Summary
During Bacillus subtilis sporulation, SpoIIIE is required for translocation of the trapped forespore chromosome across the sporulation septum, for compartmentalization of cell-specific gene expression, and for membrane fusion after engulfment. We isolated mutations within the SpoIIIE membrane domain that block localization and function. One mutant protein initially localizes normally and completes DNA translocation, but shows reduced membrane fusion after engulfment. Fluorescence recovery after photobleaching experiments demonstrate that in this mutant the sporulation septum remains open, allowing cytoplasmic contents to diffuse between daughter cells, suggesting that it blocks membrane fusion after cytokinesis as well as after engulfment. We propose that SpoIIIE catalyses these topologically opposite fusion events by assembling or disassembling a proteinaceous fusion pore. Mutants defective in SpoIIIE assembly also demonstrate that the ability of SpoIIIE to provide a diffusion barrier is directly proportional to its ability to assemble a focus at the septal midpoint during DNA translocation. Thus, SpoIIIE mediates compartmentalization by two distinct mechanisms: the SpoIIIE focus first provides a temporary diffusion barrier during DNA translocation, and then mediates the completion of membrane fusion after division to provide a permanent diffusion barrier. SpoIIIE-like proteins might therefore serve to couple the final step in cytokinesis, septal membrane fusion, to the completion of chromosome segregation.
Introduction
Membrane fusion is catalysed in eukaryotic cells and essential for such diverse processes as neuronal transmission, endocytosis and protein trafficking (Duman and Forte, 2003; Ungar and Hughson, 2003; Bonifacino and Glick, 2004; Sudhof, 2004). While most bacteria lack vesicular transport and intracellular organelles, it is likely that membrane fusion also plays an important role in bacterial cells, as at the final step of cell division, nascent daughter cells share a cytoplasmic membrane and are connected by a narrow cytoplasmic bridge. Severing this bridge requires membrane fusion at the site of this connection, to create separate membranes and allow daughter cell separation. It is unknown whether this final step of bacterial cytokinesis is catalysed, in part because of the lack of an assay for septal membrane fusion.
A unique and experimentally tractable membrane fusion event occurs during the sporulation pathway of Bacillus subtilis and its relatives (Sharp and Pogliano, 1999; Errington, 2003). Following engulfment of the smaller fore-spore by the mother cell (Fig. 1C–E), the leading edge of the engulfing membrane meets and fuses to release the forespore into the mother cell cytoplasm. This membrane rearrangement, which we will refer to as engulfment membrane fusion, requires the conserved SpoIIIE protein (Sharp and Pogliano, 1999), which has four transmembrane domains and a large cytoplasmic ATPase domain (Errington et al., 2001). The SpoIIIE membrane domain is sufficient for engulfment membrane fusion when expressed only in the cell whose membrane fuses, supporting a direct role of SpoIIIE in membrane fusion (Sharp and Pogliano, 2003). However, the multiple roles of SpoIIIE during sporulation complicates this conclusion, as SpoIIIE also acts during polar septation to translocate the trapped forespore chromosome across the sporulation septum (Wu and Errington, 1994; Fig. 1A); in the absence of SpoIIIE, only the origin-proximal third of the forespore chromosome enters the forespore (Wu and Errington, 1998). The purified ATPase domain has been demonstrated to track along DNA in an ATP-dependent manner (Bath et al., 2000). However, in cells, DNA translocation also requires the membrane domain (Wu and Errington, 1997; Sharp and Pogliano, 2002a; 2003), which localizes the protein to the septal midpoint (Bath et al., 2000). These results suggest that the cytoplasmic domain uses energy derived from ATP hydrolysis to move DNA through a channel comprised of the membrane domain (Errington et al., 2001).
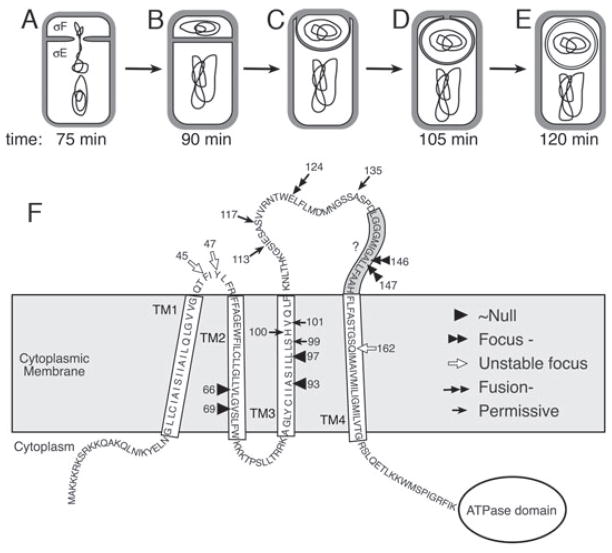
Fig. 1.
Role and predicted membrane topology of SpoIIIE.
A–E. (A) After polar septation, the forespore chromosome is bisected with the origin distal portion remaining in the mother cell. Cell-specific gene expression commences with the activation of σF in the forespore and σE in the mother cell. (B) The chromosome is translocated across the septum by SpoIIIE, and (C) engulfment commences, with the mother cell membrane migrating around the forespore, until the leading edge meets (D) and (E) fuses to release the forespore into the mother cell cytoplasm. The approximate time of sporulation at which 10% of the sporulating cells complete each step is indicated (timing data from Pogliano et al., 1999; Sharp and Pogliano, 1999).
F. Topology of SpoIIIE predicted using HMMTOP (Tusnady and Simon, 2001) and TMPRED (Hofmann and Stoffel, 1993). The shaded region indicated by the grey box is hydrophobic and has previously been proposed to be part of a long transmembrane domain 4 (Wu and Errington, 1997), although neither program predicted this region to lie within the membrane. One other region of significant ambiguity is the boundary between the first extracellular loop and TM2. Each epitope is inserted into the codon encoding the amino acid indicated by the arrow and numbers (relative to the initiation codon). Arrowheads, Class 1 (~Null); double arrowheads, Class 2 (Focus-defective); open arrows, Class 3 (Unstable focus); double arrows, Class 4 (Fusion-defective); and arrows, Class 5 (Permissive).
The role of SpoIIIE in DNA dynamics is in keeping with the roles of its homologues. For example, the Escherichia coli plasmid conjugation protein TrwB assembles a hexamer, and its cytoplasmic domain is thought to be an ATP-dependent pump that transfers DNA through a channel comprised of the TrwB membrane domain (Gomis-Ruth et al., 2001). A second SpoIIIE homologue, E. coli FtsK, is involved in cell division (Begg et al., 1995; Liu et al., 1998; Chen and Beckwith, 2001) and also aligns chromosomal dif sites and stimulates recombination during the resolution of chromosome dimers (Steiner et al., 1999). Like SpoIIIE, FtsK localizes to the division site where it assembles foci at the septal midpoint (Wang and Lutkenhaus, 1998; Yu et al., 1998a), and its soluble domain moves along DNA in an ATP-dependent manner (Aussel et al., 2002). Both FtsK and SpoIIIE are required to clear DNA trapped in vegetative septa (Sharpe and Errington, 1995; Yu et al., 1998b; Lau et al., 2003), thereby preventing DNA damage that would occur if division proceeded through an incompletely segregated chromosome. Thus, SpoIIIE homologues likely share the ability to assemble channels that translocate DNA across two cellular membranes during division or conjugation.
SpoIIIE plays a second crucial role at the stage of polar septation: immediately after polar septation, and before the completion of DNA translocation, cell-specific gene expression commences, with the activation of σF in the forespore and σE in the mother cell (Fig. 1A). However, in the absence of SpoIIIE, σF and σE activities are no longer compartmentalized (Wu and Errington, 1994; Pogliano et al., 1997), suggesting that the SpoIIIE focus might provide a diffusion barrier between the cells during DNA translocation (Wu and Errington, 1997; Hilbert et al., 2004). Interestingly, the requirement for SpoIIIE in compartmentalization can be bypassed by blocking the degradation of septal peptidoglycan during engulfment, suggesting that septal peptidoglycan provides a temporary seal between the two daughter cells (Hilbert et al., 2004). However, engulfment requires degradation of septal peptidoglycan (Abanes-De Mello et al., 2002), and when this occurs before the completion of DNA transfer, SpoIIIE appears to be required to prevent diffusion of small proteins across the open septum. Given the role of SpoIIIE in engulfment membrane fusion (Sharp and Pogliano, 1999), it is tempting to speculate that SpoIIIE-like proteins also mediate septal membrane fusion, which would result in the final separation of daughter cell membranes and cytoplasms.
It is unclear how a protein that assembles a DNA channel might also participate in membrane fusion. However, genetic and cell biological evidence suggests that SpoIIIE is involved in both processes, as DNA translocation and the entire ATPase domain are dispensable for engulfment membrane fusion (Sharp and Pogliano, 1999; 2003). Further, SpoIIIE initially localizes to the division site assembling a focus at the septal midpoint (Fig. 2A, iii), relocalizing to the pole before fusion (Fig. 2A, iv, v), and disassembling only after engulfment membrane fusion (Fig. 2A, vi; Wu and Errington, 1997; Sharp and Pogliano, 1999). We here report the localized mutagenesis of the SpoIIIE membrane domain and the isolation of mutants that block assembly and function. Importantly, we demonstrate that the ability of SpoIIIE to perform each of its functions critically depends on its ability to localize (to the septum for DNA translocation and compartmentalization, to the pole for engulfment membrane fusion). We also describe a mutant that has no effect on DNA translocation, but blocks engulfment membrane fusion. Surprisingly, this mutant also has a late compartmentalization defect, and we use fluorescence recovery after photobleaching (FRAP) to show that green fluorescent protein (GFP) diffuses from the mother cell into the forespore. We propose that this failure to seal the septum is due to incomplete septal membrane fusion and present a model for how SpoIIIE could catalyse membrane fusion after both cell division and engulfment.
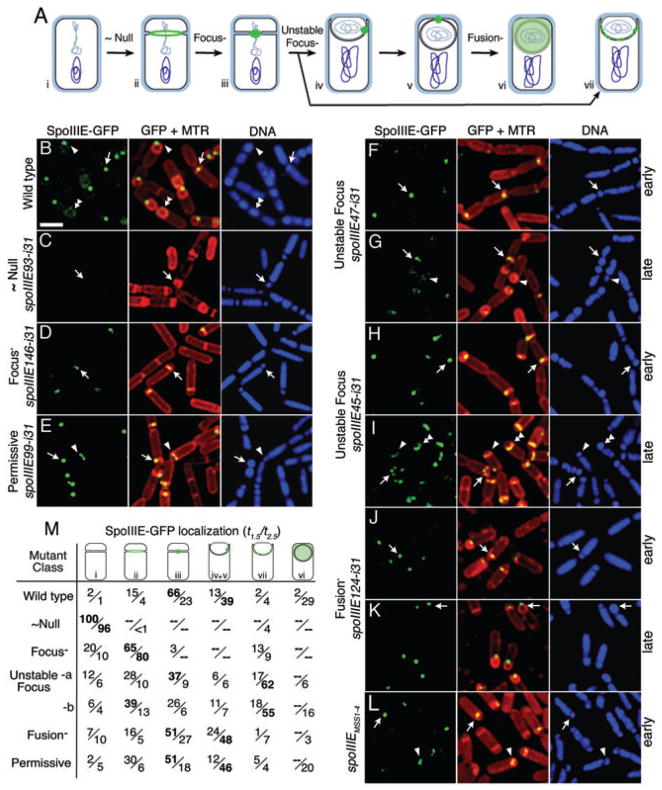
Fig. 2.
SpoIIIE-GFP assembly pathway and localization patterns of mutant proteins.
A. SpoIIIE-GFP (green) localization pathway: (i) chromosome (blue) is duplicated before division, (ii) during polar septation SpoIIIE assembles a ring, (iii) after division SpoIIIE assembles a focus and ~30% of the forespore chromosome (light blue) is trapped in the forespore, (iv and v) the SpoIIIE focus relocalizes to the cell pole during membrane migration and only after substantial DNA translocation into the forespore (Wu and Errington, 1997; Sharp and Pogliano, 1999), (vi) disassembling after fusion. Cell (vii) illustrates the localization phenotype of the Class 3 (Unstable focus) mutants. The stage at which each mutant class is blocked is indicated above the arrow.
B–L. Localization of SpoIIIE-GFP (green) with membranes stained with Mitotracker Red (MTR, red) and DNA with DAPI (blue) in live cells 2.5 h after onset of sporulation (hereafter t2.5), except that (F), (H), (J) and (L) are from t1.5 to show the early localization phenotype. (B) Wild-type SpoIIIE-GFP (KP6012). (C) Class 1 (~Null, KP6044) show no GFP (arrow). (D) Class 2 (Focus-, KP6068) mutants show a ring at the septum but few discrete foci. (E) Class 5 (Permissive, KP6095) are similar to wild type. (F and G) The Class 3 insertion spoIIIE47-i31 (Unstable focus, KP6092) is similar to wild type at t1.5 (F) but by t2.5 (G) shows diffuse fluorescence along the septum. (H and I) The Class 3 insertion spoIIIE45-i31 (Unstable focus, KP6064) is similar to wild type at t1.5 (H) but by t2.5 (I) shows either several discrete foci (arrow) or diffuse fluorescence (arrowheads) along the septum. (J and K) Class 4 (Fusion-, KP6111) mutant localizes normally early (J) in sporulation but remains as a focus even late (K) in sporulation. (L) The SpoIIIE membrane domain (spoIIIEMSS1-4, KP676) initially assembles normal foci (arrow) that disassemble during engulfment (arrowhead). Scale bar in (B) is 2 μm.
M. Quantification of SpoIIIE-GFP localization. Numbers show per cent sporangia with the indicated localization pattern (i though vii) at early (t1.5, top) and late (t2.5, bottom) times, with the predominant pattern at each time shown in bold. Unstable focus mutant ‘a’ is spoIIIE47-i31, ‘b’ is spoIIIE45-i31. An average of 203 sporangia were scored for each strain at each time point.
Results
Localization and DNA translocation by SpoIIIE mutants
We were interested in determining whether DNA translocation, cell compartmentalization and engulfment membrane fusion were genetically separable functions of SpoIIIE, and in identifying functional domains within the protein. To genetically dissect these roles, we used ISphoA/in (Manoil and Bailey, 1997) to perform epitope insertion mutagenesis targeted to the membrane domain of SpoIIIE-GFP. We isolated 16 epitope insertions at unique positions within this domain (Fig. 1F) and compared localization of the mutant proteins with wild type (Wu and Errington, 1997; Sharp and Pogliano, 1999). SpoIIIE-GFP initially localizes as a faint, transient ring at the invaginating sporulation septum (evident in the permissive mutant shown in Fig. 2E, arrowhead), and then forms a bright focus at the septal midpoint (Fig. 2B, arrow). After DNA translocation the focus moves around the forespore to the cell pole (Fig. 2B, arrowhead), disassembling after fusion with faint fluorescence around the forespore (Fig. 2B, double arrowhead).
The mutants fell into five classes. Class 1 (~Null) mutants were similar to spoIIIE null, producing few spores (≤ 115 ml−1 versus 4.7 × 108 ml−1 for wild type; Table 1), no detectable GFP and no detectable DNA translocation (Fig. 2C compared with Fig. 2B for wild type; Table 1). These Class 1 (~Null) mutants have insertions in the middle of predicted transmembrane segments (TM) 2 or 3 (Fig. 1F), likely disrupting the hydrophobic core of this domain and interfering with membrane insertion, folding or topology. The Class 5 (Permissive) mutants produced wild-type levels of spores, and had wild-type localization and DNA translocation (Fig. 2E; Table 1). These mutants have epitope insertions at the end of TM3 or on either side of the large extracellular loop (Fig. 1F), and appear to identify sites that tolerate the epitope insertion.
Table 1.
Summary of mutant phenotypes.
| Mutant class | spoIIIE allelea | DNA transferb | Membrane fusionc % (n)d | Spore titree | GFP compartmentalizationf % (n)d |
|---|---|---|---|---|---|
| Wild type | Wild type | ++ | 69 (278) | 4.7 × 108 | 100 (399) |
| Null | ΔspoIIIE | − | <1 (377) | 20 | 3 (375) |
| spoIIIE36 | −g | 5h | ND | 100 (266) | |
| spoIIIEMSS1-4 | −i | 13j | ND | 84 (490) | |
| 1. ~Null | spoIIIE66-i31 | <1 (287) | 60 | 3 (333) | |
| spoIIIE93-i31 | − | 1 (267) | 0 | 3 (346) | |
| spoIIIE69-i31 | 1 (404) | 115 | 2 (281) | ||
| spoIIIE97-i31 | 1 (356) | 30 | 5 (437) | ||
| 2. Focus- | spoIIIE147-i31 | − | 0 (331) | 100 | 7 (530) |
| spoIIIE146-i31 | 1 (211) | 20 | 1 (286) | ||
| 3. Unstable focus | spoIIIE47-i31 | 6 (514) | 2.0 × 107 | 18 (316) | |
| spoIIIE162-i31 | + | 14 (129) | 6.8 × 107 | 35 (387) | |
| spoIIIE45-i31 | 23 (195) | 1.1 × 108 | 55 (398) | ||
| 4. Fusion- | spoIIIE124-i31 | ++ | 19 (393) | 5.0 × 107 | 28 (570) |
| 5. Permissive | spoIIIE113-i31 | 76 (474) | 4.7 × 108 | 99 (454) | |
| spoIIIE135-i31 | 68 (394) | 6.0 × 108 | 99 (370) | ||
| spoIIIE117-i31 | ++ | 70 (719) | 3.8 × 108 | 98 (364) | |
| spoIIIE101-i31 | 65 (427) | 4.5 × 108 | 99 (420) | ||
| spoIIIE100-i31 | 62 (450) | 4.3 × 108 | 96 (322) | ||
| spoIIIE99-i31 | 62 (399) | 3.6 × 108 | 99 (423) |
Strains are listed in Table 2. For the i31 insertions, the allele number indicates the spoIIIE codon disrupted by the i31 insertion, numbered from the initiator methionine.
DNA translocation was assessed by DAPI staining samples from t2.5, and given three relative scores: −, no evident DNA translocation (similar to spoIIIE null);+, DNA translocation phenotype intermediate between the wild type and the null, with some sporangia appearing translocated and others not;++, DNA translocation similar to wild type in frequency and forespore chromosome size (see Fig. 4H).
Per cent sporangia that have completed membrane fusion at t3.5.
Total number of sporangia scored in assay.
Spore titre was determined by heat kill assay after 24 h of sporulation. Value represents number of heat viable spores per millilitre of culture.
Per cent of GFP expressing sporangia with soluble GFP compartmentalized to the mother cell at t3, using strains with the given spoIIIE allele plus cotE-gfp (Table 2).
Data from Wu and Errington (1994).
Data from Sharp and Pogliano (1999).
Data from Sharp and Pogliano (2003).
Data from Sharp and Pogliano (2002a).
Three mutant classes had specific defects at various stages of the SpoIIIE assembly pathway. First, Class 2 (Focus-) mutants assembled rings at the invaginating septum, but failed to assemble a focus at the septal midpoint (Fig. 2D, arrow), producing few spores (≤ 102 ml−1; Table 1) and showed no detectable DNA translocation. These Class 2 (Focus-) mutants had insertions within a hydrophobic region that is either within or before predicted TM4 (Fig. 1F), and likely produce proteins that insert into the membrane and localize to the division site. The mutant proteins might be specifically defective in focus assembly, or they might be somewhat unstable and reduce the level of SpoIIIE below some critical concentration required for focus assembly.
Second, Class 3 (Unstable focus) mutants produced spores at 4–20% of wild-type levels (Table 1). The proteins assembled foci at the septal midpoint somewhat less efficiently than wild type (Fig. 2F and H, arrows; 37–43% foci versus 79% in wild type; Fig. 2M), but 1 h later had fewer cells with single foci (Fig. 2G and I, arrows; 13–15% versus 62% in wild type; Fig. 2M). In insertion spoIIIE47-i31, SpoIIIE-GFP localized diffusely on the septum (Fig. 2G) while in spoIIIE45-i31, there were often multiple small foci along the septum (Fig. 2I, arrow). Some forespore chromosomes appeared at least partially translocated (Fig. 2G and I, arrow), while others appeared similar to chromosomes in the spoIIIE null (Fig. 2I, arrowhead, versus Fig. 2C), suggesting a decreased DNA translocation efficiency. Two Class 3 (Unstable focus) mutants had insertions within the first predicted extracellular loop, while a third had an insertion in the middle of TM4, at the end of a relatively hydrophilic patch of amino acids (Fig. 1F). These mutants appear somewhat defective in focus assembly and were unable to maintain the focus during engulfment, when the wild-type protein relocalizes to the cell pole. The Class 2 (Focus-) and Class 3 (Unstable focus) mutants together demonstrate that the ability of SpoIIIE to assemble a stable focus at the septal midpoint is closely correlated with the completion of DNA transfer.
Third, the Class 4 (Fusion-, see below) mutant produced spores at 10% of wild-type levels, and localized in a manner almost identical to wild type, assembling a GFP focus (Fig. 2J; Table 1) and relocalizing to the pole (Fig. 2K, arrow). DNA translocation appeared to be complete (Fig. 2K, arrow), but after engulfment the GFP focus remained at the pole (only 3% delocalized by t2.5, versus 29% in wild type; Fig. 2B, K and M). The Class 4 (Fusion-) mutant contained an epitope insertion in the middle of the large extracellular domain (Fig. 1F), and appeared defective in focus disassembly after engulfment.
Membrane fusion after engulfment
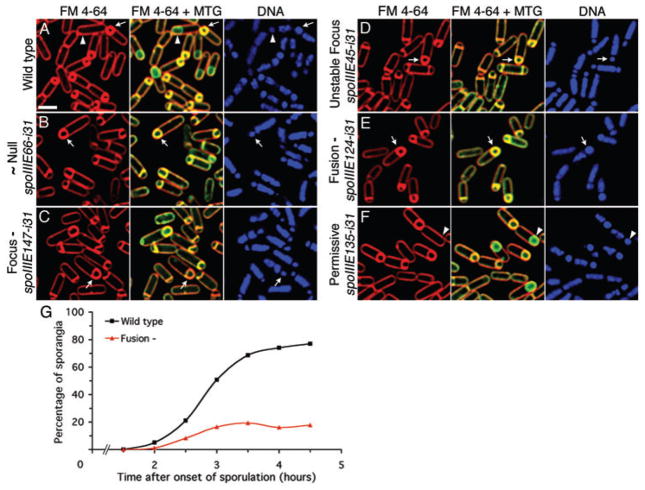
We next tested whether the mutants supported engulfment membrane fusion, using an in vivo membrane fusion assay that employs two membrane stains, the membrane-impermeable FM 4-64 and the membrane-permeable stain Mitotracker Green (Sharp and Pogliano, 1999). After engulfment membrane fusion, sporangia (here defined as a forespore and mother cell pair in any stage of engulfment) exclude FM 4-64 from the forespore membranes, resulting in a green forespore enclosed within the red mother cell (Fig. 3A, arrowhead), while unfused sporangia show FM 4-64 staining of all membranes (Fig. 3A, arrow). Samples were collected at t3.5 of sporulation, when 69% of wild-type sporangia had completed engulfment membrane fusion (Table 1). The Class 1 (~Null) and Class 2 (Focus-) mutants were similar to the spoIIIE null, with engulfment membrane fusion complete in less than 1% of sporangia (Fig. 3B and C, arrows; Table 1), while the Class 5 (Permissive) mutants fused at wild-type levels (Fig. 3F, arrow; Table 1). The Class 3 (Unstable focus) mutants showed 6–23% engulfment membrane fusion (Fig. 3D; Table 1), with a higher level of fusion occurring in spoIIIE45-i31, which also showed more stable SpoIIIE foci. Thus, the assembly of a SpoIIIE focus that relocalizes to the cell pole appears to be closely correlated with the completion of engulfment membrane fusion.
Fig. 3.
Assay for engulfment membrane fusion. Samples were collected at t3.5, and stained with DAPI (blue), FM 4-64 (red) and Mitotracker Green (MTG; green). After fusion, the membrane-impermeable stain FM 4-64 (red) is excluded from the forespore membranes, which are stained only with the membrane-permeable stain MTG (arrowheads). Before fusion, the forespore membranes stain with both FM 4-64 and MTG (arrows).
A–F. (A) Wild type (KP6012). (B) Class 1 (~Null, KP6042). (C) Class 2 (Focus-, KP6040). (D) The Class 3 insertion spoIIIE45-i31 (Unstable focus, KP6064). (E) Class 4 (Fusion-, KP6111). (F) Class 5 (Permissive, KP6066). Scale bar in (A) is 2 μm.
G. Fusion time-course of wild type (black squares) and the Class 4 (Fusion-) mutant (red triangles). Cell samples were collected every 30 min from t1.5 to t4.5. An average of 485 sporangia were scored for each strain at each time point.
The Class 4 (Fusion-) mutant was unable to support efficient engulfment membrane fusion. By t3.5, membrane fusion was complete in only 19% of sporangia compared with 69% of wild type (Table 1), although in many cases the mother cell membrane appeared to have migrated around the forespore (Fig. 3E, arrow). A time-course experiment demonstrated that engulfment membrane fusion started around t2 in both wild type and the Class 4 (Fusion-) mutant, reaching ~80% in wild type by t4.5 but more slowly in the mutant, which plateaus at ~20% by t3 (Fig. 3G). The Class 4 (Fusion-) mutant appears specifically defective in membrane fusion because it supported DNA translocation, assembled normally and relocalized to the cell pole but did not disassemble.
Cellular compartmentalization: a specific assay for diffusion across the septum
Immediately after synthesis of the sporulation septum and before the completion of DNA translocation, cell-specific gene expression commences, with the activation of σF in the forespore and σE in the mother cell (reviewed by Piggot and Losick, 2002; Errington, 2003). SpoIIIE is necessary for the compartmentalization of forespore and mother cell gene expression, and it has been suggested that this is because the SpoIIIE focus provides a diffusion barrier that prevents transcription factors from moving between cells during DNA translocation (Pogliano et al., 1997; Wu and Errington, 1997; Hilbert et al., 2004). If this is the case, then the ability of the SpoIIIE mutants to support compartmentalization should be directly correlated with their ability to assemble a focus. We therefore tested the ability of the mutants to prevent diffusion of GFP from the mother cell into the forespore, using a specific diffusion assay that does not depend on a failure to compartmentalize the activities of σF or σE. This assay employs a σE-dependent and therefore mother cell-expressed GFP reporter (cotE-GFP; Webb et al., 1995) located at the origin distal region of the chromosome, so that in the absence of DNA translocation, both copies remain in the mother cell (Fig. 4A). Thus, even in spoIIIE null mutants in which σE becomes active in both the forespore and the mother cell (Pogliano et al., 1997), GFP synthesis is restricted to the mother cell, so that forespore GFP fluorescence can occur only if GFP is able to diffuse across the septum.
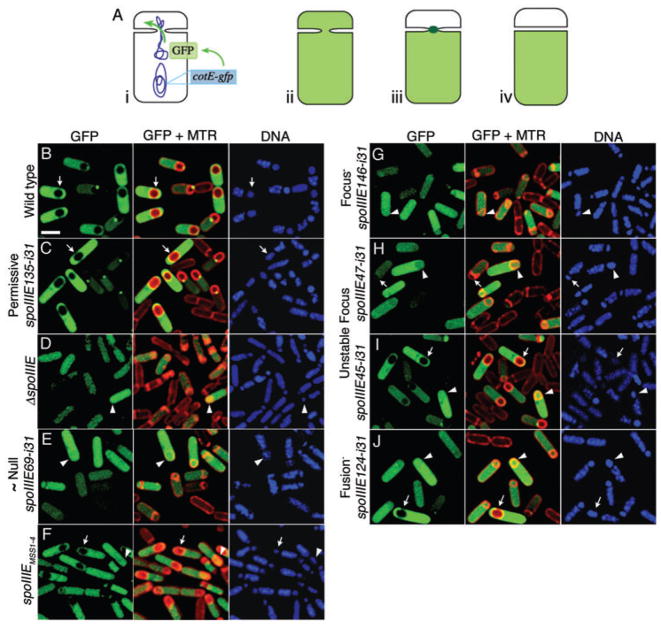
Fig. 4.
Cell compartmentalization assay.
A. Cell compartmentalization assay: (i) in DNA translocation-defective mutants, the mother cell-expressed cotE-gfp gene is trapped in the mother cell. Thus, GFP observed in the forespore must have diffused across the septum. The sporangium in (ii) shows non-compartmentalized GFP, that in (iii) shows compartmentalized GFP in the absence of septal membrane fusion but in the presence of a SpoIIIE complex at the septum (dark green), and that in (iv) shows compartmentalized GFP after septal membrane fusion.
B–J. Compartmentalization assay showing mother cell-expressed GFP (green), membranes [Mitotracker Red (MTR)] and DNA (DAPI, blue) from samples at t3. Arrows and arrowheads point to the forespores of sporangia with compartmentalized or non-compartmentalized GFP respectively. (B) The wild-type strain (KP6144) shows GFP confined to the mother cell. The cell-to-cell variability in GFP intensity is likely caused by the asynchronous initiation of sporulation and GFP synthesis. (C) Class 5 (Permissive) insertions such as spoIIIE135-i31 (KP6133) show GFP fluorescence only in the mother cell. (D) The spoIIIE null (KP725) and (E) Class 1 (~Null, KP6136) strains show GFP fluorescence in both the forespore and the mother cell. (F) The SpoIIIE membrane domain (spoIIIEMSS1-4, KP727) shows a mixture of compartmentalized (arrow) and non-compartmentalized (arrowhead) GFP. (G) Class 2 (Focus-) insertions such as spoIIIE146-i31 (KP6135) show non-compartmentalized GFP. (H and I) Class 3 (Unstable focus) insertions such as (H) spoIIIE47-i31 (KP6138) and (I) spoIIIE45-i31 (KP6131) show both compartmentalized (arrows) and non-compartmentalized (arrowheads) GFP. (J) Class 4 (Fusion-, KP6142) mutant. Scale bar in (B) is 2 μm.
In wild type and Class 5 (Permissive) sporangia, GFP produced in the mother cell remained in the mother cell (Fig. 4B and C, arrows respectively), with 100% compartmentalization (Table 1). In ΔspoIIIE, Class 1 (~Null) and Class 2 (Focus-) mutants, most sporangia showed mother cell expressed GFP in both the forespore and the mother cell (Fig. 4D, E and G, arrowheads; 3–7% compartmentalization; Table 1). In these cases, the GFP must have diffused from the mother cell into the forespore, as these mutants show little or no DNA translocation and both copies of cotE-gfp therefore remain in the mother cell. The Class 3 (Unstable focus) mutants exhibited a wider range of compartmentalization defects, ranging from 18% compartmentalization in spoIIIE47-i31 (Fig. 4H, arrow; Table 1), to 35% in spoIIIE162-i31 and 55% in spoIIIE45-i31 (Fig. 4I, arrow). There was a clear correlation between the ability of the mutant protein to assemble a stable focus (spoIIIE47-i31 < spoIIIE162-i31 < spoIIIE45-i31) and to support compartmentalization of GFP (spoIIIE47-i31 < spoIIIE162-i31 < spoIIIE45-i31), providing further evidence that the SpoIIIE focus provides a barrier to diffusion between the two cells.
Further support for this conclusion came from the examination of sporangia expressing only the membrane domain of SpoIIIE (SpoIIIEMSS1-4). The membrane domain of SpoIIIE is both necessary (Wu and Errington, 1997) and sufficient (Bath et al., 2000; Sharp and Pogliano, 2002a) for targeting the protein to the septum, but the focus it assembles appears unstable, as in engulfing sporangia a smear of GFP fluorescence is observed along the septum (Fig. 2L, arrowhead) rather than the discrete focus observed in sporangia that have not initiated engulfment (Fig. 2L, arrow). This instability during engulfment was also accompanied by a loss of diffusion barrier with time, as GFP was observed in the forespores of 8% of sporangia by t2 and 16% by t3 (Fig. 4F, arrowhead; Table 1). In this mutant, the increased GFP diffusion at later times of sporulation seems to be related to the instability of SpoIIIEMSS1-4 foci, which fall apart during engulfment (Fig. 2L, arrowhead).
Surprisingly, although the Class 4 (Fusion-) mutant assembled normally and supported DNA translocation, it showed GFP in both the forespore and mother cell in 72% of sporangia (Fig. 4J, arrowhead; Table 1). This compartmentalization defect was more pronounced late in sporulation, suggesting that the mutant protein initially assembled a functional complex at the septal midpoint, as is also indicated by its apparent ability to support DNA translocation. Thus, this mutant has a compartmentalization defect in addition to a defect in engulfment membrane fusion. This could result from a failure in septal membrane fusion, if after DNA translocation, the SpoIIIE focus relocalized to the cell pole without fusing the septal membranes, leaving membrane invaginations that retract during septal thinning. Indeed, the level of compartmentalization in this mutant (28% versus 100% in wild type) was almost identical to the per cent sporangia that complete engulfment membrane fusion (25% of wild type), suggesting that the mutation affects the ability of the protein to carry out both membrane fusion events to similar extents.
Use of FRAP to directly demonstrate diffusion across the septum
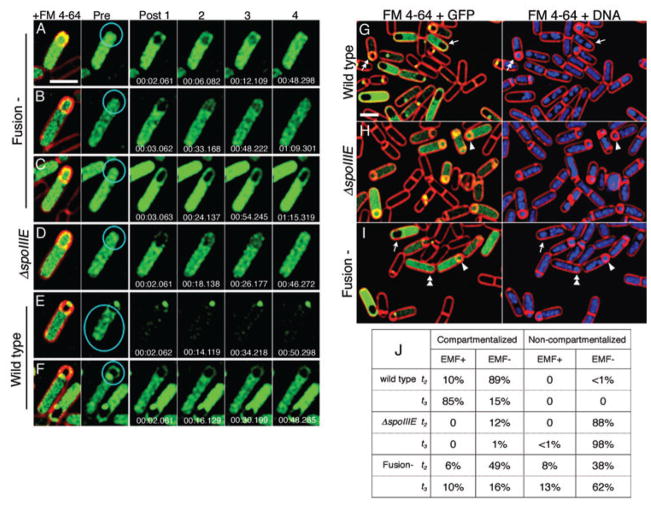
In the absence of DNA translocation, the assay described above is a specific assay for the diffusion of mother cell-produced GFP across the septum into the forespore. However, the Class 4 (Fusion-) mutant allows DNA translocation, which introduces the mother cell-specific reporter (cotE-gfp) into the forespore, where the misactivation of σE (Pogliano et al., 1997) could also produce GFP-containing forespores. To distinguish between these possibilities, we used FRAP to bleach GFP in the fore-spore and determine whether GFP from the mother cell could diffuse across the septum. We performed FRAP experiments on wild type, spoIIIE null and Class 4 (Fusion-) strains expressing cotE-gfp, choosing sporangia with moderate to bright GFP fluorescence for photobleaching. Most such sporangia have completed membrane migration and likely also DNA translocation. Control experiments with wild type demonstrated that photobleaching was irreversible and that bleaching the fore-spore did not allow GFP to diffuse across the septum (Fig. 5F). When nine GFP-containing forespores of the spoIIIE null mutant were bleached, seven allowed recovery in the forespore within 45–90 s, while two did not (Fig. 5D). It is unclear why GFP failed to diffuse across the septa in all spoIIIE null sporangia, but it is possible that the deposition of early coat proteins on the mother cell face of the septum during engulfment might create a diffusion barrier in some cells. When 18 GFP-containing forespores of the Class 4 (Fusion-) mutant were bleached, 13 allowed recovery of GFP fluorescence in the forespore cytoplasm within 45–90 s (Fig. 5A and B) while five did not (Fig. 5C). These results directly demonstrate that in the Class 4 (Fusion-) mutant, the sporulation septum remains open in most sporangia (72%), allowing diffusion of small proteins between the two cells.
Fig. 5.
FRAP assay for diffusion from the mother cell into the forespore and combined compartmentalization and engulfment membrane fusion assay.
A–F. For the FRAP assay, strains expressing cotE-gfp were harvested at t3 and stained with FM 4-64. The regions indicated by aqua circles were irreversibly bleached with a 0.05 s pulse of a 488 nm argon laser at 75% power, with subsequent images collected as quickly as possible. Times are shown as min.s.ms. (A–C) In the fusion-defective mutant (KP6142), the forespores of sporangia with GFP in both cells were bleached. Immediately after photobleaching (Post 1), GFP diffused back into the slender arms of mother cell cytoplasm around the forespore, while the forespore remained dark. (A and B) In 13/18 sporangia, GFP diffused into the forespore within 45–90 s, (C) while 5/18 failed to show recovery after 90 s. (D) In ΔspoIIIE (KP725), 7/9 sporangia recovered within 90 s. (E and F) Control experiments in wild type (KP6144), demonstrating that GFP fluorescence failed to recover when the entire mother cell was bleached (E), and that bleaching the forespore did not allow GFP to diffuse across the septum (F). In (B), (E) and (F), a focus of SpoIIIE-GFP is seen at the septum or cell pole. Scale bar in (A) is 2 μm.
G–I. Combined compartmentalization and engulfment membrane fusion assay. Cells were collected at t3 and the membranes stained with FM 4-64 (red) and DNA with DAPI (blue) to reveal fused (arrows, double arrowhead) or unfused sporangia (arrowhead, double arrow) with compartmentalized (arrows, double arrows) or non-compartmentalized CotE-GFP (arrowheads, double arrowheads). (G) Wild type (KP6144) shows compartmentalized GFP in cells before (double arrow) and after (arrow) engulfment membrane fusion. (H) ΔspoIIIE (KP725), shows no compartmentalization or engulfment membrane fusion (arrowhead). (I) Class 4 (Fusion-, KP6142) mutant. Scale bar in (G) is 2 μm.
J. Quantification of CotE-GFP compartmentalization and engulfment membrane fusion (EMF) in the combined assay at t2 and t3. Numbers indicate the per cent sporangia with the indicated phenotypes. EMF+: engulfment membrane fusion is complete, as indicated by exclusion of FM 4-64 from the forespore; EMF−: engulfment membrane fusion is not complete, as indicated FM 4-64 stained forespore membranes; Compartmentalized: GFP is restricted to the mother cell; Non-compartmentalized: GFP is in both the forespore and the mother cell. An average of 239 sporangia were scored for each strain at each time point.
Combined membrane fusion assay
The above results suggest the Class 4 (Fusion-) mutant blocks both septal membrane fusion and engulfment membrane fusion, raising the question of whether engulfment membrane fusion depends on the prior completion of septal fusion. To simultaneously visualize both events, we used the membrane-impermeable stain FM 4-64 in the GFP diffusion assay, which allowed us to assay engulfment membrane fusion by the exclusion of FM 4-64 from the forespore membrane. In wild type, 100% of sporangia expressing GFP are compartmentalized by t3 and 85% of these have completed engulfment membrane fusion (Fig. 5G, arrow, Fig. 5J). In the spoIIIE null, only 1% of sporangia show compartmentalized GFP by t3 and < 1% complete engulfment membrane fusion (Fig. 5H, arrowhead, Fig. 5J). In the Class 4 (Fusion-) mutant, approximately half the sporangia that complete engulfment membrane fusion had compartmentalized GFP, while the rest had non-compartmentalized GFP (Fig. 5I, arrow and double arrowhead, respectively; Fig. 5J). These results suggest that the ability of the fusion-defective mutant to complete engulfment membrane fusion does not depend on the completion of septal membrane fusion. The decreased compartmentalization seen in this mutant at later times (26% at t3 versus 55% at t2) suggests that compartmentalization might be lost after DNA translocation, when relocalization of the SpoIIIE focus to the pole could allow GFP to diffuse through the unfused septum.
Rescue of compartmentalization by septal thinning defects
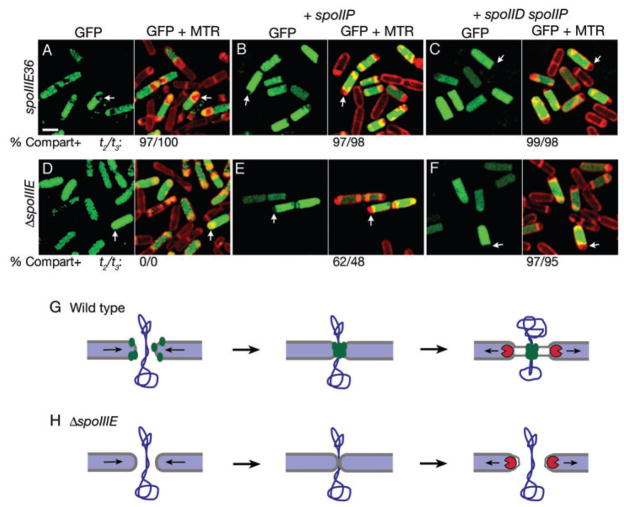
Like most bacterial septa, the sporulation septum initially contains peptidoglycan; however, during the phagocytosis-like process of engulfment, septal peptidoglycan is degraded by three mother cell-expressed proteins, SpoIID, SpoIIM and SpoIIP (Lopez-Diaz et al., 1986; Smith et al., 1993; Frandsen and Stragier, 1995; Abanes-De Mello et al., 2002). Hilbert et al. (2004) previously used a diffusion assay to determine that the absence of these engulfment proteins bypassed the requirement for SpoIIIE to prevent the diffusion of GFP across the septum. However, the GFP reporter used in this study was expressed only in sporangia that had inappropriately activated σF in the mother cell, thereby restricting the analysis to ~50% of the sporangia with a defect in the compartmentalization of σF activity. In order to determine whether blocking septal peptidoglycan degradation also rescued the compartmentalization defect in the remaining sporangia, we used the CotE-GFP-based diffusion assay, which does not depend on the miscompartmentalization of σF activity. We therefore introduced the spoIIP or spoIIP spoIID mutations into cotE-GFP strains with either the ΔspoIIIE, spoIIIE36, or our epitope insertion mutations that compromise the ability of SpoIIIE to assemble. In confirmation of previous findings (Hilbert et al., 2004), we noted that the spoIIP mutation rescued the GFP compartmentalization defect of the ΔspoIIIE strain, from 0% compartmentalization to 48% compartmentalization at t3 (Fig. 6D–E), and that the addition of both spoIIP and spoIID mutations allowed almost complete rescue, with 95% of sporangia showing normal compartmentalization at t3 (Fig. 6F). In spoIIIE36, a DNA translocation mutant that is able to assemble a focus at the septum (Wu and Errington, 1997), GFP remained compartmentalized in both the spoIIP (Fig. 6A and B) and spoIIP spoIID double mutants (Fig. 6C). The assembly-defective mutants were also rescued, and in each case the spoIIP mutation improved compartmentalization by about 50%, while the introduction of both the spoIIP and spoIID mutations rescued compartmentalization to almost wild-type levels (Fig. S1; Tables S1 and S2). These findings indicate that septal peptidoglycan is sufficient to prevent GFP from diffusing from the mother cell into the forespore in spoIIIE mutants that are either completely or partially defective in focus assembly. Thus, in such mutants, compartmentalization is likely to be lost during engulfment, when degradation of septal peptidoglycan allows the septal membranes to retract and cytoplasmic contents to diffuse between the two cells (Fig. 6H). It therefore seems likely that in wild-type sporangia, the assembly of a SpoIIIE focus at the septum helps to hold the septal membranes together during peptidoglycan hydrolysis, providing a temporary diffusion barrier during DNA translocation (Fig. 6G).
Fig. 6.
The compartmentalization defect of the spoIIIE null mutant is rescued by the absence of septal peptidoglycan degradation. Images were collected of strains expressing CotE-GFP (green) at t2 (not shown) or t3, and the membranes were stained with Mitotracker Red (MTR, red).
A–F. (A) The DNA translocation-defective mutant spoIIIE36 (KP723) shows normal compartmentalization of GFP (arrows). (B) spoIIIE36 spoIIP (KP6174). (C) spoIIIE36 spoIID spoIIP (KP6181). The numbers below indicate the per cent compartmentalization at t2 and t3. (D) The ΔspoIIIE strain (KP725) shows GFP in both cells (arrows). (E) The ΔspoIIIE spoIIP strain shows improved compartmentalization (from 0% to 48% at t3). (F) The ΔspoIIIE spoIIP spoIID strain shows almost normal compartmentalization (to 95% at t3). Scale bar in (A) is 2 μm.
G and H. Model for the relationship between septal thinning and compartmentalization. (G) In wild type, SpoIIIE (green) localizes to the leading edge of the invaginating septal membrane (grey line), ultimately assembling a SpoIIIE DNA translocation complex around the chromosome (blue). During hydrolysis of septal peptidoglycan (periwinkle) by the septal thinning machinery (red), the SpoIIIE complex remains assembled around the DNA, holding the septal membranes together and providing a diffusion barrier between the forespore and mother cell. (H) In the absence of SpoIIIE or in spoIIIE assembly mutants, the septal peptidoglycan is initially able to provide a seal between the forespore and mother cell (middle panel). However, during peptidoglycan hydrolysis, the septal membranes retract, allowing diffusion of GFP across the septum.
Discussion
Previous studies have suggested that the membrane domain of SpoIIIE is crucial for septal localization, focus assembly and membrane fusion (Wu and Errington, 1997; Sharp and Pogliano, 2003). The varied phenotypes of the mutations we have isolated that affect this domain confirm its critical role in these processes, as we have isolated mutants that block each step in the SpoIIIE localization pathway (focus assembly, relocalization to the cell pole and disassembly after engulfment) and each SpoIIIE function (DNA translocation, compartmentalization and membrane fusion). These mutants demonstrate a close correlation between SpoIIIE localization and its ability to carry out various functions. Specifically, our data demonstrate that the assembly of a stable focus at the septum is essential for DNA translocation and that this focus also provides a diffusion barrier critical for daughter cell compartmentalization. The two mutant proteins that localize to the division site but fail to assemble a focus contain epitope insertions adjacent to the site of a point mutation with a similar phenotype (Wu and Errington, 1997), suggesting that this most highly conserved part of the membrane domain (Errington et al., 2001) might be involved in either SpoIIIE–SpoIIIE or SpoIIIE–DNA (Ben-Yehuda et al., 2003) interactions required to assemble a multimeric DNA channel. Finally, SpoIIIE mutants that either fail to relocalize to the cell pole or fail to disassemble after reaching the pole show decreased membrane fusion after engulfment. These results demonstrate a close correlation between the ability of the SpoIIIE mutants to localize to a particular site and their ability to support the processes that occur at these locations.
We have also isolated a mutant that is normal in DNA translocation and relocalizes to the cell pole, but fails to support efficient membrane fusion after engulfment. This mutant, together with our previously isolated mutant in the ATPase domain that abolishes DNA translocation but not membrane fusion (Sharp and Pogliano, 1999), indicates that SpoIIIE plays genetically separable roles in engulfment membrane fusion and DNA translocation. The localization phenotype of the membrane fusion-defective mutant suggests that either disassembly of the SpoIIIE focus is required for engulfment membrane fusion or engulfment membrane fusion is required for focus disassembly. Surprisingly, FRAP analysis demonstrated that in this membrane fusion-defective mutant, GFP is able to diffuse from the mother cell into forespore. Thus, although the mutant protein assembles normally and supports DNA translocation, cytokinesis is incomplete and the septum remains open. The simplest explanation for these findings is that this mutant is defective in fusion of both the engulfing membrane and the septal membrane, as a defect in septal membrane fusion would leave the cytoplasms of nascent daughter cells connected. We therefore propose that in this mutant, DNA translocation but not septal fusion is complete, and when SpoIIIE moves to the cell pole to fuse the engulfing membrane, proteins are able to diffuse through the unfused septum (Fig. 7B).
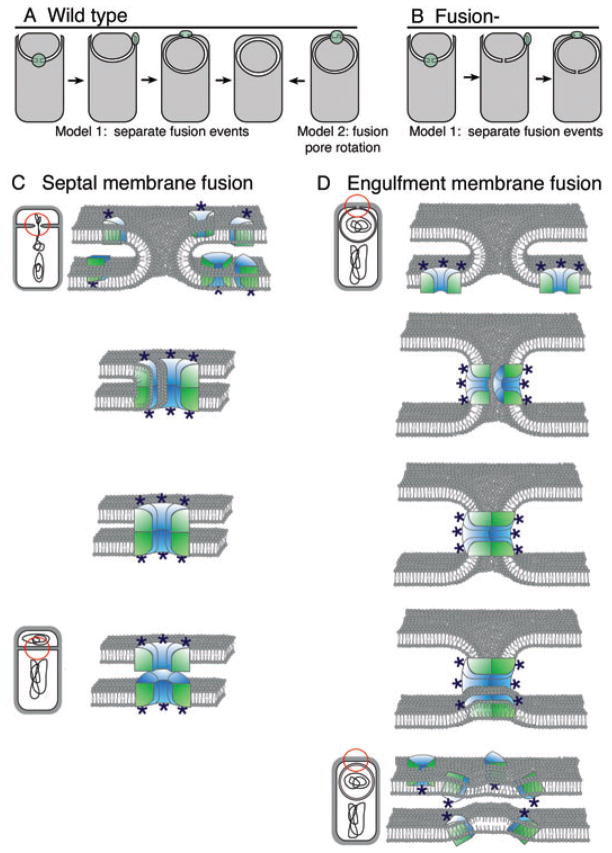
Fig. 7.
Model for SpoIIIE assembly and membrane fusion during cytokinesis and engulfment.
A. Two models for the spatial and temporal relationship between septal and engulfment membrane fusion. Model 1 proposes that septal membrane fusion and engulfment membrane fusion are spatially and temporally separate events, with septal fusion occurring after DNA translocation and before movement of a closed SpoIIIE channel (green) to the cell pole, the site of engulfment membrane fusion. Model 2 proposes that the septal fusion pore rotates to the cell pole, where the two fusion events occur in a coordinated manner.
B. Proposed membrane structure in the septal and engulfment membrane fusion-defective mutant, based on Model 1. We propose that in this mutant, the septal fusion pore is left open when the SpoIIIE channel relocalizes to the cell pole.
C and D. Model for membrane fusion during division and engulfment. The membrane domain of individual SpoIIIE subunits are shown within the lipid bilayer, with the cytoplasmic ATPase domain indicated by stars. (C) During cell division, we propose that SpoIIIE assembles a paired channel with subunits in both bilayers docking via extracellular domain interactions. Following DNA translocation (bottom), the channels undock to render the fusion event irreversible. We presume that the channel is in a closed conformation in the absence of DNA, to prevent the leakage of cytoplasmic contents. (D) During engulfment, we propose that two SpoIIIE channels within the mother cell membrane dock at the leading edge of the engulfing membrane, also via extracellular domain interactions. For clarity, the fore-spore membrane is omitted and the two channels are drawn on opposite sides of the fusion zone (top). Channel disassembly with cognate subunits remaining paired allows lipids to invade intersubunit spaces from both membranes, mediating membrane fusion (lower panel). The model is adapted from Almers (2001) and Peters et al. (2001).
It remains unclear exactly when septal membrane fusion occurs relative to engulfment membrane fusion, but we favour the hypothesis that the two membrane fusion events are both spatially and temporally separated, with septal membrane fusion occurring at the septal midpoint after DNA translocation and with engulfment membrane fusion occurring at the cell pole after membrane migration (Fig. 7A, model 1). It is also possible that the septal fusion pore rotates to the cell pole, where both fusion events might occur after membrane migration is complete (Fig. 7A, model 2). However, the latter model seems somewhat less likely, because it requires a dramatic alteration in the normal cell division pathway, such that after the sporulation septum is cleared of DNA, septal membrane fusion is delayed for the ~30 min required to complete membrane migration, rather than occurring as soon as the septum is free of DNA, as is likely the case during growth. There is also evidence that in certain situations engulfment membrane fusion can occur even when DNA remains trapped in the septum, a situation that would likely preclude septal membrane fusion. First, a mutation in the SpoIIIE ATP binding site abolishes DNA translocation but allows engulfment membrane fusion to occur in 46% of sporangia (Sharp and Pogliano, 1999). Second, the SpoIIIE membrane domain also supports engulfment membrane fusion but not DNA translocation, and elevated levels of engulfment membrane fusion occur with the expression of this domain in the mother cell (Sharp and Pogliano, 2003). However, the precise definition of the spatial and temporal relationship between septal and engulfment membrane fusion will require the development of new methods to visualize septal membrane fusion.
SpoIIIE-like proteins might be a common class of membrane fusion proteins used during bacterial cytokinesis, as most bacterial genomes contain clear SpoIIIE homologues (Errington et al., 2001). Indeed, many of these proteins, including E. coli FtsK, include a transmembrane domain similar to that of SpoIIIE, with four predicted trans-membrane domains, a glycine motif (GGGxxG) (Errington et al., 2001) and a phenylalanine-rich region after the first predicted transmembrane segment. The best-studied SpoIIIE homologue is E. coli FtsK, which assembles a focus at the septal midpoint during division (Wang and Lutkenhaus, 1998; Yu et al., 1998a). Similar to SpoIIIE, the membrane domain of FtsK is required for localization (Wang and Lutkenhaus, 1998; Yu et al., 1998a), while the cytoplasmic domain is a DNA translocase that aligns recombination sites during the resolution of chromosome dimers and termini (Yu et al., 1998b; Steiner et al., 1999; Barre et al., 2000; Aussel et al., 2002). It is possible that FtsK also participates in septal membrane fusion, as the FtsK membrane domain is required for cell division and certain ftsK mutations within the membrane domain have the late division phenotype expected for septal fusion-defective mutants (Begg et al., 1995), with chains of cells connected by membrane aggregates that likely contain peptidoglycan as well as outer and inner membranes (Diez et al., 1997). It is possible that these structures help separate the cytoplasmic contents of the daughter cells, perhaps explaining the cytoplasmic compartmentalization observed in FRAP experiments with this ftsK mutant (Goksor et al., 2003). Alternatively, compartmentalization could be maintained by septal peptidoglycan, as peptidoglycan is sufficient to mediate the initial compartmentalization of B. subtilis cells, until it is removed at the onset of engulfment. In contrast, peptidoglycan is never removed from E. coli septa, as this would cause lysis during cell separation. Thus, in bacteria with stable septal peptidoglycan, the initial compartmentalization of cytoplasmic contents could occur without septal membrane fusion, although the absence of membrane fusion might be expected to cause daughter cell lysis during cell separation.
Our results suggest that SpoIIIE participates in two membrane fusion events with opposite topology, as during septation, the cytoplasmic faces of the membrane approach one another, whereas during engulfment, the extracellular faces of the membrane approach one another, leaving opposite faces of SpoIIIE to mediate docking and/or fusion. One potential way to explain this apparent topological conundrum is based on the protein-aceous fusion pore model for eukaryotic membrane fusion. This model suggests that a pair of integral membrane channels, one in each of the fusing membranes, dock and form an intermembrane channel spanning both bilayers. The regulated lateral disassembly of each channel within the membrane while subunits in opposing membranes remain paired allows lipids to invade the channel, driving membrane fusion (Almers, 2001; Peters et al., 2001; Lindau and Alvarez de Toledo, 2003). We propose that the opposite occurs during cell division, with assembly of a paired SpoIIIE channel allowing separation of the septal membranes (Fig. 7C). Our proposed mechanism for membrane fusion during division requires a close and reversible association between the extracellular faces of the SpoIIIE membrane domain, the site of the epitope insertion in our fusion-defective mutant. The paired channel model provides a simple outline of the architecture of the SpoIIIE DNA channel, which must span two parallel bilayers to transfer DNA between daughter cells.
This view of SpoIIIE architecture during cytokinesis is in apparent contradiction with our earlier findings that the assembly of SpoIIIE during division is differentially regulated in the two cells (Sharp and Pogliano, 2002a). Specifically, we noted that while mother cell-expressed SpoIIIE assembled foci and supported wild-type levels of DNA translocation and membrane fusion, forespore-expressed SpoIIIE failed to efficiently assemble foci, and DNA was ultimately translocated out of the forespore. We suggest two possible ways to reconcile our model and cell-specific SpoIIIE assembly. First, it is possible that SpoIIIE assembles a heterologous paired channel comprised of SpoIIIE in the mother cell, and an unidentified channel-forming protein in the forespore. Second, it is possible that the membrane domain of SpoIIIE assembles a channel in the forespore, but the DNA-binding domain is held away from the channel to prevent its interaction with DNA. This proposal is in keeping with our findings that the two domains of SpoIIIE behave differently in the forespore: the cytoplasmic domain lines the septum similar to full-length SpoIIIE, while the membrane domain efficiently assembles foci at the septal midpoint (Sharp and Pogliano, 2002a). The long linker between the membrane and cytoplasmic DNA-binding domains might allow the DNA-binding domain to be held away from the DNA, perhaps via an interaction with proteins that inhibit the assembly of SpoIIIE in the forespore, such as MinCD (Sharp and Pogliano, 2002b).
After engulfment, we propose a mechanism more directly analogous to that of the proteinaceous fusion pore model (Almers, 2001; Peters et al., 2001; Lindau and Alvarez de Toledo, 2003), with two channels in the mother cell membrane pairing at the site of fusion (Fig. 7D). Importantly, this fusion event also requires interaction between the extracellular domains of SpoIIIE channels. However, here the channel subunits must disassemble within the plane of the membrane while remaining paired with their partner to allow hydrophilic regions of the lipids to invade the central hydrophilic core of the channel and the hydrophobic regions to invade the space between transmembrane domains. In possible agreement with the prediction that membrane fusion after engulfment requires channel disassembly, our membrane fusion-defective mutant maintains a stable focus at the cell pole, the site of this membrane fusion event.
The mechanism for the final separation of membranes after cell division is not yet clear for any organism. However, our model for membrane fusion during cytokinesis is the reverse of that proposed for membrane fusion mediated via a proteinaceous fusion pore (Almers, 2001; Peters et al., 2001; Lindau and Alvarez de Toledo, 2003), raising the possibility that proteins that mediate membrane fusion via a proteinaceous fusion pore or paired channel could also mediate membrane separation during cytokinesis by the de novo assembly of this structure at the division site. Thus, the list of candidates for proteins involved in the final stages of cytokinesis in eukaryotes would include the SNARE proteins, some of which have been shown to localize to the division site and to be required for the final steps of cytokinesis (Low et al., 2003). It will be interesting to determine whether eukaryotic cells have proteins that like SpoIIIE participate in two topologically opposite membrane fusion events, or whether the streamlined nature of bacterial genomes requires multifunctional proteins.
Experimental procedures
Strains and growth conditions
Bacillus subtilis strains (Table 2) were constructed by standard methods (Davidoff-Abelson and Dubnau, 1971) from wild-type strain PY79 (Youngman et al., 1984). Sporulation was induced by resuspension at 37°C and samples were collected at indicated times (tn) after the initiation of sporulation (Sterlini and Mandelstam, 1969). Heat-resistant spore titres were determined 24 h after the initiation of sporulation (Perez et al., 2000).
Table 2.
Strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| PY79 | Youngman et al. (1984) | |
| KP92 | spoIIIE36 | Wu and Errington (1994) |
| KP141 | ΔspoIIIE::spc | Pogliano et al. (1997) |
| KP514 | cotE-gfpΩkan, spoIIP::tet | This study |
| KP521 | cotE-gfpΩkan | Webb et al. (1995) |
| KP676 | spoIIIEMSS1-4-gfpΩspc | Sharp and Pogliano (2003) |
| KP723 | spoIIIE36, cotE-gfpΩkan | Sharp and Pogliano (1999) |
| KP725 | spoIIIE::spec, cotE-gfpΩkan | Sharp and Pogliano (1999) |
| KP727 | spoIIIEMSS1-4Ωspc, spoIIIE::spec, cotE-gfpΩkan | This study |
| KP6012 | amyE::spoIIIE-gfpΩcat, ΔspoIIIE::spc | This study |
| KP6040b | amyE::spoIIIE147-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc | This study |
| KP6042a | amyE::spoIIIE66-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc | This study |
| KP6044a | amyE::spoIIIE93-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc | This study |
| KP6064c | amyE::spoIIIE45-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc | This study |
| KP6065e | amyE::spoIIIE113-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc | This study |
| KP6066e | amyE::spoIIIE135-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc | This study |
| KP6067c | amyE::spoIIIE162-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc | This study |
| KP6068b | amyE::spoIIIE146-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc | This study |
| KP6090a | amyE::spoIIIE69-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc | This study |
| KP6091e | amyE::spoIIIE117-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc | This study |
| KP6092c | amyE::spoIIIE47-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc | This study |
| KP6093e | amyE::spoIIIE101-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc | This study |
| KP6094e | amyE::spoIIIE100-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc | This study |
| KP6095e | amyE::spoIIIE99-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc | This study |
| KP6111d | amyE::spoIIIE124-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc | This study |
| KP6112a | amyE::spoIIIE97-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc | This study |
| KP6128 | amyE::spoIIIE147-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc, cotE-gfpΩkan | This study |
| KP6129 | amyE::spoIIIE66-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc, cotE-gfpΩkan | This study |
| KP6130 | amyE::spoIIIE93-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc, cotE-gfpΩkan | This study |
| KP6131 | amyE::spoIIIE45-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc, cotE-gfpΩkan | This study |
| KP6132 | amyE::spoIIIE113-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc, cotE-gfpΩkan | This study |
| KP6133 | amyE::spoIIIE135-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc, cotE-gfpΩkan | This study |
| KP6134 | amyE::spoIIIE162-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc, cotE-gfpΩkan | This study |
| KP6135 | amyE::spoIIIE146-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc, cotE-gfpΩkan | This study |
| KP6136 | amyE::spoIIIE69-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc, cotE-gfpΩkan | This study |
| KP6137 | amyE::spoIIIE117-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc, cotE-gfpΩkan | This study |
| KP6138 | amyE::spoIIIE47-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc, cotE-gfpΩkan | This study |
| KP6139 | amyE::spoIIIE101-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc, cotE-gfpΩkan | This study |
| KP6140 | amyE::spoIIIE100-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc, cotE-gfpΩkan | This study |
| KP6141 | amyE::spoIIIE99-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc, cotE-gfpΩkan | This study |
| KP6142 | amyE::spoIIIE124-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc, cotE-gfpΩkan | This study |
| KP6143 | amyE::spoIIIE97-i31-‘spoIIIE-gfpΩcat, ΔspoIIIE::spc, cotE-gfpΩkan | This study |
| KP6144 | amyE::spoIIIE-gfpΩcat, ΔspoIIIE::spc, cotE-gfpΩkan | This study |
| KP6174 | spoIIIE36, cotE-gfpΩkan, spoIIP::tet | This study |
| KP6175 | ΔspoIIIE::spec, cotE-gfpΩkan, spoIIP::tet | This study |
| KP6180 | cotE-gfpΩkan, spoIIP::tet, spoIID::Tn917·mls | This study |
| KP6181 | spoIIIE36, cotE-gfpΩkan, spoIIP::tet, spoIID::Tn917·mls | This study |
| KP6182 | ΔspoIIIE::spec, cotE-gfpΩkan, spoIIP::tet, spoIID::Tn917·mls | This study |
| E. coli strains | ||
| CC118 | araD139 Δ[ara leu]7697 ΔlacX74 ΔphoA20 galE galK thi rpsE rpoB argE[am]recA1 | Calamia and Manoil (1990) |
| KP6015 |
araD139 Δ[ara leu]7697 ΔlacX74 ΔphoA20 galE galK thi rpsE rpoB argE[am] recA1 {pNJL1 (spoIIIE-gfp)} |
This study |
Class 1 (~Null).
Class 2 (Focus-).
Class 3 (Unstable focus).
Class 4 (Fusion-).
Class 5 (Permissive).
Construction of SpoIIIE-GFP plasmid pNJL1
The spoIIIE coding sequence and promoter was amplified by polymerase chain reaction (PCR) using the primers spoIIIE-5′(5′-GACTGAAGATCTGCTAGCCGAAACGTAAACCGATGATCATCC) and spoIIIE-3′ (5′-GACTGAACTAGTAGAAGAGAGCTCATCATATTTCTCTT), the product digested with BglII and SpeI and ligated into BamHI- and SpeI-digested pMDS12 (Sharp and Pogliano, 2002b). The resulting CmR plasmid encoded SpoIIIE-GFP, and could integrate at the chromosomal amyE locus by a marker replacement event. DNA sequence analysis verified that the spoIIIE region was wild type.
Mutagenesis and genetic manipulations
The TnphoA/in transposon was used to mutagenize CC118/pNJL1 (Calamia and Manoil, 1990) strain as described (Manoil and Bailey, 1997). Plasmid DNA was prepared from 12 pools and transformed into CC118, selecting for ISphoA/in element insertions on LB Amp (100 μg ml−1) Cm (40 μg ml−1) XP (40 μg ml−1) Suc (5%), without NaCl. Screening for alkaline phosphatase activity enriched for fusions to the SpoIIIE membrane domain, as APase is only active when secreted. Approximately 1000 colonies were purified and single-colony PCR was used to identify insertions within spoIIIE-gfp, using primers spoIIIE-5′ described above, and 5′-GTGCAGTAATATCGCCCTGAGCA. Seventy-six were sequenced to determine the insertion site, yielding 16 unique insertion sites. Plasmids were isolated, BamH1-digested and ligated to leave 31 codon insertions [encoding (S, P, T or A)DSYTQVASWTEPFPFSIQGDPRSDQET(V, A, D, E or G)XX; X residues determined by insertion site (Manoil and Bailey, 1997)]. The i31 insertions were given allele numbers that indicate the first codon (numbered from the initiator methionine) disrupted by the i31 insertion. For example, spoIIIE45-i31 encodes the first 44 amino acids of SpoIIIE, an amino acid generated at the junction of codon 45 and ISphoA, the amino acids encoded by the i31 insertion, a second amino acid generated at the junction of ISphoA and codon 43 and two duplicated amino acids (#44, 45) resulting from the staggered cuts made by Tn5 transposase, and the rest of SpoIIIE fused to GFP. The spoIIIE-i31 plasmids were introduced into PY79, where it integrated by a double recombination (gene replacement) event into the amyE locus. Chromosomal DNA was isolated and transformed into a spoIIIE null strain (KP141) for localization, DNA translocation, membrane fusion and spore titre analysis, and into KP725 for compartmentalization studies. These strains express only the mutagenized spoIIIE-gfp gene.
Microscopy and image analysis
Live cell images were collected at 25°C using an Applied Precision Spectris optical sectioning microscope system equipped with an Olympus IX70 microscope, an Olympus Plan Apo 100X oil immersion objective (NA 1.4), a Photometrics Cool SNAP HQ digital camera and Delta Vision standard fluorescence filters: 4′,6′-diamidino-2-phenylindole (DAPI) (excitation: UV 360/40 nm; emission: blue 457/60 nm), FITC (excitation: blue 490/20 nm; emission: green 528/38 nm) and RD-TR-PE (excitation: green 550/28 nm; emission: orange 617/73 nm). Using softWoRx software, eight optical sections, 0.15 μm apart, were collected for each sample and deconvolved using the constrained iterative deconvolution algorithm. Following deconvolution, the brightness and contrast of each fluorochrome were adjusted with softWoRx software, setting the area outside of cells to be background, and the colour balance was adjusted using Adobe Photoshop software.
Slides were prepared as previously reported (Sharp and Pogliano, 1999), except that for SpoIIIE-GFP localization and GFP diffusion assays, 500 μl of cells were collected at the desired time point and added to 5 μl each of Mitotracker Red FM (10 μg ml−1) and DAPI (70 μg ml−1) in sporulation salts, briefly pelleted and resuspended in 30 μl of the original resuspension growth medium with a final concentration of Mitotracker Red FM (0.1 μg ml−1) and DAPI (0.7 μg ml−1). The concentrated cells (3 μl) were immediately spotted onto microscope slides and firmly covered with poly L-lysine (0.1% solution, Sigma) treated coverslips. For the membrane fusion assay at the end of engulfment, 2 μl of concentrated cells (as above, without stains) were mixed with 1 μl of a stain mix containing FM 4-64 (5 μg ml−1), Mitotracker Green FM (10 μg ml−1) and DAPI (2 μg ml−1) in sporulation salts for a final concentration of FM 4-64 (1.7 μg ml−1), Mitotracker Green (3.3 μg ml−1) and DAPI (0.7 μg ml−1). For the combined compartmentalization and membrane fusion assay, 2 μl of unstained concentrated cells were mixed with 1 μl of stain mix containing FM 4-64 (1 μg ml−1) and DAPI (2 μg ml−1) for a final concentration of FM 4-64 (0.3 μg ml−1) and DAPI (0.7 μg ml−1). All stains are from Molecular Probes and stock solutions of Mitotracker Red FM (1 mg ml−1), Mitotracker Green FM (1 mg ml−1) and FM 4-64 (1 mg ml−1) were prepared in DMSO. DAPI stock (2 mg ml−1) was prepared in water.
Fluorescence recovery after photobleaching
Photobleaching experiments were performed using the same Applied Precision Spectris optical sectioning microscope system equipped with a 488 nm argon laser. Cells were collected at t3 of sporulation, concentrated and slides prepared as above. Concentrated cells (2 μl) were mixed with 1 μl of FM 4-64 (5 μg ml−1) for a final concentration of 1.7 μg ml−1. Before photobleaching, images were collected for FM 4-64 and GFP. Photobleaching was achieved using a 0.05 s pulse of a 488 nm argon laser at 75% power, and subsequent GFP images were collected as quickly as possible with exposure times limited to 2–3 s.
Supplementary Material
The following supplementary material is available for this article online:
Fig. S1. Engulfment mutants rescue the GFP diffusion defect in SpoIIIE assembly mutants. Images were collected at t3, with membranes stained with Mitotracker Red and DNA with DAPI (blue).
A. Focus-defective insertion spoIIIE147-i31 in spoIIP (KP6149).
B. Unstable focus insertion spoIIIE45-i31 in spoIIP (KP6151).
C. Unstable focus insertion spoIIIE162-i31 in spoIIP (KP6152).
D. Unstable focus insertion spoIIIE47-i31 in spoIIP (KP6153).
E. Focus-defective spoIIIE147-i31 in spoIIP spoIID (KP6176).
F. Unstable focus insertion spoIIIE45-i31 in spoIIP spoIID (KP6177).
G. Unstable focus insertion spoIIIE162-i31 in spoIIP spoIID (KP6178).
H. Unstable focus insertion spoIIIE47-i31 in spoIIP spoIID (KP6179).
For each of these mutant proteins at t3, the spoIIP mutation improved the ability of the protein to block GFP diffusion (by about 50%), while introducing both spoIIP and spoIID mutations almost completely rescued the diffusion defect (by about 97%; Table S1). Thus, septal peptidoglycan is sufficient to prevent GFP diffusion in spoIIIE mutants that are completely or partially defective in focus assembly. Scale bar in (A) is 2 μm.
Table S1. GFP diffusion in engulfment mutant backgrounds.
Table S2. Strains used in this study.
Acknowledgments
We thank Colin Manoil for providing strains, Shinobu Chiba for insightful comments on the manuscript, Rachel Larsen for her kind assistance with the FRAP experiments and the Advanced Bacterial Genetics Class of 2001 (Cold Spring Harbor Laboratory, NY), who isolated some of these epitope insertions. This research was supported by the National Science Foundation (NSF 0135955) and the National Institute of Health (GM57045).
References
- Abanes-De Mello A, Sun YL, Aung S, Pogliano K. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 2002;16:3253–3264. doi: 10.1101/gad.1039902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almers W. Fusion needs more than SNAREs. Nature. 2001;409:567–568. doi: 10.1038/35054637. [DOI] [PubMed] [Google Scholar]
- Aussel L, Barre FX, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- Barre FX, Aroyo M, Colloms SD, Helfrich A, Cornet F, Sherratt DJ. FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev. 2000;14:2976–2988. doi: 10.1101/gad.188700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath J, Wu LJ, Errington J, Wang JC. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell–prespore division septum. Science. 2000;290:995–997. doi: 10.1126/science.290.5493.995. [DOI] [PubMed] [Google Scholar]
- Begg KJ, Dewar SJ, Donachie WD. A new Escherichia coli cell division gene, ftsK. J Bacteriol. 1995;177:6211–6222. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R. Assembly of the SpoIIIE DNA translocase depends on chromosome trapping in Bacillus subtilis. Curr Biol. 2003;13:2196–2200. [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Calamia J, Manoil C. lac permease of Escherichia coli: topology and sequence elements promoting membrane insertion. Proc Natl Acad Sci USA. 1990;87:4937–4941. doi: 10.1073/pnas.87.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Beckwith J. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol Microbiol. 2001;42:395–413. doi: 10.1046/j.1365-2958.2001.02640.x. [DOI] [PubMed] [Google Scholar]
- Davidoff-Abelson R, Dubnau D. Fate of transforming DNA after uptake by competent Bacillus subtilis: failure of donor DNA to replicate in a recombination-deficient recipient. Proc Natl Acad Sci USA. 1971;68:1070–1074. doi: 10.1073/pnas.68.5.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez AA, Farewell A, Nannmark U, Nyström T. A mutation in the ftsK gene of Escherichia coli affects cell–cell separation, stationary-phase survival, stress adaptation, and expression of the gene encoding the stress protein UspA. J Bacteriol. 1997;179:5878–5883. doi: 10.1128/jb.179.18.5878-5883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman JG, Forte JG. What is the role of SNARE proteins in membrane fusion? Am J Physiol Cell Physiol. 2003;285:C237–C249. doi: 10.1152/ajpcell.00091.2003. [DOI] [PubMed] [Google Scholar]
- Errington J. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol. 2003;1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- Errington J, Bath J, Wu LJ. DNA transport in bacteria. Nat Rev Mol Cell Biol. 2001;2:538–545. doi: 10.1038/35080005. [DOI] [PubMed] [Google Scholar]
- Frandsen N, Stragier P. Identification and characterization of the Bacillus subtilis spoIIP locus. J Bacteriol. 1995;177:716–722. doi: 10.1128/jb.177.3.716-722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goksor M, Diez A, Enger J, Hanstorp D, Nystrom T. Analysis of molecular diffusion in ftsK cell-division mutants using laser surgery. EMBO Rep. 2003;4:867–871. doi: 10.1038/sj.embor.embor916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Ruth FX, Moncalian G, Perez-Luque R, Gonzalez A, Cabezon E, de la Cruz F, Coll M. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature. 2001;409:637–641. doi: 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- Hilbert DW, Chary VK, Piggot PJ. Contrasting effects of σE on compartmentalization of σF activity during sporulation of Bacillus subtilis. J Bacteriol. 2004;186:1983–1990. doi: 10.1128/JB.186.7.1983-1990.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase – a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Lau IF, Filipe SR, Soballe B, Okstad OA, Barre FX, Sherratt DJ. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- Lindau M, Alvarez de Toledo G. The fusion pore. Biochim Biophys Acta. 2003;1641:167–173. doi: 10.1016/s0167-4889(03)00085-5. [DOI] [PubMed] [Google Scholar]
- Liu G, Draper GC, Donachie WD. FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol Microbiol. 1998;29:893–903. doi: 10.1046/j.1365-2958.1998.00986.x. [DOI] [PubMed] [Google Scholar]
- Lopez-Diaz I, Clarke S, Mandelstam J. spoIID operon of Bacillus subtilis: cloning and sequence. J Gen Microbiol. 1986;132:341–354. doi: 10.1099/00221287-132-2-341. [DOI] [PubMed] [Google Scholar]
- Low SH, Li X, Miura M, Kudo N, Quinones B, Weimbs T. Syntaxin 2 and endobrevin are required for the terminal step of cytokinesis in mammalian cells. Dev Cell. 2003;4:753–759. doi: 10.1016/s1534-5807(03)00122-9. [DOI] [PubMed] [Google Scholar]
- Manoil C, Bailey J. A simple screen for permissive sites in proteins: analysis of Escherichia coli lac permease. J Mol Biol. 1997;267:250–263. doi: 10.1006/jmbi.1996.0881. [DOI] [PubMed] [Google Scholar]
- Perez AR, Abanes-De Mello A, Pogliano K. SpoIIB localizes to active sites of septal biogenesis and spatially regulates septal thinning during engulfment in Bacillus subtilis. J Bacteriol. 2000;182:1096–1108. doi: 10.1128/jb.182.4.1096-1108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters C, Bayer MJ, Buhler S, Andersen JS, Mann M, Mayer A. Trans-complex formation by proteo-lipid channels in the terminal phase of membrane fusion. Nature. 2001;409:581–588. doi: 10.1038/35054500. [DOI] [PubMed] [Google Scholar]
- Piggot PJ, Losick R. Sporulation genes and intercompartmental regulation. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and Its Relatives: From Genes to Cells. Washington, DC: American Society for Microbiology; 2002. pp. 483–518. [Google Scholar]
- Pogliano K, Hofmeister AE, Losick R. Disappearance of the σE transcription factor from the forespore and the SpoIIE phosphatase from the mother cell contributes to establishment of cell-specific gene expression during sporulation in Bacillus subtilis. J Bacteriol. 1997;179:3331–3341. doi: 10.1128/jb.179.10.3331-3341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J, Osborne N, Sharp MD, Abanes-De Mello A, Perez A, Sun YL, Pogliano K. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1149–1159. doi: 10.1046/j.1365-2958.1999.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe ME, Errington J. Postseptational chromosome partitioning in bacteria. Proc Natl Acad Sci USA. 1995;92:8630–8634. doi: 10.1073/pnas.92.19.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc Natl Acad Sci USA. 1999;96:14553–14558. doi: 10.1073/pnas.96.25.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K. Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science. 2002a;295:137–139. doi: 10.1126/science.1066274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K. MinCD-dependent regulation of the polarity of SpoIIIE assembly and DNA transfer. EMBO J. 2002b;21:6267–6274. doi: 10.1093/emboj/cdf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K. The membrane domain of SpoIIIE is required for membrane fusion during Bacillus subtilis sporulation. J Bacteriol. 2003;185:2005–2008. doi: 10.1128/JB.185.6.2005-2008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Bayer ME, Youngman P. Physical and functional characterization of the Bacillus subtilis spoIIM gene. J Bacteriol. 1993;175:3607–3617. doi: 10.1128/jb.175.11.3607-3617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner W, Liu G, Donachie WD, Kuempel P. The cytoplasmic domain of FtsK protein is required for resolution of chromosome dimers. Mol Microbiol. 1999;31:579–583. doi: 10.1046/j.1365-2958.1999.01198.x. [DOI] [PubMed] [Google Scholar]
- Sterlini JM, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. The HMMTOP trans-membrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- Ungar D, Hughson FM. SNARE protein structure and function. Annu Rev Cell Dev Biol. 2003;19:493–517. doi: 10.1146/annurev.cellbio.19.110701.155609. [DOI] [PubMed] [Google Scholar]
- Wang L, Lutkenhaus J. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol Microbiol. 1998;29:731–740. doi: 10.1046/j.1365-2958.1998.00958.x. [DOI] [PubMed] [Google Scholar]
- Webb CD, Decatur A, Teleman A, Losick R. Use of green fluorescent protein for visualization of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. J Bacteriol. 1995;177:5906–5911. doi: 10.1128/jb.177.20.5906-5911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Septal localization of the SpoIIIE chromosome partitioning protein in Bacillus subtilis. EMBO J. 1997;16:2161–2169. doi: 10.1093/emboj/16.8.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol Microbiol. 1998;27:777–786. doi: 10.1046/j.1365-2958.1998.00724.x. [DOI] [PubMed] [Google Scholar]
- Youngman P, Perkins JB, Losick R. A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol Gen Genet. 1984;195:424–433. doi: 10.1007/BF00341443. [DOI] [PubMed] [Google Scholar]
- Yu XC, Tran AH, Sun Q, Margolin W. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J Bacteriol. 1998a;180:1296–1304. doi: 10.1128/jb.180.5.1296-1304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XC, Weihe EK, Margolin W. Role of the C terminus of FtsK in Escherichia coli chromosome segregation. J Bacteriol. 1998b;180:6424–6428. doi: 10.1128/jb.180.23.6424-6428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following supplementary material is available for this article online:
Fig. S1. Engulfment mutants rescue the GFP diffusion defect in SpoIIIE assembly mutants. Images were collected at t3, with membranes stained with Mitotracker Red and DNA with DAPI (blue).
A. Focus-defective insertion spoIIIE147-i31 in spoIIP (KP6149).
B. Unstable focus insertion spoIIIE45-i31 in spoIIP (KP6151).
C. Unstable focus insertion spoIIIE162-i31 in spoIIP (KP6152).
D. Unstable focus insertion spoIIIE47-i31 in spoIIP (KP6153).
E. Focus-defective spoIIIE147-i31 in spoIIP spoIID (KP6176).
F. Unstable focus insertion spoIIIE45-i31 in spoIIP spoIID (KP6177).
G. Unstable focus insertion spoIIIE162-i31 in spoIIP spoIID (KP6178).
H. Unstable focus insertion spoIIIE47-i31 in spoIIP spoIID (KP6179).
For each of these mutant proteins at t3, the spoIIP mutation improved the ability of the protein to block GFP diffusion (by about 50%), while introducing both spoIIP and spoIID mutations almost completely rescued the diffusion defect (by about 97%; Table S1). Thus, septal peptidoglycan is sufficient to prevent GFP diffusion in spoIIIE mutants that are completely or partially defective in focus assembly. Scale bar in (A) is 2 μm.
Table S1. GFP diffusion in engulfment mutant backgrounds.
Table S2. Strains used in this study.