Abstract
Japanese encephalitis virus (JEV) consists of five genotypes (GI–V). Phylogenetic characterization of 16 JEV strains isolated from the ‘USSR’, Japan and Korea during the 1930–1970s revealed that 15 strains fell into GIII, confirming that GIII was the predominant genotype of JEV in Japan and Korea between 1935 (isolation of the prototype strain; a GIII virus) and the 1990s (when GI supplanted GIII). One of the Korean isolates fell into GII, demonstrating that GII has been circulating for at least 19 years longer than previously thought. Formerly, GII was associated with endemic disease and this genotype had never been isolated north of Southern Thailand. Additionally, the northern border of GIII prevalence was extended from Japan to the ‘USSR’.
Japanese encephalitis virus (JEV) is a mosquito-borne flavivirus that is the most significant cause of epidemic encephalitis worldwide, with 50 000 cases of encephalitis and 10 000 deaths annually (Barrett, 2008). It is transmitted in an enzootic cycle involving waterbirds, domestic pigs and rice paddy-breeding mosquitoes; humans and other non-avian vertebrates are dead-end hosts.
Encephalitis outbreaks suggestive of JE were recorded in Japan as early as 1871 and the prototype strain was isolated in Japan in 1935 (Lewis et al., 1947). Subsequently, JE has been found throughout most of Asia and the virus was most recently isolated in Papua New Guinea and Australia (Johansen et al., 2000). Genetic studies, which are limited by the predominance of newer isolates, divide JEV into five genotypes (GI–V) that exhibit geographical segregation. GI includes strains isolated in Northern Thailand, Cambodia, Korea, China, Japan, Vietnam, Taiwan and Australia between 1967 and the present, GII includes strains isolated in Southern Thailand, Malaysia, Indonesia, Papua New Guinea and Northern Australia between 1970 and 1999, GIII includes strains isolated in temperate regions of Asia from 1935 to the present, and the majority of viruses that have been studied have been characterized as members of this genotype. GIV includes strains isolated in Indonesia between 1980 and 1981 only, and GV includes a single strain isolated in Singapore in 1952 (Chen et al., 1990, 1992; Uchil & Satchidanandam, 2001). Previous studies in our laboratory suggest that JEV arose from its ancestral virus in the Indonesia–Malaysia region and evolved there into five genotypes; GIV and GV, the most divergent genotypes, remained confined to the Indonesia–Malaysia region, whereas GI, GII and GIII, the most recent genotypes spread across Asia (Solomon et al., 2003).
There is little genetic information on JEV strains isolated prior to the 1970s and the majority are in GIII. Furthermore, the geographical range of GI endemicity has expanded over the past 25 years, and GI has recently replaced GIII as the most frequently isolated virus genotype in a number of East and South-east Asian countries (Nga et al., 2004; Nitatpattana et al., 2008). The absence of genetic information on older strains has limited our understanding of the phylogeography, evolution and epidemiology of JEV. We hypothesized that genetic characterization of older strains of JEV will identify the extent of genetic variation among and within the virus genotypes. Therefore, we examined 16 JEV strains obtained from the Walter Reed Army Institute of Research (WRAIR), which were isolated from specimens collected in the ‘USSR’, Japan and Korea during the 1930s through the 1970s; and 10 strains obtained from the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA), which were isolated in Korea during the 1940s through the 1990s (Table 1).
Table 1.
Details of isolates used for analysis in this study
All isolates are JEV, except for MVE-1-51 which is a strain of Murray Valley encephalitis virus.
| Strain | Origin | Year of isolation | Host | Passage no. | GenBank accession no(s). |
|---|---|---|---|---|---|
| 05734 | India | 2005 | Human | EF623988 | |
| 691004 | Sri Lanka | 1969 | Human | Z34097 | |
| Autumn 4* | ‘USSR’ | 1943 | Human | 4 | FJ515932 |
| Beijing-1 | China | 1949 | Mosquito | L48961 | |
| Bennett* | Korea | Before 1951 | Human | 5 | FJ515927, FJ872376 |
| CH1392 | Taiwan | 1990 | Mosquito | AF254452 | |
| CNS138-11 | Malaysia | 1999 | Human | AY184213 | |
| CTS | China | 1955 | Human | AY243830 | |
| Equine* | Japan | 1947 | Equid | 27 | FJ515929, FJ872378 |
| FU | Australia | 1995 | Human | AF217620 | |
| GP78 | India | 1978 | Human | AF075723 | |
| HV1 | Taiwan | 1958 | Human | AF098735 | |
| Ishikawa | Japan | 1999 | Mosquito | AB051292 | |
| JaGAr01 | Japan | 1959 | Mosquito | AF069076 | |
| JaOArS982 | Japan | 1982 | Mosquito | NC_001437 | |
| JaOH0566 | Japan | 1966 | Human | AY508813 | |
| JaTH160 | Japan | 1960 | Human | AB269326 | |
| JE-82† | Korea | 1982 | Mosquito | Unknown | GQ415347 |
| JE-83† | Korea | 1983 | Mosquito | Unknown | GQ415348 |
| JE-84† | Korea | 1984 | Mosquito | Unknown | GQ415349 |
| JE-85† | Korea | 1985 | Mosquito | Unknown | GQ415350 |
| JE-86† | Korea | 1986 | Mosquito | Unknown | GQ415351 |
| JE-87† | Korea | 1987 | Mosquito | Unknown | GQ415352 |
| JE-88† | Korea | 1988 | Mosquito | Unknown | GQ415353 |
| JE-89† | Korea | 1989 | Mosquito | Unknown | GQ415354 |
| JE-91† | Korea | 1991 | Mosquito | Unknown | GQ415355 |
| JEV # 40783* | Korea | Before 1971 | Human | 8 | FJ515923 |
| JEV/sw/Mie/40/2004 | Japan | 2004 | Swine | AB241118 | |
| JKT5441 | Indonesia | 1981 | Mosquito | U70406 | |
| JKT6468 | Indonesia | 1981 | Mosquito | AY184212 | |
| K-29† | Korea | 1949 | Human | Unknown | GQ415356 |
| K87P39 | Korea | 1987 | Mosquito | AY585242 | |
| K94P05 | Korea | 1994 | Mosquito | AF045551 | |
| Kalinina* | Japan | 1935 | Human | 5 | FJ515928 |
| ‘Korea Jap B’* | Korea | Before 1950 | 7 | FJ515926, FJ872379 | |
| KV1899 | Korea | 1999 | Swine | AY316157 | |
| Ling | Taiwan | 1965 | Human | L78128 | |
| LYZ | China | 1957 | Human | AY243834 | |
| Matsunaga* | Japan | 1939 | Human | 3 | FJ515930, FJ872381 |
| MVE-1-51 | Australia | 1951 | Human | AF161266 | |
| Nakayama | Japan | 1935 | Human | EF571853 | |
| Nakayama mb-1* | Japan | 1935 | Human | 3 | FJ515931 |
| P20778 | India | 1958 | Human | AF080251 | |
| P3 | China | 1950 | Mosquito | U47032 | |
| Roum* | Korea | 1946 | Human | 5 | FJ515922, FJ872377 |
| SA14 | China | 1954 | Mosquito | JEU14163 | |
| T1P1 | Taiwan | 1997 | Mosquito | AF254453 | |
| Taira* | Japan | 1948 | Human | 5 | FJ515933, FJ872384 |
| V9-3182* | Japan | 1949 | Human | 5 | FJ515924 |
| V9-3702* | Japan | 1949 | Human | 9 | FJ515934 |
| V9-3901* | Japan | Before 1950 | Human | 8 | FJ515935, FJ872382 |
| V9-3902* | Japan | Before 1950 | Human | 8 | FJ515936, FJ872383 |
| V9-4358* | Japan | Before 1950 | Human | 6 | FJ515925 |
| V9-4399* | Japan | Before 1950 | Human | 7 | FJ515937, FJ872380 |
| WTP-70-22 | Malaysia | 1970 | Mosquito | U70421 | |
| XJ69 | China | 2007 | Mosquito | EU880214 |
*WRAIR isolates sequenced in this study.
†WRCEVA isolates sequenced in this study.
The viruses were amplified once in Aedes albopictus C6/36 mosquito cells. Culture supernatants were harvested and viral RNA was extracted using the QIAamp Viral RNA Mini kit (Qiagen). The NS5 gene/3′ UTR of the 16 WRAIR strains was amplified using the Titan One Tube RT-PCR System (Roche) and the universal mosquito-borne flavivirus primer pair EMF1/VD8 (Pierre et al., 1994) (for primer sequences see Supplementary Table S1, available in JGV Online). This primer pair was selected not only because it can amplify small amounts of RNA and be used to differentiate among mosquito-borne flaviviruses, but also because the 3′ UTR of flaviviruses is hypervariable and is thus considered to be a good marker for predicting genetic relatedness and viral variation (Shurtleff et al., 2001). However, EMF1/VD8 only amplifies a 562 nt region of the virus genome, and therefore amplicons contain relatively few phylogenetically informative sites, which can result in low bootstrap values for some branches of the phylogeny. Hence, the envelope (E) gene (1500 nt in length) of nine of the 16 WRAIR strains and the 10 WRCEVA strains was amplified by RT-PCR and one of two sets of primer pairs: (i) F879/R2570 or (ii) 940S/1720A, 1598S/2171A and 2115S/2624A. The E gene of seven of the 16 WRAIR strains could not be amplified with either E gene-targeting primer sets due to extremely low quantities of viral RNA. The PCR products were purified and sequenced directly by standard methods using the primer sets: (i) EMF1 and VD8 and (ii) F879, R2570, F1468 and R2088 or (iii) 940S, 1720A, 1598S, 2171A, 2115S and 2624A. The WRAIR NS5 gene/3′ UTR nucleotide sequence file was assembled using ContigExpress and then aligned, using AlignX (Vector NTI; Invitrogen), with 22 geospatiotemporally distinct strains of JEV genotypes I–IV and one strain of Murray Valley encephalitis virus that were retrieved from GenBank. Two E gene nucleotide sequence files (WRAIR and WRAIR/WRCEVA) were assembled using ContigExpress and then aligned, using AlignX, with 26 geospatiotemporally distinct strains of JEV genotypes I–IV and one strain of Murray Valley encephalitis virus that were retrieved from GenBank. The sequence alignment files were transferred to BioEdit (Hall, 1999) to manually align gaps in the 3′ UTR in the NS5 gene/3′ UTR file and to translate the WRAIR E gene nucleotide sequence alignment file to produce a WRAIR E protein amino acid sequence alignment file. Neighbour-joining, maximum-likelihood and maximum-parsimony phylogenetic analyses were performed on the NS5 gene/3′ UTR and E gene datasets using the phylip package (Felsenstein, 1989). All trees were drawn using FigTree software (http://tree.bio.ed.ac.uk/software/figtree/).
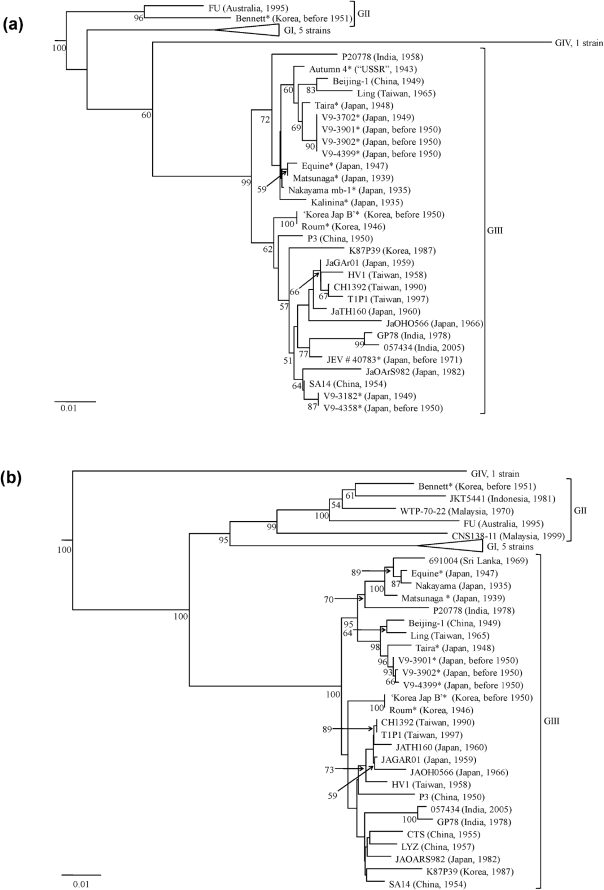
In comparison to the prototype Nakayama strain, nucleotide sequence divergence between the various WRAIR strains ranged from 0.4 to 13.0 % in the NS5 gene/3′ UTR and from 0.6 to 17.0 % in the E gene. Phylogenetic trees produced from the NS5 gene/3′ UTR and the E gene nucleotide sequence files identified four major clades (representing GI–IV) and revealed similar virus groupings (Fig. 1a, b, c). Furthermore, all three methods of phylogenetic analyses (neighbour-joining, maximum-likelihood and maximum-parsimony) performed on the sequence files identified four major clades (representing GI–IV) and revealed similar tree topologies differing only in terminal branching. JEV genotypes in this study were defined as viruses that differed in nucleotide composition by at least 7.2 % in the NS5 gene/3′ UTR and by more than 9.2 % in the E gene.
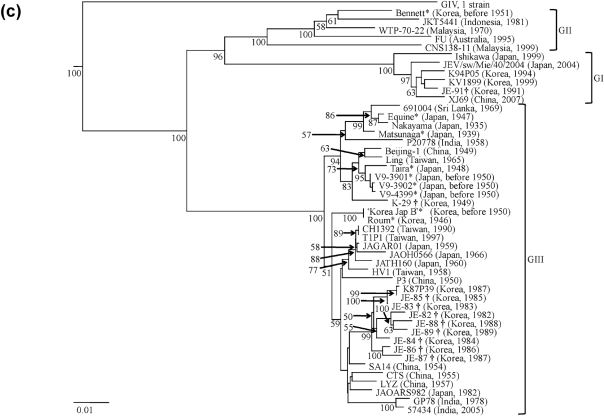
Fig. 1.
Neighbour-joining phylogenies based on sequence information derived from (a) a 562 nt region spanning the NS5 gene/3′ UTR of the WRAIR isolates, (b) the E gene of the WRAIR isolates and (c) the E gene of the WRAIR/WRCEVA isolates. The trees were rooted using Murray Valley encephalitis virus (MVE-1-51), which is a member of the Japanese encephalitis serogroup, but has been removed to allow for better visualization of branch lengths. Horizontal branch lengths are proportional to the genetic distance between isolates and the scale at the lower-left of each tree indicates the number of nucleotide substitutions per site. Genotypes (GI–IV) are represented to the right of each tree. Numbers to the lower-left of the nodes represent per cent bootstrap values based on 1000 replicates. Only bootstrap values greater than 50 % are indicated. WRAIR isolates sequenced in this study are indicated by ‘*’ and WRCEVA isolates sequenced in this study are indicated by ‘†’.
Fifteen of the WRAIR isolates fell into GIII, while one of the isolates fell into GII [Bennett (Korea, 1951)], demonstrating that GII has been circulating for at least 19 years longer than previously thought (Fig. 1a, b).
In Japan, the first outbreak suggestive of JE was recorded in 1871 and major epidemics occurred in 1924, 1935 and 1948. As expected, two viruses (Nakayama mb-1 and Kalinina) isolated from humans in the 1935 epidemic in Tokyo group closely within GIII (Fig. 1a) (Kudo et al., 1937). Matsunaga was isolated from a human in Japan in 1939 and groups with Equine, which was isolated from a horse in Japan in 1947 (Fig. 1a, b). Taira was isolated during the 1948 epidemic in Japan and shares a common ancestor with four strains (V9-3702, V9-3901, V9-3902 and V9-4399) isolated between 1949 and 1950 in Japan (Fig. 1a, b). Interestingly, two strains (V9-3182 and V9-4358) isolated during the same time period in Japan formed a different cluster within GIII and differed by 19 nt within the amplified NS5 gene/3′ UTR, suggesting that two genetically distinct strains of JEV were co-circulating (Fig. 1a).
A JE epidemic was reported for the first time in restricted seacoast areas of the ‘USSR’ in 1938, reappeared in epidemic form in 1939 and occurred sporadically in subsequent years (Grascenkov, 1964). Autumn 4 was isolated from a human in the ‘USSR’ in 1943 and represents the first genetically characterized strain of JEV from this geographical region (Fig. 1a).
Major outbreaks of JE were recorded in Korea among American soldiers in 1946, in the civilian population in 1949 and among American military personnel in 1950. JE had been suggested to occur in Korea as early as 1926, but was never proven. For this reason the American forces stationed in Korea were not given JE vaccine during 1946 as were those stationed in Japan. Consequently, three cases of severe encephalitis, resulting in one fatality, occurred among 1500 American soldiers stationed in Kunsan, Korea in 1946. JEV was isolated from the fatal case and it is likely that this is the Roum strain (Sabin et al., 1947). Roum and the ‘Korea Jap B’ strain, which was isolated in Korea in 1950, group together (Fig. 1a, b). The isolation of ‘Korea Jap B’ coincides with an epidemic of JE, resulting in 17 deaths that occurred among a camp of 300 American soldiers stationed to the 1949–1950 defence ‘perimeter’ about Pusan, Korea (Lincoln & Sivertson, 1952). Interestingly, the Bennett strain was isolated in Korea during the same period of time (before 1951) as ‘Korea Jap B’. However, Bennett fell into GII (Fig. 1a, b), indicating two genotypes were in Korea concurrently. Bennett was isolated at least 19 years prior to the oldest published GII strain, WTP-70-22, which was isolated in Malaysia in 1970 (Chen et al., 1990). A sequence alignment revealed that Bennett had the same unique 11 nt deletion immediately following the stop codon in the 3′ UTR as all other GI and GII strains used in this analysis (data not shown).
To determine whether GII circulated in Korea or the Bennett strain represented a single imported case, an E gene phylogeny containing three Korean strains retrieved from GenBank, three Korean WRAIR strains and 10 Korean strains obtained from WRCEVA was constructed (Fig. 1c). Of the 16 Korean strains examined, 12 belong to GIII, three belong to GI and the Bennett strain represents the only isolate of a GII virus (Fig. 1c). However, there is no nucleotide sequence information available on Korean strains isolated between 1951 and 1971. Therefore, it remains unknown whether the isolation of the Bennett strain coincided with a GII epidemic focus that quickly died off, or GII circulated in Korea for a period of time prior to its extinction, or the Bennett strain represented a single imported case from another region. The latter is improbable given that almost all American ground forces arrived to the Korean War zone by sea after a journey devoid of culicine mosquito exposure for days, and there was little or no rest and recuperation, which occurred in Japan, for American ground forces assigned to the Korean War zone (Hastings, 1987). Thus, the possibility of importation is unlikely, and if importation did occur the origin of infection was nearby Japan. The Bennett strain was most likely isolated from an American soldier bitten by an infective mosquito in Korea, as minimum infection rates of mosquitoes even during intense JEV outbreaks among susceptible humans are low (Gingrich et al., 1992). Therefore, it is likely that there was at least transient transmission occurring locally among amplifying hosts to generate the infective mosquito that transmitted the Bennett strain.
A WRAIR E protein amino acid sequence alignment was generated using sequence information derived from one GI isolate (Ishikawa), two GII isolates (Bennett and FU), four GIII isolates [Nakayama, Matsunaga, V9-3901 and ‘Korea Jap B (representative of distinct nodes within the WRAIR E gene phylogeny)] and one GIV isolate (JKT6468) (Fig. 2). The range of intergenotypic amino acid sequence divergence among the viruses included in Fig. 2 is as follows: GI versus GII (2.2–3.4 %), GI versus GIII (2.8–4.0 %), GI versus GIV (6.6 %), GII versus GIII (0.6–2.6 %), GII versus GIV (4.4–5.6 %) and GIII versus GIV (4.4–5.6 %). The amino acid composition of the two GII strains shown in the E protein alignment, Bennett and FU, differed by 1.2 %. The FU strain possessed six amino acid substitutions within the E protein that differentiated it from the Bennett strain (F108S, S208P, K307N, F308S, A311R and A366S). The E protein of flaviviruses is composed of three domains: domain III is the receptor-binding domain, domain II is the dimerization domain and domain I acts as a hinge between domains II and III (Rey et al., 1995). The predicted three-dimensional structure of the E protein of the Bennett and FU strains were modelled onto the crystal structure of West Nile virus (a flavivirus that is closely related to JEV) (Nybakken et al., 2006), illustrating that all six of the amino acid substitutions were surface exposed. Four of the amino acid substitutions (K307N, F308S, A311R and A366S) mapped to domain III of the E protein; while two of the substitutions mapped to domain II of the E protein (F108S and S208P), with F108S specifically mapping to the fusion peptide of domain II (Williams et al., 2000). Five of these amino acid substitutions (S208P, K307N, F308S, A311R and A366S) are unique to the FU strain and three other GII strains (M15, NO and M40) that were also isolated in Australia in 1995 (data not shown). One of the amino acid substitutions (F108S) is also found in the three Australian strains isolated in 1995, as well as in JKT5441 which was isolated in Indonesia in 1981 (data not shown). This evidence supports the hypothesis that the E protein amino acid substitutions unique to the four virus strains isolated in Australia in 1995 have evolved recently. The intragenotypic divergence of GIII within the E protein ranged from 0.0 to 1.8 %. Matsunaga, V9-3901 and ‘Korea Jap B’ exhibited six amino acid substitutions when compared with the prototype Nakayama strain (V51S, K83E, T176I, P223S, N276S and R290K). In addition to the six amino substitutions, ‘Korea Jap B’ possessed three unique amino acid mutations when compared with the Nakayama strain (S64L, T146A and K336R). The E protein amino acid sequence alignment illustrated the genetic diversity exhibited among early isolates of GIII of JEV.
Fig. 2.
Alignment of JEV GI–IV E protein amino acid sequences relative to the prototype Nakayama strain. Dots indicate consensus. The letter X represents amino acids that are unidentified.
Overall, the results of the current study confirm that GIII was the predominant genotype of JEV in Japan and Korea between 1935 (isolation of the prototype strain; a GIII virus) and the 1990s (GI supplanted GIII) (Nga et al., 2004; Nitatpattana et al., 2008). The reasons underlying the initial disproportionate expansion and establishment of GIII as the most widely distributed genotype of JEV throughout East and South-east Asia, as well as the recent replacement of GIII by GI are uncertain and have been discussed previously (Solomon et al., 2003). JEV utilizes a broad range of reservoir hosts and vector species across Asia and differences in host and vector availability may explain the observed patterns of genotype expansion and establishment. In addition, these observed patterns could also be due to genotype-defining viral molecular determinants that relate to differences in host preference. This study also revealed that GIII existed as far north as the ‘USSR’ [Autumn 4 (1943)]. Genetic characterization of the Bennett strain (Korea, before 1951) demonstrated that GII has been circulating for at least 19 years longer than previously thought. Prior to this study, GII was associated with endemic disease and this genotype had never been isolated north of Southern Thailand. Although the possibility of the Bennett strain representing a single imported case from nearby Japan cannot be excluded, it is more likely that the isolation of this strain coincided with a GII epidemic focus that quickly became extinct or GII became endemic in Korea for a period of time and subsequently disappeared.
Supplementary Material
Acknowledgments
We thank the WRAIR for donating the older JEV strains. This work was supported in part by an NIH grant AI 067847 (to A. D. T. B.) and an NIH contract N01-AI 30027 (to R. B. T.). A. J. S. is supported by an NIH T32 training grant AI 60549.
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the sequences described in this study are FJ515922–FJ515937, FJ872376–FJ872384 and GQ415347–GQ415356.
A supplementary table of primer sequences is available with the online version of this paper.
References
- Barrett, A. D. T. (2008). Japanese encephalitis virus. In Encyclopaedia of Virology, 3rd edn, vol. 5, pp. 182–188. Edited by B. W. J. Mahy & M. V. H. van Regenmortel. Oxford: Elsevier.
- Chen, W. R., Tesh, R. B. & Rico-Hesse, R. (1990). Genetic variation of Japanese encephalitis virus in nature. J Gen Virol 71, 2915–2922. [DOI] [PubMed] [Google Scholar]
- Chen, W. R., Rico-Hesse, R. & Tesh, R. B. (1992). A new genotype of Japanese encephalitis virus from Indonesia. Am J Trop Med Hyg 47, 61–69. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1989). phylip – phylogeny inference package (version 3.2). Cladistics 5, 164–166. [Google Scholar]
- Gingrich, J. B., Nisalak, A., Latendresse, J. R., Sattabongkot, J., Hoke, C. H., Pomsdhit, J., Chantalakana, C., Satayaphanta, C., Uechiewcharnkit, K. & Innis, B. L. (1992). Japanese encephalitis virus in Bangkok: factors influencing vector infections in three suburban communities. J Med Entomol 29, 436–444. [DOI] [PubMed] [Google Scholar]
- Grascenkov, N. I. (1964). Japanese encephalitis in the USSR. Bull World Health Organ 30, 161–172. [PMC free article] [PubMed] [Google Scholar]
- Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41, 95–98. [Google Scholar]
- Hastings, M. (1987). The Korean War. New York: Simon and Schuster.
- Johansen, C. A., van den Hurk, A. F., Ritchie, S. A., Zborowski, P., Nisbet, D. J., Paru, R., Bockarie, M. J., Macdonald, J., Drew, A. C. & other authors (2000). Isolation of Japanese encephalitis virus from mosquitoes (Diptera: Culicidae) collected in the Western Province of Papua New Guinea, 1997–1998. Am J Trop Med Hyg 62, 631–638. [DOI] [PubMed] [Google Scholar]
- Kudo, M., Uraguchi, K., Matsuda, S. & Hashimoto, H. (1937). Experimental studies on encephalitis virus of 1935 epidemic in Tokyo with special reference to its serological difference from the virus of St. Louis encephalitis. J Immunol 32, 129–135. [Google Scholar]
- Lewis, L., Taylor, H. G., Sorem, M. B., Norcross, J. W. & Kindsvatter, V. H. (1947). Japanese B encephalitis. Arch Neurol Psychiatry 57, 430–463. [PubMed] [Google Scholar]
- Lincoln, A. F. & Sivertson, S. E. (1952). Acute phase of Japanese B encephalitis; two hundred and one cases in American soldiers, Korea, 1950. J Am Med Assoc 150, 268–273. [DOI] [PubMed] [Google Scholar]
- Nga, P. T., del Carmen Parquet, M., Cuong, V. D., Ma, S. P., Hasebe, F., Inoue, S., Makino, Y., Takagi, M., Nam, V. S. & Morita, K. (2004). Shift in Japanese encephalitis virus (JEV) genotype circulating in northern Vietnam: implications for frequent introductions of JEV from Southeast Asia to East Asia. J Gen Virol 85, 1625–1631. [DOI] [PubMed] [Google Scholar]
- Nitatpattana, N., Dubot-Peres, A., Gouilh, M. A., Souris, M., Barbazan, P., Yoksan, S., de Lamballerie, X. & Gonzalez, J. P. (2008). Change in Japanese encephalitis virus distribution, Thailand. Emerg Infect Dis 14, 1762–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken, G. E., Nelson, C. A., Chen, B. R., Diamond, M. S. & Fremont, D. H. (2006). Crystal structure of the West Nile virus envelope glycoprotein. J Virol 80, 11467–11474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre, V., Drouet, M. T. & Deubel, V. (1994). Identification of mosquito-borne flavivirus sequences using universal primers and reverse transcription/polymerase chain reaction. Res Virol 145, 93–104. [DOI] [PubMed] [Google Scholar]
- Rey, F. A., Heinz, F. X., Mandl, C., Kunz, C. & Harrison, S. C. (1995). The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375, 291–298. [DOI] [PubMed] [Google Scholar]
- Sabin, A. B., Schlesinger, R. W. & other authors (1947). Japanese B encephalitis in American soldiers in Korea. Am J Hyg 46, 356–375. [DOI] [PubMed] [Google Scholar]
- Shurtleff, A. C., Beasley, D. W., Chen, J. J., Ni, H., Suderman, M. T., Wang, H., Xu, R., Wang, E., Weaver, S. C. & other authors (2001). Genetic variation in the 3′ non-coding region of dengue viruses. Virology 281, 75–87. [DOI] [PubMed] [Google Scholar]
- Solomon, T., Ni, H., Beasley, D. W., Ekkelenkamp, M., Cardosa, M. J. & Barrett, A. D. (2003). Origin and evolution of Japanese encephalitis virus in southeast Asia. J Virol 77, 3091–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchil, P. D. & Satchidanandam, V. (2001). Phylogenetic analysis of Japanese encephalitis virus: envelope gene based analysis reveals a fifth genotype, geographic clustering, and multiple introductions of the virus into the Indian subcontinent. Am J Trop Med Hyg 65, 242–251. [DOI] [PubMed] [Google Scholar]
- Williams, D. T., Wang, L. F., Daniels, P. W. & Mackenzie, J. S. (2000). Molecular characterization of the first Australian isolate of Japanese encephalitis virus, the FU strain. J Gen Virol 81, 2471–2480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.