Abstract
In prokaryotes, the transfer of DNA between cellular compartments is essential for the segregation and exchange of genetic material. SpoIIIE and FtsK are AAA+ ATPases responsible for intercompartmental chromosome translocation in bacteria. Despite functional and sequence similarities, these motors were proposed to use drastically different mechanisms: SpoIIIE was suggested to be a unidirectional DNA transporter that exports DNA from the compartment in which it assembles, whereas FtsK was shown to establish translocation directionality by interacting with highly skewed chromosomal sequences. Here we use a combination of single-molecule, bioinformatics and in vivo fluorescence methodologies to study the properties of DNA translocation by SpoIIIE in vitro and in vivo. These data allow us to propose a sequence-directed DNA exporter model that reconciles previously proposed models for SpoIIIE and FtsK, constituting a unified model for directional DNA transport by the SpoIIIE/FtsK family of AAA+ ring ATPases.
The segregation and exchange of genetic material are central to the processes of cell division and evolution. Although mechanisms of genetic transfer between cellular compartments are diverse, all require the movement of DNA across cellular membranes. A dramatic example of intercellular transmembrane DNA transport is the segregation of chromosomes during sporulation in Bacillus subtilis. During this process, newly replicated chromosomes are rearranged into an elongated structure, or axial filament, in which the origin of replication of each chromosome is tethered to opposite cell poles1,2. Next, the division plane is relocated to one pole, creating two asymmetric cellular compartments (the forespore and the mother cell) and trapping the origin-proximal 30% of one chromosome within the forespore3,4. The septally bound DNA translocase SpoIIIE assembles on the trapped chromosome and uses the energy of ATP hydrolysis to rapidly transfer the remaining ~3 megabases (Mb) of DNA from the mother cell into the forespore5–7. Genetic evidence suggests that SpoIIIE also functions during vegetative growth to clear DNA from division septa8–10. Both functions require that SpoIIIE mobilize DNA directionally.
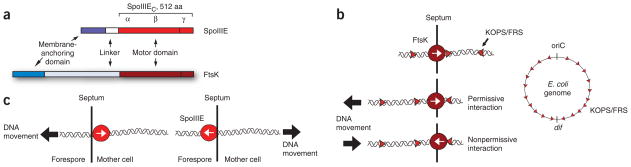
FtsK, a closely related DNA translocase from Escherichia coli, is involved in the coordination of chromosome segregation and cell division11,12. FtsK brings the sites of chromosome dimer resolution (dif sites) into close proximity by translocating DNA directionally13. SpoIIIE and FtsK are composed of an N-terminal transmembrane domain involved in septal localization14–16, a putatively unstructured linker and a C-terminal motor domain involved in DNA translocation (Fig. 1a), which itself is composed of three separate subdomains: α, β and γ7,16,17. Genetic and single-molecule studies showed that the translocation directionality of FtsK is dictated by specific chromosomal DNA sequence motifs GGGNAGGG (FtsK orienting polar sequence, KOPS)18 or GNGNAGGG (FtsK recognition sequence, FRS)19. The interaction of FtsK with KOPS is orientation specific: when encountering KOPS from the 3′ end of the G-rich strand (nonpermissive direction) FtsK pauses and frequently reverses its translocation direction, but it passes unobstructed when encountering KOPS from the opposite direction (permissive orientation)18–20 (Fig. 1b, left). The KOPS distribution is highly skewed (defined as the percentage of occurrences of the sequence on the leading versus the lagging strand of DNA replication) in the E. coli chromosome and switches strand and orientation at dif (Fig. 1b, right). Thus, this orientation-specific recognition mechanism and the asymmetric chromosomal distribution of KOPS direct FtsK translocation toward dif from any location along the chromosome.
Figure 1.
Architecture of SpoIIIE and FtsK monomers and models for DNA translocation directionality. (a) Schematic depicting the architecture of the SpoIIIE and FtsK monomers (above and below, respectively), showing the N-terminal transmembrane domain (blue), the central linker domain (white) and the motor domain consisting of α, β and γ subdomains (red). (b) Sequence-directed model for DNA translocation directionality of FtsK. Above left, FtsK moves along DNA in the direction of the white arrow. The encounter of FtsK with KOPS in the permissive direction (rightward-pointing red arrowheads) does not affect the translocation direction (middle left). FtsK can reverse translocation direction (below left) when encountering KOPS in the nonpermissive orientation (leftward-pointing red arrowheads). KOPS are highly skewed in the chromosome of E. coli and switch strand and orientation at dif (right). (c) Simple exporter model for DNA translocation directionality by SpoIIIE. In this model, the relative abundance of SpoIIIE (red circle, arrows indicating its relative orientation) in the mother cell or other cell-specific factors favor the assembly of a unidirectional DNA exporter in the mother cell that determines the directionality of transfer (left). Expression of SpoIIIE in the forespore after septation leads to DNA transfer into the mother cell21 (right).
The high sequence homology and similar functions of FtsK and SpoIIIE imply a common translocation mechanism. However, fluorescence microscopy studies suggested that SpoIIIE is a unidirectional DNA transporter that exports DNA out of the compartment in which it is expressed21 (the unidirectional exporter model, shown in Figure 1c). This model proposed that the directionality of DNA export is regulated by the asymmetric partitioning and cell-specific assembly of SpoIIIE21,22. The fact that SpoIIIE is expressed throughout vegetative growth implied the existence of another unidentified factor that regulates translocation polarity in the absence of cellular asymmetry21,22. Recent work showed that, in a racA soj-spo0J mutant background, the misorientation of the chromosome relative to the septum caused SpoIIIE to translocate the chromosome out of the forespore23. This result suggests that SpoIIIE directional DNA transfer may also be regulated by the polarity of the chromosome itself. However, there is no direct evidence for a DNA sequence–directed mechanism for SpoIIIE translocation directionality. To clarify these conflicting models, we used single-molecule, bioinformatics and fluorescence microscopy methods to demonstrate that specific interactions between the γ-domain of SpoIIIE and specific, highly skewed chromosomal DNA sequences (SpoIIIE recognition sequence or SRS) regulate the compartment-specific activation of a mother-cell SpoIIIE complex responsible for DNA export into the forespore. This sequence-directed DNA exporter model unifies conflicting sequence-directed and exporter models and explains the establishment of directional DNA translocation by the FtsK/SpoIIIE family of DNA translocases.
RESULTS
DNA translocation by SpoIIIEC does not require SpoIIIE-γ
Previous measurements of DNA translocation by SpoIIIE have relied on indirect bulk methods or static microscopy6,7. Here, we used a simplified in vitro system to observe directly the dynamics of DNA translocation by single SpoIIIE motors using magnetic tweezers. A single nicked DNA molecule is tethered between a magnetic particle and a glass surface, and DNA translocation by the soluble C-terminal motor domain of SpoIIIE7 (SpoIIIEC; Fig. 1a) is monitored by the shortening of the DNA extension due to the formation of a DNA loop (Supplementary Fig. 1a and Supplementary Methods online). In these experiments, SpoIIIEC translocated DNA rapidly and processively at rates of 4 ± 1 kb s−1 (Supplementary Fig. 1b,d), notably faster than estimated translocation velocities in vivo (~1.5 kb s−1 per chromosomal arm)24–26. This difference could be due to protein or DNA roadblocks, or larger loads opposing translocation in vivo. Recent findings showed that FtsK-γ is required for KOPS recognition27,28. On the basis of the sequence similarity between FtsK-γ and SpoIIIE-γ, we predicted that SpoIIIE-γ may be involved in regulating the directionality of DNA transfer during sporulation but not in the mechanical process of DNA translocation itself. To test these predictions, we first examined the in vitro motor properties of SpoIIIEC-Δγ, a truncation mutant of SpoIIIEC lacking the γ-domain (Supplementary Methods). SpoIIIEC-Δγ was able to translocate DNA at rates that were only slightly slower than SpoIIIEC (3 ± 1 kb s–1; Supplementary Fig. 1c,e), showing that SpoIIIE-γ is not essential for the motor activity of SpoIIIE in vitro.
SpoIIIE-γ is required for directional DNA translocation in vivo
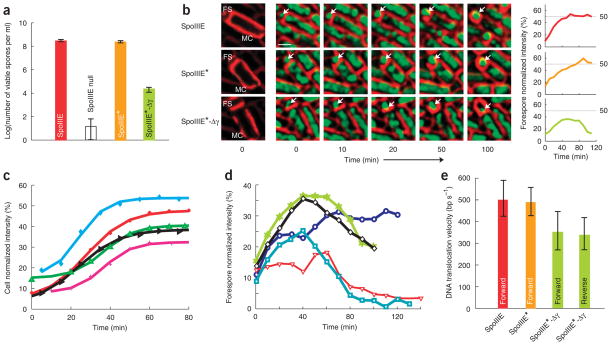
We investigated the role of SpoIIIE-γ in vivo during sporulation by testing the ability of SpoIIIE-Δγ to rescue a spoIIIE-null strain and produce viable spores (Methods). The genes encoding wild-type SpoIIIE or SpoIIIE-Δγ under control of the spoIIIE promoter were integrated into spoIIIE-null strains (hereafter referred to as SpoIIIE★ and SpoIIIE★-Δγ, respectively; Supplementary Methods). The ability of the SpoIIIE★ strain to produce viable spores was similar to that of a wild-type strain21, but the SpoIIIE★-Δγ strain produced 10,000-fold fewer viable spores than SpoIIIE★ (Fig. 2a and Supplementary Table 1 online). Thus, SpoIIIE-γ is not necessary for DNA translocation in vitro but is necessary for efficient sporulation in vivo.
Figure 2.
SpoIIIE-γ is necessary for efficient sporulation but does not affect motor function. (a) Log of viable spore titers for wild-type (red), SpoIIIE null (white), SpoIIIE★ (orange) and SpoIIIE★-Δγ (green) cells. Error bars indicate s.d. (b) Timelapse microscopic images of wild-type (above), SpoIIIE★ (middle) and SpoIIIE★-Δγ (below) cells. Membrane images (red) at the left are enhanced to indicate the location and orientation of the cell in each frame, and the forespore and mother cell are indicated (FS and MC, respectively). A time-lapse image series is shown, with arrows indicating the forespore DNA (green). The scale bar of 1 μm is shown. Plots to the right of each series show the normalized forespore intensity versus time. (c) Normalized DNA fluorescence intensity in the forespore versus that in the mother cell from time-lapse fluorescence images of SpoIIIE★ strains permits quantification of the percentage of DNA translocated into the forespore as a function of time. (d) Normalized DNA fluorescence intensity traces of individual cells (different colors shown) of the SpoIIIE★-Δγ strain. (e) In vivo velocities of DNA translocation for SpoIIIE (red), SpoIIIE★ (orange) and SpoIIIE★-Δγ (green) obtained from DNA fluorescence intensity traces. Error bars indicate s.d.
To understand this sporulation defect triggered by the lack of SpoIIIE-γ, we used a time-lapse fluorescence microscopy technique23 to characterize quantitatively the dynamics of chromosome translocation in vivo by SpoIIIE. Synchronized sporulating cultures were grown on agar pads and imaged at regular intervals by fluorescence microscopy. To measure DNA translocation velocities and to observe the behavior of various SpoIIIE mutants in vivo, we quantified the fluorescence intensity of DNA in the forespore versus total fluorescence intensity in the cell at different time points during sporulation (Supplementary Methods). Wild-type B. subtilis and SpoIIIE★ strains translocated DNA exclusively from the mother cell to the forespore (n = 243; Fig. 2b,c and Supplementary Video 1 online) at velocities of 500 ± 80 bp s−1 per chromosomal arm. The slightly slower DNA translocation velocity in these experiments versus previous estimates may be attributed to differences in our experimental conditions, such as temperature (30 °C versus 37 °C) and the presence of intercalated fluorescent dyes (Methods) that could affect SpoIIIE velocities in vivo.
In contrast, SpoIIIE★-Δγ cells showed a marked defect in directional chromosome transfer, as at least 40% of the cells were observed to reverse-translocate DNA out of the forespore, and none completed forward chromosome translocation (n = 349; Supplementary Video 2 online). Quantitative analysis of these events revealed a frequent DNA translocation defect in which forward translocation was followed by a pause and a subsequent reversal of the translocation direction out of the forespore (Fig. 2b,d and Supplementary Fig. 2a–d online). The mean distance translocated by SpoIIIE-Δγ before pausing was 0.75 ± 0.3 Mb per chromosome arm, and the mean pause length was 20 ± 10 min (Supplementary Fig. 3b,c online). The velocities of forward and reverse DNA translocation in these events were indistinguishable (Fig. 2e) and consistently ~25% slower than in wild-type and SpoIIIE★ strains (n = 77, P-value = 0.0001; Fig. 2e), mimicking the difference in translocation rates measured in single-molecule in vitro experiments between SpoIIIEC and SpoIIIEC-Δγ (Supplementary Fig. 1). A control strain expressing only SpoIIIE36, an ATPase mutant of SpoIIIE5, did not show any forward or reverse translocation (data not shown). Previous studies showed that FtsK-γ is necessary for sequence recognition in vitro27,28 and in vivo in a plasmid resolution assay28. Our data show that the SpoIIIE-γ is necessary for regulating the directionality of chromosomal transport in vivo. In particular, the frequent in vivo translocation direction reversals observed in the SpoIIIE★-Δγ strain show that the translocation directionality imparted by SpoIIIE is not simply due to its partitioning to the mother cell before septation (Fig. 1c) and suggest that the substrate polarity is also involved in maintaining the directionality of transfer. The long pause durations required for reversing translocation direction in vivo (~20 min) suggest that the mechanism responsible for translocation reversal is slow, possibly involving the recruitment of SpoIIIE and the assembly of the forespore complex. This observation, together with the high processivity of SpoIIIE-Δγ in vivo, is also inconsistent with a strictly sequence-directed model (Fig. 1b).
Identification and characterization of SRS
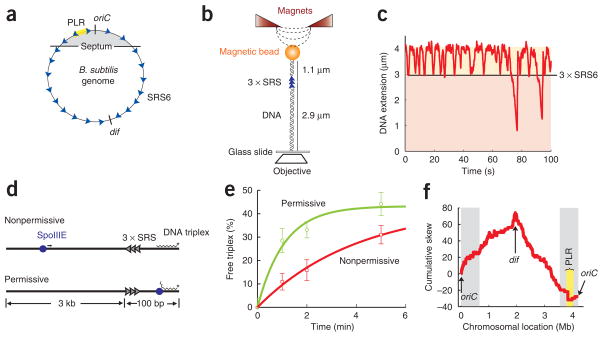
The role of SpoIIIE-γ in regulating directional DNA transfer and the function of FtsK-γ in KOPS recognition27,28 raised the possibility that orientation-specific interactions between SpoIIIE and highly skewed SRS sequences could control directional DNA transfer during sporulation (Fig. 3a, Supplementary Fig. 3a and Supplementary Methods). Sequence similarity between FtsK-γ and SpoIIIE-γ, and the high skew of KOPS along the chromosome of B. subtilis (84%, P-value = 1 × 10−27), suggested that KOPS may guide directional translocation by SpoIIIE. To test the effect of KOPS on SpoIIIE DNA translocation, we used an in vitro magnetic tweezers assay for sequence recognition (Fig. 3b and Methods) that was previously used to detect specific KOPS-FtsK interactions18,27,29. In this assay, we quantified the recognition probability of a triple tandem repeat of KOPS (3 × KOPS, or 3 × SRS1) by measuring the reversal frequency of SpoIIIEC at DNA extensions corresponding to the location of the test sequence on the molecule (Supplementary Methods). Previous studies determined that FtsK recognizes KOPS with a probability of ~40%19. Notably, we found that SpoIIIEC recognizes KOPS with a probability of only 3 ± 3% per sequence (Supplementary Table 2 online), showing that at least this instance of KOPS cannot guide SpoIIIE during sporulation.
Figure 3.
Identification and characterization of a SpoIIIE recognition sequence (SRS). (a) Schematic depicting the orientation of SRS6 sequences (blue arrowheads) along the B. subtilis chromosome (circle). The replication origin (oriC), the dif sequence and the polar localization region (PLR) are indicated. Gray area marks the region of the chromosome initially trapped by the septum before DNA translocation by SpoIIIE commences. (b) Schematic depicting the magnetic tweezers setup for DNA sequence recognition. A naked double-stranded DNA molecule (black helix) is tethered between a glass slide and magnetic particle. A repeat of three SRS candidates to be tested (blue arrowheads) is located near the top, 1.1 μm from the bead and 2.9 μm from the glass surface. Tension in the DNA is introduced using magnets (red triangles). SpoIIIEC-induced DNA looping shortens the DNA extension, monitored by the change in height of the magnetic particle using an inverted objective (white rhombus). Specific interactions between SpoIIIEC and SRS are observed as pauses and translocation reversals at extensions corresponding to the location of the tandem repeat of three SRS sequences (3 × SRS). (c) Representative trace of the change in DNA extension due to single-molecule SpoIIIEC activity on a DNA molecule containing a 3 × SRS6 at the test location (black line). (d) Schematic of DNA triplex displacement assay. SRS repeats (arrowheads) in the nonpermissive or the permissive orientations are located on a linear duplex DNA between a 21-bp triplex (jagged line) and a 3-kb ‘antenna’ region. SpoIIIE (blue sphere) often binds within the antenna region because of its relative length. Translocation of SpoIIIE into the nonpermissive SRS region frequently reverses SpoIIIE, protecting the triplex (above). SpoIIIE translocation into the permissive SRS region leads to displacement of the triplex (below). (e) SpoIIIEC triplex displacement reactions on DNA substrates with permissive (green circles) or nonpermissive (red squares) SRS6 orientations. The percentage of free triplex is plotted as a function of time, and error bars indicate s.d. Data were normalized for the initial percentage of free triplex and fit to an exponential function. The ratio of displacement on nonpermissive versus permissive substrates is 2.5 ± 0.5. (f) Step plot indicating the locations and orientations of SRS6 along the B. subtilis genome (see text). Cumulative skew is shown as a function of chromosomal location (Supplementary Methods). The replication origin (oriC), the dif site and the PLR are indicated. Gray area marks the region of the chromosome initially trapped by the septum before DNA segregation by SpoIIIE commences.
To identify other candidate SRS sequences, we conducted a bio-informatics search for highly skewed octameric DNA sequences in the B. subtilis chromosome that switched strand at dif (Supplementary Table 2, Supplementary Fig. 4 and Supplementary Methods online). The role of SpoIIIE in vegetative growth10 and the location of dif at the center of the terminus of replication in B. subtilis30,31 suggest that the dif region is the last chromosomal fragment to be translocated into the forespore during sporulation. We tested six candidate sequences identified with this approach (SRS1–6) using the magnetic tweezers assay for specific DNA sequence recognition. We observed similar low levels of recognition (~5% per SRS instance) for SRS1–5, despite their high chromosomal skews and low P-values (that is, the probability that their asymmetric chromosomal distributions occurred by chance; see Supplementary Fig. 5a and Supplementary Table 2 online). Notably, on DNA molecules containing 3 × SRS6 (GAGAAGGG), SpoIIIEC frequently paused and reversed at DNA extensions corresponding to the SRS6 repeats with a frequency of 29 ± 5% per instance (Fig. 3c, Supplementary Table 2). However, to guarantee directional DNA movement, SpoIIIE must be able to recognize SRS6 in an orientation-specific manner. We implemented a DNA triplex-displacement assay in which orientation specificity is detected by differences in triplex-displacement rates by SpoIIIEC on DNA substrates containing permissive or nonpermissive SRS19 (Fig. 3d). Consistent with the lack of KOPS recognition, triplex-displacement rates by SpoIIIEC were similar on substrates containing 3 × SRS1 (KOPS) in permissive or nonpermissive orientations (Supplementary Fig. 5b). In contrast, displacement rates on substrates containing SRS6 showed a considerable difference (2.5 ± 0.5–fold) between permissive and nonpermissive orientations (Fig. 3e).
This robust and orientation-specific recognition of SRS6 in vitro prompted us to examine its relevance in vivo by using a genetic assay for sequence recognition (Supplementary Figs. 6a,b and Supplementary Methods online). This assay relies on a kinetic competition between SpoIIIE translocation and the Cre-mediated recombination of a chromosomal kanamycin-resistance cassette flanked by loxP32. Recognition of a tandem repeat of SRS6 that disrupts forward translocation by SpoIIIE increases the frequency of recombination of the kanamycin-resistance gene in the mother cell, and thus the frequency of kanamycin-sensitive spores. Using this assay, we measured an 8% increase in kanamycin-sensitive spores in the nonpermissive versus the permissive SRS6 strain (n = 24, P-value = 0.02), suggesting that SpoIIIE recognizes SRS6 specifically in vivo during sporulation.
SRS6 is a member of AIMS (architecture imparting sequences)33. On the B. subtilis chromosome, SRS6 is highly skewed (81%, P-value = 1 × 10−15) and switches strand at dif. Surprisingly, SRS6 does not switch at oriC but at a locus approximately 250 kb to the left of oriC (Fig. 3a,f). Interestingly, this is the location of the polar localization region (PLR), the centromere-like sequence in B. subtilis (extending from −150 kb to −300 kb) required to segregate the oriC region to the cell pole during sporulation34. It is likely that SRS6 is an instance of a degenerate sequence family or motif, such as that represented by KOPS19,29. In a preliminary search for a consensus SRS, we found that GAG(C/A)AGGG is the most likely SRS motif (Supplementary Results and Supplementary Table 3 online). This putative family of sequences has a skew of 82%, a significantly higher density and thus a lower P-value (1 × 10−26) than SRS6.
SpoIIIE-γ recognizes SRS6 specifically in vitro and in vivo
The orientation-specific recognition of SRS6 by SpoIIIE and the requirement of SpoIIIE-γ for directional DNA transfer in vivo suggest that SpoIIIE-γ specifically mediates SRS6 interactions. To test this prediction, we introduced a single mutation in the putative DNA binding region of SpoIIIE-γ (SpoIIIEC-R773A)28 and measured the ability of this protein to recognize SRS6 in vitro and to support sporulation in vivo. In the magnetic tweezers assay, SRS6 recognition by SpoIIIEC-R773A was reduced by approximately two-fold (Supplementary Fig. 5c and Supplementary Table 2). Consistent with this reduction, SpoIIIE★-R773A produced three-fold fewer viable spores than SpoIIIE★ (Supplementary Table 1), indicating that SpoIIIE-γ mediates direct interactions with SRS6 in vivo. This modest defect in spore titers in vivo agrees with calculations that predicted that large changes in sequence-recognition probability produce minor changes in the efficiency of directional DNA translocation19.
FtsK-γ and SpoIIIE-γ are exchangeable modules
The sequence-directed model for DNA translocation directionality predicts that SpoIIIE-γ–SRS6 interactions are modular. Thus, the exchange of SpoIIIE-γ by FtsK-γ should switch the recognition specificity of SpoIIIE from SRS to KOPS. We tested this prediction by replacing SpoIIIE-γ with FtsK-γ to create a chimera termed SpoIIIEC-SK (Fig. 4a), and assayed its interaction with KOPS using the magnetic tweezers and triplex-displacement assays. In magnetic tweezers experiments, SpoIIIEC-SK reversed at DNA extensions corresponding to the location of 3 × KOPS with a probability of ~35% per KOPS instance (Fig. 4b, Supplementary Fig. 5e and Supplementary Table 2) and showed a pattern of sequence recognition similar to that of FtsK50C27. Recognition of KOPS by SpoIIIEC-SK is orientation specific, as triplex-displacement rates on permissive 3 × KOPS substrates were 1.8 ± 0.25–fold faster than on nonpermissive substrates (Fig. 4c). Notably, the turnaround probability of SpoIIIEC-SK at SRS6 was only 3% (Supplementary Figs. 2 and 5d), indicating that SpoIIIEC-SK can recognize KOPS but not SRS in vitro.
Figure 4.
γ-domains are modular and can be switched between species to confer altered DNA sequence specificities. (a) Schematic depicting the architecture of the SpoIIIE-SK chimera containing the γ-domain of FtsK (dark red) fused to the motor domain of SpoIIIE (light red). (b) Representative trace of SpoIIIEC-SK chimera-induced changes in DNA extension versus time in the magnetic tweezers on a DNA substrate containing a triple repeat of KOPS (3 × SRS1) at the test location (black line). (c) SpoIIIEC-SK chimera DNA triplex displacement reactions on substrates containing either a triple KOPS sequence in permissive (green circles) or nonpermissive (red squares) orientations. Error bars indicate s.d. Data were normalized for the initial percentage of free triplex and fit to an exponential function. Ratio of displacement on permissive versus nonpermissive substrates is 1.8 ± 0.25. (d) Log of heat-resistant spore titers for wild-type SpoIIIE★ (orange), SpoIIIE★-Δγ (green) and SpoIIIE★-SK chimera (red) strains. Error bars indicate s.d. (e) Normalized DNA fluorescence intensity traces of individual SpoIIIE★-SK cells (different colors shown). Velocities of DNA translocation in vivo were 520 ± 100 bp s−1 (n = 16), similar to those of SpoIIIE★.
The efficient, orientation-specific KOPS recognition by SpoIIIEC-SK in vitro and the high skew of KOPS along the chromosome of B. subtilis suggested that the translocation directionality of SpoIIIEC-SK could be guided by KOPS in vivo. To test this hypothesis, we constructed strains lacking wild-type spoIIIE, but expressing either SpoIIIE★, SpoIIIE★-Δγ or SpoIIIE★-SK from the spoIIIE promoter (Supplementary Methods) and examined their ability to translocate DNA directionally in vivo by measuring the number of viable spores per ml produced by each strain and the ability of SpoIIIE★-SK to translocate DNA directionally. Notably, we found that SpoIIIE★-SK rescued sporulation to near wild-type levels (Fig. 4d) and translocated DNA in vivo at wild-type velocities (data not shown). In contrast to SpoIIIE★-Δγ, less than 3% of SpoIIIE★-SK cells showed reversals of DNA translocation direction (n = 72, Fig. 4e), and the remaining 97% completed DNA translocation into the forespore. These experiments reveal that the γ-domains of the FtsK/SpoIIIE family of DNA translocases are functional modules that seem to have evolved for the recognition of specific DNA sequences and can be interchanged between motors to confer different sequence-recognition properties.
SpoIIIE-γ regulates the compartmental localization of SpoIIIE
So far, our data suggest that directional DNA transfer during sporulation is regulated by orientation-specific interactions between SpoIIIE and highly skewed chromosomal sequences that switch orientation at dif, and not only due to the compartment-specific assembly of SpoIIIE, as proposed by the simple exporter model. The long pauses required for reversing translocation direction in vivo (~20 min), however, are inconsistent with a strictly sequence-directed model (Fig. 1b) and suggest that the translocation reversal mechanism possibly involves the assembly of a forespore-specific SpoIIIE complex. To further discern between these two models, we studied the role of SpoIIIE-γ in compartment-specific SpoIIIE assembly. The simple exporter model predicts that, in either the presence or the absence of SpoIIIE-γ, SpoIIIE should assemble a complex only in the mother cell. In contrast, the sequence-directed model suggests that SpoIIIE may assemble on both sides of the sporulation septum.
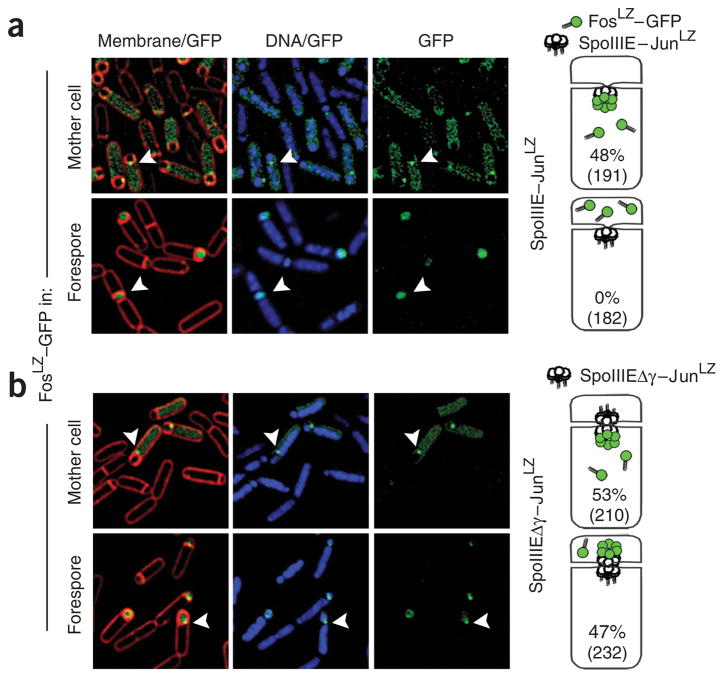
Previous methods used to localize SpoIIIE complexes in vivo, such as GFP-fusion localization, cannot distinguish SpoIIIE localization on one side of the septum from localization on the other21. Here we used a cell-specific GFP-tagging method to visualize SpoIIIE complexes assembled at the mother-cell or forespore side of the sporulation septum23. In this method, the leucine zipper domains of Jun and Fos (JunLZ and FosLZ)35 are used as specific tags to link SpoIIIE and GFP. GFP–FosLZ was expressed in either the forespore or mother-cell compartments after septation. SpoIIIE–JunLZ complexes that assemble in the compartment expressing GFP–FosLZ were visualized by the appearance of a septal GFP focus due to the binding of GFP–FosLZ molecules to the SpoIIIE–JunLZ complex (Fig. 5a, right). SpoIIIE★–JunLZ and SpoIIIE★-Δγ–JunLZ were expressed from the native SpoIIIE promoter and locus, and the frequency of focus formation in either forespore or mother-cell compartments was measured during sporulation. SpoIIIE★–JunLZ strains produced foci exclusively in the mother cell, as 48% of cells contained a visible mother-cell GFP focus (Fig. 5a, above, n = 191) and 0% of cells produced forespore foci (n = 182). In contrast, SpoIIIE★-Δγ–JunLZ strains produced GFP foci with approximately equal frequency in both mother-cell (53%, n = 210) and forespore compartments (47%, Fig. 5b, n = 232). Thus, these results conflict with both the simple exporter and the strictly sequence-directed models. In the next section, we will present a unified model for the mechanism of directional DNA transfer by SpoIIIE/FtsK that is consistent with the available data.
Figure 5.
Cell-specific GFP tagging indicates that SpoIIIE-Δγ assembles on both sides of the sporulation septum with equal frequency. Cartoons show the experimental setup (right). FosLZ–GFP (green circles) was expressed after septation in the forespore or mother-cell compartments in strains expressing SpoIIIE–JunLZ or SpoIIIE–Δγ–JunLZ (white circles). The location of SpoIIIE complexes was observed by the appearance of a GFP focus (green cluster) at the septal midpoint. Fluorescence microscopic images show a membrane and GFP overlay (left), a DNA and GFP overlay (center) and GFP only (right). GFP foci are indicated by white arrowheads. (a) SpoIIIE assembles only on the mother-cell side of the septum during DNA translocation. SpoIIIE–JunLZ foci were observed on the mother-cell side of the septum in 48% of the cells (n = 191, above), whereas no SpoIIIE–JunLZ foci were observed in the forespore compartment (n = 182, below). (b) SpoIIIE–Δγ assembles on both sides of the septal membrane with equal frequency. Assembly of SpoIIIE–Δγ–JunLZ foci was observed in the mother cell (53%, n = 210, above) and forespore (47%, n = 232, below) sides of the septum.
DISCUSSION
Here we advance a sequence-directed DNA exporter model for SpoIIIE translocation directionality that is consistent with both our data and previous experiments. Structural and fluorescence microscopy evidence suggests that during sporulation each chromosome arm is translocated through an independent SpoIIIE channel17,23,36. Our compartment-specific SpoIIIE-tagging experiments showed that during sporulation SpoIIIE is exclusively assembled on the mother-cell side of the septum, whereas SpoIIIE-Δγ is assembled on both sides with equal frequency. Furthermore, our fluorescence microscopy results demonstrated that SpoIIIE-Δγ can translocate DNA in both forward and reverse directions in vivo. These results are consistent with a model in which each chromosome arm can be initially translocated by opposing unidirectional complexes, one on each side of the newly formed division septum (forespore and mother-cell exporters, Fig. 6a). Our study reveals that SpoIIIE-γ not only mediates orientation-specific SpoIIIE–SRS interactions in vitro, but is also required for the disassembly of the forespore exporter and the maintenance of persistent directional DNA transfer in vivo. The long pause durations required for reversing translocation direction by SpoIIIE-Δγ in our in vivo DNA translocation assay (~20 min) further suggest that the mechanism giving rise to translocation reversals is slow and may involve the reassembly of the forespore complex. Recent studies determined that the ability of SpoIIIE to hydrolyze ATP is required for the disassembly of the forespore SpoIIIE complex and that the orientation of the chromosome after septation determines the direction of DNA translocation23. Taken together, these results suggest that translocation by the forespore exporter leads to specific interactions between SpoIIIE-γ and nonpermissive SRS that lead to the inactivation and subsequent dissociation of the forespore complex (Fig. 6b,c), a process that ultimately decides the direction of DNA transfer. This model explains the observation that SpoIIIE expressed in the forespore after septation is severely deficient in DNA export into the mother cell21, as the forespore complex may be frequently disassembled by interactions with nonpermissive SRS. The idea that SpoIIIE may not be required on the forespore side of the septum for forward translocation from the mother cell is supported by two observations: (i) the expression of SpoIIIE only in the mother cell after septation was shown to rescue viable spore production to near wild-type levels21; and (ii) highly homologous motors of the SpoIIIE/FtsK family involved in conjugative transfer (such as the Tra protein from pIJ101 in Streptomyces lividans37) are present only in the donor-cell side of the communicating cell wall. Thus, compartment-specific localization may be relevant to the mechanisms for directional DNA transfer during cell division, sporulation and conjugation.
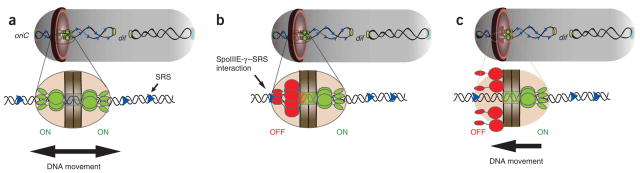
Figure 6.
Model for SpoIIIE sequence-directed DNA export during sporulation. (a) Schematic of a sporulating B. subtilis cell (gray) at the onset of DNA translocation, in which each chromosomal arm (black ribbon) is bound by two opposing active unidirectional complexes (green) that assemble on each side of the division septum (brown disc). Below, an enlarged view of a single chromosomal arm (helix), division septum (gray) and SRS sequences (blue triangles) are represented. Black arrows depict the overall direction of DNA transport. We assume that during translocation DNA is transported in the direction from SpoIIIE-β to SpoIIIE-α29; (b) SpoIIIE-γ–mediated interactions with nonpermissive SRS in the forespore leads to the inactivation of the forespore complex (red). (c) The inactivation of the forespore SpoIIIE complex converts the bidirectional channel into a DNA exporter, leading to unidirectional DNA transport into the forespore.
Previous studies proposed that the oriented loading of FtsK on KOPS29, or the reversal of translocation direction of FtsK upon encountering nonpermissive KOPS19,29,38 or both could act as the mechanism for setting the direction of DNA translocation. The ability of SpoIIIE to translocate DNA substrates lacking SRS (Supplementary Fig. 5a), the interaction of SpoIIIE with nonpermissive SRS repeats distant from the SpoIIIE initiation site, the requirement for the ATPase activity of SpoIIIE to disassemble the forespore complex23, and the low density of SRS and KOPS along the chromosome suggest these sequences are not simply loading sites for SpoIIIE/FtsK in vivo. The detailed molecular mechanism by which SpoIIIE-γ–SRS and FtsK-γ–KOPS interactions specifically affect translocation directionality and the process by which in vitro reversibility is related to the mechanism of SpoIIIE/FtsK in vivo remain unclear and will be the subject of our future studies.
Our sequence-directed DNA exporter model predicts that, in the absence of SpoIIIE-γ, the initial direction of DNA translocation early in sporulation would partition between forward and reverse orientations with equal probabilities. However, quantitative analysis of our live time-lapse microscopy data showed that SpoIIIE-Δγ commences translocation predominantly in the forward direction (>96%, n = 32). These results imply the existence of additional mechanisms that bias the initial direction of DNA segregation. Several possible DNA sequence–independent processes could contribute to the initial inactivation of the forespore exporter and thus produce the observed bias of the initial DNA translocation direction. The frequent reverse translocations by SpoIIIE in minCD-null strains22 suggest that MinCD may influence chromosome architecture or inactivate forespore SpoIIIE complexes. Alternative mechanisms may involve forespore-specific DNA–protein complexes that could obstruct DNA translocation by the forespore SpoIIIE exporter by creating DNA roadblocks or membrane anchors. These complexes could include the ParB homolog Spo0J, which compacts the origin-proximal region into filamentous protein-DNA architectures1,39,40, and RacA, which tightly tethers the chromosomal origin to the cell pole40–42. The late appearance of translocation reversal events could be related to the release of oriC tethering later in the process of sporulation. Finally, other unknown forespore-specific factors, asymmetric concentrations of SpoIIIE between compartments or early structural asymmetry in the sporulation septum may bias the activation state of the forespore exporter at the onset of DNA translocation.
Despite these possible overlapping mechanisms, our data demonstrate that DNA sequence recognition is both robust and necessary for directional DNA translocation by SpoIIIE. Our model for sequence-directed DNA export may also explain the directional chromosomal transfer by FtsK and SpoIIIE during vegetative growth, as DNA sequence asymmetry alone could bias the activation state of exporter complexes in the absence of compartmental asymmetry. Overall, the data presented here reconcile previous contradictory models for FtsK and SpoIIIE, and suggest a new sequence-directed DNA exporter model for the establishment of translocation polarity of the FtsK/SpoIIIE/Tra family of DNA transporters.
METHODS
SpoIIIEC cloning and purification
SpoIIIE expression constructs were derived from pJB103 (ref. 7), which contains the C-terminal motor domain of SpoIIIE with an N-terminal OmpA tag and a C-terminal hexahistidine tag (plasmid provided by J. Bath, Oxford University). PredictProtein43 was used to create a multiple sequence alignment and to predict conserved regions, secondary structure and unstructured regions. Point mutations were introduced using the QuickChange Method (Stratagene). All clones were propagated in E. coli DH5α cells and verified by sequencing. For further details see Supplementary Methods. SpoIIIEC was expressed and purified as described7. Pure fractions (>95% pure by SDS-PAGE and Coomasie blue staining) were pooled, dialyzed in storage buffer (50 mM HEPES, 100 mM NaCl, 0.1 mM EDTA, 30% (v/v) glycerol, pH 7.5) and quantified using optical density and the Bradford method.
Triplex displacement assays
Fluorescent triplex substrates were designed as described in Levy et al.19 and prepared following the method developed by McClelland et al.44. Displacement reactions were conducted at 25 °C in 50 mM Tris (pH 7.5), 5 mM MgCl2, 3 mM ATP, 0.1 mg ml−1 BSA, 10 nM 5′-6-rhodamine triplex substrate and 20–50 nM SpoIIIE or 100nM SpoIIIE-SK chimera. Reactions were quenched by the addition of an equal volume of 250 mM MES (pH 5.5), 3% (w/v) SDS, 15% (w/v) glucose45 and analyzed on 1.5% agarose gels (40 mM Tris acetate (pH 5.5), 5 mM sodium acetate, 1 mM MgCl2 at 10 V cm−1 at 4 °C8. Results were visualized using Typhoon Imager and the proportion of free oligonucleotide to triplex substrate was determined by using IMAGEQUANT (GE Healthcare). The percentage of free triplex at zero time was subtracted from each time point, and the percentage of triplex displaced versus time was fitted to A0(1 − e−kt), where the rate (k) and the baseline (A0) were floated. Further details can be found in the Supplementary Methods.
Magnetic tweezers experiments
DNA tethers were prepared as previously described27, using a plasmid containing a 12.2-kb segment of the E. coli genome devoid of any FRS or KOPS family motifs18,19. SRS repeats were synthesized as 5′-phosphorylated duplex oligonucleotides and cloned into the desired location (Supplementary Methods). To create DNA tethers, the plasmid described above was digested with XbaI and KpnI to create inserts of 12.2 kb with the SRS repeat located 3 kb from the KpnI end. These molecules were ligated to biotin- and digoxygenin-modified PCR products. DNA tethers were oriented between a biotin-BSA–straptavidin-coated glass surface and a 1-μm diameter anti-digoxigenin antibody coated bead (Dynal, My One Beads). Tweezers experiments were conducted on a magnetic tweezers instrument described46. SpoIIIE reactions contained: 1–3 nM SpoIIIEC, 50 mM Tris-HCl (pH 7.5), 3mM ATP, 1–3 nM SpoIIIE and 0.1 mg ml−1 BSA at constant forces between 5–8 pN. See Supplementary Methods for additional information.
Spore titer assay and sporulation time-lapse microscopy
Spore titers were determined as described47. Briefly, strains were inoculated into Difco sporulation medium and incubated at 37 °C and 2,600 × g for 24 h. Cultures were heated to 80 °C for 10 min, diluted in 1× TBase and plated onto LB agar plates for determining colony numbers. Time-lapse microscopy experiments were performed as described23. Briefly, exponentially growing cultures were induced to sporulate by resuspension48 at 30 °C and grown for a further 2 h in the presence of 0.5 μg ml−1 FM4-64 (Molecular Probes/Invitrogen) and 0.5 μM SytoxGreen (Invitrogen). Sporulation supernatants were collected and an additional 0.5 μg ml−1 of FM4-64 was added. Molten agarose was added to 1% final, poured into shallow-welled microscope slides (VWR; 18 mm × 1.75 mm), and allowed to solidify. Cultures were applied to the agarose pads and imaged at 30 °C using an Applied Precision Spectris microscope with a WeatherStation temperature control chamber. Exposure times varied between 100 ms and 300 ms with neutral density filter settings of 32% for both FM4-64 and SytoxGreen images. Image files were deconvolved using SoftWoRx (15 iterations on conservative setting). The ‘polygon’ function of ImageJ software (http://rsb.info.nih.gov/ij/) was used to quantitate forespore and mother-cell fluorescence signals.
SRS in vivo recognition experiments
Strains were grown in DSM medium47 + 1.5 μg ml−1 kanamycin at 37 °C for 24 h, heat killed for 20 min at 80 °C, diluted in 1× TBase47 and plated onto LB and LB + 15 μg ml−1 kanamycin plates. Strains containing the tet gene in both orientations produced no measurable difference in kanamycin sensitive spore frequency (data not shown). Further details can be found in Supplementary Methods.
Cell-specific GFP tagging
Cell-specific JunLZ- and FosLZ-tagging experiments were performed as described23. Briefly, exponentially growing cultures were induced to sporulate in the presence of 0.5 μg ml−1 FM4-64 (Invitrogen). Cultures were adjusted to 0.2 μg ml−1 SYTOX Green dye, applied to poly–L-lysine–coated coverslips and imaged using and Applied Precision microscope. Images were deconvolved using SoftWoRx v3.3.6 (Applied Precision).
Strain construction, cloning in vivo constructs and culture conditions
All strains (Supplementary Table 2) are derived from B. subtilis PY79 and were created using standard methods49. Sporulation was induced by resuspension48 at 30 °C. Strains, cloning and construct design details can be found in Supplementary Methods.
Supplementary Material
Acknowledgments
The authors would like to dedicate this work to our friend and colleague Nicholas R. Cozzarelli, who passed away during completion of this research. We thank J. Berger for his continuous advice during this research and N. Crisona for critical reading. This work was supported by National Institutes of Health Grants GM31655 (to N.R.C.) and the Human Frontier Science Program (M.N.).
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
J.L.P. and M.N. planned, collected and interpreted single-molecule, biochemical and fluorescence data; E.C.B. performed time-lapse fluorescence studies of SpoIIIE-SK chimera and compartment-specific GFP-tagging experiments; J.L.P., M.N., E.C.B., K.P. and C.B. wrote the paper.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Ryter A. Morphologic study of the sporulation of Bacillus subtilis. Ann Inst Pasteur (Paris) 1965;108:40–60. [PubMed] [Google Scholar]
- 2.Thomaides HB, Freeman M, El Karoui M, Errington J. Division site selection protein DivIVA of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation. Genes Dev. 2001;15:1662–1673. doi: 10.1101/gad.197501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levin PA, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 1996;10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- 4.Pogliano J, Sharp MD, Pogliano K. Partitioning of chromosomal DNA during establishment of cellular asymmetry in Bacillus subtilis. J Bacteriol. 2002;184:1743–1749. doi: 10.1128/JB.184.4.1743-1749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu LJ, Errington J. Bacillus subtilis spoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- 6.Wu LJ, Lewis PJ, Allmansberger R, Hauser PM, Errington J. A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes Dev. 1995;9:1316–1326. doi: 10.1101/gad.9.11.1316. [DOI] [PubMed] [Google Scholar]
- 7.Bath J, Wu LJ, Errington J, Wang JC. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science. 2000;290:995–997. doi: 10.1126/science.290.5493.995. [DOI] [PubMed] [Google Scholar]
- 8.Sharpe ME, Errington J. Postseptational chromosome partitioning in bacteria. Proc Natl Acad Sci USA. 1995;92:8630–8634. doi: 10.1073/pnas.92.19.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Britton RA, Grossman AD. Synthetic lethal phenotypes caused by mutations affecting chromosome partitioning in Bacillus subtilis. J Bacteriol. 1999;181:5860–5864. doi: 10.1128/jb.181.18.5860-5864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemon KP, Grossman AD. Effects of replication termination mutants on chromosome partitioning in Bacillus subtilis. Proc Natl Acad Sci USA. 2001;98:212–217. doi: 10.1073/pnas.011506098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu G, Draper GC, Donachie WD. FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol Microbiol. 1998;29:893–903. doi: 10.1046/j.1365-2958.1998.00986.x. [DOI] [PubMed] [Google Scholar]
- 12.Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capiaux H, Lesterlin C, Perals K, Louarn JM, Cornet F. A dual role for the FtsK protein in Escherichia coli chromosome segregation. EMBO Rep. 2002;3:532–536. doi: 10.1093/embo-reports/kvf116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu LJ, Errington J. Septal localization of the SpoIIIE chromosome partitioning protein in Bacillus subtilis. EMBO J. 1997;16:2161–2169. doi: 10.1093/emboj/16.8.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Lutkenhaus J. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol Microbiol. 1998;29:731–740. doi: 10.1046/j.1365-2958.1998.00958.x. [DOI] [PubMed] [Google Scholar]
- 16.Yu XC, Tran AH, Sun Q, Margolin W. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J Bacteriol. 1998;180:1296–1304. doi: 10.1128/jb.180.5.1296-1304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massey TH, Mercogliano CP, Yates J, Sherratt DJ, Lowe J. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol Cell. 2006;23:457–469. doi: 10.1016/j.molcel.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 18.Bigot S, et al. KOPS: DNA motifs that control E. coli chromosome segregation by orienting the FtsK translocase. EMBO J. 2005;24:3770–3780. doi: 10.1038/sj.emboj.7600835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy O, et al. Identification of oligonucleotide sequences that direct the movement of the Escherichia coli FtsK translocase. Proc Natl Acad Sci USA. 2005;102:17618–17623. doi: 10.1073/pnas.0508932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pease PJ, et al. Sequence-directed DNA translocation by purified FtsK. Science. 2005;307:586–590. doi: 10.1126/science.1104885. [DOI] [PubMed] [Google Scholar]
- 21.Sharp MD, Pogliano K. Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science. 2002;295:137–139. doi: 10.1126/science.1066274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharp MD, Pogliano K. MinCD-dependent regulation of the polarity of SpoIIIE assembly and DNA transfer. EMBO J. 2002;21:6267–6274. doi: 10.1093/emboj/cdf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker EC, Pogliano K. Cell-specific SpoIIIE assembly and DNA translocation polarity are dictated by chromosome orientation. Mol Microbiol. 2007;66:1066–1079. doi: 10.1111/j.1365-2958.2007.05992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Partridge SR, Errington J. The importance of morphological events and intercellular interactions in the regulation of prespore-specific gene expression during sporulation in Bacillus subtilis. Mol Microbiol. 1993;8:945–955. doi: 10.1111/j.1365-2958.1993.tb01639.x. [DOI] [PubMed] [Google Scholar]
- 25.Lewis PJ, Partridge SR, Errington J. Sigma factors, asymmetry, and the determination of cell fate in Bacillus subtilis. Proc Natl Acad Sci USA. 1994;91:3849–3853. doi: 10.1073/pnas.91.9.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pogliano J, et al. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1149–1159. doi: 10.1046/j.1365-2958.1999.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ptacin JL, Nollmann M, Bustamante C, Cozzarelli NR. Identification of the FtsK sequence-recognition domain. Nat Struct Mol Biol. 2006;13:1023–1025. doi: 10.1038/nsmb1157. [DOI] [PubMed] [Google Scholar]
- 28.Sivanathan V, et al. The FtsK γ domain directs oriented DNA translocation by interacting with KOPS. Nat Struct Mol Biol. 2006;13:965–972. doi: 10.1038/nsmb1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bigot S, Saleh OA, Cornet F, Allemand JF, Barre FX. Oriented loading of FtsK on KOPS. Nat Struct Mol Biol. 2006;13:1026–1028. doi: 10.1038/nsmb1159. [DOI] [PubMed] [Google Scholar]
- 30.Griffiths AA, Wake RG. Search for additional replication terminators in the Bacillus subtilis 168 chromosome. J Bacteriol. 1997;179:3358–3361. doi: 10.1128/jb.179.10.3358-3361.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sciochetti SA, Piggot PJ, Blakely GW. Identification and characterization of the dif site from Bacillus subtilis. J Bacteriol. 2001;183:1058–1068. doi: 10.1128/JB.183.3.1058-1068.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becker E, et al. DNA segregation by the bacterial actin AlfA during Bacillus subtilis growth and development. EMBO J. 2006;25:5919–5931. doi: 10.1038/sj.emboj.7601443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hendrickson H, Lawrence JG. Selection for chromosome architecture in bacteria. J Mol Evol. 2006;62:615–629. doi: 10.1007/s00239-005-0192-2. [DOI] [PubMed] [Google Scholar]
- 34.Wu LJ, Errington J. A large dispersed chromosomal region required for chromosome segregation in sporulating cells of Bacillus subtilis. EMBO J. 2002;21:4001–4011. doi: 10.1093/emboj/cdf393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohler JJ, Schepartz A. Kinetic studies of FosJunDNA complex formation: DNA binding prior to dimerization. Biochemistry. 2001;40:130–142. doi: 10.1021/bi001881p. [DOI] [PubMed] [Google Scholar]
- 36.Burton BM, Marquis KA, Sullivan NL, Rapoport TA, Rudner DZ. The ATPase SpoIIIE transports DNA across fused septal membranes during sporulation in Bacillus subtilis. Cell. 2007;131:1301–1312. doi: 10.1016/j.cell.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettis GS, Cohen SN. Unraveling the essential role in conjugation of the Tra protein of Streptomyces lividans plasmid pIJ101. Antonie Van Leeuwenhoek. 2001;79:247–250. doi: 10.1023/a:1012079707140. [DOI] [PubMed] [Google Scholar]
- 38.Bigot S, Sivanathan V, Possoz C, Barre FX, Cornet F. FtsK, a literate chromosome segregation machine. Mol Microbiol. 2007;64:1434–1441. doi: 10.1111/j.1365-2958.2007.05755.x. [DOI] [PubMed] [Google Scholar]
- 39.Sharpe ME, Errington J. The Bacillus subtilis soj-spo0J locus is required for a centromere-like function involved in prespore chromosome partitioning. Mol Microbiol. 1996;21:501–509. doi: 10.1111/j.1365-2958.1996.tb02559.x. [DOI] [PubMed] [Google Scholar]
- 40.Wu LJ, Errington J. RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol Microbiol. 2003;49:1463–1475. doi: 10.1046/j.1365-2958.2003.03643.x. [DOI] [PubMed] [Google Scholar]
- 41.Ben-Yehuda S, Rudner DZ, Losick R. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science. 2003;299:532–536. doi: 10.1126/science.1079914. [DOI] [PubMed] [Google Scholar]
- 42.Ben-Yehuda S, et al. Defining a centromere-like element in Bacillus subtilis by identifying the binding sites for the chromosome-anchoring protein RacA. Mol Cell. 2005;17:773–782. doi: 10.1016/j.molcel.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 43.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McClelland SE, Dryden DT, Szczelkun MD. Continuous assays for DNA translocation using fluorescent triplex dissociation: application to type I restriction endonucleases. J Mol Biol. 2005;348:895–915. doi: 10.1016/j.jmb.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 45.Firman K, Szczelkun MD. Measuring motion on DNA by the type I restriction endonuclease EcoR124I using triplex displacement. EMBO J. 2000;19:2094–2102. doi: 10.1093/emboj/19.9.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strick TR, Allemand JF, Bensimon D, Croquette V. Behavior of supercoiled DNA. Biophys J. 1998;74:2016–2028. doi: 10.1016/S0006-3495(98)77908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perez AR, Abanes-De Mello A, Pogliano K. SpoIIB localizes to active sites of septal biogenesis and spatially regulates septal thinning during engulfment in Bacillus subtilis. J Bacteriol. 2000;182:1096–1108. doi: 10.1128/jb.182.4.1096-1108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sterlini JM, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoch JA. Genetic analysis in Bacillus subtilis. Methods Enzymol. 1991;204:305–320. doi: 10.1016/0076-6879(91)04015-g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.