Summary
SpoIIIE and FtsK are related proteins that translocate chromosomes across septa. Previous results suggested that SpoIIIE exports DNA and that translocation polarity is governed by the cell-specific regulation of its assembly, but that FtsK is a reversible motor for which translocation polarity is governed by its DNA substrate. Seeking to reconcile these conclusions, we used cell-specific GFP tagging to demonstrate that SpoIIIE assembles a complex only in the mother cell, from which DNA is exported, but that DNA translocation-defective SpoIIIE proteins assemble in both cells. Altering chromosome architecture by soj-spo0J and racA soj-spo0J mutations allowed wild-type SpoIIIE to assemble in the forespore and export the forespore chromosome. Combining LacI-CFP tagging of oriC with time-lapse microscopy, we demonstrate that the chromosome is exported from the forespore when oriC fails to be trapped in the forespore. Thus, the position of oriC after septation determines which cell will receive the chromosome and which will assemble SpoIIIE.
Introduction
The architectural simplicity of bacterial cells, which have a circular chromosome compacted within the cytoplasm, exposes the chromosome to a significant risk of damage each time the cell divides. This is because the circular chromosome frequently multimerizes by catenation (the interlinking of the two circles) and dimerization (resulting from an odd number of recombination events between chromosomes). Division traps these interlinked daughter chromosomes in the septum and if cell separation proceeds, one or both chromosomes could receive potentially lethal damage (reviewed by Sherratt, 2003; Wu, 2004; Thanbichler et al., 2005; Cozzarelli et al., 2006). To avert such damage, bacterial cells employ proteins that couple the final stages of cell division to the resolution of multimeric chromosomes, the conserved SpoIIIE/FtsK family of DNA translocases (reviewed by Errington et al., 2001). These proteins contain an N-terminal membrane anchor that directs the protein to the septum (Wang and Lutkenhaus, 1998; Yu et al., 1998; Bath et al., 2000; Wang et al., 2006) and which, in Bacillus subtilis, also participates in the final step of cytokinesis, membrane fission (Sharp and Pogliano, 1999; Liu et al., 2006). The membrane anchor is connected by a variable ‘linker’ domain of unknown function to a C-terminal domain that translocates along DNA in an ATP-dependent manner (Bath et al., 2000; Aussel et al., 2002). DNA translocation is used to align the recombination sites at which multimerized chromosomes are resolved and to move chromosomes out of septa (Barre et al., 2000; Corre and Louarn, 2002; Lau et al., 2003; Yates et al., 2006), likely through an oligomeric channel comprised of the membrane domain (Errington et al., 2001). Remarkably, FtsK-like proteins interact with enzymes that resolve both catenated and dimeric chromosomes, recruiting Topoisomerase IV to the septum to mediate decatenation (Espeli et al., 2003; Wang et al., 2006) and stimulating the XerC/D recombinase that mediates resolution of dimeric chromosomes (Barre et al., 2000; Aussel et al., 2002). Coupling the activity of these enzymes to FtsK likely ensures that they act only on appropriate substrates (chromosomes trapped in septa), and that their activity is subject to precise temporal and spatial control.
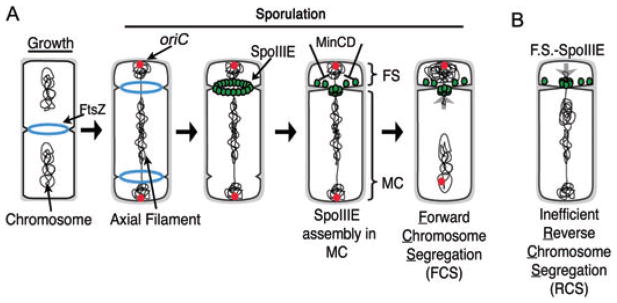
While FtsK-like proteins usually translocate a relatively small region of the chromosome, the B. subtilis SpoIIIE protein translocates 70% of the chromosome across the septum (~3 Mb) during B. subtilis sporulation (Wu and Errington, 1994; Wu et al., 1995). This is because during sporulation, a septum is formed at the extreme cell pole to generate a smaller cell (the future spore or forespore) containing only the origin-proximal 30% of the chromosome (Fig. 1A). The completion of chromosome segregation requires SpoIIIE, which first localizes as a ring at nascent division sites (Liu et al., 2006) and ultimately assembles a focus at the septal midpoint (Fig. 1A; Wu and Errington, 1997; Sharp and Pogliano, 1999). These foci likely represent active translocation complexes, as assembly defective SpoIIIE mutants demonstrate a correlation between the frequency of focus assembly and the amount of DNA moved into the forespore (Liu et al., 2006). After the completion of forespore chromosome segregation, SpoIIIE mediates separation of the forespore and mother cell membranes (Liu et al., 2006). It also moves around the forespore during engulfment and mediates separation of the mother cell cytoplasmic membrane and the outer forespore membrane, thereby releasing the forespore into the mother cell cytoplasm (Sharp and Pogliano, 1999; Liu et al., 2006).
Fig. 1.
Chromosome segregation and SpoIIIE assembly during growth and development.
A. During growth, FtsZ rings (Z-rings) assemble at midcell between separated chromosomes. During sporulation, Z-rings assemble at both poles and the chromosome assembles an axial filament with both origins (red spots) at the poles. During septation, SpoIIIE (green) assembles a ring then a focus. SpoIIIE appears to assemble preferentially in the mother cell from which it exports DNA. FS, forespore; MC, mother cell.
B. When expressed in the forespore, SpoIIIE slowly assembles and exports the trapped chromosome out of the forespore. We call this process reverse chromosome segregation (RCS), because it moves the partially segregated forespore chromosome back into the mother cell, thereby producing one anucleate daughter cell (the forespore) and one daughter cell (the mother cell) containing two chromosomes.
Both FtsK and SpoIIIE translocate DNA in a vectoral manner, showing net movement towards the chromosomal terminus or dif site from a more origin-proximal site at which they initially bind the chromosome. There are currently two contrasting models for how translocation polarity is determined by these proteins. The first is that the polarity of the DNA substrate determines translocation polarity. This model is supported both by genetic experiments (Corre and Louarn, 2002) and by single molecule experiments that demonstrate FtsK to behave as a reversible motor in which the polarity of translocation is determined by skewed octameric DNA sequences (FRS or KOPS) that are over-represented on the leading strands of DNA replication (Bigot et al., 2005; Levy et al., 2005; Pease et al., 2005) and recognized by the C-terminal γ-domain (Bigot et al., 2006; Ptacin et al., 2006; Sivanathan et al., 2006). These motifs cause FtsK to pause and reverse direction when encountered from the non-permissive direction (which in cells would correspond to movement of FtsK towards the origin), but have no effect on proteins approaching from the permissive direction (movement towards the terminus). Thus, for FtsK, DNA translocation polarity is dictated by the polarity of the DNA substrate itself.
The second model is supported by our studies of SpoIIIE during B. subtilis sporulation, which demonstrated that when SpoIIIE was expressed specifically in one or the other daughter cell (rather than before septation), DNA was exported from the cell in which SpoIIIE was expressed (Sharp and Pogliano, 2002a). This suggested that in living cells SpoIIIE acted as a DNA exporter, rather than a reversible DNA translocase. SpoIIIE also failed to efficiently assemble foci when expressed in the forespore, suggesting that the direction of DNA translocation was controlled by the cell-specific inhibition of SpoIIIE assembly (Sharp and Pogliano, 2002a). We subsequently provided evidence that the MinCD proteins that spatially regulate cell division were required for the forespore-specific repression of SpoIIIE assembly (Fig. 1A), and that in the absence of these proteins DNA was exported from the forespore in about 8% of all sporangia (Sharp and Pogliano, 2002b). It appeared that the asymmetric distribution of SpoIIIE across the septum trumped any potential impact of DNA substrate polarity in determining the direction of DNA translocation, as expression of SpoIIIE in the forespore reversed the direction of chromosome segregation. However, we were unable to achieve efficient export of the forespore chromosome even when SpoIIIE was expressed in the forespore in the absence of MinCD, suggesting the existence of an additional determinant of SpoIIIE translocation polarity, such as substrate polarity (Sharp and Pogliano, 2002b).
In an attempt to reconcile these two models, we developed new methods to investigate the mechanism by which SpoIIIE translocation polarity is established in B. subtilis cells. We first developed a method (cell-specific GFP tagging) that allows the specific localization of proteins made before septation (such as SpoIIIE) in the forespore and mother cell. We find that wild-type SpoIIIE assembles a focus only in the mother cell, supporting our previous conclusion that it exports DNA. However, translocation-defective SpoIIIE mutants assembled complexes in both cells, suggesting that DNA translocation is required to prevent assembly of a stable complex in the forespore, as might be expected if SpoIIIE assembly is dictated by the polarity of its DNA substrate. In keeping with this proposal, mutants that disrupt chromosome orientation (soj-spo0J) allow assembly of SpoIIIE in the forespore. We next developed an efficient time-lapse method for following chromosome segregation in living cells, and found that disrupting chromosome orientation also allows export of the forespore chromosome, specifically in sporangia in which the origin fails to be trapped in the forespore. These results suggest that in living cells, SpoIIIE behaves as a DNA exporter whose cell-specific assembly and hence the polarity of chromosome segregation are governed by the polarity of its DNA substrate.
Results
Cell-specific GFP tagging to probe the distribution of SpoIIIE across the sporulation septum
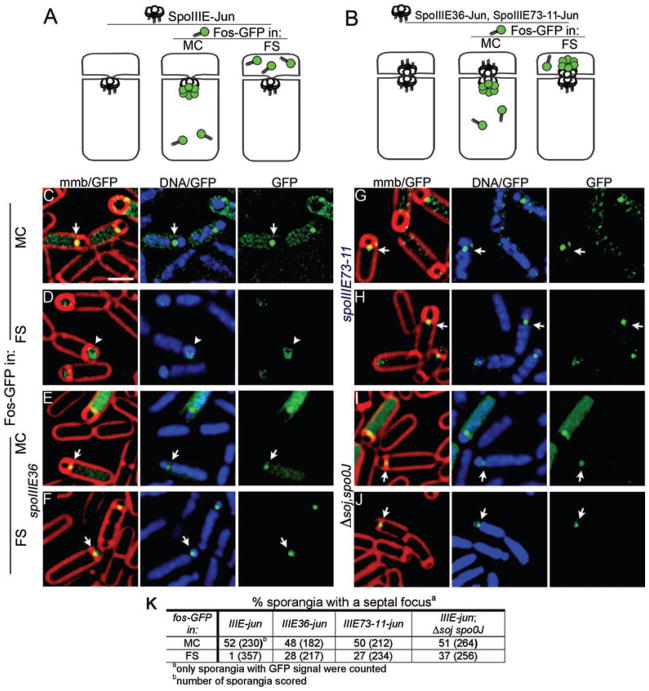
Our previous data suggested that in vivo SpoIIIE acts as a DNA exporter that assembles a stable translocation complex only in the mother cell (Sharp and Pogliano, 2002a), thereby exporting DNA from the mother cell into the forespore. This model was based on the observation that forespore-expressed SpoIIIE–GFP assembled inefficiently and moved DNA out of the forespore (thereby reversing chromosome segregation), whereas mother cell-expressed SpoIIIE–GFP efficiently assembled and moved the trapped chromosome into the forespore (Fig. 1A and B; Sharp and Pogliano, 2002a). However, SpoIIIE is normally expressed prior to polar septation, leaving open the possibility that it normally assembles on both faces of the sporulation septum and that the direction it moved DNA in the cell is controlled by another mechanism. We therefore developed a method (cell-specific GFP tagging) to allow SpoIIIE synthesized from its native promoter to be specifically tagged with GFP in either the mother cell or forespore. This method takes advantage of the high-affinity interaction (Kd = 8.7 nM; Kohler and Schepartz, 2001) between the leucine zipper domains of cFos (FosLZ) and cJun (JunLZ). We fused JunLZ to the C-terminus of SpoIIIE (SpoIIIE–JunLZ) and FosLZ to the amino terminus of GFP (FosLZ–GFP). The GFP fusion protein was then expressed specifically in the forespore or the mother cell using cell-specific promoters (PspoIIR and PspoIID respectively). If SpoIIIE assembles a translocation complex only in the mother cell, then a GFP focus should be observed when FosLZ–GFP is expressed in the mother cell but not when it is expressed in the forespore (Fig. 2A). However, if SpoIIIE assembles a translocation complex in both cells, GFP foci should be observed when FosLZ–GFP is expressed in either cell (Fig. 2B). It is important to note that we do not expect to detect foci in all sporangia in such experiments, due to the asynchrony of the sporulating culture, the transient assembly of the SpoIIIE focus and the continued synthesis of excess FosLZ–GFP, which will ultimately display bright cytoplasmic fluorescence and mask the SpoIIIE focus.
Fig. 2.
Cell-specific GFP tagging to investigate the distribution of proteins across the sporulation septum.
A. If SpoIIIE–JunLZ (white circles) assembles only in the mother cell, FosLZ–GFP (green) will form a septal focus when expressed in the mother cell, but not when expressed in the forespore.
B. If it assembles in both cells, FosLZ–GFP (green) will form a focus when expressed in either cell.
C and D. Cell-specific GFP tagging of wild-type SpoIIIE–JunLZ FosLZ–GFP forms foci when expressed in the mother cell (EBS488) (C; arrow), but not in the forespore (EBS494) (D; arrowhead).
E and F. Translocase-defective SpoIIIE36–JunLZ mutant forms foci when FosLZ–GFP is expressed in either the mother cell (EBS773) (E) or forespore (EBS775) (F).
G and H. Translocase-defective SpoIIIE73-11–JunLZ demonstrates that foci are assembled when FosLZ–GFP is expressed in either the mother cell (EBS490) (G) or forespore (EBS492) (H).
I and J. SpoIIIE–JunLZ in the soj-spo0J mutant. Foci are assembled when FosLZ–GFP is expressed in either the mother cell (EBS726) (I) or forespore (EBS729) (DAPI channel is overexposed to show DNA in the forespore) (J).
K. Frequency of focus formation.
Scale bar (C), 1 μm.
As expected, FosLZ–GFP displayed diffuse cytoplasmic fluorescence when expressed in cells without a JunLZ fusion protein (not shown). However, when FosLZ–GFP was expressed in the mother cell of SpoIIIE–JunLZ sporangia, a GFP focus was detected in 52% of sporangia at t3 (Fig. 2C, arrows, Fig. 2K). Foci were observed in early sporangia and before completion of DNA translocation, indicating that cell-specific GFP tagging rapidly labels the target protein (likely < 20 min after polar division, as chromosome translocation requires 30–40 min at 30°C). However, when FosLZ–GFP was expressed in the forespore of SpoIIIE–JunLZ cells, septal foci were detected in only 1% of sporangia, and most forespores exhibited diffuse cytoplasmic GFP fluorescence (Fig. 2D, arrowheads, Fig. 2K). Thus SpoIIIE–JunLZ efficiently assembles a complex that can be visualized using FosLZ–GFP tagging in the mother cell but not in the forespore (Fig. 2A). As discussed below, in some strains we can visualize SpoIIIE–JunLZ in the forespore, so the failure to detect wild-type SpoIIIE in the forespore likely reflects either the absence of a SpoIIIE complex in the forespore or a major conformational change that prevents the C-terminal JunLZ tag from interacting with FosLZ or prevents visualization of this complex (see Fig. S1). Thus, the assembly of the SpoIIIE motor domain is differentially regulated on the two faces of the septum such that in living cells SpoIIIE assembles a complex accessible to FosLZ–GFP only in the cell from which DNA is exported, the mother cell. These results suggest that the SpoIIIE motor domain fails to assemble a stable translocation complex in the forespore.
DNA translocation-defective SpoIIIE proteins assemble on both faces of the sporulation septum
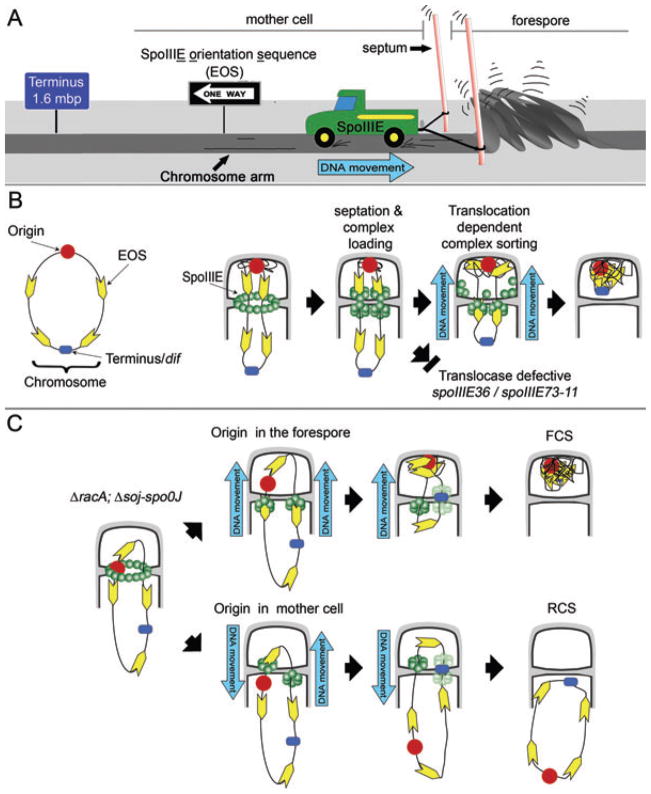
Single-molecule experiments indicate that the polarity with which FtsK moves along DNA is determined by FRS sequences that are highly skewed to one DNA strand and direct the FtsK towards dif sites by causing the motor to pause and reverse direction when it hits the sequence from the incorrect direction (Bigot et al., 2005; Levy et al., 2005). In contrast, our in vivo results indicate that SpoIIIE exports DNA and that the polarity with which SpoIIIE moves DNA in cells is regulated at the level of complex assembly, which is repressed in the forespore. These results could be reconciled if B. subtilis has similar polarity-determining DNA sequences that promote the disassembly of SpoIIIE complexes moving towards the origin (as would be the case for complexes exporting DNA from the forespore during reverse chromosome segregation or RCS), but have no activity on complexes moving towards the terminus or dif site (i.e. those exporting DNA from the mother cell during forward chromosome segregation or FCS; Fig. 3A and B). If this were the case, then translocation-defective SpoIIIE proteins would assemble similar complexes on both faces of the sporulation septum.
Fig. 3.
Models for the establishment of SpoIIIE polarity.
A. Cartoon showing that although SpoIIIE (truck) travels in the direction dictated by the SpoIIIE orientation sequences (EOS; one-way sign) the fact that it is tethered to the septal membrane (posts) demands that the chromosome (road) moves in the opposite direction.
B. We propose that either after or during division, SpoIIIE complexes (green) in each daughter cell (or nascent daughter cell) begin moving along DNA in their preferred direction relative to the septum (to promote DNA export). However, the forespore complex (moving towards the origin) encounters the skewed EOS (yellow arrows) in the non-permissive direction, resulting in the disassembly of the forespore complex. In contrast the mother cell complex (moving towards the terminus) encounters these sequences in the permissive direction, allowing continued DNA export into the forespore. It remains unclear how the final portion of the circular chromosome enters the forespore, because each chromosome arm is likely translocated through independent channels, which poses a topological barrier to moving the final chromosome loop across the septum. We speculate that moving this loop requires either the transient linearization of the forespore chromosome or the consolidation of the two channels into a single channel to accommodate the looped DNA.
C. This model for segregation polarity predicts that in ΔracA soj-spoOJ sporangia the EOS skew ensures that sporangia that trap oriC in the forespore after septation will always complete forespore chromosome segregation (top), while those that do not will always export the forespore chromosome by rotating the terminus into the forespore. It is unclear what happens when the terminus reaches the septum, as at this point, the EOS skew might allow SpoIIIE to assemble in both cells. We here represent this step with transparent SpoIIIE complexes to represent this uncertainty. Sporangia that trap the terminus in the forespore after polar septation will assemble two forespore complexes and rapidly export the chromosome from the forespore (not shown). However, the strains we here examine rarely trap the terminus in the absence of SpoIIIE DNA translocase activity (< 2% of all sporangia) because they do not completely invert chromosome orientation within the cell but rather more subtly shift the region trapped in forespore away from the origin (Sharpe and Errington, 1996; Lee et al., 2003; Wu and Errington, 2003).
To test this hypothesis, cell-specific GFP tagging experiments were performed on two mutant SpoIIIE proteins that are translocation-defective but localize to the septum, SpoIIIE36 (Wu et al., 1995; Wu and Errington, 1997) and SpoIIIE73-11 (Sharp and Pogliano, 1999). SpoIIIE36 contains three amino acid substitutions (Wu et al., 1995), but the reason for its translocation defect is unclear, while SpoIIIE73-11 contains an amino acid substitution in the Walker A ATP binding site that is predicted to abolish ATP hydrolysis but not DNA binding (G467S; Sharp and Pogliano, 1999). In contrast to the wild type, both proteins assembled foci detected when FosLZ–GFP was expressed in either the mother cell (Fig. 2E and G) or the forespore (Fig. 2F and H). Quantification of the fluorescence intensity revealed that forespore-expressed Fos–GFPLZ assembled foci that were at least as intense as mother cell-expressed Fos–GFPLZ (Fig. S2), suggesting that septation traps sufficient wild-type SpoIIIE in the forespore to be detected by cell-specific GFP tagging. Thus, although wild-type SpoIIIE assembles only in the mother cell, DNA translocation-defective proteins assemble on both faces of the sporulation septum (Fig. 2B).
SpoIIIE assembles on both faces of the septum in soj-spo0J sporangia
The above results suggested that SpoIIIE might move along DNA until it recognizes some polarity-determining feature of the chromosome that either dissociates complexes moving in the non-permissive direction (towards the origin) or prevents their visualization by cell-specific GFP tagging. If this were the case, then mutants that disrupt chromosome orientation and fail to trap the origin in the forespore should disrupt the cell specificity of SpoIIIE assembly, as some forespore complexes will be moving towards the terminus region and therefore remain stably assembled while some mother cell complexes will be moving towards the origin and therefore will not remain stably assembled (Fig. 3C). To test this prediction, we used cell-specific GFP tagging to investigate the polarity of SpoIIIE assembly in a strain deleted for the chromosome-partitioning proteins Soj-Spo0J. The absence of these proteins disrupts chromosome architecture, so that the origin region fails to be trapped in the forespore in ~20% of sporangia and a variable amount of DNA is trapped in the forespore (Sharpe and Errington, 1996; Lee et al., 2003; Wu and Errington, 2003). In contrast to the wild type, in soj-spo0J cells we detected foci when FosLZ–GFP was expressed in either the forespore (37%; Fig. 2J and K), or the mother cell (51%; Fig. 2I and K). The latter represents a > 30× increase in the frequency of SpoIIIE foci in the forespore than in wild type (37% versus 1%). Thus, when chromosome architecture is perturbed by the absence of Soj and Spo0J, SpoIIIE assembles in both cells.
Reversed chromosome segregation in sporangia defective in chromosome positioning
The above results suggest that mutants that disrupt chromosome architecture allow SpoIIIE to assemble in the forespore. If SpoIIIE exports DNA from cells in which it assembles, then such mutants should frequently move the trapped chromosome out of the forespore, thereby reversing chromosome segregation to produce mother cells with two chromosomes and anucleate forespores. To test this hypothesis, we scored the frequency of RCS in strains lacking one or both of the chromosome-partitioning systems known to operate during sporulation, Soj-Spo0J (discussed above) and RacA, which anchors the chromosomal origin of replication to the cell pole (Ben-Yehuda et al., 2003; Wu and Errington, 2003). In contrast to mutants lacking Soj-Spo0J, the absence of RacA mainly affects the amount of DNA trapped in the forespore while chromosome orientation is unchanged (Ben-Yehuda et al., 2003; Wu and Errington, 2003). The RacA and Soj-Spo0J systems likely act together to condense the origin-proximal portion of the chromosome and partition it to the forespore, as the combined absence of the two systems results in a more severe defect in chromosome orientation than predicted based on their individual defects (Wu and Errington, 2003).
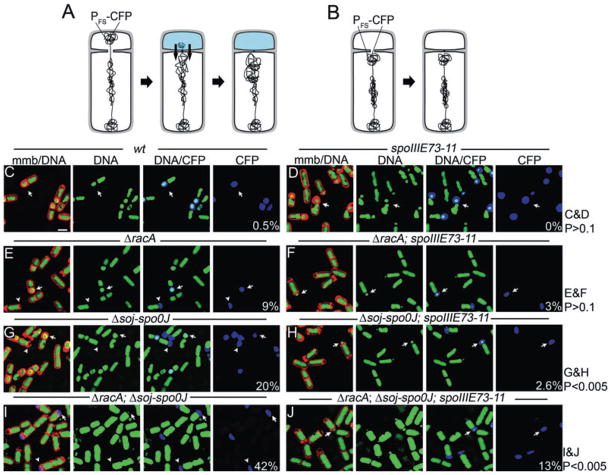
We used a liquid culture assay to determine if strains lacking either RacA or Soj-Spo0J or both systems produced anucleate forespores by SpoIIIE-mediated RCS. Cultures were grown in the presence of vital nucleic acid and membrane stains (CYTOX-green and FM 4-64 respectively) and visualized by fluorescence microscopy. A CFP reporter of forespore-specific gene expression was integrated into a chromosomal region that is normally trapped in the forespore after division (amyE), but which is a substantial distance from oriC (327 kb), so that some sporangia might trap the reporter but not oriC in the forespore, and therefore export the chromosome. Anucleate cells lacking CFP are either forespores that never contained the CFP reporter gene or minicells resulting from polar placement of vegetative septa (Fig. 4B); such cells were not scored in this assay. Small anucleate cells with CFP could be one of two classes. First, they could have initially contained DNA, activated forespore gene expression and undergone RCS (Fig. 4A). Second, they could have trapped the CFP reporter with just a small amount of chromosomal DNAthat could not be detected with CYTOX-green staining. To discriminate between these latter two classes, we intentionally overexposed the CYTOX-green images, so that smaller amounts of DNA could be visualized. We next compared the frequency of anucleate, CFP+ cells in strains with wild-type SpoIIIE and in the DNA translocation-defective SpoIIIE73-11 strain, to determine if SpoIIIE DNA translocation significantly increased the frequency of anucleate cells, as would only be the case if they arose by SpoIIIE-mediated RCS.
Fig. 4.
Visualization of reverse chromosome segregation during sporulation. Membranes and DNA were stained with FM 4-64 (red) and CYTOX-green (respectively) and cells harvested at t4. CFP (blue) is produced in the forespore from the spoIIQ promoter. Arrows indicate forespores containing a chromosome; arrowheads indicated anucleate forespores (small polar cells containing CFP but lacking DNA).
A. Septation traps the origin-proximal region of the chromosome and the CFP reporter of forespore gene expression (PFS-CFP) in the forespore. Small anucleate cells containing CFP are either anucleate forespores resulting from RCS or they contain a small and undetectable amount of DNA.
B. Small anucleate cells without CFP are either minicells or forespores that failed to trap the CFP reporter gene.
C–J. (C) Wild type (EBS527), (D) spoIIIE73-11 (EBS575), (E) ΔracA (EBS546), (F) ΔracA, spoIIIE73-11 (EBS550), (G) Δsoj-spo0J (EBS548), (H) Δsoj-spo0J, spoIIIE73-11 (EBS544), (I) ΔracA, Δsoj-spo0J (EBS561), (J) ΔracA, Δsoj-spo0J, spoIIIE73-11 (EBS563).
Scale bar (C), 1 μm. The frequency of apparently anucleate forespores for each strain is shown in the lower right-hand corner as a per cent of all sporangia (n > 200 for each strain). A χ2 test was performed to calculate the probability that the differences in the number of anucleate forespores in the designated data sets occurred by chance, yielding the P-values shown on the right.
The absence of the RacA chromosome-anchoring protein caused a variability in the amount of DNA trapped by polar septation (as expected; Ben-Yehuda et al., 2003; Wu and Errington, 2003), producing 9% apparently anucleate forespores in spoIIIE+ cells and 3% in the spoIIIE73-11 strain (Fig. 4E and F, arrowheads). This decrease was not significant (P-value > 0.1), suggesting that many or all of these apparently anucleate forespores actually contain an undetectable amount of chromosomal DNA (which must include the amyE region encoding the CFP reporter). We conclude that RacA-mediated polar chromosome anchoring does not significantly contribute to the polarity of SpoIIIE-mediated chromosome segregation. The absence of the Soj Spo0J chromosome-partitioning proteins caused the production of 20% apparently anucleate forespores in an otherwise wild-type strain, but only 2.6% in the spoIIIE73-11 strain (Fig. 4G and H, arrowheads). This significant difference (P < 0.005) indicates that most of these anucleate forespores result from SpoIIIE-mediated RCS. In a strain lacking both systems (racA soj-spo0J), 42% anucleate forespores were observed in an otherwise wild-type strain, but only 13% in the spoIIIE73-11 strain (Fig. 4I and J, arrowheads). This difference is significant (P < 0.005) and indicates that most of these anucleate forespores result from SpoIIIE-mediated RCS. Thus, disrupting chromosome positioning by inactivating the two known machineries that orient the chromosome, or simply inactivating the Soj-Spo0J system, causes SpoIIIE to move DNA out of the forespore.
Direct visualization of RCS by time-lapse fluorescence microscopy
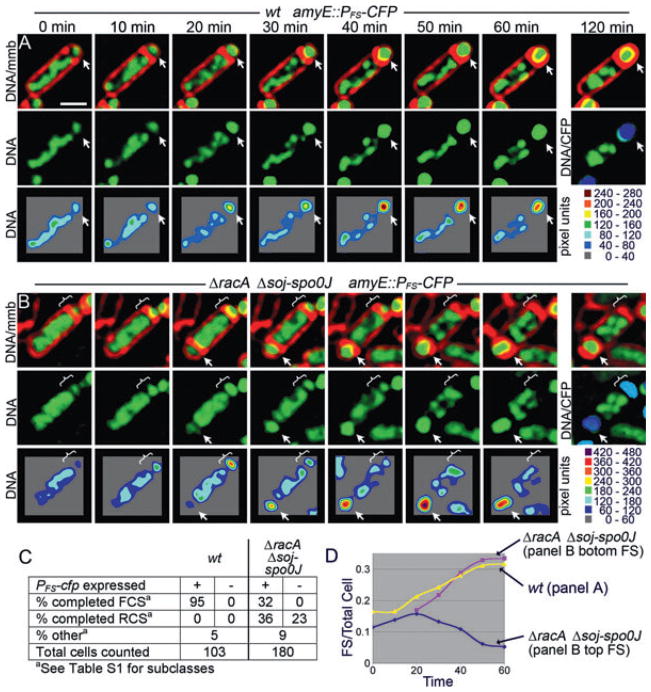
The above method probably underestimates RCS events as it requires that the CFP reporter of forespore positioning (amyE::PFS-cfp) initially be present in the forespore in mutants that have aberrant chromosomal architecture and in which many forespores fail to trap oriC (Fig. S3). This would produce many undetected RCS events. We therefore developed a method to visualize chromosome segregation using time-lapse fluorescence microscopy. Sporulating cells were applied to agarose pads in chambered slides 2 h after the initiation of sporulation at 30°C (t2; Experimental procedures), and the FM 4-64-stained membranes and CYTOX-green-stained DNA were visualized at 10 min intervals. Expression of CFP from PFS-cfp was visualized after the last time point (120 min), to reduce phototoxicity. This protocol allowed robust sporulation and 95% of wild-type sporangia completed forespore chromosome segregation 30–40 min after septation (Fig. 5A and C, Movies S1 and S2). Most sporangia also completed engulfment with little evidence of lysis after 2 h of microscopy. This represents a substantial improvement in sporulation efficiency during time-lapse microscopy (Pogliano et al., 2002), likely because our new method employs agarose pads containing culture supernatant (which contains factors necessary for the initiation of sporulation) and air reservoirs (O2 is necessary for sporulation).
Fig. 5.
Direct visualization of forward and reverse chromosome segregation using time-lapse fluorescence microscopy.
A and B. Membranes are stained with FM 4-64 (red), DNA with CYTOX-green (green). Sporulation was performed at 30°C, with a single CFP image (blue) collected after the experiment (t120 min). Bottom panels of each time-lapse consist of two-dimensional graphs of the raw pixel intensity data for the fluorescent DNA stain. (A) Wild type (EBS527) synthesizes a sporulation septum (arrow) and completes forespore chromosome segregation (FCS) within 40 min. See Movies S1 and S2. (B) A ΔracA Δsoj-spoOJ triple mutant (EBS561) switching from forward (between 0 and 20 min) to reverse chromosome segregation (between 20 and 50 min, brackets). The same sporangium synthesizes a second sporulation septum between 10 and 20 min (arrow) and completes chromosome segregation into this forespore within 40 min. See Movies S3 and S4.
C. Quantification of wild type (wt) and the ΔracA Δsoj-spoOJ triple mutant showing the number of cells that completed FCS or RCS with or without CFP in the forespore.
D. A graph depicting the relative increase in forespore DNA fluorescence (the ratio of forespore/total cell fluorescence) as a function of time for cells shown in (A) and (B).
Scale bar (A), 1 μm.
The racA soj-spo0J sporangia showed a variety of aberrant chromosome translocation events in experiments using the PFS-cfp reporter integrated at the origin-proximal gene amyE (Fig. 5C, Movies S3 and S4). First, only 32% of sporangia completed forespore chromosome segregation (Fig. 5B, arrows), compared with 95% of wild type (Fig. 5C). Second, movement of DNA out of the forespore (RCS) was directly observed in 36% of forespores that contained CFP at the end of the experiment (Fig. 5B and C). These cells had a variable amount of DNA in the forespore at the beginning of the experiment but were anucleate by the end of the experiment (Fig. 5B, bracket). Third, RCS was observed in 23% of small cells that failed to show subsequent CFP expression. These cells could be minicells or forespores in which the PFS-cfp reporter either was not trapped in the forespore or was translocated out of the forespore before being expressed. If we include these events, then in the absence of the Soj-Spo0J and RacA proteins, RCS occurs in 59% of all cells (36% + 23%) with small cellular compartments (minicells or forespores). We also clearly visualized sporangia (7%) in which the direction of DNA translocation reversed during the experiment, with net movement of DNA into the forespore immediately after septation followed by the export of the forespore chromosome to produce an anucleate forespore (Fig. 5B, bracket, Fig. 5D, Table S1). Thus SpoIIIE is able to reverse the direction it moves DNA in living cells in which chromosome architecture is perturbed.
Correlation between the location of oriC in individual cells and the polarity of chromosome segregation
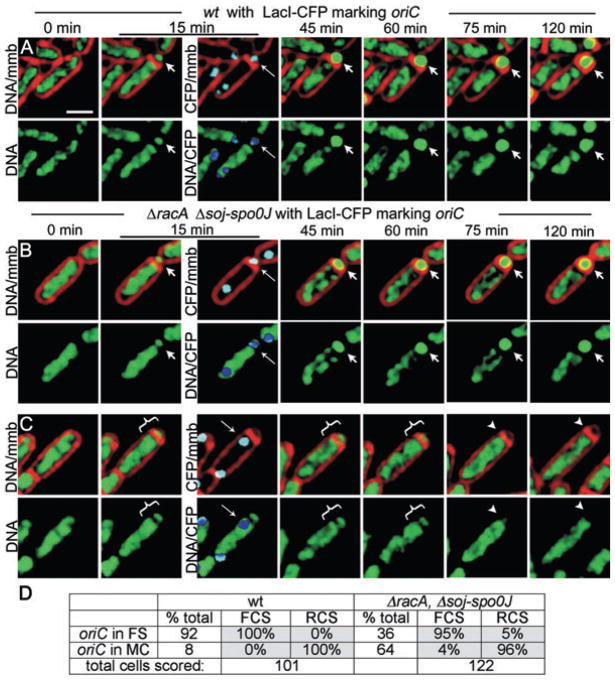
We were interested in determining if in individual racA soj-spo0J cells, the polarity of DNA segregation was directly correlated with the position of oriC after polar septation (as depicted in Fig. 3C). We therefore used a strain in which the lacO array was integrated at the soj-spo0J locus (Webb et al., 1997), which is 10 kb away from oriC, and expressed LacI-CFP early in sporulation (from the spoIIE promoter). The position of the lacO array was determined early in the time-course and the polarity of chromosome segregation monitored by time-lapse fluorescence microscopy. In wild-type cells, the lacO array was trapped in the forespore in 92% of all sporangia (n = 101), all of which completed forespore chromosome segregation (FCS; Fig. 6A, Movie S5). Surprisingly, we noted that 8% of wild-type sporangia failed to trap the lacO array in the forespore (Movie S6, Fig. 6D). We suspect that the lacO array subtly interferes with the normal function of the soj-spo0J locus at which it integrates (Webb et al., 1997), resulting in a partial defect in origin trapping. Importantly, all of the wild-type sporangia that failed to trap the origin in the forespore exported the forespore chromosome to produce anucleate forespores (Fig. 6D). There was also a close correlation between origin positioning and chromosome segregation polarity in the racA soj-spo0J mutant (Movie S7). For example, 64% of all sporangia failed to trap oriC in the forespore (n = 122) and 96% of these underwent RCS, with just 4% moving the chromosome into the forespore (Fig. 6C and D). In contrast, 95% of the sporangia that trapped oriC in the forespore completed forespore chromosome segregation, with just 5% RCS (Fig. 6B and D).
Fig. 6.
Localization of oriC during sporulation.
A–C. Chromosome segregation visualized using time-lapse microscopy of cells harbouring a lacO cassette near the origin of replication and producing LacI-CFP. Sporangia are stained with FM 4-64 (red), CYTOX-green (green). LacI-CFP (cyan) bound to the lacO cassette near the origin form foci (thin arrows) visualized 15 min after the start of the experiment. Overlapping LacI-CFP (cyan) and DNA (green) signals are pseudocoloured dark blue for maximal contrast. (A) Wild type (EBS897) synthesizes a sporulation septum trapping oriC in the forespore (thin arrow, cyan spot) and completes forespore chromosome segregation (arrows). See Movies S5 and S6. (B) ΔracA Δsoj-spoOJ sporangium (EBS887) traps oriC in the forespore (thin arrow, cyan spot) and completes forespore chromosome segregation. (C) ΔracA Δsoj-spoOJ sporangium traps oriC in the mother cell (thin arrow, cyan spot) and undergoes RCS. See Movie S7.
D. Quantification of the frequency of RCS and FCS in sporangia with oriC in the forespore (FS) or mother cell (MC).
Scale bar (A), 1 μm.
These results demonstrate a clear correlation between the position of oriC in individual cells and the polarity of SpoIIIE-mediated chromosome segregation. Indeed for at least 95% of the cells undergoing chromosome translocation, the forespore chromosome was partitioned to the cell that initially contained the origin-proximal lacO array (Fig. 6D). The few cells in which lacO position was not correlated with translocation polarity could be due to the ~10 kb distance between the lacO array and oriC, which could allow the two loci to be on opposite sites of the septum, as just 1 kb of extended B-form DNA is 3.3 μm in length (approximately the length of the entire sporangium). Clearly the position of oriC after polar septation is a critical determinant of chromosome segregation polarity.
oriC does not enter the forespore during RCS
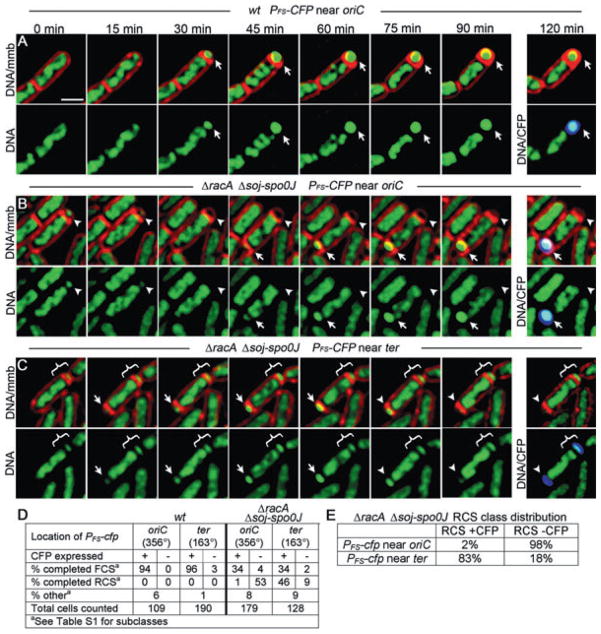
Although lacO/LacI-CFP tagging method used in the previous section effectively correlated origin positioning with the polarity of chromosome segregation, it has potential limitations. For example, an otherwise wild-type strain harbouring the lacO/LacI-CFP tagging construct produces 8% anucleate forespores, which are rarely produced by the wild-type strain PY79 (0.5% in the liquid culture assay, Fig. 4C; never detected during time-lapse microscopy, Table S1). Furthermore, binding of LacI-CFP to the lacO array might block chromosome segregation if SpoIIIE is incapable of removing LacI from DNA. Indeed, in the absence of inducer, LacI binds tightly enough to DNA to block replication (Possoz et al., 2006), so it seems reasonable to suspect that it could also block SpoIIIE-mediated DNA translocation. We therefore sought to confirm our findings using a less disruptive method to correlate chromosome orientation with the polarity of DNA translocation. Towards this aim, we integrated the CFP reporter of forespore gene expression (PFS-cfp) very close to either the origin (cotF, 48 kb from oriC) or the terminus (cotC, 37 kb from dif), and followed chromosome segregation polarity using time-lapse microscopy. If the direction that SpoIIIE moves DNA is correlated with the position of oriC after septation (in or out of the forespore), then forespore chromosome export should not occur in sporangia in which CFP is expressed from the origin-proximal cotF locus. However, if the direction in which SpoIIIE pumps the chromosome is not correlated with origin position, then FCS and RCS would occur equally in sporangia expressing CFP from origin- and terminus-proximal locations.
As expected, wild-type strains containing PFS-cfp integrated near oriC or ter were indistinguishable, with 94% and 96% of sporangia completing forespore chromosome segregation and expressing CFP respectively (Fig. 7A and D). For racA soj-spo0J strains, 34% completed forespore chromosome segregation and expressed CFP from both loci (Fig. 7B, arrow, Fig. 7D). However, in the strain with PFS-cfp integrated near oriC, 98% of the sporangia that underwent RCS showed no CFP fluorescence (Fig. 7B, arrowhead, Fig. 7E), indicating that the origin region had not been in the forespore. Conversely, for the racA soj-spo0J strain with PFS-cfp integrated near the terminus, 83% of sporangia that underwent RCS expressed CFP (Fig. 7C, arrowhead, Fig. 7E). Thus, most sporangia that undergo RCS fail to express CFP from origin-proximal locations, while most express CFP from terminus-proximal positions. These results, together with our lacO/LacI-CFP tagging experiments, clearly demonstrate that the direction that SpoIIIE moves DNA is closely correlated with the position of the origin, and that the chromosome is moved into the cell that initially contains the origin after septation.
Fig. 7.
Localizing origin and terminus regions using a reporter of forespore gene expression during time-lapse microscopy. Time-lapse microscopy was performed as in Fig. 4.
A. Wild-type sporangia (EBS850) with the CFP reporter of forespore expression located near the origin (356°) undergoing FCS.
B. ΔracA Δsoj-spoOJ sporangium with origin-proximal CFP reporter of forespore expression (strain EBS833) undergoing both RCS to produce anucleate forespores (arrowhead) and FCS to complete forespore chromosome segregation (arrow).
C. ΔracA Δsoj-spoOJ sporangium with reporter of forespore expression near the terminus (162°, EBS607) with forespore that does not appear to undergo significant translocation (bracket) and a second forespore undergoing FCS (15–60 min) and RCS (60–120 min) (arrow). See Movie S8.
D. Quantification of the frequency of RCS and FCS in sporangia showing CFP expressed from forespore promoter integrated near either oriC or ter.
E. Per cent ΔracA Δsoj-spoOJ sporangia that underwent RCS and expressed CFP from near either oriC or ter.
Scale bar (A), 1 μm.
Evidence for the assembly of independent SpoIIIE complexes for each chromosome arm
Interestingly, in the absence of SpoIIIE DNA translocase activity, only 2% of racA soj-spo0J sporangia trap ter in the forespore (Fig. S3; Wu and Errington, 2003), while in the presence of active SpoIIIE, 83% of the anucleate forespores contain CFP expressed from ter-proximal locations. This indicates that the ter region enters the forespore by SpoIIIE-mediated DNA translocation. However, examination of the time-lapse images for such sporangia indicated that the racA, soj-spo0J sporangia that ultimately expressed CFP from ter-proximal regions often showed only a small amount of net forward DNA translocation before abruptly exporting the chromosome (Movie S8). The model that translocation polarity is dictated by DNA sequences predicts that this would be observed when a single chromosome arm is trapped in the septum with both oriC and ter remaining in the mother cell. In this case, the chromosome arm would direct one SpoIIIE complex to assemble in the mother cell to move the ter-proximal region of the arm into the forespore and another to assemble in the forespore to move the ori-proximal region into the mother cell (Fig. 3C). This suggests that SpoIIIE might be capable of assembling translocation complexes of opposite polarity in the same cell, although further experiments are required to test this proposal more directly.
Discussion
Our results demonstrate that the position of oriC after septation dictates the polarity with which SpoIIIE moves DNA, because 95% of the cells that initially contain oriC ultimately receive the forespore chromosome. This suggests that the B. subtilis chromosome is decorated with skewed SpoIIIE orientation sequences (EOS) that are present on both chromosome arms and lead from oriC towards ter (Fig. 3B). Our results also support previous observations that SpoIIIE exports DNA and further indicate that blocking DNA translocation or disrupting chromosome architecture allows SpoIIIE to assemble in the forespore. To account for these results, we propose the following model for the establishment of DNA translocation polarity in intact cells (Fig. 3B). First, we propose that in living cells, the polarity with which SpoIIIE assembles relative to the septum is constrained such that it specifically exports DNA (i.e. that SpoIIIE hexamers assemble with a defined polarity relative to the septum). This proposal is consistent with our previous findings that SpoIIIE exports DNA from the cell in which it is expressed (Sharp and Pogliano, 2002a) and with cell-specific GFP tagging, which indicates that SpoIIIE assembles a complex accessible to GFP tagging in the mother cell but not the forespore. Second, we propose that the cell in which SpoIIIE assembles a stable and accessible complex is determined by the polarity of the DNA substrate, which normally directs SpoIIIE to move towards the terminus (Fig. 3). It therefore seems likely that SpoIIIE recognizes specific sequences that are similar to the FRS/KOPS recognized by FtsK, and that these dictate the polarity with which SpoIIIE moves DNA.
Our results seem most consistent with the model that DNA polarity is recognized after the initial assembly of complexes in both cells that begin exporting DNA until they encounter the EOS. These would have no effect on SpoIIIE complexes moving with the correct polarity (towards the terminus) but would promote disassembly of SpoIIIE complexes moving in the non-permissive direction (towards the origin). This is supported by our observation that translocation-defective SpoIIIE proteins assemble in both cells, suggesting that DNA translocation is required to prevent assembly SpoIIIE in the forespore. The scanning of the chromosome that determines the polarity of SpoIIIE assembly could occur either during or immediately after cell division, accounting for our failure to detect forespore foci in wild-type cells using cell-specific GFP tagging, which requires the onset of cell-specific gene expression.
DNA substrate-mediated translocation polarity would provide a mechanism to allow cells to cope with unanticipated consequences of aberrant division or chromosome segregation events. For example, it would allow the precise resolution of chromosomes that pass through the membrane several times, which likely occur in mutants defective in chromosome condensation (Niki et al., 1991; Britton and Grossman, 1999), and which could require the chromosome to be ‘snaked’ through the membrane. Interestingly, the septa of growing Escherichia coli cells are inherently polarized in a manner nearly identical to the B. subtilis sporulation septum, as both termini often lie in just one of two daughter cells immediately after cell division (Lau et al., 2003). It is therefore tempting to speculate that in living cells FtsK also acts as a DNA exporter whose polarity of assembly is determined by the location of the terminus after division.
Experimental procedures
Strains and growth conditions
Sporulation was induced by re-suspension (Sterlini and Mandelstam, 1969), except that the bacteria were grown in 20% LB prior to re-suspension, rather than CH medium. Strains (Table S2) are PY79 derivatives (Youngman et al., 1984) and were constructed using standard methods (Hoch, 1991). PCR used Pfu DNA polymerase (Stratagene) with oligonucleotides (Table S3) from Integrated DNA Technologies. New England Biolabs supplied restriction endonucleases and T4 DNA ligase, and cloning was performed in E. coli DH5α. Details of plasmid construction, including the sequence of all oligonucleotides are found in Supplementary material.
Microscopy
For Jun/Fos tagging experiments, cells were sporulated in the presence of 0.5 μg ml−1 FM 4-64 (Invitrogen), harvested by centrifugation, concentrated 10-fold in culture supernatant. Two microlitres of cells were applied to a slide, mixed with 1 μl of 0.6 μg ml−1 DAPI (Invitrogen) and immobilized with poly L-lysine-treated coverslips. Images were collected using an Applied Precision optical sectioning microscope (Liu et al., 2006) and deconvolved using softWoRx v3.3.6 (Applied Precision), assembling figures with Photoshop v7.0 (Adobe). For counting anucleate forespores, the sporulation salts contained 0.5 μg ml−1 FM 4-64 and the nucleotide stain 0.5 μM CYTOX-green (Invitrogen). This method allows forespore chromosomes to be visualized up to 6 h after the onset of sporulation without the loss of fluorescence previously observed using DAPI (Setlow et al., 1991). Five 0.2 μm optical sections were collected. Pixel data from all five optical sections were summed using softWoRx Z-projection and the summed data used for scoring anucleate forespores.
Time-lapse fluorescence microscopy
Time-lapse microscopy of sporulating cultures used agarose pads (Stewart et al., 2005) prepared as follows. Cells were re-suspended in 2 ml of sporulation salts containing 0.5 μg ml−1 FM 4-64 (and 0.5 μM CYTOX-green when nucleic acids were visualized), and incubated in a small glass tube in a 30°C roller. After 2 h, 1 ml of culture was centrifuged and the supernatant collected added to 2 ml of melted 3.5% agarose solution in A+B. The resulting 1.2% solution of molten agar/culture supernatant was supplemented with 0.5 μg ml−1 FM 4-64, added to the well of a culture slide (wells 18 mm diameter × 1.75 mm depth) and covered with a glass slide. After cooling, the cover glass was removed and two air pockets cut out of the agarose with a 15 ml tube, leaving a 3–5 mm agar bridge in the centre of the well. Seven microlitres of the remaining culture was spread over the agar, partially dried and sealed with cover glass. Slides were equilibrated in an environmental chamber at 30°C (Precision Control Weather Station) for 30 min prior to visualization. Single sections and short exposures times (< 0.2 s) were used to minimize phototoxicity.
Supplementary Material
Acknowledgments
We thank Eric Stewart for providing advice on the preparation of agarose pads for time-lapse microscopy and Jerod Ptacin and Marcelo Nollmann for many helpful discussions. This research was supported by the National Science Foundation (NSF0135955) and by the National Institute of Health (GM57045 to K.P. and Postdoctoral Fellowship GM65692-03 to E.C.B.).
Footnotes
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2958.2007.05992.x
(This link will take you to the article abstract).
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Aussel L, Barre FX, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- Barre FX, Aroyo M, Colloms SD, Helfrich A, Cornet F, Sherratt DJ. FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev. 2000;14:2976–2988. doi: 10.1101/gad.188700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath J, Wu LJ, Errington J, Wang JC. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science. 2000;290:995–997. doi: 10.1126/science.290.5493.995. [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science. 2003;299:532–536. doi: 10.1126/science.1079914. [DOI] [PubMed] [Google Scholar]
- Bigot S, Saleh OA, Lesterlin C, Pages C, El Karoui M, Dennis C, et al. KOPS: DNA motifs that control E. coli chromosome segregation by orienting the FtsK translocase. EMBO J. 2005;24:3770–3780. doi: 10.1038/sj.emboj.7600835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot S, Saleh OA, Cornet F, Allemand JF, Barre FX. Oriented loading of FtsK on KOPS. Nat Struct Mol Biol. 2006;13:1026–1028. doi: 10.1038/nsmb1159. [DOI] [PubMed] [Google Scholar]
- Britton RA, Grossman AD. Synthetic lethal phenotypes caused by mutations affecting chromosome partitioning in Bacillus subtilis. J Bacteriol. 1999;181:5860–5864. doi: 10.1128/jb.181.18.5860-5864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corre J, Louarn JM. Evidence from terminal recombination gradients that FtsK uses replichore polarity to control chromosome terminus positioning at division in Escherichia coli. J Bacteriol. 2002;184:3801–3807. doi: 10.1128/JB.184.14.3801-3807.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzarelli NR, Cost GJ, Nollmann M, Viard T, Stray JE. Giant proteins that move DNA: bullies of the genomic playground. Nat Rev Mol Cell Biol. 2006;7:580–588. doi: 10.1038/nrm1982. [DOI] [PubMed] [Google Scholar]
- Errington J, Bath J, Wu LJ. DNA transport in bacteria. Nat Rev Mol Cell Biol. 2001;2:538–545. doi: 10.1038/35080005. [DOI] [PubMed] [Google Scholar]
- Espeli O, Lee C, Marians KJ. A physical and functional interaction between Escherichia coli FtsK and topoisomerase IV. J Biol Chem. 2003;278:44639–44644. doi: 10.1074/jbc.M308926200. [DOI] [PubMed] [Google Scholar]
- Hoch JA. Genetic analysis in Bacillus subtilis. Methods Enzymol. 1991;204:305–320. doi: 10.1016/0076-6879(91)04015-g. [DOI] [PubMed] [Google Scholar]
- Kohler JJ, Schepartz A. Kinetic studies of Fos. Jun DNA complex formation: DNA binding prior to dimerization. Biochemistry. 2001;40:130–142. doi: 10.1021/bi001881p. [DOI] [PubMed] [Google Scholar]
- Lau IF, Filipe SR, Soballe B, Okstad OA, Barre FX, Sherratt DJ. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- Lee PS, Lin DC, Moriya S, Grossman AD. Effects of the chromosome partitioning protein Spo0J (ParB) on oriC positioning and replication initiation in Bacillus subtilis. J Bacteriol. 2003;185:1326–1337. doi: 10.1128/JB.185.4.1326-1337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy O, Ptacin JL, Pease PJ, Gore J, Eisen MB, Bustamante C, Cozzarelli NR. Identification of oligonucleotide sequences that direct the movement of the Escherichia coli FtsK translocase. Proc Natl Acad Sci USA. 2005;102:17618–17623. doi: 10.1073/pnas.0508932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu NJ, Dutton RJ, Pogliano K. Evidence that the SpoIIIE DNA translocase participates in membrane fusion during cytokinesis and engulfment. Mol Microbiol. 2006;59:1097–1113. doi: 10.1111/j.1365-2958.2005.05004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H, Jaffe A, Imamura R, Ogura T, Hiraga S. The new gene mukB codes for a 177 kD protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 1991;10:183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease PJ, Levy O, Cost GJ, Gore J, Ptacin JL, Sherratt D, et al. Sequence-directed DNA translocation by purified FtsK. Science. 2005;307:586–590. doi: 10.1126/science.1104885. [DOI] [PubMed] [Google Scholar]
- Pogliano J, Sharp MD, Pogliano K. Partitioning of chromosomal DNA during establishment of cellular asymmetry in Bacillus subtilis. J Bacteriol. 2002;184:1743–1749. doi: 10.1128/JB.184.4.1743-1749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possoz C, Filipe SR, Grainge I, Sherratt DJ. Tracking of controlled Escherichia coli replication fork stalling and restart at repressor-bound DNA in vivo. EMBO J. 2006;25:2596–2604. doi: 10.1038/sj.emboj.7601155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacin JL, Nollmann M, Bustamante C, Cozzarelli NR. Identification of the FtsK sequence-recognition domain. Nat Struct Mol Biol. 2006;13:1023–1025. doi: 10.1038/nsmb1157. [DOI] [PubMed] [Google Scholar]
- Setlow B, Magill N, Febbroriello P, Nakhimovsky L, Koppel DE, Setlow P. Condensation of the forespore nucleoid early in sporulation of Bacillus species. J Bacteriol. 1991;173:6270–6278. doi: 10.1128/jb.173.19.6270-6278.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc Natl Acad Sci USA. 1999;96:14553–14558. doi: 10.1073/pnas.96.25.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K. Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science. 2002a;295:137–139. doi: 10.1126/science.1066274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K. MinCD-dependent regulation of the polarity of SpoIIIE assembly and DNA transfer. EMBO J. 2002b;21:6267–6274. doi: 10.1093/emboj/cdf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe ME, Errington J. The Bacillus subtilis soj-spo0J locus is required for a centromere-like function involved in prespore chromosome partitioning. Mol Microbiol. 1996;21:501–509. doi: 10.1111/j.1365-2958.1996.tb02559.x. [DOI] [PubMed] [Google Scholar]
- Sherratt DJ. Bacterial chromosome dynamics. Science. 2003;301:780–785. doi: 10.1126/science.1084780. [DOI] [PubMed] [Google Scholar]
- Sivanathan V, Allen MD, de Bekker C, Baker R, Arciszewska LK, Freund SM, et al. The FtsK gamma domain directs oriented DNA translocation by interacting with KOPS. Nat Struct Mol Biol. 2006;13:965–972. doi: 10.1038/nsmb1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlini JM, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart EJ, Madden R, Paul G, Taddei F. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 2005;3:e45. doi: 10.1371/journal.pbio.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Wang SC, Shapiro L. The bacterial nucleoid: a highly organized and dynamic structure. J Cell Biochem. 2005;96:506–521. doi: 10.1002/jcb.20519. [DOI] [PubMed] [Google Scholar]
- Wang L, Lutkenhaus J. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol Microbiol. 1998;29:731–740. doi: 10.1046/j.1365-2958.1998.00958.x. [DOI] [PubMed] [Google Scholar]
- Wang SC, West L, Shapiro L. The bifunctional FtsK protein mediates chromosome partitioning and cell division in Caulobacter. J Bacteriol. 2006;188:1497–1508. doi: 10.1128/JB.188.4.1497-1508.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CD, Teleman A, Gordon S, Straight A, Belmont A, Lin DC, et al. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- Wu LJ. Structure and segregation of the bacterial nucleoid. Curr Opin Genet Dev. 2004;14:126–132. doi: 10.1016/j.gde.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Septal localization of the SpoIIIE chromosome partitioning protein in Bacillus subtilis. EMBO J. 1997;16:2161–2169. doi: 10.1093/emboj/16.8.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LJ, Errington J. RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol Microbiol. 2003;49:1463–1475. doi: 10.1046/j.1365-2958.2003.03643.x. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Lewis PJ, Allmansberger R, Hauser PM, Errington J. A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes Dev. 1995;9:1316–1326. doi: 10.1101/gad.9.11.1316. [DOI] [PubMed] [Google Scholar]
- Yates J, Zhekov I, Baker R, Eklund B, Sherratt DJ, Arciszewska LK. Dissection of a functional interaction between the DNA translocase, FtsK, and the XerD recombinase. Mol Microbiol. 2006;59:1754–1766. doi: 10.1111/j.1365-2958.2005.05033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P, Perkins JB, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- Yu XC, Tran AH, Sun Q, Margolin W. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J Bacteriol. 1998;180:1296–1304. doi: 10.1128/jb.180.5.1296-1304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.