREV-TET-ON (Clontech). Each plasmid was transfected into 293T cells together with the pEQPAM3 packaging plasmid (Persons et al., 1998). Retroviruses were collected after 48 and 72 h, and added to cells with 6 μg polybrene ml−1. Triply transduced cells were selected with puromycin+hygromycin+G418.
Viruses.
The luciferase coding sequence plus polyadenylation signal was removed from pGL4.10 (Promega) by digestion with BglII/SalI and cloned into the BamHI/SalI sites of pSP73, downstream of a 500 bp MuHV-4 M3 promoter (May et al., 2005a). M3-luciferase-polyA was then excised with BglII/SalI, blunted with Klenow fragment DNA polymerase and cloned into the blunted MfeI site (genomic co-ordinate 77 176, GenBank accession no. U97553) of a BglII MuHV-4 genomic clone (co-ordinates 75 338–78 717), again in pSP73. The expression cassette plus genomic flanks was subcloned into the BamHI site of the pST76K-SR shuttle vector and recombined into a MuHV-4 bacterial artificial chromosome (BAC; Adler et al., 2000). An ORF50-deficient derivative was made by cloning a HincII genomic fragment (co-ordinates 63 844–70 433) into the HincII site of pUC9 (New England Biolabs), with the BamHI site of pUC9 at the 70 433 end of the insert. This was cut with BsmI (67 792) and ClaI (69 177) to remove most of ORF50 exon 2 (67 661–69 376), blunted and dephosphorylated with Antarctic alkaline phosphatase (New England Biolabs). The eGFP coding sequence from pEGFP-N3 (Clontech) was ligated in place of the removed fragment. EGFP plus its genomic flanks was then excised using a genomic KpnI site (66 120) and the BamHI site in pUC9, cloned into the BamHI/KpnI sites of pST76K-SR, and recombined into the M3-LUC BAC. M3-LUC virus was recovered by transfecting BAC DNA into BHK-21 cells. For in vivo experiments, its loxP-flanked BAC/eGFP cassette was removed by passage through NIH-3T3-CRE cells. Virus stocks were grown in BHK-21 cells (Coleman et al., 2003). ORF50−M3-LUC virus was recovered by transfecting BAC DNA into NIH-3T3-TET50 cells and propagated by treating the cells with doxycycline.
Viral infectivity assays.
Virus stocks were titrated by plaque assay on BHK-21 cells (de Lima et al., 2004), or on doxycycline-treated NIH-3T3-TET50 cells for ORF50−M3-LUC. Cell monolayers were incubated with virus (2 h, 37 °C), overlaid with 0.3 % carboxymethylcellulose, and 4 days later fixed and stained for plaque counting. Infectious virus in lungs was measured by freeze–thawing the lungs and homogenizing them in 1 ml complete medium prior to plaque assay. Latent virus was measured by infectious centre assay (de Lima et al., 2004): spleen cells were co-cultured with BHK-21 cells, then fixed and stained for plaque counting after 4 days. Plaque assay titres of freeze–thawed lymphoid homogenates were always <1 % of infectious centre assay titres, so the latter essentially measured reactivable latent virus.
Viral genome quantification.
Viral genome loads were measured by real-time PCR (Gaspar et al., 2008). DNA from organs (50–80 ng) was used to amplify MuHV-4 genomic co-ordinates 4166–4252 (Rotor Gene 3000; Corbett Research). The PCR products were quantified by hybridization with a TaqMan probe (genomic co-ordinates 4218–4189) and converted to genome copies by comparison with a standard curve of cloned plasmid template amplified in parallel. Cellular DNA was quantified in parallel by amplifying part of the adenosine phosphoribosyl transferase gene (forward primer 5′-GGGGCAAAACCAAAAAAGGA-3′, reverse primer 5′-TGTGTGTGGGGCCTGAGTC-3′, probe 5′-TGCCTAAACACAAGCATCCCTACCTCAA-3′).
Southern blotting.
Viral DNA was extracted by alkaline lysis (Coleman et al., 2003), digested, electrophoresed and transferred to nylon membranes (Roche Diagnostics). A [32P]dCTP-labelled probe (APBiotech) was generated by random primer extension (DECAprime II kit; Ambion). Membranes were hybridized with probe (65 °C, 18 h), washed in 30 mM sodium chloride, 3 mM sodium citrate, 0.1 % SDS solution at 65 °C and exposed to X-ray film.
In vitro luciferase assays.
Cells were washed twice in PBS, then lysed in 1 % Triton X-100 (15 min, 4 °C). Cell debris was pelleted (13 000 g, 5 min) and supernatants incubated in 20 mM glycylglycine, 20 mM potassium phosphate buffer (pH 7.8) with 1 mM dithiothreitol, 10 mM MgSO4, 3 mM EGTA, 2.5 mM ATP and 100 μM luciferin. The light emission of triplicate samples was detected by luminometry (Hewlett Packard).
ELISA.
MuHV-4 virions were recovered from infected cell supernatants by ultracentrifugation, disrupted with 0.05 % Triton X-100 in 50 mM sodium carbonate buffer (pH 8.5), and coated onto MaxiSorp ELISA plates (Nunc). The plates were washed three times in PBS containing 0.1 % Tween 20, blocked with 2 % BSA in PBS 0.1 % Tween 20, then incubated with threefold serum dilutions from MuHV-4-exposed mice (1 h, 23 °C). The plates were then washed four times in PBS 0.1 % Tween 20, incubated (1 h, 23 °C) with alkaline phosphatase-conjugated goat anti-mouse IgG-Fc pAb (Sigma), washed five times, and developed with nitrophenylphosphate (Sigma). Absorbance was measured at 405 nm (Bio-Rad).
RESULTS
Generation and in vitro analysis of MuHV-4 expressing luciferase
An ideal luciferase reporter would mark both lytically and latently infected cells. However, high level latent gene expression is probably incompatible with normal host colonization for a gammaherpesvirus: most EBV-infected cells express no latency genes at all (Thorley-Lawson, 2001); both EBV and MuHV-4 limit antigen presentation from their episome maintenance proteins by low turnover (Yin et al., 2003; Bennett et al., 2005); and bypassing this evasion severely attenuates MuHV-4 latency (Bennett et al., 2005). Autonomous promoters such as that of the human cytomegalovirus (HCMV) IE1 gene (Rosa et al., 2007; Smith et al., 2007) may therefore expose latently infected cells to unphysiological CD8+ T-cell recognition. This would explain why HCMV IE1-driven reporter gene expression is associated with marked in vivo MuHV-4 attenuation (Adler et al., 2001).
We therefore aimed for lytic reporter gene expression, using as a promoter an ectopic copy of the 500 bp upstream of the MuHV-4 M3, an abundant early/late lytic gene (van Berkel et al., 1999). In situ detection of M3 mRNA in infected lymphoid tissue (Simas et al., 1999) and a latency establishment deficit of MuHV-4 M3 mutants (Bridgeman et al., 2001) suggest that M3 might also be transcribed in latency. However, other early lytic transcripts are detectable in spleens (Marques et al., 2003) – B cells even drive a substantial lytic antigen-specific CD8+ T-cell response (Stevenson et al., 1999a) – and the latency deficit of M3 mutants may simply reflect that lytically infected cells no longer secrete M3 to provide bystander protection for their latently infected neighbours (Rice et al., 2002; Stevenson, 2004).
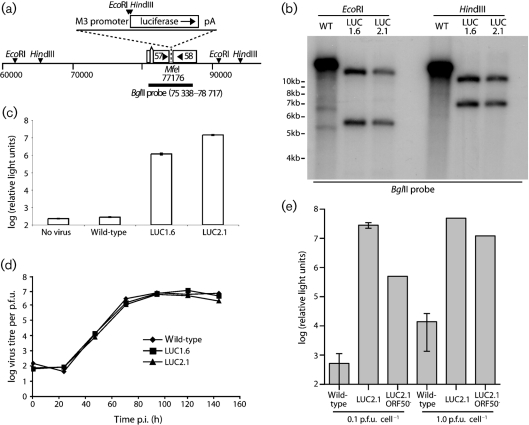
The luciferase expression cassette was inserted between the polyadenylation signals of ORFs 57 and 58 (Fig. 1a). Southern blotting (Fig. 1b) confirmed the predicted structures of recombinant viral genomes. Infected cells showed strong luciferase expression (Fig. 1c). M3-LUC viruses showed no in vitro lytic replication defect (Fig. 1d). Since ORF57 is essential for lytic replication (Song et al., 2005) and ORF58 contributes to inter-cellular viral spread (May et al., 2005b), luciferase expression appeared not to compromise the functions of neighbouring genes. Gp48 expression, which is ORF58-dependent, was also normal (data not shown).
Fig. 1.
Generation of MuHV-4 expressing luciferase. (a) A 2 kb luciferase-polyA cassette was placed downstream of a 500 bp M3 promoter, in a MfeI restriction site between ORFs 57 and 58. Relevant restrictions sites are shown. (b) Viral DNA was digested with EcoRI or HindIII and probed with the 75 338–78 717 BglII clone shown in (a). The luciferase expression cassette changes a 14.9 kb EcoRI band to 5.5 kb+12.0 kb, and a 14.5 kb HindIII band to 6.8 kb+10.1 kb. M3-LUC1.6 and M3-LUC2.1 are independently generated recombinant viruses. (c) BHK-21 cells were left uninfected or infected overnight (1 p.f.u. cell−1), then lysed and assayed for luciferase expression. Each bar shows the mean±sd of triplicate cultures. (d) BHK-21 cells were infected with wild-type or M3-LUC MuHV-4 (0.01 p.f.u. cell−1, 2 h), washed with PBS to remove unbound virions, then incubated at 37 °C. The infectious virus in each culture was measured by plaque assay. (e) BHK-21 cells were infected with wild-type MuHV-4, the M3-LUC2.1 recombinant or its ORF50− derivative. Luciferase expression was assayed 18 h later by luminometry. Each bar shows mean±sd of five replicate infections. The ORF50− cultures contained no replication-competent virus by plaque assay.
We tested whether M3-LUC luciferase expression depended on the ORF50 lytic transactivator by generating an ORF50-deficient derivative (Fig. 1e). This was propagated in complementing NIH-3T3-TRE50 cells and then tested for luciferase expression in non-complementing BHK-21 cells. At 0.1 p.f.u. cell−1, ORF50 disruption reduced the luciferase signal by 98–99 %. At 1 p.f.u. cell−1 the reduction was 80 %. Inhibiting viral DNA replication with phosphonoacetic acid had no effect on luciferase expression (data not shown). Thus, luciferase expression corresponded mainly to early lytic gene expression, with some additional ORF50-independent expression at high multiplicity infection. In vivo cells may differ from transformed cell lines in viral transcription, but it seemed unlikely that luciferase expression would reveal latent infection. We considered it to mark early lytic gene expression.
In vivo analysis of MuHV-4 expressing luciferase
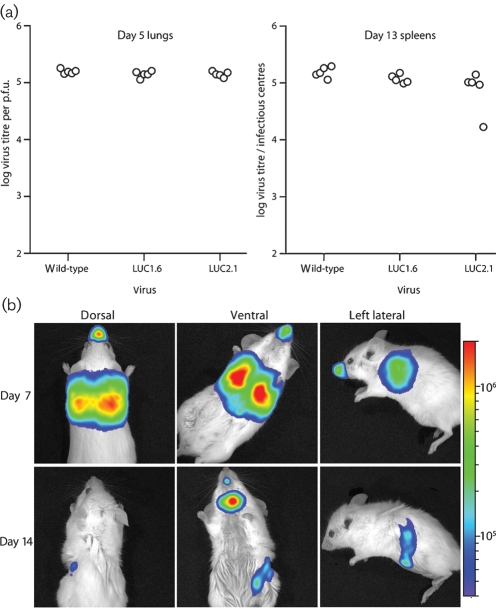
We have previously shown that M3 promoter-driven ovalbumin or eGFP expression has a minimal effect on MuHV-4 replication in vivo (Smith et al., 2006). M3-LUC viruses similarly showed normal replication after intranasal inoculation (Fig. 2a). We selected M3-LUC2.1 for further analysis. We infected anaesthetized mice intranasally, then monitored luciferase expression by luciferin injection and charge-coupled-device camera scanning (Fig. 2b). In preliminary experiments, removing the fur from mice prior to imaging had little effect on the signal obtained. Therefore in subsequent experiments no fur was removed. At the peak of lytic replication (5–7 days p.i.), a strong signal was visible in the thorax and the nose. At the peak of latency amplification (13–14 days p.i.), luciferase signals were weak or undetectable in the thorax and nose, but strong in the neck. Some mice also showed a weaker abdominal signal.
Fig. 2.
In vivo infection by luciferase-expressing MuHV-4. (a) M3-LUC1.6 and M3-LUC2.1 were compared with wild-type MuHV-4 for their capacity to colonize mice after intranasal infection. Lytic replication was tested by plaque assay of lungs after 5 days. Latency establishment was tested by infectious centre assay of spleens after 13 days. Each point shows the titre of one mouse. There was no significant difference between each virus. (b) Mice were infected intranasally (104 p.f.u.) with M3-LUC2.1 MuHV-4 under general anaesthesia, and then injected with luciferin and imaged every 3–4 days. Images show a representative mouse at day 7 and 14 p.i. The signal in the mouth was atypical and probably corresponds to the strong neck signal reflecting off the incisors. The scale bar shows photons s−1 cm−2 sr−1.
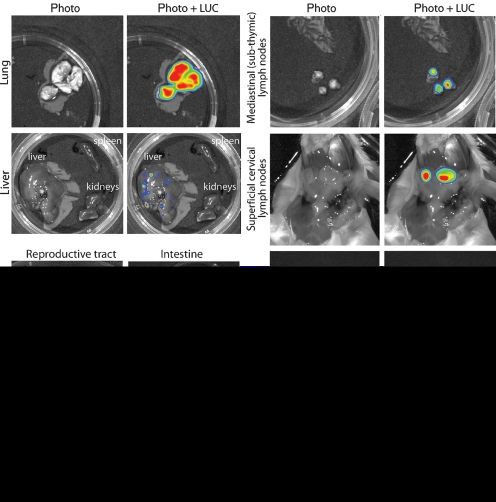
Ex vivo imaging of dissected organs (Fig. 3) established the sources of the live images. The thoracic signal came from the lungs; the abdominal signal came from the spleen; and the neck signal came from the SCLN, particularly the most rostral node that lies alongside the salivary glands. The mediastinal (subthymic) and deep cervical LN showed weaker signals. Despite strong signals from nose and lungs, we saw no signal from the intervening trachea or main bronchi. Although MuHV-4 has been reported to infect mice after inoculation into the stomach (Peacock & Bost, 2000), and at least some of a 30 μl intranasal inoculum is likely to be swallowed, we saw no sign of intestinal infection. A signal was occasionally observed (10–20 % of mice) in the liver at day 7 (Fig. 3) and in abdominal LN at day 10–14 (data not shown). This could have reflected some intestinal infection. But it could also have reflected contiguous spread from the lungs and diaphragm or systemic virus spread via B cells. Other abdominal organs, such as the kidneys and reproductive tract, were consistently negative.
Fig. 3.
Luciferase signals from isolated organs after intranasal MuHV-4 infection. Mice equivalent to those in Fig. 2(b) were dissected and their organs imaged ex vivo. Each image is representative of data from at least five mice, and shows either a standard photograph (Photo) or that photograph overlaid with the luciferase signal (Photo+LUC). The colour scheme for relative signal intensity is as for Fig. 2.
Quantification of luciferase signals
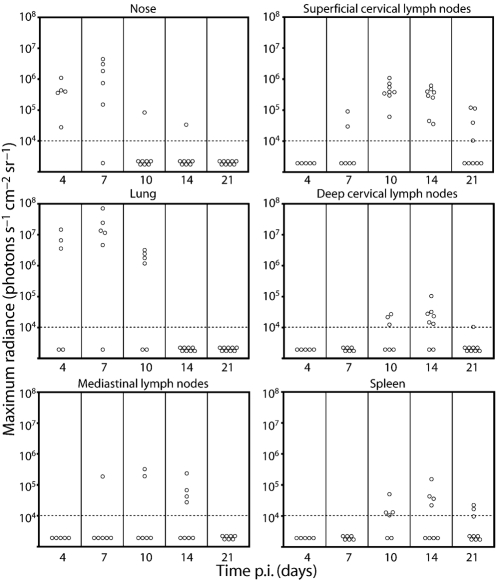
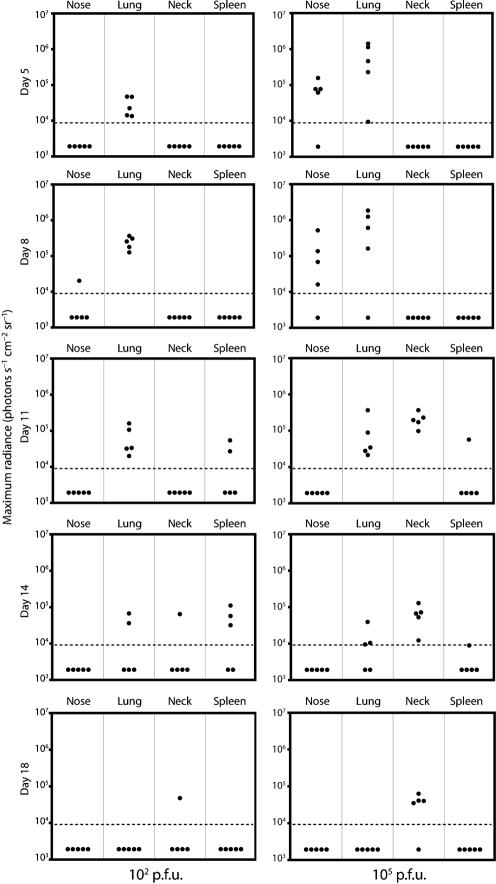
Luciferase signals were quantified as the maximum radiance (photons s−1 cm−2 sr−1) over a region of interest – typically one dissected organ. Fig. 4 shows a time-course after intranasal infection. Occasional mice showed no signal even at peak average time points. Tracking individual mice showed that none remained uninfected. Rather the kinetics of luciferase expression varied; total signals over the whole time-course were similar. This possibly reflected variation in inoculum dose to the lung: some virus may have been trapped in nasal sinuses or coughed up after recovery from anaesthesia. SCLN signals were the most long-lived, sometimes persisting even at day 25 p.i. But generally all sites were negative by day 30.
Fig. 4.
Quantification of luciferase signals from isolated organs after intranasal MuHV-4 infection. Mice were infected intranasally (104 p.f.u.) with M3-LUC. At each time point, at least five mice per group were dissected for ex vivo organ imaging. Each point shows the maximum radiance value for one mouse. The horizontal dashed lines mark an arbitrary sensitivity threshold, chosen to minimize the chance of artefactual signals such as secondary light reflections.
Several caveats apply to live imaging. Overlying tissues clearly reduced light transmission, as neck signals were evident ventrally but not dorsally, and the dissected organs signals were stronger than live images. This limits the interpretation of fine quantitative differences. Mediastinal LN and deep cervical LN signals were only evident after dissection because they were obscured by those of the lungs and SCLN. Very strong signals sometimes gave secondary reflections. For example, in the day 14 ventral view (Fig. 2b), the neck signal is reflecting off the incisors: after dissection, no mouth signal was observed. However, there was generally a good correlation between live images and dissected organ signals.
Since luciferase expression corresponded to early lytic gene expression, a lack of signal did not necessarily imply a lack of infection: tightly maintained viral latency might be missed. The lytic antigen-specific CD8+ T-cell responses generated by 7–10 days p.i. (Stevenson & Doherty, 1998) would also be expected to suppress luciferase expression. Nevertheless, luciferase signals appeared to match quite well the results of other assays (Sunil-Chandra et al., 1992): an acute lytic infection in the lung (day 4–10) progressed to a subacute infection of lymphoid tissue (day 10–25), and both were largely resolved by day 30. Although MuHV-4 does not show significant productive infection in lymphoid tissue after intranasal inoculation (Nash & Sunil-Chandra, 1994), luciferase signals here were entirely consistent with evidence of early lytic gene expression (Stevenson et al., 1999a; Liu et al., 1999; Marques et al., 2003). Thus, early lytic luciferase expression revealed both productive lytic replication and acute lymphoid colonization.
Intraperitoneal infection
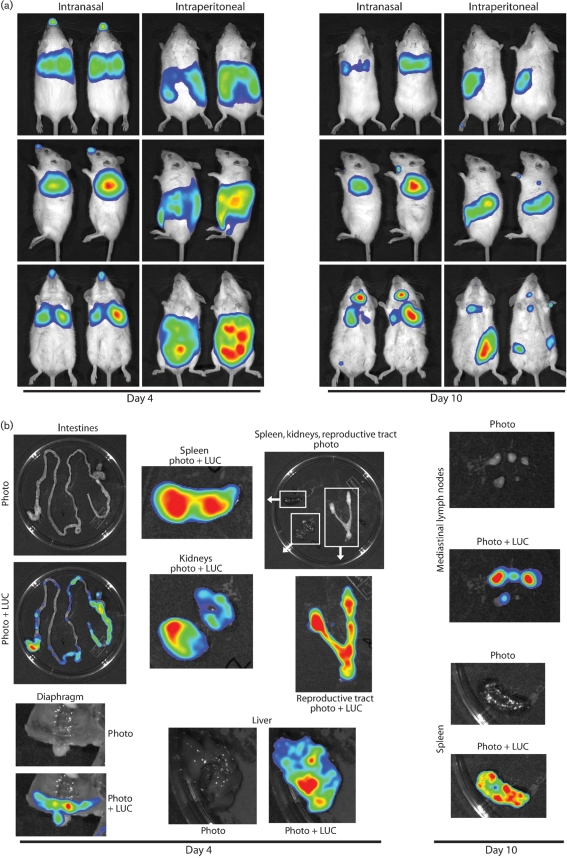
Many MuHV-4 pathogenesis studies have used intraperitoneal infection rather than intranasal (Speck & Virgin, 1999). Understanding the relationship between these infections is therefore important for integrating existing pathogenesis data into a coherent whole. Luciferase expression patterns after intraperitoneal or intranasal M3-LUC inoculations were markedly different (Fig. 5a). At day 4 after intraperitoneal inoculation, there was a strong signal from the abdominal cavity and none from the nose or lungs. At the same time after intranasal inoculation, there was a strong signal from the lungs and none from the abdomen. By 10 days after intraperitoneal inoculation, there was still no signal from the lungs. The abdominal signal had decreased, and the only spread was to the mediastinal LN, which receive lymphatic drainage from the peritoneal cavity; the strong SCLN signal associated with intranasal infection remained weak or absent.
Fig. 5.
Comparison of intranasal and intraperitoneal MuHV-4 infections. (a) Mice were inoculated intranasally or intraperitoneally with 104 p.f.u. of M3-LUC, then monitored for luciferase expression. Representative pairs of mice are shown. (b) Mice were dissected at 4 or 10 days after intraperitoneal virus inoculation to identify the source of the live imaging signals. The colour scheme for relative signal intensity is as for Fig. 2.
Ex vivo imaging (Fig. 5b) showed that the abdominal signal of intraperitoneal infection had many sources, including the liver, spleen, kidneys and reproductive tract. The gut was also patchily positive, presumably reflecting serosal infection. Therefore, although MuHV-4 reached lymphoid tissue by either route, the distributions of lytic infection were almost mutually exclusive; in neither case did infected B cells appear to seed major new non-lymphoid sites of lytic gene expression. Even the spleen signals differed: intraperitoneal infection showed strong luciferase expression early on, consistent with productive infection (Weck et al., 1996), while intranasal infection showed later, weaker expression, consistent with a predominantly latent infection. These data explained why intraperitoneal and intranasal infections have given such different phenotypes, for example with M11 knockouts (Gangappa et al., 2002; de Lima et al., 2005).
Different infection doses
We next tested different inoculation doses via the intranasal route with general anaesthesia (Fig. 6). High dose infection (105 p.f.u.) gave a luciferase signal in the lung that peaked at 4–7 days. After low dose infection (102 p.f.u.) the signal peaked at 8–11 days and was generally less extensive. Both infections led to lymphoid colonization. Thus, beyond more virus giving more extensive early lytic replication, there was little difference between low and high doses, consistent with infectivity assays (Tibbetts et al., 2003). Luciferase signals in the nose were uncommon after low dose infection, presumably because all the infectious particles ended up in the lungs. SCLN signals were also weak, implying that this site is colonized principally via the upper respiratory tract.
Fig. 6.
High dose and low dose intranasal infections. Mice were infected intranasally with M3-LUC (102 or 105 p.f.u.) under general anaesthesia, then live-imaged for luciferase expression every 3–5 days. Each point corresponds to one mouse. The horizontal dashed lines mark an arbitrary sensitivity threshold.
Intranasal infection without anaesthesia
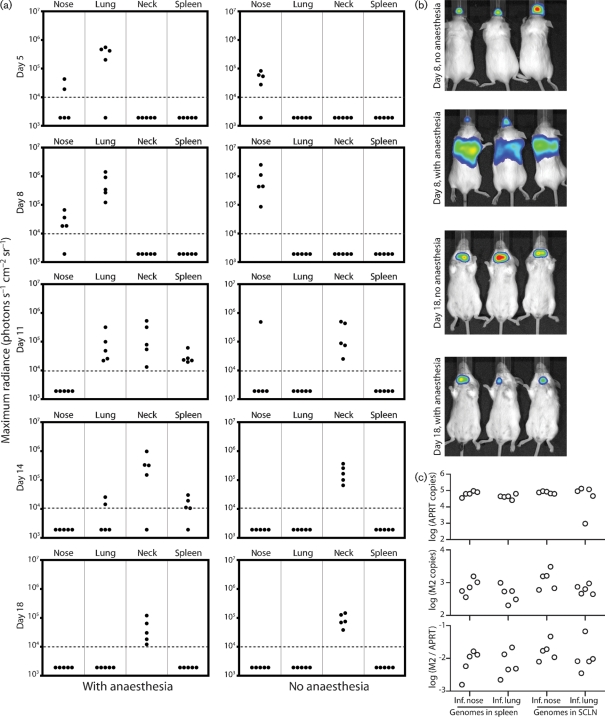
The prominent luciferase signals in noses after intranasal inoculation suggested that host entry might occur via the upper respiratory tract as well as via the lungs, where it has mostly been studied to date. We tested this by intranasal infection without anaesthesia (Fig. 7). As before, the anaesthetized controls showed strong luciferase signals in lungs and less consistent signals in the nose (Fig. 7a, b). Infection then spread to the SCLN and spleen. Non-anaesthetized mice given the same virus dose showed a strong signal in the nose and none in the lungs. Thus, infection did not reach the lungs without anaesthesia. Non-anaesthetized mice also showed no luciferase signal in the spleen. However, their SCLN signals were at least as strong as those of anaesthetized mice. The lack of spleen signal presumably reflected that it was colonized relatively late in infection, when lytic antigen-specific CD8+ T-cell responses were strong. Real-time PCR quantification of viral genomes (Fig. 7c) established that virus delivered to the nose still established normal persistence in both the SCLN and spleen.
Fig. 7.
Intranasal infection with and without anaesthesia. (a) Mice were infected intranasally with M3-LUC (104 p.f.u.), either awake or while anaesthetized, then imaged for luciferase expression every 3–5 days. Each point corresponds to one mouse. The horizontal dashed lines mark an arbitrary sensitivity threshold. (b) Representative images from the data summarized in (a). (c) At 1 month p.i., mice were analysed for viral genome loads in the spleen and SCLN by real-time PCR. The top panel shows the cellular control (APRT), the middle panel the MuHV-4 M2 gene, and the bottom panel the M2 load normalized by APRT. Each point corresponds to one mouse. There was no statistically significant difference between mice given anaesthesia (Inf. lung) or not (Inf. nose).
Oral infection
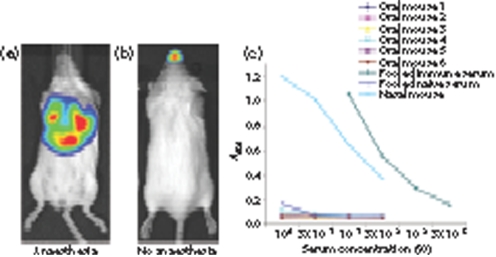
We also tested oral infection (Fig. 8). A 104 p.f.u. inoculum with general anaesthesia gave no luciferase signal in 3/5 mice. What mice were infected showed the typical lung-dominated pattern of intranasal infection (Fig. 8a), suggesting aspiration of the oral inoculum. A 106 p.f.u. inoculum without anaesthesia gave luciferase signals in 5/8 mice. This time, the infected mice had the typical pattern of nose infection (Fig. 8b), suggesting that here some of the oral inoculum reached the nose. Thus, there was no evidence of host entry by the oral route: normal infection was established if virions reached the nose or lung, but there was no infection if they did not.
Fig. 8.
Oral infection. (a) Representative image of a luciferase-positive mouse 8 days after oral infection under general anaesthesia. (b) Representative image of a luciferase-positive mouse 7 days after oral infection without anaesthesia. (c) Sera from mice 1 month after oral inoculation without anaesthesia (50 p.f.u.) were analysed for MuHV-4-specific IgG by ELISA, along with pooled naive mouse serum, pooled immune serum and serum from a mouse infected (10 p.f.u.) intranasally without anaesthesia.
Lower dose infections without anaesthesia – reducing the chance of inadvertent respiratory tract contamination – gave no luciferase signal (0/10 mice for 103 p.f.u.; 0/24 mice for 50 p.f.u.), even when we imaged dissected organs. In contrast, a 10 p.f.u. intranasal inoculum without anaesthesia gave strong luciferase signals in 12/12 mice. A lack of infection by 50 p.f.u. of oral virus was confirmed by ELISA for virus-specific serum IgG at 1 month post-exposure (Fig. 8c). PCR of spleen cell DNA for viral genomes at 15 days after oral inoculation was also completely negative in 6/6 mice (<5 viral genome copies per 80 ng DNA). Thus in adult mice, oral MuHV-4 was poorly infectious.
DISCUSSION
How do herpesviruses enter their hosts? We compared different modes of experimental MuHV-4 infection, aiming to establish a plausible correlate of natural transmission for adult mice. For an anatomically complete view, we monitored infection by luciferase expression from the viral genome. Analysis of the standard intranasal infection model validated this approach. Thus, strong luciferase expression in the lung, weaker expression in lymphoid tissue, then quiescence, matched the consensus picture from other assays of productive viral replication in the lung, latency amplification with some lytic gene expression in lymphoid tissue, then immune control. Different modes of virus inoculation gave markedly different patterns of luciferase expression. Intranasal inoculation under general anaesthesia gave strong expression in the nose and lungs and weaker expression in lymphoid tissue; intraperitoneal inoculation gave strong expression in multiple abdominal organs, including the spleen, and none in the respiratory tract; intranasal inoculation without anaesthesia gave expression in just the nose and draining lymphoid tissue; and oral inoculation gave no expression at all. Normal transmission is presumably mucosal. The nose – but not the lung – is therefore a likely point of normal host entry.
The poor infectivity of oral MuHV-4 was surprising, as human gammaherpesviruses are thought to transmit orally via saliva. MuHV-4 may use nasal entry because noses feature more prominently in murine social life. The nasopharynx may also be important for human gammaherpesvirus transmission: there is little evidence for a specifically oral EBV or KSHV entry route, and EBV infection notably predisposes to nasopharyngeal carcinoma.
The limited lytic spread of MuHV-4 from the nose might be seen as suboptimal for host colonization. However, gammaherpesvirus epidemiology indicates that transmission depends more on long-term virus shedding than primary lytic infection, and this correlates with the latent load (Yao et al., 1985). MuHV-4 latency is relatively independent of the extent of primary lytic infection (Stevenson et al., 1999c; Coleman et al., 2003). It depends much more on latency-associated lymphoproliferation (May et al., 2004). Extensive primary lytic spread might even be counter-productive by providing a powerful immune stimulus and by predisposing the host to disease. Thus, gammaherpesviruses may have evolved to infect their hosts without extensive lytic spread.
The apparent failure of intranasal MuHV-4 to infect the oropharynx or trachea, and of oral MuHV-4 to infect anywhere, suggest that incoming virions cross epithelia by specialized routes. Although MuHV-4 virions readily infect most epithelial cells in vitro, their strong dependence on heparan sulphate for cell binding (Gillet et al., 2008) raises questions about epithelial infection in vivo, as here heparan sulfate is predominantly basolateral rather than apical (Hayashi et al., 1987). In vitro epithelial infection may correspond more to host exit, when virions would emerge from B cells to infect basolaterally. Notably, MuHV-4 infects confluent, polarized in vitro epithelial monolayers much less well than subconfluent monolayers (our unpublished data).
An important task now is to identify the cell types targeted in the nose. This is not necessarily straightforward: high dose inocula may reach non-physiological sites, low dose inocula are inherently hard to track, and once there is viral replication, histology may fail to distinguish host entry from exit. Entry via the nose may be qualitatively different to that via the lungs or the peritoneal cavity. The latter both contain abundant macrophage populations without an epithelial barrier, and peritoneal macrophages at least are quite readily infected (Rosa et al., 2007). A related question is how incoming virions reach B cells. This may occur submucosally or in lymph nodes. The strong luciferase signals in SCLN following nasal infection argued for a significant lymphatic transport of infectious virions to this site. One precedent for such transport is normal immune priming (Belz et al., 2007). A role for dendritic cells in B-cell infection would therefore not be surprising. The present study provides a basis for further understanding by identifying the nose as an entry point and the SCLN as the major associated lymphoid target.
Acknowledgments
R. M., S. M., M. A. and J. P. S. are supported by the Portuguese Foundation for Science and Technology (POCTI/ESP/467378/2002). L. G. is a Postdoctoral Researcher of the Fonds National Belge de la Recherche Scientifique. This work was also supported by CR-UK (C19612/A6189 to P. G. S. and S. E.), the UK Medical Research Council (G0701185 to P. G. S. and G9800903 to P. G. S., S. E. and others) and the Wellcome Trust (GR076956MA to P. G. S.).
Footnotes
Published online ahead of print on 15 October 2008 as DOI 10.1099/vir.0.2008/006569-0.
References
- Adler, H., Messerle, M., Wagner, M. & Koszinowski, U. H. (2000). Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J Virol 74, 6964–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler, H., Messerle, M. & Koszinowski, U. H. (2001). Virus reconstituted from infectious bacterial artificial chromosome (BAC)-cloned murine gammaherpesvirus 68 acquires wild-type properties in vivo only after excision of BAC vector sequences. J Virol 75, 5692–5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belz, G. T., Wilson, N. S., Kupresanin, F., Mount, A. M. & Smith, C. M. (2007). Shaping naive and memory CD8+ T cell responses in pathogen infections through antigen presentation. Adv Exp Med Biol 590, 31–42. [DOI] [PubMed] [Google Scholar]
- Bennett, N. J., May, J. S. & Stevenson, P. G. (2005). Gamma-herpesvirus latency requires T cell evasion during episome maintenance. PLoS Biol 3, e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasdell, K., McCracken, C., Morris, A., Nash, A. A., Begon, M., Bennett, M. & Stewart, J. P. (2003). The wood mouse is a natural host for murid herpesvirus 4. J Gen Virol 84, 111–113. [DOI] [PubMed] [Google Scholar]
- Blaskovic, D., Stancekova, M., Svobodova, J. & Mistrikova, J. (1980). Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol 24, 468. [PubMed] [Google Scholar]
- Blaskovic, D., Stanekova, D. & Rajcani, J. (1984). Experimental pathogenesis of murine herpesvirus in newborn mice. Acta Virol 28, 225–231. [PubMed] [Google Scholar]
- Boname, J. M. & Stevenson, P. G. (2001). MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 15, 627–636. [DOI] [PubMed] [Google Scholar]
- Boname, J. M., de Lima, B. D., Lehner, P. J. & Stevenson, P. G. (2004). Viral degradation of the MHC class I peptide loading complex. Immunity 20, 305–317. [DOI] [PubMed] [Google Scholar]
- Bridgeman, A., Stevenson, P. G., Simas, J. P. & Efstathiou, S. (2001). A secreted chemokine binding protein encoded by murine gammaherpesvirus-68 is necessary for the establishment of a normal latent load. J Exp Med 194, 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastel, C., Beaucournu, J. P., Chastel, O., Legrand, M. C. & Le Goff, F. (1994). A herpesvirus from an European shrew (Crocidura russula). Acta Virol 38, 309. [PubMed] [Google Scholar]
- Coleman, H. M., de Lima, B., Morton, V. & Stevenson, P. G. (2003). Murine gammaherpesvirus 68 lacking thymidine kinase shows severe attenuation of lytic cycle replication in vivo but still establishes latency. J Virol 77, 2410–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima, B. D., May, J. S. & Stevenson, P. G. (2004). Murine gammaherpesvirus 68 lacking gp150 shows defective virion release but establishes normal latency in vivo. J Virol 78, 5103–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima, B. D., May, J. S., Marques, S., Simas, J. P. & Stevenson, P. G. (2005). Murine gammaherpesvirus 68 bcl-2 homologue contributes to latency establishment in vivo. J Gen Virol 86, 31–40. [DOI] [PubMed] [Google Scholar]
- Flano, E., Kim, I. J., Woodland, D. L. & Blackman, M. A. (2002). γ-herpesvirus latency is preferentially maintained in splenic germinal center and memory B cells. J Exp Med 196, 1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa, S., van Dyk, L. F., Jewett, T. J., Speck, S. H. & Virgin, H. W. (2002). Identification of the in vivo role of a viral bcl-2. J Exp Med 195, 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar, M., Gill, M. B., Loesing, J. B., May, J. S. & Stevenson, P. G. (2008). Multiple functions for ORF75c in murid herpesvirus-4 infection. PLoS One 3, e2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet, L., Adler, H. & Stevenson, P. G. (2007a). Glycosaminoglycan interactions in murine gammaherpesvirus-68 infection. PLoS One 2, e347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet, L., May, J. S. & Stevenson, P. G. (2007b). Post-exposure vaccination improves gammaherpesvirus neutralization. PLoS One 2, e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet, L., Colaco, S. & Stevenson, P. G. ( (2008). The murid herpesvirus-4 gH/gL binds to glycosaminoglycans. PLoS ONE 3, e1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, K., Hayashi, M., Jalkanen, M., Firestone, J. H., Trelstad, R. L. & Bernfield, M. (1987). Immunocytochemistry of cell surface heparan sulfate proteoglycan in mouse tissues. A light and electron microscopic study. J Histochem Cytochem 35, 1079–1088. [DOI] [PubMed] [Google Scholar]
- Hutchens, M. & Luker, G. D. (2007). Applications of bioluminescence imaging to the study of infectious diseases. Cell Microbiol 9, 2315–2322. [DOI] [PubMed] [Google Scholar]
- Kapadia, S. B., Molina, H., van Berkel, V., Speck, S. H. & Virgin, H. W. (1999). Murine gammaherpesvirus 68 encodes a functional regulator of complement activation. J Virol 73, 7658–7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozuch, O., Reichel, M., Lesso, J., Remenová, A., Labuda, M., Lysy, J. & Mistríková, J. (1993). Further isolation of murine herpesviruses from small mammals in southwestern Slovakia. Acta Virol 37, 101–105. [PubMed] [Google Scholar]
- Laichalk, L. L., Hochberg, D., Babcock, G. J., Freeman, R. B. & Thorley-Lawson, D. A. (2002). The dispersal of mucosal memory B cells: evidence from persistent EBV infection. Immunity 16, 745–754. [DOI] [PubMed] [Google Scholar]
- Liu, L., Flaño, E., Usherwood, E. J., Surman, S., Blackman, M. A. & Woodland, D. L. (1999). Lytic cycle T cell epitopes are expressed in two distinct phases during MHV-68 infection. J Immunol 163, 868–874. [PubMed] [Google Scholar]
- Lybarger, L., Wang, X., Harris, M. R., Virgin, H. W. & Hansen, T. H. (2003). Virus subversion of the MHC class I peptide-loading complex. Immunity 18, 121–130. [DOI] [PubMed] [Google Scholar]
- Marques, S., Efstathiou, S., Smith, K. G., Haury, M. & Simas, J. P. (2003). Selective gene expression of latent murine gammaherpesvirus 68 in B lymphocytes. J Virol 77, 7308–7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, J. S., Coleman, H. M., Smillie, B., Efstathiou, S. & Stevenson, P. G. (2004). Forced lytic replication impairs host colonization by a latency-deficient mutant of murine gammaherpesvirus-68. J Gen Virol 85, 137–146. [DOI] [PubMed] [Google Scholar]
- May, J. S., Coleman, H. M., Boname, J. M. & Stevenson, P. G. (2005a). The murine gammaherpesvirus-68 ORF28 encodes a non-essential virion glycoprotein. J Gen Virol 86, 919–928. [DOI] [PubMed] [Google Scholar]
- May, J. S., de Lima, B. D., Colaco, S. & Stevenson, P. G. (2005b). Intercellular gamma-herpesvirus dissemination involves co-ordinated intracellular membrane protein transport. Traffic 6, 780–793. [DOI] [PubMed] [Google Scholar]
- Mistrikova, J., Kozuch, O., Klempa, B., Kontsekova, E., Labuda, M. & Mrmusova, M. (2000). New findings on the ecology and epidemiology of murine herpes virus isolated in Slovakia. Bratisl Lek Listy 101, 157–162 (in Slovak). [PubMed] [Google Scholar]
- Nash, A. A. & Sunil-Chandra, N. P. (1994). Interactions of the murine gammaherpesvirus with the immune system. Curr Opin Immunol 6, 560–563. [DOI] [PubMed] [Google Scholar]
- Nash, A. A., Dutia, B. M., Stewart, J. P. & Davison, A. J. (2001). Natural history of murine γ-herpesvirus infection. Philos Trans R Soc Lond B Biol Sci 356, 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry, C. M., Simas, J. P., Smith, V. P., Stewart, C. A., Minson, A. C., Efstathiou, S. & Alcami, A. (2000). A broad spectrum secreted chemokine binding protein encoded by a herpesvirus. J Exp Med 191, 573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock, J. W. & Bost, K. L. (2000). Infection of intestinal epithelial cells and development of systemic disease following gastric instillation of murine gammaherpesvirus-68. J Gen Virol 81, 421–429. [DOI] [PubMed] [Google Scholar]
- Persons, D. A., Mehaffey, M. G., Kaleko, M., Nienhuis, A. W. & Vanin, E. F. (1998). An improved method for generating retroviral producer clones for vectors lacking a selectable marker gene. Blood Cells Mol Dis 24, 167–182. [DOI] [PubMed] [Google Scholar]
- Raslova, H., Berebbi, M., Rajcani, J., Sarasin, A., Matis, J. & Kudelova, M. (2001). Susceptibility of mouse mammary glands to murine gammaherpesvirus 72 (MHV-72) infection: evidence of MHV-72 transmission via breast milk. Microb Pathog 31, 47–58. [DOI] [PubMed] [Google Scholar]
- Rice, J., de Lima, B., Stevenson, F. K. & Stevenson, P. G. (2002). A γ-herpesvirus immune evasion gene allows tumor cells in vivo to escape attack by cytotoxic T cells specific for a tumor epitope. Eur J Immunol 32, 3481–3487. [DOI] [PubMed] [Google Scholar]
- Rosa, G. T., Gillet, L., Smith, C. M., de Lima, B. D. & Stevenson, P. G. (2007). IgG Fc receptors provide an alternative infection route for murine gamma-herpesvirus-68. PLoS One 2, e560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simas, J. P., Swann, D., Bowden, R. & Efstathiou, S. (1999). Analysis of murine gammaherpesvirus-68 transcription during lytic and latent infection. J Gen Virol 80, 75–82. [DOI] [PubMed] [Google Scholar]
- Smith, C. M., Rosa, G. T., May, J. S., Bennett, N. J., Mount, A. M., Belz, G. T. & Stevenson, P. G. (2006). CD4+ T cells specific for a model latency-associated antigen fail to control a gammaherpesvirus in vivo. Eur J Immunol 36, 3186–3197. [DOI] [PubMed] [Google Scholar]
- Smith, C. M., Gill, M. B., May, J. S. & Stevenson, P. G. (2007). Murine gammaherpesvirus-68 inhibits antigen presentation by dendritic cells. PLoS One 2, e1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, M. J., Hwang, S., Wong, W. H., Wu, T. T., Lee, S., Liao, H. I. & Sun, R. (2005). Identification of viral genes essential for replication of murine γ-herpesvirus 68 using signature-tagged mutagenesis. Proc Natl Acad Sci U S A 102, 3805–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck, S. H. & Virgin, H. W. (1999). Host and viral genetics of chronic infection: a mouse model of gamma-herpesvirus pathogenesis. Curr Opin Microbiol 2, 403–409. [DOI] [PubMed] [Google Scholar]
- Stevenson, P. G. (2004). Immune evasion by gamma-herpesviruses. Curr Opin Immunol 16, 456–462. [DOI] [PubMed] [Google Scholar]
- Stevenson, P. G. & Doherty, P. C. (1998). Kinetic analysis of the host immune response to a murine gammaherpesvirus. J Virol 72, 943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson, P. G. & Efstathiou, S. (2005). Immune mechanisms in murine gammaherpesvirus-68 infection. Viral Immunol 18, 445–456. [DOI] [PubMed] [Google Scholar]
- Stevenson, P. G., Belz, G. T., Altman, J. D. & Doherty, P. C. (1999a). Changing patterns of dominance in the CD8+ T cell response during acute and persistent murine γ-herpesvirus infection. Eur J Immunol 29, 1059–1067. [DOI] [PubMed] [Google Scholar]
- Stevenson, P. G., Cardin, R. D., Christensen, J. P. & Doherty, P. C. (1999b). Immunological control of a murine gammaherpesvirus independent of CD8+ T cells. J Gen Virol 80, 477–483. [DOI] [PubMed] [Google Scholar]
- Stevenson, P. G., Belz, G. T., Castrucci, M. R., Altman, J. D. & Doherty, P. C. (1999c). A γ-herpesvirus sneaks through a CD8+ T cell response primed to a lytic-phase epitope. Proc Natl Acad Sci U S A 96, 9281–9286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson, P. G., Boname, J. M., de Lima, B. & Efstathiou, S. (2002a). A battle for survival: immune control and immune evasion in murine γ-herpesvirus-68 infection. Microbes Infect 4, 1177–1182. [DOI] [PubMed] [Google Scholar]
- Stevenson, P. G., May, J. S., Smith, X. G., Marques, S., Adler, H., Koszinowski, U. H., Simas, J. P. & Efstathiou, S. (2002b). K3-mediated evasion of CD8+ T cells aids amplification of a latent γ-herpesvirus. Nat Immunol 3, 733–740. [DOI] [PubMed] [Google Scholar]
- Sunil-Chandra, N. P., Efstathiou, S., Arno, J. & Nash, A. A. (1992). Virological and pathological features of mice infected with murine gammaherpesvirus 68. J Gen Virol 73, 2347–2356. [DOI] [PubMed] [Google Scholar]
- Terry, L. A., Stewart, J. P., Nash, A. A. & Fazakerley, J. K. (2000). Murine gammaherpesvirus-68 infection of and persistence in the central nervous system. J Gen Virol 81, 2635–2643. [DOI] [PubMed] [Google Scholar]
- Thorley-Lawson, D. A. (2001). Epstein-Barr virus: exploiting the immune system. Nat Rev Immunol 1, 75–82. [DOI] [PubMed] [Google Scholar]
- Tibbetts, S. A., Loh, J., Van Berkel, V., McClellan, J. S., Jacoby, M. A., Kapadia, S. B., Speck, S. H. & Virgin, H. W. (2003). Establishment and maintenance of gammaherpesvirus latency are independent of infective dose and route of infection. J Virol 77, 7696–7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berkel, V., Preiter, K., Virgin, H. W. & Speck, S. H. (1999). Identification and initial characterization of the murine gammaherpesvirus 68 gene M3, encoding an abundantly secreted protein. J Virol 73, 4524–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berkel, V., Barrett, J., Tiffany, H. L., Fremont, D. H., Murphy, P. M., McFadden, G., Speck, S. H. & Virgin, H. W. (2000). Identification of a gammaherpesvirus selective chemokine binding protein that inhibits chemokine action. J Virol 74, 6741–6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weck, K. E., Barkon, M. L., Yoo, L. I., Speck, S. H. & Virgin, H. W. (1996). Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J Virol 70, 6775–6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, Q. Y., Rickinson, A. B. & Epstein, M. A. (1985). A re-examination of the Epstein-Barr virus carrier state in healthy seropositive individuals. Int J Cancer 35, 35–42. [DOI] [PubMed] [Google Scholar]
- Yin, Y., Manoury, B. & Fahraeus, R. (2003). Self-inhibition of synthesis and antigen presentation by Epstein-Barr virus-encoded EBNA1. Science 301, 1371–1374. [DOI] [PubMed] [Google Scholar]