Abstract
Expression of p185Bcr-Abl in Ba/F3 cells inhibits chemotactic response of these cells to SDF1α. A mutant p185Bcr-Abl with deletion of amino acids from 176 to 426 (p185Δ176–426) is deficient in suppressing SDF1α-stimulated chemotaxis. Comparison of the gene expression profiles among parental Ba/F3 cells and the cells transformed by p185Bcr-Abl and p185Δ176–426 reveals that class II phosphoinositide 3-kinase γ (PI3KC2γ) expression is markedly down-regulated by p185Bcr-Abl but not p185Δ176–426. Furthermore, knockdown of PI3KC2γ expression in p185Δ176–426 cells is sufficient to suppress SDF1α-stimulated chemotaxis and to promote infiltration of these cells into liver. Together, these studies suggest that inhibition of PI3KC2γ expression may represent a mechanism by which Bcr-Abl suppresses SDF1α-induced chemotaxis and induces abnormal homing of leukemic cells.
Keywords: PI3KC2γ, p185Bcr-Abl, SDF-1α, chemotaxis
INTRODUCTION
Bcr-Abl oncoproteins, generated by a chromosomal translocation event which leads to the fusion of c-Abl gene with Bcr gene, are causative agents for most human chronic myelogenous leukemia and a subset of acute lymphocytic leukemia [1, 2]. The Bcr-Abl-positive leukemia is characterized by the pre-mature release of lymphoid and myeloid progenitor cells into peripheral blood and massive expansion of these cells in peripheral hematopoietic tissues such as blood and spleen, as well as infiltration of these cells into the liver and lung [1]. Hematopoietic cells isolated from patients with Bcr-Abl-positive leukemia exhibit multiple abnormalities of cytoskeletal function [3]. Among these is a reduced response to stromal cell derived factor 1α (SDF1α), a chemokine important for hematopoietic cell homing [4–6]. It has been suggested that a reduced response of Bcr-Abl-positive hematopoietic cells to SDF1α may contribute to leukemogenesis, as it may promote pre-mature release of lymphoid and myeloid progenitor cells from bone marrow, where the SDF1α concentration is high, to peripheral hematopoietic tissues such as blood, spleen, and thymus [4–9]. This hypothesis, however, needs to be experimentally tested in vivo.
How Bcr-Abl suppresses chemotaxis of hematopoietic progenitor cells to SDF1α remains largely unknown. The Bcr-Abl transformation of hematopoietic cells results in the activation of multiple signaling pathways including the one mediated by phosphoinositide 3-kinase (PI3K) [1]. The PI3K proteins are involved in regulation of many fundamental cellular processes such as proliferation, migration, and survival [10,11]. Deregulation of PI3K-mediated pathways is implicated in the development of many diseases including leukemia [12–14]. The PI3K family proteins are divided into three classes (I–III) based on their protein domain structure and substrate specificity [15]. Among the three classes of PI3K proteins, class I is the best characterized, whereas class II has received increasing attention only recently. While numerous studies have shown that class I PI3K is a downstream target of Bcr-Abl oncoproteins, it is not clear whether the class II PI3K is also involved in Bcr-Abl-induced leukemogenesis. There are three members of class II PI3K, PI3KC2α, PI3KC2β, and PI3KC2γ [15]. Although several lines of evidence suggest that PI3KC2α and PI3KC2β are linked to diverse receptor-mediated signaling processes that control cell migration, trafficking, and survival [16–19], little is known about the activation, regulation, and function of PI3KC2γ which was identified by PCR cloning strategy and data mining of genome sequencing projects [20,21]. In this report we show that the expression of PI3KC2γ is repressed in p185Bcr-Abl-transformed Ba/F3 cells (p185wt cells). The markedly reduced expression of PI3KC2γ in p185wt cells correlates with a decrease of SDF1α-stimulated chemotaxis in these cells. By mutagenesis analysis and short hairpin RNA (shRNA)-mediated gene silencing studies, we present data here to support a role of PI3KC2γ in regulation of SDF1α-stimulated chemotaxis as well as in Bcr-Abl-induced leukemogenesis. Identification of PI3KC2γ as a novel target of Bcr-Abl may help to shed new light on therapy of Bcr-Abl-positive leukemia.
MATERIALS AND METHODS
Cell Lines And Reagents
Ba/F3 cells were grown in RPMI-1640 (Mediatech, Manassas, VA, USA) containing 10% fetal bovine serum (FBS) (Sigma, St. Louis, MO, USA) and 15% conditioned medium collected from WEHI3 cell cultures as a source of IL3. The Ba/F3 cell lines expressing the wild type and mutant forms of p185Bcr-Abl were cultured in RPMI-1640 containing 10%FBS. Retroviral packaging cell line GP2-293 (ClonTech, Mountain View, CA, USA) was grown in DMEM (Mediatech, Manassas, VA, USA) containing 10% FBS. The antibodies against tyrosine-phosphorylated proteins were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA) and the monoclonal antibodies for Abl were obtained from BD Pharmingen (San Diego, CA, USA). The protease inhibitor cocktail was purchased from Sigma (Catalog number P8340, St. Louis, MO, USA). The recombinant human SDF1α was purchased from R&D Systems. (Minneapolis, MN, USA).
Plasmids And Retroviral Infection
Transfection of Ba/F3 cells with pSRαp185wt and pSRαp185Δ176–426 was performed as previously described [22]. Construction of MSCV-based retroviral vectors expressing p185wt, p185ΔSH3, p185ΔC, and p185ΔSH3ΔC has been described previously [23]. To generate the retroviral vector expressing p185Δ176–426, the full length cDNA encoding p185Δ176–426 was released from pSRαp185Δ176–426 by restriction digestion with EcoR1. The purified cDNA was subsequently ligated to MSCV at EcoR1 sites. Two MSCV-based pSM2 retroviral vectors containing cDNAs encoding short hairpin RNAs (shRNAs) that specifically target PI3KC2γ transcript and a control vector encoding non-silencing shRNA were purchased from Open Biosystems (Huntsville, AL, USA). The sequences in PI3KC2γ that are targeted by two shRNAs are as the follows: shRNA P1: TGT GGA AAG GAC GAA ACA CC; shRNA P2: TGT GGA AAG GAC GAA ACA CC. The expression of shRNAs was driven by U6 promoter. Amplification and purification of plasmid DNAs were performed as specified by manufacturer’s instruction. Retrovirus production and infection of Ba/F3 cells and the Ba/F3 cells transformed by wild type and mutant forms of p185Bcr-Abl were performed as described previously [23]. The mass populations of cells that were stably transformed were selected by puromycin (2µg/ml) and used for subsequent experiments.
DNA Microarray Analysis
Total RNAs were isolated from Ba/F3 cells transduced with control retrovirus or the retroviruses expressing either p185wt or p185Δ176–426 using RNeasy mini kit (QIAGEN, Valencia, CA, USA). The concentration of total RNA was determined by NanoDrop ND-3300 Fluorospectrometer and the integrity of RNA was examined using an Experion analyzer (Bio-Rad, Hercules, CA, USA) with Experion RNA HighSens Analysis Kit. Five-microgram of total RNA isolated from each cell line was individually converted to double-stranded cDNA (ds-cDNA) using the Superscript Choice system (Life Technologies, Rockville, MD, USA). In vitro transcription and biotin-labeling were then performed with an RNA transcript labeling kit (Enzo Biochem, Inc., New York, NY, USA). Synthesized cRNA was used to probe Affymetrix Mouse Genome 430 2.0 Array that represented over 39,000 transcripts and variants from over 34,000 well-characterized mouse genes (Affymetrix Inc., Santa Clara, CA, USA). After washing and staining with streptavidin-phycoerythrin, the probed arrays were scanned using a Gene-array scanner (Hewlett Packard, Palo Alto, CA, USA). The data were analyzed using MicroArray Suite Version 4.0 software (Affymetrix Inc.). Detailed protocols for data analysis of Affymetrix microarrays and extensive documentation of the sensitivity and quantitative aspects of the method have been described (Affymetrix Microarray Suite, version 4.0, User’s guide).
Biochemical Assays
Quantitative reverse transcription-PCR was performed using mouse PI3KC2γ primers (forward: 5’- ACTGCACCTTAGGGCTCTTGTC-3’; reverse: 5’- AGTCACTACGTGGAACAGGTGCTA -3’) and SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA). Briefly, total RNAs were isolated from cells using RNeasy mini kit (QIAGEN, Valencia, CA, USA) and the cDNAs were subsequently generated using SuperScript III First-strand Synthesis System (Invitrogen, Carlsbad, CA, USA). The PCR reactions began with 10 min at 95°C for AmpliTaq Gold activation, followed by 40 cycles at 95°C for 15 s for denature and then 60°C for 1 min for annealing/extension. The RT-PCR was performed on MyiQ single color real-time PCR detection system (Bio-Rad, Hercules, CA, USA). Relative quantification was done using Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an endogenous housekeeping transcript control (forward primer: 5’-AACGACCCCTTCATTGAC-3’, Reverse primer: 5’-TCCACGACATACTCAG CAC-3’).
Immunoprecipitation and Western blot analyses were performed as previously described [24]. Cells were lysed in lysis buffer (20 mM Hepes, pH7.2; 150 mM NaCl, 1% Triton X-100, and 10% glycerol) and incubated with indicated antibodies bound to Sepharose beads. The immunoprecipitates were separated on SDS-PAGE, transferred to nitrocellulose, and immunoblotted with appropriate antibodies.
Migration Assays
The transwell cell migration assay was performed as described previously [23]. The inserts of transwell plates (8-µm pores, Corning Costar Corp., Cambridge, MA) were coated with human fibronectin (Sigma, St. Louis, MO, USA). The control Ba/F3 cells and the Ba/F3 cells expressing the wild type or mutant forms of p185Bcr-Abl, as well as their derivative lines expressing PI3KC2γ shRNA or scrambled control shRNA, were added into inserts (1×105 cells/insert in 0.1 ml RPMI containing 0.1% bovine serum albumin). The cells were incubated in 5% CO2 incubator at 37° C for 6–8 hours to allow migration to the bottom chamber containing 0.6 ml RPMI/0.1% BSA medium with or without SDF1α (100 ng/ml). At the end of incubation the cells migrated to the bottom chamber were counted and the percentage of migrated cells against total cells was calculated. The chemotactic index was determined by dividing the number of cells that migrated in response to SDF-1α with the number of the cells that migrated spontaneously.
In Vivo Leukemogenesis Studies
1×106 Ba/F3 cells and Ba/F3 cells expressing p185wt, p185Δ176–426, or p185Δ176–426 plus control or PI3KC2γ shRNA were injected into 6–8 week old female BALB/c mice through tail vein as previously described (n=5 for each group) [24]. The mice were followed for disease development, as judged by symptoms such as abnormal gait and labored breathing. Moribund animals were sacrificed by CO2 asphyxiation and were examined for tumors or other visible abnormalities. Collection and weighing of livers, spleens and other tissues was performed immediately after sacrifice. Some tissues were fixed in 10% buffered formalin solution and analyzed by pathological/histological examination. All protocols used were approved by Institutional Animal Review Committee at the Texas Tech University Health Sciences Center.
Statistical Analysis
Descriptive statistics were generated for all quantitative data with presentation of average ± SDs. Significance of comparisons between experimental groups was tested using the Student's t test. P<0.05 was considered as statistically significant.
RESULTS
The SDF1α-Stimulated Chemotaxis Requires PI3 Kinases And Is Inhibited By p185Bcr-Abl
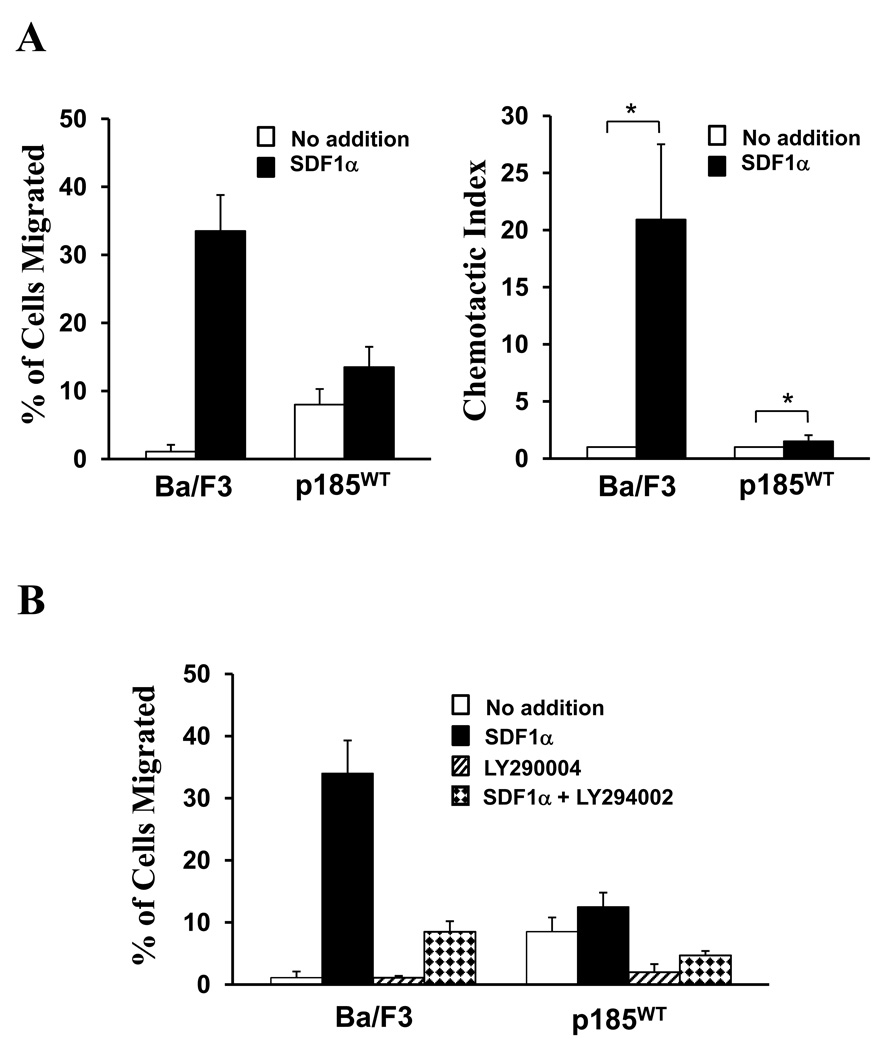
Ba/F3 is a murine pro-B cell line whose growth is IL-3-dependent. Previous studies have shown that Ba/F3 cells express CXCR4, the membrane receptor for SDF1α [4] In a transwell migration assay, these cells exhibited low spontaneous migration to the bottom chamber containing medium alone. Addition of SDF1α into the bottom chamber stimulated Ba/F3 cell migration by 20.9 +/− 6.6 fold (*P < 0.05), indicating that these cells have strong chemotactic response to SDF1α (Figure 1A). Consistent with previous report [23], expression of p185Bcr-Abl in Ba/F3 cells significantly increased spontaneous cell migration (Figure 1A). However, the chemotactic response to SDF1α was dramatically reduced by p185Bcr-Abl transformation. As shown in Figure 1A, addition of SDF1α resulted in only a slight increase (1.5 fold, *P < 0.05) of cell migration in BaF3 cells transformed by p185Bcr-Abl (p185wt cells). This is in contrast to control Ba/F3 cells, in which SDF1α induced an increase of migration by 21 fold.
Figure 1. The chemotactic response of Ba/F3 cells to SDF1α is suppressed by p185Bcr-Abl transformation and pharmacological inhibition of PI3 kinases.
A. Transformation of Ba/F3 cell by p185Bcr-Abl impaired SDF1α-stimulated chemotaxis of Ba/F3 cell. The left panel represents one of nine transwell cell migration experiments. The vertical axis shows the percentage of migrated cell and is expressed as the average +/− S.D. of triplicate wells. The chemotactic index shown in the right panel is expressed in vertical axis as the average +/− S.D (*P < 0.05) calculated from 9 independent experiments. B. Inhibition of PI3K pathway impaired the SDF1α-stimulated chemotaxis of both Ba/F3 cell and p185wt cell. The Ba/F3 cells and p185wt cells were pre-incubated with or without 50µM LY294002, as indicated, for 5 hours. 1×105 cells were tested in trans-well migration assay at the presence or absence of SDF1α (100 ng/ml) in bottom chamber. The data represents one of three independent migration experiments. The percentage of migrated cells is shown in vertical axis as the average +/− S.D. calculated from triplicate wells.
It has been shown that the PI3K pathway is a downstream target of Bcr-Abl and plays a key role in regulation of cell migration and chemotaxis [2,5]. We therefore examined the effect of PI3K inhibitor, LY294002, on spontaneous migration as well as SDF1α-stimulated chemotaxis in Ba/F3 cells and p185wt cells. Consistent with a role of PI3K in Bcr-Abl-induced spontaneous cell migration, treatment of p185wt cells with LY294002 alone inhibited spontaneous cell migration of these cells (Figure 1B). Furthermore, addition of LY294002 dramatically reduced the SDF1α-stimulated chemotaxis in both Ba/F3 cells and p185wt cells (Figure 1B), suggesting that the activation of PI3K pathway is required for SDF1α-stimulated chemotaxis of these cells.
Deletion Of Bcr Sequences From Amino Acids 176 To 426 Abrogated The Ability Of The Mutant p185Bcr-Abl To Inhibit SDF1α-Stimulated Chemotaxis
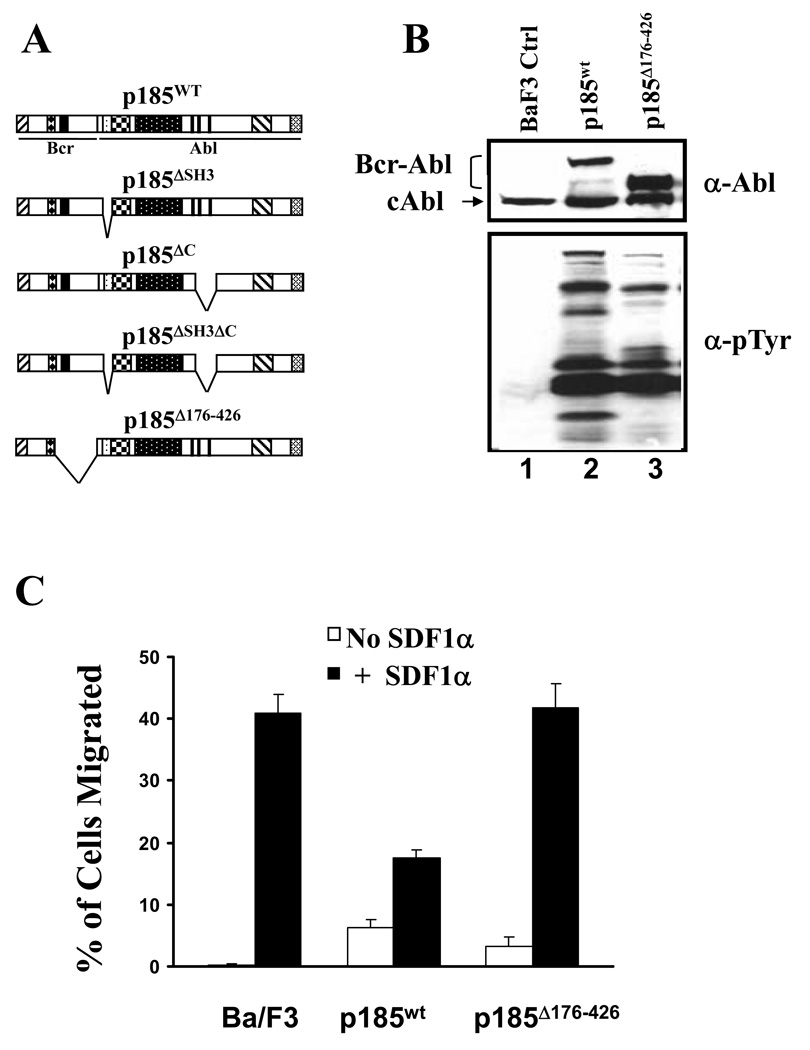
To determine the sequences in p185Bcr-Abl that contribute to the inhibition of SDF1α-stimulated chemotaxis, we examined several mutant forms of p185Bcr-Abl for their abilities to inhibit SDF1α-stimulated chemotaxis. These include the mutant forms of p185 with deletion of SH3 domain (p185ΔSH3), C-terminal proline-rich sequences (p185ΔC), both SH3 and C-terminal proline-rich sequences (p185ΔSH3ΔC), as well as Bcr sequences from amino acids 176 to 426 (p185Δ176–426, Figure 2A). Previous studies have shown that these sequences are critical for Bcr-Abl to activate signaling pathways important for cell adhesion and migration [22,23,25]. These mutant forms of p185Bcr-Abl were introduced into Ba/F3 cells by retrovirus-mediated gene transfer. The deletions in these mutant forms of Bcr-Abl did not affect their tyrosine kinase activity, as the wild type and mutant forms of p185Bcr-Abl were all able to stimulate protein tyrosine phosphorylation in BaF3 cells [23] (Figure 2B, compare lanes 2 and 3 with lane 1). We noticed that the profile of protein tyrosine phosphorylation stimulated by mutant forms of Bcr-Abl differs from that stimulated by wild type p185Bcr-Abl (Figure 2B, compare lane 3 with lane 2). It is possible that deletions in mutant forms of Bcr-Abl may prevent them from interacting with some downstream substrates [23] and therefore alter the protein tyrosine phosphorylation profile. Like wild type p185Bcr-Abl, all mutant forms of Bcr-Abl were capable of inducing IL3-independent growth when expressed in Ba/F3 cells. To determine if the deletions affect the abilities of these mutant forms of Bcr-Abl to inhibit SDF1α-stimulated chemotaxis, the cells expressing mutant Bcr-Abl proteins were tested for their responsiveness to SDF1α in a transwell migration assay. Deletion of the SH3 domain, C-terminal proline-rich sequences, or both SH3 and C-terminal proline-rich sequences did not affect the ability of the mutant Bcr-Abl to inhibit chemotaxis (data not shown). However, deletion of Bcr sequences from amino acids 176 to 426 abolished the Bcr-Abl-induced inhibition of chemotaxis (Figure 2C). Thus, our data indicate that Bcr sequences from amino acids 176 to 426 are essential for Bcr-Abl to exert its inhibitory effect on SDF1α-stimulated chemotaxis.
Figure 2. A mutant Bcr-Abl with deletion in Bcr sequences failed to inhibit SDF1α-stimulated chemotaxis.
A. Schematic representation of the wild type (p185wt) and mutant forms of p185Bcr-Abl. B. Profiles of protein tyrosine phosphorylation in Ba/F3 cells transduced with control retrovirus (BaF3 Ctrl) or retroviruses expressing either p185wt or p185Δ176–426, as indicated. The cells were starved in RPMI-1640 medium containing 0.1% bovine serum albumin for 6 hours. Total lysates from 1×106 cells were subjected to Western blot analysis with anti-Abl antibody (α-Abl) and anti-phosphotyrosine (α-Tyr) antibodies. The positions of the wild type and mutant form of p185Bcr-Abl as well as endogenous c-Abl are indicated. C. The p185Δ176–426 failed to inhibit SDF1α-stimulated chemotaxis in Ba/F3 cells. The data represents one of three independent cell migration experiments. The vertical axis shows the percentage of migrated cells and is expressed as the average +/− S.D. calculated from triplicate wells.
Expression Of P185wt, But Not P185Δ176–426, In Ba/F3 Cells Down-Regulates PI3 Kinase C2γ Expression
To determine the mechanism by which p185wt inhibits chemotactic response of Ba/F3 cells to SDF1α, we compared the gene expression profile of p185wt cells to that of control Ba/F3 cells as well as p185Δ176–426 cells by genome-wide microarray analysis. Among a list of genes that are either up-regulated or down-regulated by p185wt transformation, PIK3C2G, which encodes for PI3KC2γ, came to our attention because: 1) the expression of this gene was down-regulated dramatically (approximately 20-fold) in p185wt cells as compared to Ba/F3 control cells; 2) the down-regulation of this gene was not observed in cells transformed by p185Δ176–426, the mutant Bcr-Abl that is defective in inhibiting chemotaxis; and 3) our studies have shown that PI3K activity is required for SDF1α-stimulated chemotaxis (Figure 1B).
The down-regulation of PI3KC2γ expression by p185wt was confirmed by RT-PCR (Figure 3A) as well as quantitative real time PCR (Figure 3B). To determine if the down-regulation of PI3KC2γ expression requires the tyrosine kinase activity of p185wt, we treated p185wt cells with imatinib mesylate, a selective inhibitor of Bcr-Abl tyrosine kinase. As shown in Figure 3C, the treatment of p185wt cells with 5 µM imatinib mesylate completely inhibited Bcr-Abl-induced protein tyrosine phosphorylation. The treatment of p185wt cells with 5 µM imatinib mesylate also abrogated Bcr-Abl-induced down-regulation of PI3KC2γ (Figure 3D), suggesting that the tyrosine kinase activity is required for Bcr-Abl to down-regulate PI3KC2γ. Furthermore, the quantitative real time PCR analysis revealed that the p185Δ176–426 is deficient in suppressing PI3KC2γ expression (Figure 3B). Therefore, the results suggest that Bcr sequences from amino acids 176 to 426 are essential for p185wt to induce downregulation of PI3KC2γ.
Figure 3. The p185wt, but not p185Δ176–426, down-regulates the expression of PI3KC2γ.
A. Downregulation of PI3KC2γ expression in p185wt cells. Total RNAs isolated from the Ba/F3 cells transduced with the control retrovirus (Ba/F3) or retrovirus expressing p185Bcr-Abl (p185wt) were subjected to RT-PCR analysis using specific primers for PI3KC2γ and GAPDH, respectively. The DNA fragments amplified from PI3KC2γ and GAPDH are indicated. B. Quantitative real time PCR (qRT-PCR) analysis of PI3KC2γ expression in Ba/F3 cells transduced with control retrovirus (Ba/F3) or retroviruses expressing p185wt or p185Δ176–426, as indicated. The PI3KC2γ mRNA levels, expressed as the fold relative to that of GAPDH, are shown in vertical axis as the average +/− S.D calculated from triplicates of a representative experiment. Three independent experiments were performed. *P < 0.05. C. Inhibition of tyrosine kinase activity of p185wt by Imatinib Mesylate (IM). The p185wt cells were treated with Imatinib Mesylate at the indicated concentration and cell lysates were analyzed by western blot using anti-phosphotyrosine antibodies (α-pTyr), as indicated. D. The tyrosine kinase activity is required for p185wt to repress the expression of PI3KC2γ. The p185wt cells were treated with or without 5µM Imatinib Mesylate (IM), as indicated, for 6 hours. Total RNAs were isolated and subjected to qRT-PCR analysis as described in Materials and Methods. The PI3KC2γ mRNA levels relative to that of GAPDH were determined and the folds increased in IM-treated cells over untreated cells are shown in vertical axis as the average +/− S.D. calculated from triplicates of a representative experiment. Three independent experiments were performed. *P < 0.05.
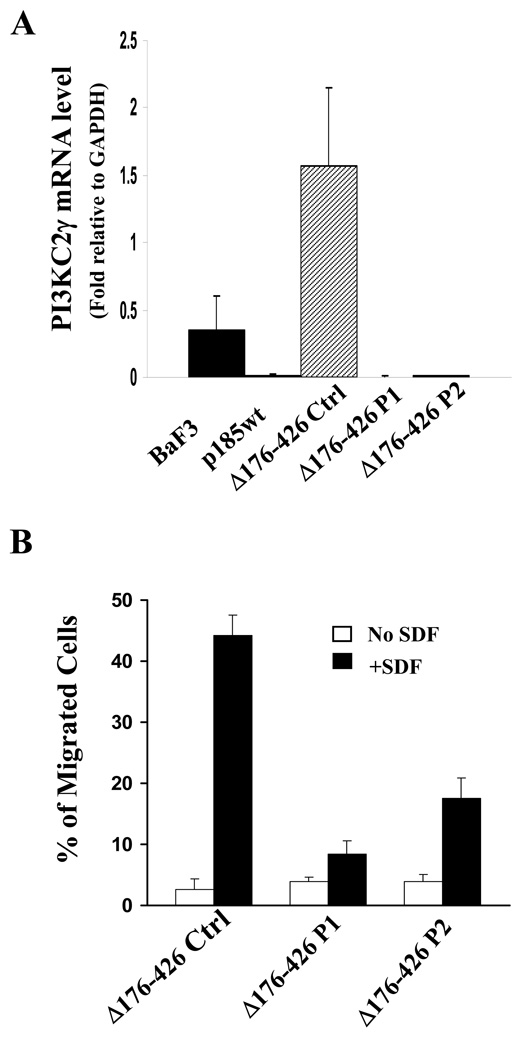
Knockdown Of PI3KC2γ Expression In P185Δ176–427 Cells Inhibited The Chemotaxis Of These Cells To SDF1α
It is notable that the levels of PI3kC2γ expression in Ba/F3, p185wt, and p185Δ176–426 cells correlate well with the chemotactic responsiveness of these cells to SDF1α. This raised the question as to whether the defect of p185Δ176–426 in suppressing SDF1α-stimulated chemotaxis is due to its inability to down-regulate PI3KC2γ expression. If this is the case we reason that forced down-regulation of PI3KC2γ in p185Δ176–426 cells may result in the inhibition of the chemotaxis. To test this, the retroviral constructs expressing a scrambled control shRNA (control) or the shRNAs that specifically target different region of mouse PI3KC2γ transcript (PI3KC2γ P1 and P2) were introduced into p185Δ176–426 cells. As shown in Figure 4A, the expression of PI3KC2γ in p185Δ176–426 cells transduced with retroviruses expressing PI3KC2γ shRNAs was drastically decreased as compared to the p185Δ176–426 cells transduced with the retrovirus expressing control shRNA. More importantly, transduction of the p185Δ176–426 cells with retroviruses expressing PI3KC2γ shRNAs resulted in an inhibition of chemotactic response to SDF1α (Figure 4B). Together, these results are consistent with the hypotheses that the PI3KC2γ may play a role in SDF1α-stimulated chemotaxis and that down-regulation of PI3KC2γ in p185wt cells may represent a mechanism by which p185wt inhibits SDF1α-stimulated chemotaxis.
Figure 4. Knockdown of the PI3KC2γ expression in p185Δ176–426 cells inhibited SDF1α-stimulated chemotaxis.
A. Knockdown of PI3KC2γ expression in p185Δ176–426 cells. Total RNAs isolated from Ba/F3, p185wt, and p185Δ176–426 cells transduced with control retrovirus (p185Δ176–426 Ctrl) or retroviruses expressing two shRNAs that specifically target different region of PI3KC2γ mRNA (p185Δ176–426 P1 and p185Δ176–426 P2) were subjected to qRT-PCR analysis. The vertical axis shows the PI3KC2γ mRNA levels relative to that of GAPDH. The data represents the average +/− S.D. calculated from triplicates of a representative experiment. Two independent experiments were performed. B. Knockdown of PI3KC2γ expression impaired the SDF1α-stimulated chemotaxis of p185Δ176–426 cells. 1×105 p185Δ176–426 cells transduced with indicated retroviruses were tested in trans-well migration assay for their abilities to migrate spontaneously (no SDF) or in response to SDF1α (+SDF, 100ng/ml). The vertical axis shows the percentage of migrated cells. The data represents the average +/− S.D. calculated from triplicate wells of a representative experiment. Two independent experiments were performed.
PI3KC2γ Gene Silencing Alters The Phenotype Of Leukemic Disease Induced By P185Δ176–426
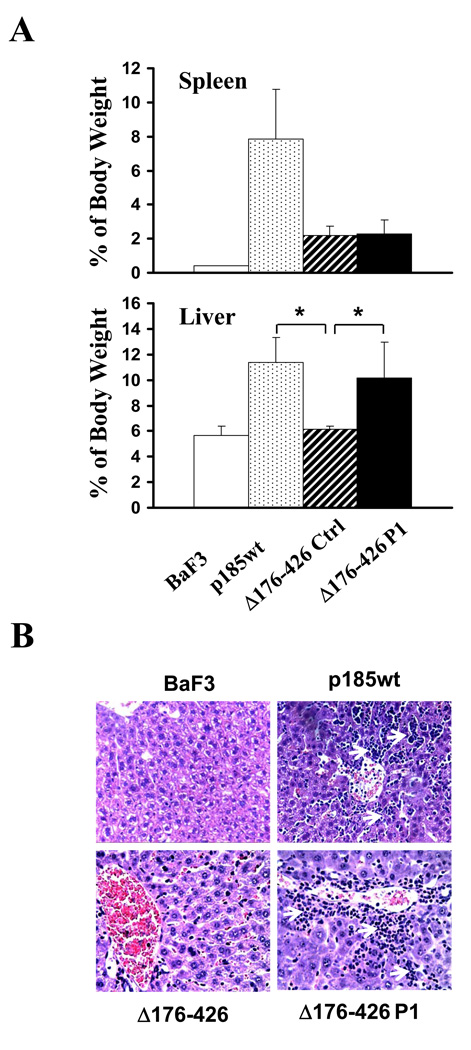
Previous studies have shown that deletion of Bcr sequences from amino acids 176 to 426 severely impaired transforming activity of the p185Δ176–426 [26]. The finding that the p185Δ176–426 is also defective in inhibiting SDF1α-stimulated chemotaxis prompted us to test if this defect of the p185Δ176–426 contributes to the inhibition of leukemogenic activity. We made use of the p185Δ176–426 cells transduced with the retrovirus expressing PI3KC2γ shRNA (p185Δ176–426 P1 cells). We have shown that PI3KC2γ expression in these cells was down-regulated to a level comparable to that of p185wt cells (Figure 4A). Like p185wt cells, the p185Δ176–426 P1 cells also displayed a reduced chemotactic response to SDF-1α (Figure 4B). The p185Δ176–426 P1 cells as well as Ba/F3, p185wt, and the p185Δ176–426 cells transduced with scrambled shRNA (p185Δ176–426 control cells) were injected intravenously into BALB/c mice, from which the parental Ba/F3 cells were derived. The mice injected with p185wt cells (p185wt mice) all developed leukemia, as evidenced by massive expansion of leukemic cells in spleens and livers that lead to destruction of normal cytoarchitecture of these tissues (Figure 5A and 5B). These mice died with an average survival of 28 days, while the mice injected with Ba/F3 cells stayed healthy for more than 90 days. The mice injected with p185Δ176–426 control cells (Δ176–426 mice) also developed leukemic disease. However, consistent with previous reports, these mice survived longer (average 60 days) compared to the mice injected with p185wt cells. The spleens of these mice were enlarged only moderately compared to p185wt mice and the animals showed no sign of hepatomegaly (Figure 5A). Despite the moderate leukemic cell accumulation and infiltration, no obvious destruction of cytoarchitecture was observed in spleens from p185Δ176–426 mice. Interestingly, the mice injected with p185Δ176–426 P1 cells (Δ176–426 P1 mice) developed a leukemic disease that is different from that observed in either p185wt mice or p185Δ176–426 mice. Similar to p185Δ176–426 mice, these mice developed only moderate splenomegaly (Figure 5A) and survived longer (average 66.75 days) compared to p185wt mice. Histopathology analysis showed no apparent difference between the spleen from p185Δ176–426 mouse and that from p185Δ176–426 P1 mouse. However, in contrast to p185Δ176–426 mice which had no sign of hepatomegaly, the p185Δ176–426 P1 mice displayed hepatomegaly that is similar to that observed in p185wt mice (Figure 5A). The destruction of normal cytoarchitecture due to massive invasion of leukemic cells in livers was observed in both p185wt mice and p185Δ176–426 P1 mice, whereas no apparent histological abnormality was observed in the livers of p185Δ176–427 mice (Figure 5B). Taken together, these results suggest that knock-down of PI3KC2γ expression in p185Δ176–427 cells altered the phenotype of the disease induced by p185Δ176–427, possibly through the alteration of the leukemic cell homing.
Figure 5. Pathology analysis of the mice injected with Ba/F3 cells, p185wt cells, and p185Δ176–426 cells transduced with control retrovirus or the retrovirus expressing PI3KC2γ shRNA.
A. Spleen (upper panel) and liver (lower panel) weights of the mice injected with Ba/F3 cells, p185wt cells, and p185Δ176–426 cells transduced with control shRNA (Δ176–426 Ctrl) or PI3KC2γ shRNA (Δ176–426 P1). The spleen and liver weights are expressed as the percentage of total body weights (average +/− S.D., n=5). *P<0.05. B. Histology of livers of mice receiving the indicated cells. The liver tissue sections were stained by Haematoxylin and Eosin. Arrows indicate the infiltrated leukemic cells.
DISCUSSION
In this report, the inhibition of SDF1α-stimulated chemotaxis by oncogenic Bcr-Abl kinase was studied in Ba/F3 cells by a combination of mutagenesis analysis, gene expression profile analysis, and functional analysis. The mutagenesis analysis showed that Bcr sequences from amino acids 176 to 426 are essential for p185Bcr-Abl to inhibit chemotactic response of Ba/F3 cells to SDF1α. Comparison of gene expression profiles among parental Ba/F3 cells and the Ba/F3 cells transformed by p185wt and p185Δ176–426, a mutant form of p185Bcr-Abl defective in suppressing chemotaxis, revealed a 20-fold decrease of the expression of PI3KC2γ in p185wt-transformed Ba/F3 cells compared to parental Ba/F3 cells and p185Δ176–426 cells. This was confirmed by RT-PCR as well as quantitative real time PCR analyses. Because PI3KC2γ expression is repressed only in p185wt cells in which SDF1α-stimulated chemotaxis is inhibited, we investigated the role of this signaling molecule in SDF1α-stimulated chemotaxis. We found that knockdown of PI3KC2γ expression in p185Δ176–426 cells resulted in a significant reduction in chemotactic response of these cells to SDF1α. A similar result was also obtained with Ba/F3 cells (data not shown). To examine the role of PI3KC2γ in Bcr-Abl-induced leukemogenesis, we injected mice with p185Δ176–426 cells expressing PI3KC2γ shRNA and compared the leukemogenesis in these mice to those injected with p185wt cells or p185Δ176–426 control cells. Our results show that in contrast to p185Δ176–426 cells, which could not induce hepatomegaly when injected into mice, the p185Δ176–426 cells expressing PI3KC2γ shRNA induced hepatomegaly as what p185wt cells did. Taken together, these studies identify PI3KC2γ as a novel downstream target of p185Bcr-Abl and suggest that repression of PI3KC2γ expression by p185Bcr-Abl may contribute to impaired chemotaxis and aberrant homing of Bcr-Abl-positive leukemic cells.
SDF1α is an important chemokine that regulates the homing, growth, and differentiation of hematopoietic stem/progenitor cells in bone marrow microenvironment [7,9,27]. Given the importance of SDF1α signaling in regulating hematopoietic progenitor cell homing, it is conceivable that Bcr-Abl-induced suppression of chemotactic response to SDF1α may contribute to premature release of Bcr-Abl-positive progenitor cells from bone marrow and facilitate the homing of these cells to peripheral hematopoietic tissues. In line with this hypothesis, we found that the Ba/F3 cells transformed by p185Δ176–426, the mutant that is deficient in suppressing chemotaxis, failed to induce hepatomegaly in mouse model. However, when the expression of PI3KC2γ was knocked down in these cells the chemotactic response to SDF-1α was reduced to the level comparable to that in p185wt-transformed cells. In correlation with the decrease in chemotactic responsiveness to SDF-1α, these cells re-gain the ability to induce hepatomegaly when injected into mouse. Thus, our studies provide the first evidence that the experimental manipulation of the chemotactic responsiveness to SDF1α in hematopoietic cells transformed by a mutant Bcr-Abl may alter homing of these leukemic cells in vivo.
Ptasznik et al have shown that Src family kinase Lyn is activated by SDF1α and is required for SDF1α-stimulated chemotaxis in hematopoietic cells [5]. They found that class I PI3K is activated by Lyn upon SDF1α stimulation and the expression of Bcr-Abl in hematopoietic cells constitutively activates Lyn/class I PI3 kinase pathway [5]. Consistent with a role of PI3 kinases in regulation of SDF-stimulated chemotaxis, we also found that pharmacological inhibitor of PI3 kinases blocked SDF1α-stimulated chemotaxis. Based on our studies and those by Ptasznik et al, at least two hypotheses may be proposed to explain how Bcr-Abl suppresses SDF1α-stimulated chemotaxis. One hypothesis, as proposed by Ptasznik et al, is that the constitutive activation of class I PI3K by Bcr-Abl prevents these enzymes from being further activated by SDF1α. As a consequence, this leads to reduced chemotactic response to SDF1α [5]. Another hypothesis, as suggested by our studies, is that PI3KC2γ plays a role in SDF1α-stimulated chemotaxis and that Bcr-Abl inhibits the SDF1α-stimulated chemotaxis by, at least in part, repressing the expression of PI3KC2γ. In support of the latter hypothesis, we found that a mutant Bcr-Abl (p185Δ176–426) defective in repressing PI3KC2γ expression is also deficient in inhibiting chemotaxis. Furthermore, we show that knockdown of the expression of PI3KC2γ in the Ba/F3 cells transformed by p185Δ176–426 is sufficient to inhibit SDF1α-stimulated chemotaxis. It should be pointed out that these two hypotheses do not have to be mutually exclusive. It is possible, for example, that inhibition of chemotaxis by Bcr-Abl is the result of an additive effect of the constitutive activation of class 1 PI3K, which prevents it from being further activated by SDF, and the repression of PI3KC2γ expression. In this regard, it is notable that expression of Bcr-Abl in Ba/F3 cells stimulated spontaneous cell migration which could be attenuated by pharmacological inhibition of PI3K (Figure 1A). The increase of spontaneous cell migration induced by Bcr-Abl in p185wt cells is much less than that stimulated by SDF1α in Ba/F3 cells (Figure 1B). Furthermore, addition of SDF1α failed to further stimulate p185wt cell migration to a level comparable to that stimulated by SDF1α in Ba/F3 cells. One possible explanation for these observations is that, in normal Ba/F3 cells SDF1α stimulates not only class I PI3K but also PI3KC2γ and thereby induces maximal increase of cell migration. In p185wt cells, on the other hand, while Bcr-Abl constitutively activates class I PI3K and thereby stimulates basal cell migration, it also suppresses the expression of PI3KC2γ, which is required for maximal stimulation of cell migration. Thus, the insensitiveness of p185wt cells to SDF1α may be due in part to an additive effect of constitutive activation of class 1 PI3K and repression of PI3KC2γ.
The sequences from aimno acids 176 to 426 in Bcr-Abl have been shown to be important for activation of several signaling pathways. These sequences are important for activation of class I PI3K, as deletion of this region severely impaired interaction of Bcr-Abl with p85 subunit of PI3 kinase [25]. Also, the tyrosine residue 177 in this region is constitutively phosphorylated in Bcr-Abl [28,29]. The phosphorylated tyrosine 177 binds to adaptor proteins such as Grb2 and, by doing so, links Bcr-Abl to Ras and Map kinase pathway [28,29]. In addition, the sequences of amino acids 176–426 in Bcr-Abl are also involved in the interaction with its own SH2 domain as well as Src family kinases [22,30]. Therefore, further mutagenesis analysis in this region will provide insight into the mechanism by which Bcr-Abl represses PI3KC2γ expression and inhibits SDF-stimulated chemotaxis.
ACKNOWLEDGEMENTS
We thank Drs. Yingzhu Li and Vishakha Kale for assistance in migration assay, Dr. Bifeng Gao for DNA microarray analysis, and Dr. William C. Gilmore for pathology/histology analysis. This work was supported in part by a grant from National Cancer Institute (R01 CA094921, Z. Dai), a grant from the When Everyone Survives Foundation (Z Dai), and a grant from National Institute of Diabetes and Digestive and Kidney Diseases (grant K01 DK067191, Y. Tao).
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
REFERENCES
- 1.Wong S, Witte ON. The BCR-ABL story: bench to bedside and back. Annu Rev Immunol. 2004;22:247–306. doi: 10.1146/annurev.immunol.22.012703.104753. [DOI] [PubMed] [Google Scholar]
- 2.Melo JV, Deininger MW. Biology of chronic myelogenous leukemia--signaling pathways of initiation and transformation. Hematol Oncol Clin North Am. 2004;18:545–568. doi: 10.1016/j.hoc.2004.03.008. vii–viii. [DOI] [PubMed] [Google Scholar]
- 3.Salgia R, Li JL, Ewaniuk DS, et al. BCR/ABL induces multiple abnormalities of cytoskeletal function. J Clin Invest. 1997;100:46–57. doi: 10.1172/JCI119520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salgia R, Quackenbush E, Lin J, et al. The BCR/ABL oncogene alters the chemotactic response to stromal-derived factor-1alpha. Blood. 1999;94:4233–4246. [PubMed] [Google Scholar]
- 5.Ptasznik A, Urbanowska E, Chinta S, et al. Crosstalk between BCR/ABL oncoprotein and CXCR4 signaling through a Src family kinase in human leukemia cells. J Exp Med. 2002;196:667–678. doi: 10.1084/jem.20020519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geay JF, Buet D, Zhang Y, et al. p210BCR-ABL inhibits SDF-1 chemotactic response via alteration of CXCR4 signaling and down-regulation of CXCR4 expression. Cancer Res. 2005;65:2676–2683. doi: 10.1158/0008-5472.CAN-04-2152. [DOI] [PubMed] [Google Scholar]
- 7.Durig J, Rosenthal C, Elmaagacli A, et al. Biological effects of stroma-derived factor-1alpha on normal and CML CD34+ haemopoietic cells. Leukemia. 2000;14:1652–1660. doi: 10.1038/sj.leu.2401875. [DOI] [PubMed] [Google Scholar]
- 8.Peled A, Hardan I, Trakhtenbrot L, et al. Immature leukemic CD34+CXCR4+ cells from CML patients have lower integrin-dependent migration and adhesion in response to the chemokine SDF-1. Stem Cells. 2002;20:259–266. doi: 10.1634/stemcells.20-3-259. [DOI] [PubMed] [Google Scholar]
- 9.Cashman J, Clark-Lewis I, Eaves A, et al. Stromal-derived factor 1 inhibits the cycling of very primitive human hematopoietic cells in vitro and in NOD/SCID mice. Blood. 2002;99:792–799. doi: 10.1182/blood.v99.3.792. [DOI] [PubMed] [Google Scholar]
- 10.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 11.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steelman LS, Pohnert SC, Shelton JG, et al. JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia. 2004;18:189–218. doi: 10.1038/sj.leu.2403241. [DOI] [PubMed] [Google Scholar]
- 13.Martelli AM, Nyakern M, Tabellini G, et al. Phosphoinositide 3-kinase/Akt signaling pathway and its therapeutical implications for human acute myeloid leukemia. Leukemia. 2006;20:911–928. doi: 10.1038/sj.leu.2404245. [DOI] [PubMed] [Google Scholar]
- 14.Kharas MG, Fruman DA. ABL oncogenes and phosphoinositide 3-kinase: mechanism of activation and downstream effectors. Cancer Res. 2005;65:2047–2053. doi: 10.1158/0008-5472.CAN-04-3888. [DOI] [PubMed] [Google Scholar]
- 15.Traer CJ, Foster FM, Abraham SM, et al. Are class II phosphoinositide 3-kinases potential targets for anticancer therapies? Bull Cancer. 2006;93:E53–E58. [PubMed] [Google Scholar]
- 16.Maffucci T, Cooke FT, Foster FM, et al. Class II phosphoinositide 3-kinase defines a novel signaling pathway in cell migration. J Cell Biol. 2005;169:789–799. doi: 10.1083/jcb.200408005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arcaro A, Zvelebil MJ, Wallasch C, et al. Class II phosphoinositide 3-kinases are downstream targets of activated polypeptide growth factor receptors. Mol Cell Biol. 2000;20:3817–3830. doi: 10.1128/mcb.20.11.3817-3830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Domin J, Gaidarov I, Smith ME, et al. The class II phosphoinositide 3-kinase PI3K-C2alpha is concentrated in the trans-Golgi network and present in clathrin-coated vesicles. J Biol Chem. 2000;275:11943–11950. doi: 10.1074/jbc.275.16.11943. [DOI] [PubMed] [Google Scholar]
- 19.Gaidarov I, Smith ME, Domin J, et al. The class II phosphoinositide 3-kinase C2alpha is activated by clathrin and regulates clathrin-mediated membrane trafficking. Mol Cell. 2001;7:443–449. doi: 10.1016/s1097-2765(01)00191-5. [DOI] [PubMed] [Google Scholar]
- 20.Rozycka M, Lu YJ, Brown RA, et al. cDNA cloning of a third human C2-domain-containing class II phosphoinositide 3-kinase, PI3K-C2gamma, and chromosomal assignment of this gene (PIK3C2G) to 12p12. Genomics. 1998;54:569–574. doi: 10.1006/geno.1998.5621. [DOI] [PubMed] [Google Scholar]
- 21.Misawa H, Ohtsubo M, Copeland NG, et al. Cloning and characterization of a novel class II phosphoinositide 3-kinase containing C2 domain. Biochem Biophys Res Commun. 1998;244:531–539. doi: 10.1006/bbrc.1998.8294. [DOI] [PubMed] [Google Scholar]
- 22.Pendergast AM, Muller AJ, Havlik MH, et al. BCR sequences essential for transformation by the BCR-ABL oncogene bind to the ABL SH2 regulatory domain in a non-phosphotyrosine-dependent manner. Cell. 1991;66:161–171. doi: 10.1016/0092-8674(91)90148-r. [DOI] [PubMed] [Google Scholar]
- 23.Dai Z, Kerzic P, Schroeder WG, et al. Deletion of the Src homology 3 domain and C-terminal proline-rich sequences in Bcr-Abl prevents Abl interactor 2 degradation and spontaneous cell migration and impairs leukemogenesis. J Biol Chem. 2001;276:28954–28960. doi: 10.1074/jbc.M101170200. [DOI] [PubMed] [Google Scholar]
- 24.Yu W, Sun X, Clough N, et al. Abi1 gene silencing by short hairpin RNA impairs Bcr-Abl-induced cell adhesion and migration in vitro and leukemogenesis in vivo. Carcinogenesis. 2008;29:1717–1724. doi: 10.1093/carcin/bgn098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skorski T, Bellacosa A, Nieborowska-Skorska M, et al. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. Embo J. 1997;16:6151–6161. doi: 10.1093/emboj/16.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cortez D, Kadlec L, Pendergast AM. Structural and signaling requirements for BCR-ABL-mediated transformation and inhibition of apoptosis. Mol Cell Biol. 1995;15:5531–5541. doi: 10.1128/mcb.15.10.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lataillade JJ, Clay D, Bourin P, et al. Stromal cell-derived factor 1 regulates primitive hematopoiesis by suppressing apoptosis and by promoting G(0)/G(1) transition in CD34(+) cells: evidence for an autocrine/paracrine mechanism. Blood. 2002;99:1117–1129. doi: 10.1182/blood.v99.4.1117. [DOI] [PubMed] [Google Scholar]
- 28.Pendergast AM, Quilliam LA, Cripe LD, et al. BCR-ABL-induced oncogenesis is mediated by direct interaction with the SH2 domain of the GRB-2 adaptor protein. Cell. 1993;75:175–185. [PubMed] [Google Scholar]
- 29.Puil L, Liu J, Gish G, et al. Bcr-Abl oncoproteins bind directly to activators of the Ras signalling pathway. Embo J. 1994;13:764–773. doi: 10.1002/j.1460-2075.1994.tb06319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanglmaier M, Warmuth M, Kleinlein I, et al. The interaction of the Bcr-Abl tyrosine kinase with the Src kinase Hck is mediated by multiple binding domains. Leukemia. 2003;17:283–289. doi: 10.1038/sj.leu.2402778. [DOI] [PubMed] [Google Scholar]