Abstract
Activation of the host gene egr1 is essential for the lytic replication of Epstein–Barr virus (EBV). egr1 is activated by Zta (BZLF1, ZEBRA). Zta interacts directly with DNA through a series of closely related Zta-response elements (ZREs). Here we dissect the mechanism used by Zta to interact with the egr1 promoter and identify a weak interaction with egr1ZRE that is dependent on the distal part of egr1ZRE. Furthermore, we demonstrate that the ability of Zta to interact with egr1ZRE is enhanced at least tenfold by methylation. The ability of Zta to transactivate a reporter construct driven by the egr1 promoter can be enhanced by methylation. As the ability of Zta to interact with a methylated ZRE in the EBV genome correlates with its ability to activate the expression of the endogenous viral gene BRLF1, this suggests that Zta may also have the capability to overturn epigenetic control of egr1.
Following infection, Epstein–Barr virus (EBV) establishes long-lived latency in memory B-lymphocytes (Thorley-Lawson et al., 1996) and other lineages (Chaganti et al., 2008; Conacher et al., 2005). Reactivation from latency can occur from both memory B-lymphocytes and tumour cells, induced by a variety of physiological stimuli (Miller, 1989). The key regulator of EBV lytic replication, Zta (BZLF1, Zta, ZEBRA) is a virally encoded transcription and replication factor which interacts directly with DNA through its basic region both in Zta-regulated promoters and within the origin of lytic replication (Feederle et al., 2000; Miller et al., 2007; Sinclair, 2003, 2006; Sinclair & Farrell, 1992; Speck et al., 1997). Zta also interacts indirectly with DNA and has the potential to alter gene expression through its interactions with some host transcription factors (Gutsch et al., 1994; Sista et al., 1993, 1995; Swenson et al., 2001; Wu et al., 2003; Zhang et al., 1994).
Zta interacts with a subset of ZREs that contain CpG motifs (Bhende et al., 2004). Methylation of transcription factor binding sites at CpG motifs is normally either inhibitory for binding (BSAP, ATF, YY1, EPO80) or neutral (SP1, RFX) (Falzon & Kuff, 1991; Holler et al., 1988; Iguchi-Ariga & Schaffner, 1989; Mancini et al., 1998; Tierney et al., 2000). Methylated DNA is generally associated with non-expressed genes and the inability of some transcription factors to interact with methylated DNA may play a role in either silencing, or maintaining the inhibition of, gene expression. In contrast, Zta has the unusual property of displaying enhanced binding to methylated ZREs (Bhende et al., 2004, 2005; Karlsson et al., 2008a, b; Petosa et al., 2006; Wang et al., 2005).
The host gene egr1 encodes a transcription factor which is induced following EBV lytic cycle activation, through the action of Zta (Chang et al., 2006). egr1 is involved in diverse biological functions (Sukhatme et al., 1988). Mutational analysis of the promoter suggests that activation by Zta occurs through a serum response element (SRE) that is flanked by two Ets response elements and through a tandem pair of potential ZREs (Fig. 1). Zta activates Erk, a member of the MAP kinase family, resulting in transactivation through the Ets response elements (Chang et al., 2006). Indeed, mutation of either the Ets or ZRE sites impacts upon Zta activation in vivo (Chang et al., 2006), suggesting that both direct and indirect mechanisms are used to activate expression through Zta.
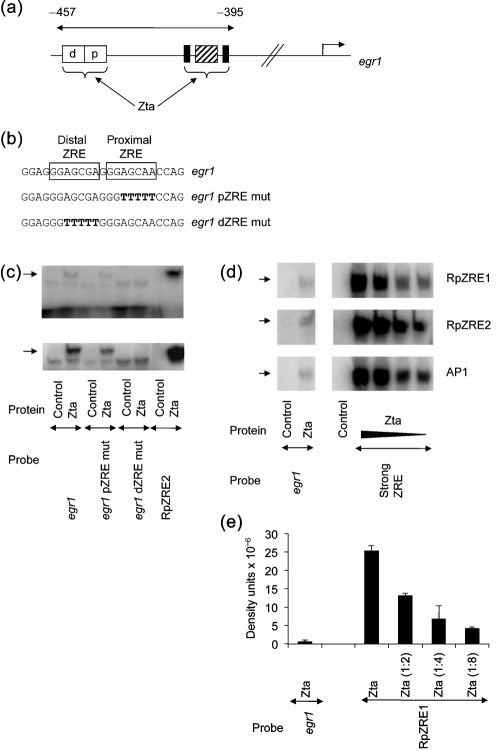
Fig. 1.
Interaction of Zta with egr1ZREs. (a) The organization of the egr1 promoter is shown schematically. The numbers represent distance from the transcription initiation site. The Ets response elements are shown by filled boxes, with the SRE hatched. The open boxes represent the tandem ZREs, proximal (p) and distal (d). (b) The nucleotide sequence of one strand of the oligonucleotide spanning the ZREs is shown. egr1 represents the wild-type sequence, egr1 dZRE mut, the mutation in the distal ZRE and egr1 pZRE mut, the mutation in the proximal ZRE. Double-strand versions of these sequences were used as probes for EMSA. (c) EMSA analysis of the interaction between Zta and egr1, egr1 pZRE mut, egr1 dZREmut and RpZRE2 was undertaken as described previously (Schelcher et al., 2005, 2007). Probes were labelled to approximately equivalent specific activities and the relative amounts used in the reactions are: egr1 (1.0), egr1 pZRE mut (0.5), egr1 dZRE mut (0.7) and RpZRE2 (0.3). In these experiments the probe is in excess, as shown by the dose response in (d) and (e). Zta protein was generated in a wheatgerm translation system, and its interaction with a labelled probe was compared with an unprogrammed lysate as control. For each panel, complexes were separated on the same gel by electrophoresis and visualized by phosphoimaging. The exposure of the bottom panel was ten times longer than that shown in the top panel to visualize the weak Zta–egr1 protein–DNA complexes. The experiment was repeated with similar results. (d) EMSA analysis of the interaction between Zta and egr1, compared with known strong ZREs, RpZRE1 (GATCTCTTTTATGAGCCATTGGCA), RpZRE2 (GATCAAGCTTATGAGCGATTTTTAT) (Bhende et al., 2004) and AP1 (GATCCATGACTCAGAGGAAAACATACG) (Hicks et al., 2003; Kouzarides et al., 1991), were undertaken as in (c). The relative amounts of probes used in the reactions are equivalent. The experiment was repeated with similar results. (e) Quantification of the complexes for RpZRE1 and egr1. These were detected by phosphoimaging in two experiments. Error bars indicate sd.
The egr1 promoter contains tandem copies of potential ZREs. A mutation covering both sites was shown to decrease Zta transactivation in vivo (Chang et al., 2006); however, the precise sequence elements required for Zta to interact with the egr1 promoter have not been mapped. In order to investigate the molecular mechanism by which Zta can directly activate egr1 expression, we questioned the contribution from each ZRE for the interaction with Zta.
A schematic diagram of the egr1 promoter region is shown in Fig. 1. The ability of Zta to interact with the egr1ZRE was assessed using electrophoretic mobility shift assays (EMSA). A Zta–DNA complex was readily detected between Zta and a previously characterized ZRE from the EBV BRLF1 promoter (RpZRE2) (Fig. 1c). The presence of a Zta complex with the egr1ZRE probe, which spans both ZREs, was difficult to detect in comparison, although long exposures revealed a weak protein–DNA complex. From titration experiments we demonstrate that Zta forms a complex with the egr1ZRE probe that is at least 20-fold weaker than that formed with three examples of previously characterized ZREs: RpZRE2, RpZRE1 and AP1 (Fig. 1d). The individual mutations of the tandem ZRE sequences reveal that both contribute to complex formation (Fig. 1c); mutation of the proximal site results in a modest reduction in binding, whereas mutation of the distal site ablates Zta complex formation under these conditions.
Intriguingly, the distal egr1ZRE contains a CpG motif (Fig. 2a). Zta has the unusual property of being able to recognize response elements when methylated at CpG motifs; indeed, the interaction between Zta and two previously characterized ZREs from the BRLF1 promoter, RpZRE2 and RpZRE3, is strongly enhanced by methylation (Bhende et al., 2004, 2005; Karlsson et al., 2008b). egr1 is methylated in B-cells, yet a combination of demethylation agents and physiological stimuli is able to induce egr1 expression (Seyfert et al., 1990). This demonstrates that methylation can play a role in the control of egr1 gene expression.
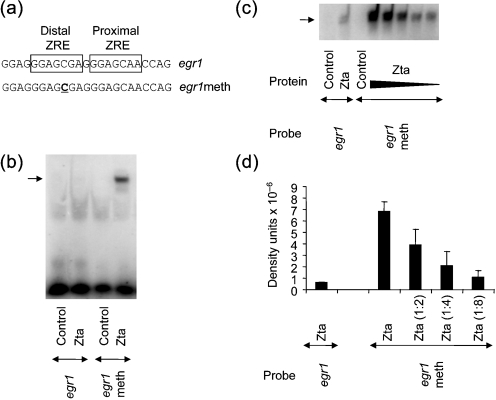
Fig. 2.
Interaction of Zta with methylated egr1ZRE. (a) The nucleotide sequence of one strand of the oligonucleotide spanning the ZREs is shown, together with the methylated version (underlined). Double-strand versions of these sequences were used as probes in EMSA. (b) EMSA analysis of the indicated proteins with the egr1 probes was carried out as described in Fig. 1. Probes were labelled to approximately equivalent specific activities and the relative amounts used in each comparison are egr1 (1.0) and egr1meth (0.72). The experiment was repeated with similar results. (c) EMSA analysis of the interaction between Zta and egr1, compared with the methylated egr1 was undertaken using serial dilutions of Zta protein. The experiment was repeated with similar results. (d) Quantification of the complexes for egr1meth and egr1 (density units×10−6) after adjustment for the relative amounts of probes used for each EMSA. These were detected by phosphoimaging in two experiments. Error bars indicate sd.
To assess whether methylation has any effect on the interaction between Zta and the egr1ZRE, we generated unmethylated and methylated egr1ZRE probes and assessed the interaction of each with Zta by EMSA (Fig. 2). Methylation of the single CpG motif in the distal ZRE clearly enhanced DNA binding and titration experiments revealed that at least tenfold more Zta–DNA complex is formed with the methylated egr1ZRE compared with the unmethylated egr1ZRE.
Molecular modelling predicts that the methylated cytosines of RpZRE2 and RpZRE3 make contact with Zta amino acid residues S186, C189 and R190 as indicated in Fig. 3 (Bhende et al., 2005; Karlsson et al., 2008a; Petosa et al., 2006). Indeed, S186 and C189 have been shown to aid binding to methylated RpZRE2 and RpZRE3 experimentally (Bhende et al., 2005; Karlsson et al., 2008a, b; Wang et al., 2005). In addition to their relevance for interaction with methylated ZREs, mutations of C189 and S186 promote differential binding to some non-CpG-containing ZREs (summarized by Heston et al., 2006).
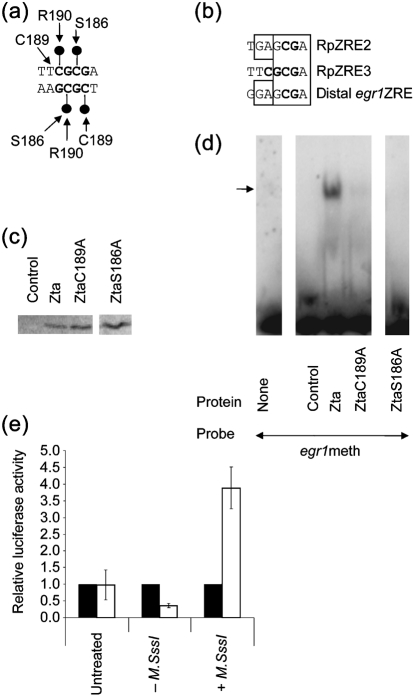
Fig. 3.
Relevance of serine 186 and cysteine 189 for interaction with methylated egr1ZRE and effect of methylation on Zta activation of promoter. (a) Schematic diagram summarizing the known interactions between serine 186, cysteine 189 and arginine 190 with methylated cytosines (indicated as balls) and between cysteine 189, and with the methyl group of a thymidine residue in RpZRE3. (b) Alignment of RpZRE2, RpZRE3 and the distal egr1ZRE. Nucleotides conserved between the distal egr1ZRE with either RpZRE2 or RpZRE3 are highlighted with boxes. (c) Zta, ZtaC189A and ZtaS186A (Karlsson et al., 2008a) were generated in a wheatgerm translation system, together with trace levels of [35S]methionine, and fractionated by SDS-PAGE. The expression levels were determined and equivalent amounts were used for EMSA analysis. (d) The interaction of the indicated Zta proteins, or unprogrammed lysate as control, with the methylated egr1ZRE probe was determined by EMSA. The experiment was repeated with similar results. (e) HeLa cells were transfected with the indicated plasmids using Effectene (Qiagen). After 48 h cell extracts were prepared and assayed for luciferase activity using the luciferase assay system (Promega). The data for each type of plasmid [luciferase activity (μg protein)−1] are expressed relative to the activity levels seen in the absence of Zta, together with the sd, which was derived from at least two experiments. The reporter plasmid used was egr1 (−504/+9)LUC (Chang et al., 2006). The plasmid was transfected in an untreated form, or it underwent a mock methylation reaction (−M.SssI), or it went through a methylation reaction with the methyl transferase M.SssI (New England Biolabs) (+M.SssI), both overnight, followed by purification on a QIAprep column (Qiagen). Prior to transfection, the extent of methylation was evaluated by a diagnostic digestion with the methylation-sensitive restriction enzyme BstUI. The open bars represent transfections undertaken with pBabe BZLF1 (Hicks et al., 2003), while the filled bars represent transfections undertaken with the ‘empty’ pBabe vector.
The distal egr1ZRE, containing the CpG motif, has strong homology with RpZRE2 (6/7 of the heptad core), and significant homology with RpZRE3 (4/7 of the heptad core), suggesting that the same amino acid residues may contribute to interaction with the methylated CpG motif. As only the rightwards CpG motif of RpZRE3 is conserved within the distal egr1ZRE, it is relevant to question the contribution of amino acids C189 and S186 to the DNA-binding function. To assess this, we used single point mutants of Zta, ZtaC189A and ZtaS186A, to investigate their impact on the interaction of Zta with methylated egr1ZRE. The full-length version of each protein was generated in an in vitro translation system (Fig. 3) and the interaction of equivalent amounts of each with methylated egr1ZRE probe was analysed by EMSA. Mutation of either S186 or C189 greatly reduced the ability of Zta to interact with the methylated egr1ZRE.
To assess whether methylation of the egr1 promoter affects the ability of Zta to activate it, we co-expressed Zta and a previously characterized egr1 promoter construct (Chang et al., 2006) in two epithelial cell lines. A comparison was undertaken between methylated and non-methylated versions of the promoter construct (Fig. 3e). In HeLa cells, Zta did not activate expression of the untreated plasmid, yet Zta activated the methylated reporter construct approximately fourfold. This clearly demonstrates that Zta can preferentially activate the methylated egr1 promoter construct.
We note that the Zta activation occurs against a background of decreased Zta-induced activity of the egr1 promoter plasmid that had undergone a ‘mock-methylation’ reaction. We also noticed a reduction in the basal activity of plasmids that have been methylated and a partial reduction in basal activity of those plasmids that have been through the mock-methylation reaction in all cell types tested (data not shown). This mirrors what has been described previously for the BRLF1 and BZLF1 promoters in 293T and DG75 cells (Bhende et al., 2004, 2005).
In contrast, in U2OS cells, Zta activation of expression of the egr1 promoter plasmid was approximately twofold for both the untreated and the methylated plasmids, suggesting no methylation-dependent enhancement (Supplementary Fig. S1, available in JGV Online). This analysis suggests that the enhanced activation by Zta from the methylated egr1 promoter is cell-line dependent.
Taken together, these data show that Zta is able to interact with the egr1ZRE through the distal ZRE and furthermore that methylation of the CpG motif in the distal element does not prevent the interaction with Zta, but rather enhances binding. The methylation sensitivity of the interaction of Zta with egr1ZRE and the dependence on S186 and C189 resembles its binding with RpZRE3 (Karlsson et al., 2008a, b). The BRLF1 promoter is methylated during viral latency (Bhende et al., 2005) and the ability of Zta to interact with methylated RpZRE3 correlates with its ability to activate expression of BRLF1 in a B-cell line (Karlsson et al., 2008a, b). The demonstration that activation of the egr1 promoter by Zta can be enhanced by methylation suggests that Zta may be able to activate the egr1 promoter in vivo, when it is methylated. Thus the potential arises that Zta is able to overturn the epigenetic silencing of both a viral promoter (BRLF1) and a host promoter (egr1).
Supplementary Material
Acknowledgments
This research was supported by the Medical Research Council and the Wellcome Trust. We are grateful to Professor Ching-Hwa Tsai and colleagues (National Taiwan University, Taipei, Taiwan) for the generous gift of the pEgr-1(−504/+9)-LUC plasmid.
Footnotes
A supplementary figure showing the effect of methylation and Zta expression on the activity of the erg1 promoter in U2OS cells is available with the online version of this paper.
References
- Bhende, P. M., Seaman, W. T., Delecluse, H. J. & Kenney, S. C. (2004). The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat Genet 36, 1099–1104. [DOI] [PubMed] [Google Scholar]
- Bhende, P. M., Seaman, W. T., Delecluse, H. J. & Kenney, S. C. (2005). BZLF1 activation of the methylated form of the BRLF1 immediate-early promoter is regulated by BZLF1 residue 186. J Virol 79, 7338–7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti, S., Ma, C. S., Bell, A. I., Croom-Carter, D., Hislop, A. D., Tangye, S. G. & Rickinson, A. B. (2008). Epstein–Barr virus persistence in the absence of conventional memory B cells: IgM+IgD+CD27+ B cells harbor the virus in X-linked lymphoproliferative disease patients. Blood 112, 672–679. [DOI] [PubMed] [Google Scholar]
- Chang, Y., Lee, H. H., Chen, Y. T., Lu, J., Wu, S. Y., Chen, C. W., Takada, K. & Tsai, C. H. (2006). Induction of the early growth response 1 gene by Epstein–Barr virus lytic transactivator Zta. J Virol 80, 7748–7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conacher, M., Callard, R., McAulay, K., Chapel, H., Webster, D., Kumararatne, D., Chandra, A., Spickett, G., Hopwood, P. A. & Crawford, D. H. (2005). Epstein-Barr virus can establish infection in the absence of a classical memory B-cell population. J Virol 79, 11128–11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzon, M. & Kuff, E. L. (1991). Binding of the transcription factor EBP-80 mediates the methylation response of an intracisternal A-particle long terminal repeat promoter. Mol Cell Biol 11, 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feederle, R., Kost, M., Baumann, M., Janz, A., Drouet, E., Hammerschmidt, W. & Delecluse, H. J. (2000). The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J 19, 3080–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsch, D. E., Holley-Guthrie, E. A., Zhang, Q., Stein, B., Blanar, M. A., Baldwin, A. S. & Kenney, S. C. (1994). The bZIP transactivator of Epstein-Barr virus, BZLF1, functionally and physically interacts with the p65 subunit of NF-κB. Mol Cell Biol 14, 1939–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heston, L., El-Guindy, A., Countryman, J., Dela Cruz, C., Delecluse, H. J. & Miller, G. (2006). Amino acids in the basic domain of Epstein-Barr virus ZEBRA protein play distinct roles in DNA binding, activation of early lytic gene expression, and promotion of viral DNA replication. J Virol 80, 9115–9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks, M. R., Al-Mehairi, S. S. & Sinclair, A. J. (2003). The zipper region of Epstein-Barr virus bZIP transcription factor Zta is necessary but not sufficient to direct DNA binding. J Virol 77, 8173–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler, M., Westin, G., Jiricny, J. & Schaffner, W. (1988). Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev 2, 1127–1135. [DOI] [PubMed] [Google Scholar]
- Iguchi-Ariga, S. M. & Schaffner, W. (1989). CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev 3, 612–619. [DOI] [PubMed] [Google Scholar]
- Karlsson, Q. H., Schelcher, C., Verrall, E., Petosa, C. & Sinclair, A. J. (2008a). Methylated DNA recognition during the reversal of epigenetic silencing is regulated by cysteine and serine residues in the Epstein-Barr virus lytic switch protein. PLoS Pathog 4, e1000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson, Q. H., Schelcher, C., Verrall, E., Petosa, C. & Sinclair, A. J. (2008b). The reversal of epigenetic silencing of the EBV genome is regulated by viral bZIP protein. Biochem Soc Trans 36, 637–639. [DOI] [PubMed] [Google Scholar]
- Kouzarides, T., Packham, G., Cook, A. & Farrell, P. J. (1991). The BZLF1 protein of EBV has a coiled coil dimerization domain without a heptad leucine repeat but with homology to the C/EBP leucine zipper. Oncogene 6, 195–204. [PubMed] [Google Scholar]
- Mancini, D. N., Rodenhiser, D. I., Ainsworth, P. J., O'Malley, F. P., Singh, S. M., Xing, W. & Archer, T. K. (1998). CpG methylation within the 5′ regulatory region of the BRCA1 gene is tumor specific and includes a putative CREB binding site. Oncogene 16, 1161–1169. [DOI] [PubMed] [Google Scholar]
- Miller, G. (1989). The switch between EBV latency and replication. Yale J Biol Med 62, 205–213. [PMC free article] [PubMed] [Google Scholar]
- Miller, G., El-Guindy, A., Countryman, J., Ye, J. & Gradoville, L. (2007). Lytic cycle switches of oncogenic human gammaherpesviruses. Adv Cancer Res 97, 81–109. [DOI] [PubMed] [Google Scholar]
- Petosa, C., Morand, P., Baudin, F., Moulin, M., Artero, J. B. & Muller, C. W. (2006). Structural basis of lytic cycle activation by the Epstein-Barr virus ZEBRA protein. Mol Cell 21, 565–572. [DOI] [PubMed] [Google Scholar]
- Schelcher, C., Valencia, S., Delecluse, H. J., Hicks, M. & Sinclair, A. J. (2005). Mutation of a single amino acid residue in the basic region of the Epstein-Barr virus (EBV) lytic cycle switch protein Zta (BZLF1) prevents reactivation of EBV from latency. J Virol 79, 13822–13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelcher, C., Al Mehairi, S., Verrall, E., Hope, Q., Flower, K., Bromley, B., Woolfson, D. N., West, M. J. & Sinclair, A. J. (2007). Atypical bZIP domain of viral transcription factor contributes to stability of dimer formation and transcriptional function. J Virol 81, 7149–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfert, V. L., McMahon, S., Glenn, W., Cao, X. M., Sukhatme, V. P. & Monroe, J. G. (1990). Egr-1 expression in surface Ig-mediated B cell activation. Kinetics and association with protein kinase C activation. J Immunol 145, 3647–3653. [PubMed] [Google Scholar]
- Sinclair, A. J. (2003). bZIP proteins of human gammaherpesviruses. J Gen Virol 84, 1941–1949. [DOI] [PubMed] [Google Scholar]
- Sinclair, A. J. (2006). Unexpected structure of Epstein-Barr virus lytic cycle activator Zta. Trends Microbiol 14, 289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair, A. J. & Farrell, P. J. (1992). Epstein-Barr virus transcription factors. Cell Growth Differ 3, 557–563. [PubMed] [Google Scholar]
- Sista, N. D., Pagano, J. S., Liao, W. & Kenney, S. (1993). Retinoic acid is a negative regulator of the Epstein-Barr virus protein (BZLF1) that mediates disruption of latent infection. Proc Natl Acad Sci U S A 90, 3894–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sista, N. D., Barry, C., Sampson, K. & Pagano, J. (1995). Physical and functional interaction of the Epstein-Barr virus BZLF1 transactivator with the retinoic acid receptors RARα and RXRα. Nucleic Acids Res 23, 1729–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck, S. H., Chatila, T. & Flemington, E. (1997). Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol 5, 399–405. [DOI] [PubMed] [Google Scholar]
- Sukhatme, V. P., Cao, X. M., Chang, L. C., Tsai-Morris, C. H., Stamenkovich, D., Ferreira, P. C., Cohen, D. R., Edwards, S. A., Shows, T. B. & other authors (1988). A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell 53, 37–43. [DOI] [PubMed] [Google Scholar]
- Swenson, J. J., Holley-Guthrie, E. & Kenney, S. C. (2001). Epstein-Barr virus immediate-early protein BRLF1 interacts with CBP, promoting enhanced BRLF1 transactivation. J Virol 75, 6228–6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorley-Lawson, D. A., Miyashita, E. M. & Khan, G. (1996). Epstein-Barr virus and the B-cell: that's all it takes. Trends Microbiol 4, 204–208. [DOI] [PubMed] [Google Scholar]
- Tierney, R. J., Kirby, H. E., Nagra, J. K., Desmond, J., Bell, A. I. & Rickinson, A. B. (2000). Methylation of transcription factor binding sites in the Epstein-Barr virus latent cycle promoter Wp coincides with promoter down-regulation during virus-induced B-cell transformation. J Virol 74, 10468–10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P., Day, L., Dheekollu, J. & Lieberman, P. M. (2005). A redox-sensitive cysteine in Zta is required for Epstein-Barr virus lytic cycle DNA replication. J Virol 79, 13298–13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F. Y., Chen, H., Wang, S. E., ApRhys, C. M., Liao, G., Fujimuro, M., Farrell, C. J., Huang, J., Hayward, S. D. & Hayward, G. S. (2003). CCAAT/enhancer binding protein α interacts with ZTA and mediates ZTA-induced p21CIP-1 accumulation and G1 cell cycle arrest during the Epstein-Barr virus lytic cycle. J Virol 77, 1481–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Gutsch, D. & Kenney, S. (1994). Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol Cell Biol 14, 1929–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.